Abstract
Sustained TCR stimulation is required for maintaining germinal center T follicular helper cells (GC-TFH). Paradoxically, TCR activation induces interleukin-2 receptor (IL-2R) expression and IL-2 production, thereby initiating a feedback loop of IL-2 signaling that normally inhibits TFH cells. It is unclear how GC-TFH cells can receive prolonged TCR signaling without succumbing to the detrimental effects of IL-2. Using an influenza infection model, we show here that GC-TFH cells secreted large amounts of IL-2 but responded poorly to it. Importantly, to maintain their IL-2-hyporesponsiveness, GC-TFH cells required intrinsic interleukin-6 (IL-6) signaling. Mechanistically, we found that IL-6 inhibited upregulation of IL-2Rβ (CD122) by preventing association of STAT5 with the Il2rb locus, thus allowing GC-TFH cells to receive sustained TCR signaling and produce IL-2 without initiating a TCR/IL-2-inhibitory feedback loop. Collectively, our results identify a regulatory mechanism that controls the generation of GC-TFH cells.
ONE SENTENCE SUMMARY
IL-6-mediated inhibition of CD122 allows TFH cells to receive TCR signaling without initiating an inhibitory TCR/IL-2 loop.
INTRODUCTION
T follicular helper (TFH) cells are a subset of CD4+ T cells that provide survival and differentiation signals for the development and maintenance of the germinal centers (GCs) (1, 2). TFH cells are primed outside of B cell follicles by antigen (Ag)-bearing dendritic cells (DCs) (3–6). This early stage of the TFH cell response is independent of the presence of B cells (3, 4, 7) and is termed the “DC-phase.” Following their initial interaction with DCs, CXCR5 guides TFH cells into the B/T cell border, where engagement of ICOS and PD-1 by bystander B cells directs TFH cells into B cell follicles (3, 8, 9). Once inside the follicles, the interaction of GC-TFH cells with activated B cells leads to the formation of GCs (1, 2), where persistent Ag presentation by GC B cells sustains the TFH cell response (4, 5, 10, 11).
Interleukin-2 (IL-2) signaling inhibits TFH cell differentiation by repressing Bcl-6 expression via STAT5 (12–14). Consequently, TFH cell responses fail to develop in high-IL-2 environments (14–16). Strikingly, TFH cells produce large amounts of IL-2 upon in vitro re-stimulation (17), and a recent study indicates that IL-2-producing cells are the precursors of TFH cells (18). This is particularly intriguing since prolonged TCR stimulation, which is required for normal TFH cell responses (5, 10), normally promotes IL-2R expression, thereby initiating a positive-feedback loop of IL-2/STAT5 signaling that results in increased IL-2 responsiveness (19, 20). Thus, the exact mechanisms that allow TFH cells to receive sustained TCR stimulation without responding to IL-2 are unclear.
Whereas IL-2 inhibits Bcl-6 expression, IL-6 signaling via STAT3 transiently induces Bcl-6 up-regulation (21, 22). The role of IL-6 in TFH cells is, however, puzzling. Although antiviral TFH cell responses are normally initiated in the absence of IL-6/IL-6R interactions (23–25), intrinsic IL-6 signaling is critical for sustaining TFH cell responses during the late stages of chronic viral infections (24). These data suggest that IL-6 signaling is not absolutely required for the initiation of the TFH cell program but is essential for supporting antiviral TFH responses during the GC-phase. The mechanisms by which IL-6 signaling contribute to the development of GC-TFH cells are unknown.
Using an influenza infection model, we show here that IL-6 was dispensable for the initial priming of influenza-specific TFH cells but was critical for the generation of GC-TFH cells. Our results demonstrate that fully differentiated GC-TFH cells produced large amounts of IL-2 and that intrinsic IL-6 signaling was required for maintaining their IL-2 hyporesponsiveness. Mechanistically, IL-6 negatively regulated CD122 expression, thus preventing the initiation of a negative TCR/IL-2-feedback loop that inhibits the generation of GC-TFH cells during the non-GC to GC-TFH transition phase.
RESULTS
GC-TFH cells require intrinsic IL-6 signaling
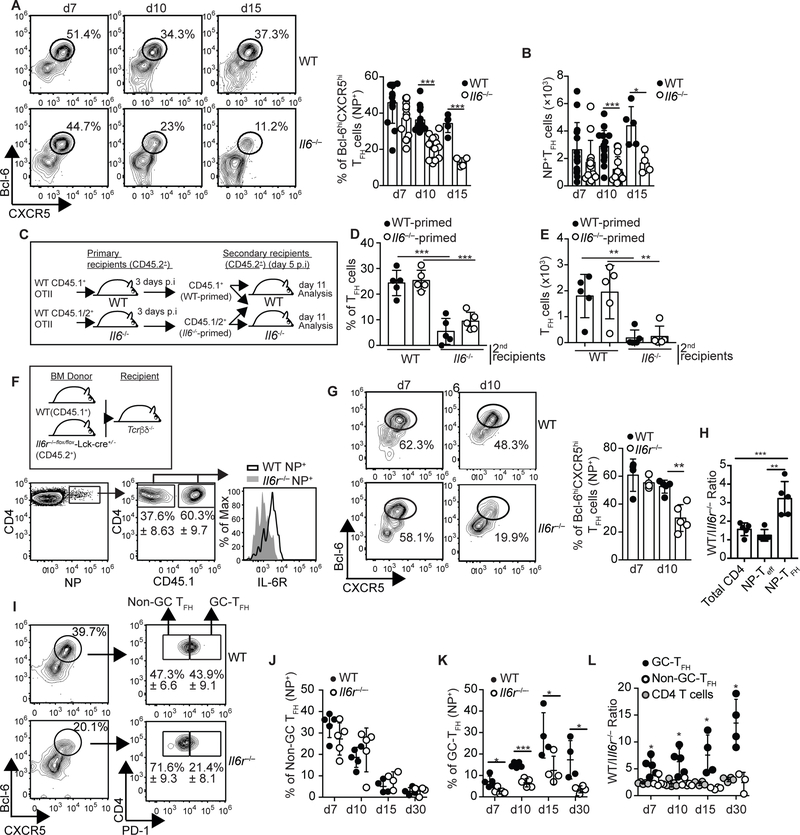
To study the role of IL-6 in the influenza-specific TFH cell response, we infected C57BL/6 (WT) and C57BL/6.Il6−/− (Il6−/−) mice with influenza and followed the kinetics of the nucleoprotein (NP)-specific TFH cells in the lung-draining mediastinal lymph node (mLN) (Fig. 1A,B). The frequency (Fig. 1A) and number (Fig. 1B) of Bcl-6hiCXCR5hi NP-specific TFH cells were similar in WT and Il6−/− mice early after infection (d7). However, the NP-specific TFH cell response prematurely contracted in Il6−/− mice (Fig. 1A,B). Similar results were obtained in WT mice treated with anti-IL6 + anti-IL-6R (anti-IL6/R) blocking Abs (Fig. S1A).
Figure 1. Late IL-6 signaling is required for the accumulation of influenza-specific GC-TFH cells.
(a-b) C57BL/6 (WT) and C57BL/6.Il6−/− (Il6−/−) mice were infected with PR8 and the frequency (a) and number (b) of NP-specific CD4+ T cells with a Bcl-6hiCXCR5hi TFH cell phenotype were evaluated in the mLN at the indicated time points. Representative plots gated on NP-specific CD4+ T cells are shown. Data were pooled from three independent experiments from a total of five independent experiments (Data are shown as the mean ± SD, n=4–5 mice per experiment). *P < 0.05, **P < 0.01, ***P < 0.001. P values were determined using a two-tailed Studentś t-test. (c-e) Equivalent numbers of CD45.1+CD45.2- and CD45.1+CD45.2+ OTII cells were respectively transferred into CD45.1-CD45.2+ WT and CD45.1-CD45.2+ Il6−/− primary recipient mice. One day later, the recipient mice were infected with PR8-OTII influenza virus. Three days after infection, CD4+ T cells from the mLN of WT and Il6−/− recipient mice were purified and mixed so that the mixture contained equivalent numbers of CD45.1+CD45.2- (WT-primed) and CD45.1+CD45.2+ (Il6−/−-primed) OTII cells. Cell numbers were then normalized to the concentration of OTII cells and 2×103 cells of the 1:1 mixture of WT-primed and Il6−/−-primed OTII cells were adoptively transferred into WT and Il6−/−mice that were previously infected with PR8-OTII influenza 5 days earlier. The frequency (d) and number (e) of WT-primed and Il6−/−-primed OTII cells with a PD-1hiCXCR5hi TFH phenotype were determined on day 11 after infection in the mLN. Data are representative of three independent experiments. Data are shown as the mean ± SD (n=5 mice). *P < 0.05, **P < 0.01, ***P < 0.001. P values were determined using a two-tailed Studentś t-test. (f-l) Tcrb−/−Tcrd−/− mice were irradiated and reconstituted with a 50:50 mix of BM from CD45.1+ C57BL/6 (WT) and CD45.2+ Il6raflox/flox-lck-cre−/+ (Il6r/−) donors. Eight weeks later, reconstituted mice were infected with PR8 and cells from the mLN were analyzed at the indicated time points. (f) Expression of CD126 (IL-6Ra) in the CD45.1+ and CD45.2+ NP-specific CD4+ T cell compartments. Representative plots on day 7 are shown. (g) Frequency of CD45.1+ and CD45.2+ NP-specific CD4+ T cells with a Bcl-6-hiCXCR5hi phenotype. Representative plots are shown. Data in the graph are shown as the mean ± SD (n=5 mice/time point). (h) The ratio of WT to Il6r−/− CD4+ T cells, NP-TEFF (Bcl-6-loCXCR5lo) and NP-TFH (Bcl-6-hiCXCR5hi) cells on day 10 were calculated (mean ± SD, n=5 mice). (i) PD-1 expression within the CD45.1+ and CD45.2+ NP-specific TFH cell compartments. Representative plots on day 10 are shown. Frequency of GC-TFH (Bcl-6hiCXCR5hiPD-1hi) (j) and non-GC-TFH (Bcl-6hiCXCR5hiPD-1lo) (k) cells within the CD45.1+ and CD45.2+ NP-specific CD4+ T cell compartments. (l) The ratio of WT to Il6r−/− NP-specific GC-TFH, NP-specific non-GC-TFH, and total CD4+ T cells were calculated (mean ± SD, n=5 mice). Data are representative of three independent experiments. Data are shown as the mean ± SD (n=4–5 mice/time point). *P < 0.05, **P < 0.01, ***P < 0.001. P values were determined using a two-tailed Studentś t-test.
To test whether IL-6 was required beyond the priming phase, we transferred CD45.1+ and CD45.1+/2+ OTII cells into congenically different WT or Il6−/−recipient mice (Fig. 1C). One day later, the recipient mice were infected with an engineered PR8 influenza virus expressing the OVA323–339 peptide (PR8-OTII) (26). Three days after infection, CD4+ T cells from WT and Il6−/− recipients were purified and mixed so that the mixture contained equivalent numbers of CD45.1+ (WT-primed) and CD45.1+/2+ (Il6−/−-primed) OTII cells. As expected, OTII cells recovered from WT and Il6−/− primary recipient mice contained similar frequencies of Bcl-6hiCXCR5hi TFH cells (Fig. S1B). Cell numbers were then normalized to the concentration of OTII cells and a 1:1 mixture of WT and Il6−/−-primed OTII cells was transferred into day 5 infected WT or Il6−/−secondary recipient mice. We found that regardless of whether OTII cells were initially primed in a WT or Il6−/− environment, nearly 30% of the donor OTII cells were TFH cells in the WT mice on day 11 after infection (Fig. 1D,E and S1C). In contrast, less than 12% of the WT and Il6−/−-primed OTII cells displayed a TFH cell phenotype in the Il6−/− recipient mice (Fig. 1D,E and Fig. S1C). These results indicated that TFH cell responses at the peak of the infection were compromised in the absence of IL-6, regardless of whether CD4+ T cells were primed in an IL-6-sufficient environment.
T follicular regulatory (Tfr) cells suppress TFH cell responses in some models (27). The frequency and phenotype of Tfr cells was, however, similar in WT and IL-6−/− mice (Fig. S1D and E). Hence, changes in Tfr cells were not likely responsible for the diminished TFH cell response observed in the Il-6−/− mice. We next assessed whether the requirement for IL-6 signaling was T cell-intrinsic. To do this, we infected WT/Il6r−/− mixed bone marrow (BM) chimeras (Fig. 1F) with influenza and determined the frequency of TFH cells within the WT and Il6r−/− NP-specific CD4+ T cell compartments at different times after infection (Fig. 1G). The frequency of NP-specific TFH cells was similar at day 7 (Fig. 1G). In contrast, NP-specific TFH cells failed to accumulate in the Il6r−/− compared to WT compartment at day 10 after infection (Fig. 1G and H). These results indicated that IL-6 signaling beyond the initial priming phase was intrinsically required for the TFH cell response to influenza. This conclusion was consistent with the presence of high levels of IL-6 during the post-priming phase of the primary response (Fig. S1F).
We next separated Bcl-6hiCXCR5hi NP-specific TFH cells into PD-1hi cells (GC-TFH) and PD-1lo cells (non-GC-TFH) in our WT/Il6r−/− chimeras (Fig. 1I). The frequency of NP-specific Tfh cells with a non-GC-TFH cell phenotype peaked at day 7 after infection and was similar in the WT and Il6r−/− donors at all time points analyzed (Fig. 1J). In contrast, the frequency of NP-specific Tfh cells with a GC-TFH phenotype peaked between days 10 and 15 and was significantly diminished in the Il6r−/− relative to the WT compartment (Fig. 1K). As a result, the WT: Il6r−/− ratio of GC-TFH was increased compared to the ratio of non-GC-TFH or total CD4+ T cells (Fig. 1L). These results indicated that the diminished TFH cell response observed in the absence of IL-6 signaling was due to the lack of influenza-specific GC-TFH cells. In agreement with this, whereas the frequency of extrafollicular CD138+ antibody-secreting cells (ASCs) was similar in WT and Il6−/− mice (Fig. S1G), the GC B cell response was reduced in Il6−/− relative to WT mice at the peak of the infection (Fig. S1H and I). Furthermore, though the number of GC B cells was similar in WT and Il6−/− mice at later time points (Fig. S1I), we observed a significant reduction in the long-term influenza-specific IgG2b and IgG2c levels in Il6−/− mice compared to WT mice (Fig. SJ). Our data suggested that IL-6 was dispensable for the priming on non-GC-TFH cells but was intrinsically required for the generation of GC-TFH cells during the non-GC to GC-TFH cell transition.
Late IL-6 prevents IL-2 responsiveness of GC-TFH cells
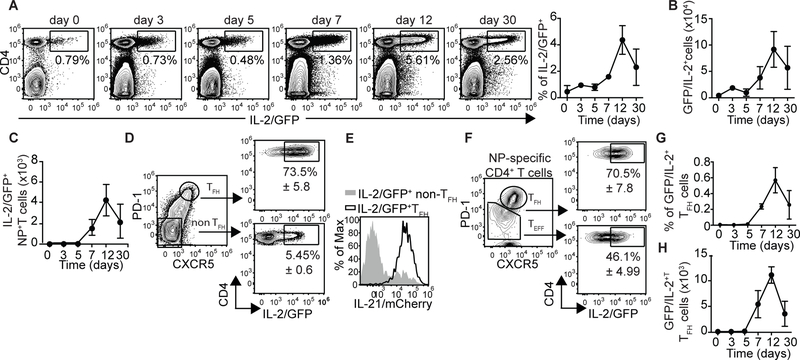
Given that IL-2 inhibits TFH cell responses (12–14) and that IL-6 antagonizes IL-2 signaling in some models (28, 29), we decided to investigate whether the late requirement for IL-6 signaling correlated with changes in IL-2 production. Using Il21-mCherry-Il2-emGFP dual-reporter mice (30), we found that the vast majority of IL-2 producing cells were CD4+ T cells (Fig. 2A). Although IL-2 producing CD4+ T cells were detected early after infection, the frequency (Fig. 2A) and number (Fig. 2B) of these cells peaked at day 12 post-infection. The same results were obtained when IL-2-producing NP-specific CD4+ T cells were enumerated (Fig. 2C), or when we assessed IL-2 production by intracellular staining (Fig. S2A). We also found that close to 80% of the GC-TFH cells were GFP/IL-2+ at day 12 (Fig. 2D). As expected, GFP/IL-2+ TFH cells were also mCherry/IL-21+ (Fig. 2E). The capacity of GC-TFH cells to secrete IL-2 was further confirmed by intracellular staining (Fig. S2B). Similar results were obtained when we analyzed IL-2 production by NP-specific Tfh cells (Fig. 2F). Importantly, while IL-2+ GC-TFH cells were already detectable at day 7, they largely accumulated during the peak of the response (day 12) (Fig. 2G and H). These results indicated that GC-TFH cells produce large amounts of IL-2 during the peak of the infection.
Figure 2. GC-TFH cells produce large amounts of IL-2 at the peak of the infection.
(a-c) Il21-mCherry-Il2-emGFP reporter mice were infected with PR8 and cells from the mLN were analyzed at the indicated time-points. (a) Frequency and (b) number of IL2/GFP+CD4+ T cells. (c) Number of IL2/GFP+ NP-specific CD4+ T cells. Representative plots are shown. Data in the graphs are shown as the mean ± SD (n=4 mice/time point). Data are representative of three independent experiments. (d-g) Il21-mCherry-Il2-emGFP reporter mice were infected with PR8 and the frequency of IL2/GFP+ cells within the PD-1hiCXCR5hi (TFH) and PD-1loCXCR5lo (non-TFH) CD4+ T cell populations were calculated on day 12 after infection. Representative plots gated on CD19-CD4+ T cells. (Data are shown as the mean ± SD, n=4 mice/time point). (e) Expression of IL-21/mCherry in IL2/GFP+ GC-TFH and IL2/GFP+ non-TFH cells. Data are representative of three independent experiments (f) Frequency of IL2/GFP+ cells within the PD-1hiCXCR5hi and PD-1loCXCR5lo NP-specific CD4+ T cell populations on day 12 post-infection. (Data are shown as the mean ± SD, n=4) mice. (g-h) Frequency (g) and number (h) of IL2/GFP+ GC-TFH cells at the indicated time points. Data are representative of three independent experiments. Data are shown as the mean ± SD (n=4 mice/time point)
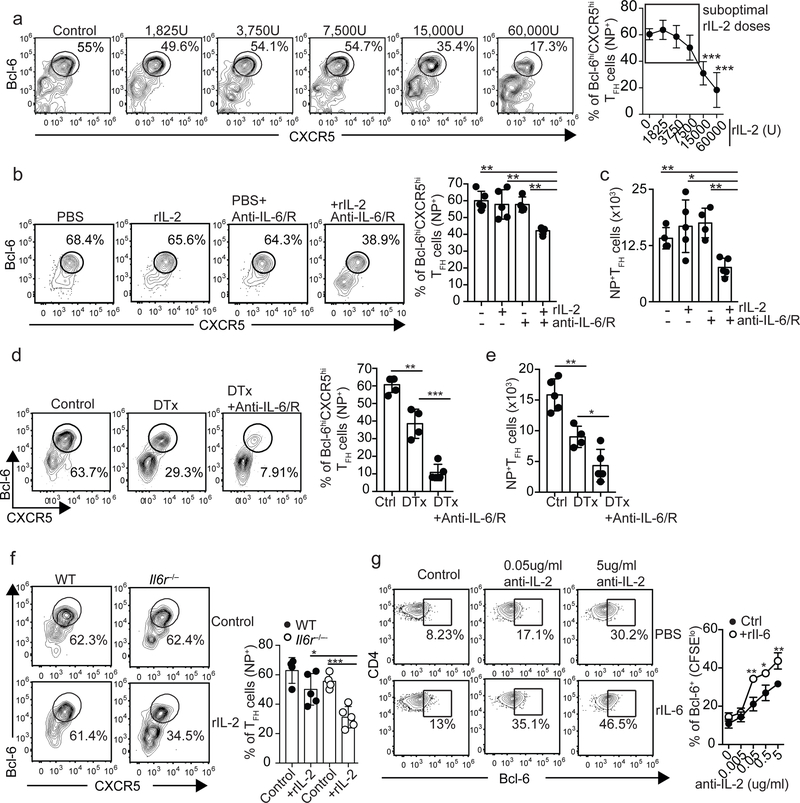
We next decided to increase IL-2 availability early after infection, a time during which TFH cells normally developed in the absence of IL-6. Importantly, treatment of WT mice with recombinant IL-2 (rIL-2) doses ≥ 15,000 units (U) prevented Tfh cell responses (Fig. 3A). In contrast, no differences were detected with doses under the 15,000U threshold, which we will refer to as suboptimal doses (Fig. 3A). Thus, we treated influenza-infected WT mice with a suboptimal dose of rIL-2 or PBS, administered either control or anti-IL6/R Ab, and evaluated the NP-specific TFH cell response on day 7 (Fig. 3B,C). Whereas rIL-2 or anti-IL-6/R Abs alone did not significantly affect the NP-specific TFH cell response, NP-specific TFH cells failed to accumulate in mice that received rIL-2 in combination with anti-IL-6/R Abs. Similar results were obtained in WT and Il6−/− mice (Fig. S3A). These results suggested that the lack of IL-6 signaling lowered the threshold of IL-2 required for suppressing TFH cell responses, thereby rendering TFH cells more responsive to IL-2.
Figure 3. IL-6 signaling is required for sustaining TFH cells developing in a high-IL-2 environment.
(a) B6 mice were infected with PR8 and treated daily with the indicated doses of rIL-2 starting one day after infection. The frequency of NP-specific cells with Bcl-6hiCXCR5hi TFH cell phenotype was determined in the mLN on day 7 after infection. Data in the graph are shown as the mean ± SD (n=4 mice per group). Data are representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. P values were determined using a two-tailed Studentś t-test. (b-c) B6 mice were infected with PR8, treated daily with either 7,500U of rIL-2 or PBS alone, or in combination with 250 μg of a mix of anti-IL-6+anti- IL-6/R Abs. The frequency (b) and number (c) of Bcl-6hiCXCR5hi NP-specific TFH cells were determined in the mLN on day 7 after infection. Representative plots are shown. Data in the graph are shown as the mean ± SD (n=4–5 mice per group). Data are representative of two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. P values were determined using a two-tailed Studentś t-test. (d-e) Foxp3-DTR–GFP mice were infected with PR8 and treated with either PBS (control) or DT on day 3 after infection. A third group was treated with DT and administered 250 μg of a mix of anti-IL-6+anti-IL-6/R Abs on days 0, 2 and 4 after infection. The frequency (d) and number (e) of Bcl-6hiCXCR5hi NP-specific TFH cells in the mLN were calculated on day 7 after infection. Representative plots gated on NP-specific CD4+ T cells are shown. Data in the graph are shown as the mean ± SD (n=4–5 mice per group). Data are representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. P values were determined using a two-tailed Studentś t-test. (f) Irradiated Tcrb−/−Tcrd−/− mice were reconstituted with a 50:50 mix of BM from CD45.1+ C57BL/6 (WT) and CD45.2+ Il6raflox/flox-lck-cre−/+ (Il6r−/−) donors. Two months later, chimeric mice were infected with PR8, treated daily with 7,500U of rIL-2 or PBS starting on day 1 after infection, and the frequency of Bcl-6hiCXCR5hi cells within the WT and Il6r−/− NP-specific CD4+ T cell compartments calculated on day 7 post-infection in the mLN. Representative plots are shown. Data in the graph are shown as the mean ± SD (n=4–5 mice per group). Data are representative of two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. P values were determined using a two-tailed Studentś t-test. (g) CFSE-labeled CD4+ T cells from the spleen of naïve B6 mice were activated in vitro with plate-bound anti-CD3/CD28 Abs in the presence of the indicated concentration of anti-IL-2 Abs (JES6–1A12+S4B6) and either 10ng/ml of rIL-6 or PBS was added to the cultures. The expression of Bcl-6 in CFSElowCD4+ T cells was assessed at 48h by flow cytometry. Data are representative of four independent experiments. All values were obtained in triplicate and the data are shown as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001P. P values were determined using a two-tailed Studentś t-test.
CD25+FoxP3+Treg cells consume IL-2 early after infection (31–34), thereby lowering the IL-2 environment and helping TFH cell differentiation (15). Thus, we considered the possibility that IL-6 was dispensable early after infection because IL-2 consumption by Treg cells was sufficient for lowering the IL-2 availability below the necessary threshold for TFH cell suppression. To test this hypothesis, we infected FoxP3-DTR mice with influenza, depleted Treg cells to increase IL-2 availability (31–34), and assessed whether early TFH cell responses developed in the absence of IL-6 signaling (Fig. 3D,E). In agreement with our previous studies (15), the frequency (Fig. 3D) and number (Fig. 3E) of NP-specific TFH cells were significantly reduced in Treg-depleted relative to control mice. Importantly, however, the NP-specific TFH cell response was further reduced in Treg-depleted mice that received anti-IL6/R Abs (Fig. 3D,E). These data suggest that, when developing in a high IL-2 environment, IL-6 was required for preventing IL-2 mediated suppression of the TFH cell response.
To examine whether IL-6 signaling was intrinsically required to limit IL-2 responsiveness of TFH cells, we infected WT/Il6r−/− chimeras with influenza, treated them daily with a suboptimal dose of rIL-2 or control PBS, and analyzed WT and Il6r−/− TFH cells on day 7 after infection (Fig. 3F), a time during which WT and Il6r−/− TFH cells similarly accumulated (Fig. 1G). As expected, the frequency of NP-specific TFH cells within the WT and Il6r−/− compartments were comparable in the PBS-treated mice (Fig. 3F). In contrast, the frequency of Il6r−/− NP-specific TFH cells was significantly diminished in the rIL-2 treated mice (Fig. 3F). These results indicated that intrinsic IL-6 signaling protected TFH cells from IL-2-mediated suppression.
To confirm these conclusions, we carried out an in vitro study to test whether the presence of IL-6 affected the threshold of IL-2 required to prevent Bcl-6 expression. Carboxyfluorescein succinimidyl ester (CFSE)-labeled CD4+ T cells were activated in vitro in the presence of increasing concentrations anti-IL-2 neutralizing Abs and either rIL-6 or control PBS. Bcl-6 expression in proliferating (CFSElo) CD4+ T cells was measured two days later by flow cytometry (Fig. 3G). Proliferating CD4+ T cells cultured in the absence of anti-IL-2 neutralizing Abs (i.e. high IL-2 conditions) expressed low levels of Bcl-6 (Fig. 3G). Bcl-6 expression, however, gradually increased with progressively higher concentrations of anti-IL-2 Abs (i.e. low IL-2 conditions) (Fig. 3G). Importantly, in the presence of IL-6, a lower concentration of anti-IL-2 Ab was required to promote Bcl-6 up-regulation (Fig. 3G). CD4+ T cells proliferated at a similar rate in all conditions (Fig. S3B). These data suggested that intrinsic IL-6 signaling prevented IL-2 responsiveness of TFH cells, which was required for sustaining TFH cell responses when present in a high-IL-2 environment.
IL-6 prevents IL-2 responsiveness by inhibiting CD122 expression
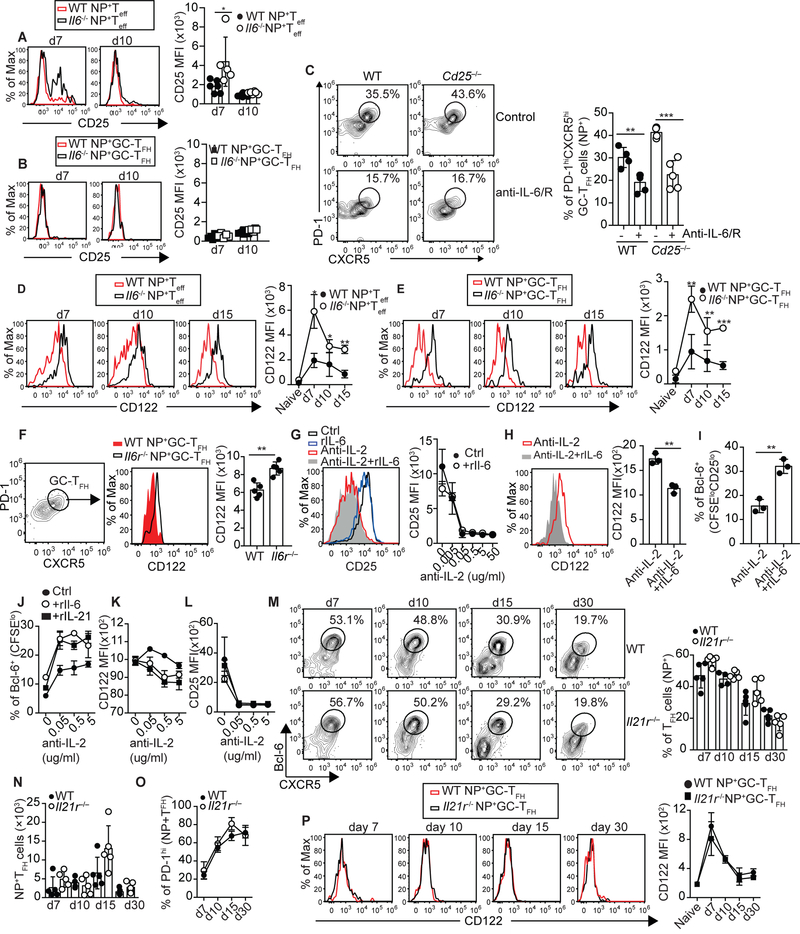
TCR stimulation, which is required for the maintenance of the GC-TFH cells (4, 5, 10, 11), induces CD25 expression. Given that IL-6 signaling negatively regulates CD25 expression (28, 35, 36), we considered the possibility that IL-6 signaling inhibited IL-2 responsiveness of GC-TFH cells by preventing CD25 upregulation. To test this idea, we infected WT and Il6−/− mice with influenza and assessed the expression of CD25 on NP-specific GC-TFH cells and NP-specific TEFF cells at different times after infection (Fig. 4A,B). Il6−/− NP-specific TEFF cells expressed higher amounts of CD25 relative to WT controls early after infection (Fig. 4A). We found, however, that CD25 was virtually undetectable in both WT and Il6−/− NP-specific GC-TFH cells (Fig. 4B). We next infected WT/Cd25−/− mixed BM chimeras, treated them with control or anti-IL-6/R Abs, and enumerated WT and Cd25−/− NP-specific GC-TFH cells on day 10 after infection (Fig. 4C). The NP-specific CD4+ T cells developed comparably from WT and Cd25−/− donors in both groups (Fig. S4A). As expected, WT GC-TFH cells failed to accumulate in the anti-IL-6/R treated mice relative to control counterparts (Fig. 4C). However, treatment with anti-IL-6/R Abs also prevented the accumulation of Cd25−/− NP-specific GC-TFH cells (Fig. 4C). These results suggested that IL-6 controlled the threshold of IL-2 responsiveness of TFH cells by a CD25-independent mechanism
Figure 4. IL-6 signaling limits IL-2 responsiveness by preventing CD122 expression.
(a-b) WT and Il6−/− mice were infected with PR8 and the expression of CD25 in PD-1loCXCR5lo NP-specific Teff cells (a) and PD-1hiCXCR5hi NP-specific GC-TFH cells (b) was examined in the mLN at the indicated time points. Data are representative of three independent experiments. Data in the graph are shown as the mean ± SD (n=5 mice). *P < 0.05, **P < 0.01, ***P < 0.001. P values were determined using a two-tailed Studentś t-test. (c) Irradiated Tcrb−/−Tcrd−/− mice were reconstituted with a 50:50 mix of BM from CD45.1+ C57BL/6 (WT) and CD45.2+ C57BL/6.Cd25−/− (Cd25−/−) donors. Two months later, chimeric mice were infected with PR8, treated or not with 250 μg of a mix of anti-IL-6 + anti-IL-6/R Abs on days 0, 2, 4, 6 and 8 after infection, and the frequency of PD-1hiCXCR5hi GC-TFH cells within the WT and Cd25−/− NP-specific CD4+ T cells were examined on day 10 after infection. Representative plots are shown. Data in the graph are shown as the mean ± SD (n=4–5 mice per group). Data are representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. P values were determined using a two-tailed Studentś t-test. (d-e) Kinetics demonstrating the expression of CD122 in PD-1loCXCR5lo NP-specific Teff cells (d) and PD-1hiCXCR5hi NP-specific GC-TFH cells (e) from PR8-infected WT and Il6−/− mice. Data are representative of three independent experiments. Data in the graph are shown as the mean ± SD (n=5 mice). *P < 0.05, **P < 0.01, ***P < 0.001. P values were determined using a two-tailed Studentś t-test. (f) WT/ Il6r−/− mixed BM chimeras were infected with PR8 and expression of CD122 in WT and Il6r−/− PD-1hiCXCR5hi NP-specific GC-TFH cells was determined on day 10 after infection. Data are representative of two independent experiments. Data in the graph are shown as the mean ± SD (n=5). *P < 0.05, **P < 0.01, ***P < 0.001. P values were determined using a two-tailed Studentś t-test. (g-h) CFSE-labeled CD4+ T cells from the spleen of naïve B6 mice were activated in vitro with plate-bound anti-CD3/CD28 Abs in the presence of 0.5μg/ml of anti-IL-2 Abs (JES6–1A12+S4B6) and either 10ng/ml of rIL-6 or PBS was added to the cultures. (g) Expression of CD25 in CFSEloCD4+ T cells at 48h. (h) Expression of CD122 in CFSEloCD25lo CD4+ T cells. (i) Expression of Bcl-6 in CFSEloCD25lo CD4+ T cells. Data are representative of three independent experiments. All values were obtained in triplicate and the data are shown as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001P. P values were determined using a two-tailed Studentś t-test. (j-l) CFSE-labeled CD4+ T were activated in vitro in the presence of anti-IL-2 Abs (JES6–1A12+S4B6) and rIL-6 (10ng/ml), rIL-21 (50ng/ml), or PBS was added to the cultures. The expression of Bcl-6 (j), CD122 (k), and Cd25 (l) in CFSElowCD4+ T cells was assessed at 48h. Data are representative of two independent experiments. All values were obtained in triplicate and the data are shown as the mean ± SD. (m-o) WT and Il21r–/– mice were infected with PR8 and cells from the mLN were analyzed at the indicated time points. Frequency (m) and number (n) of Bcl-6hiCXCR5hi NP-specific TFH cells. (o) Frequency of PD-1hi GC-TFH cells within the NP-specific TFH cell population. (p) Expression of CD122 in NP-specific GC-TFH cells. Data are representative of two independent experiments. Data in the graph are shown as the mean ± SD (n=5).
We next studied whether IL-6 deficiency resulted in changes in CD122 levels. We found that the expression of CD122 was increased in both Il6−/− NP-specific TEFF (Fig. 4D) and NP-specific GC-TFH cells (Fig. 4E and Fig. S4B) relative to WT counterparts. Similar results were obtained when we analyzed total GC-TFH cells (Fig. S4C). To determine whether IL-6 signaling was intrinsically required for preventing CD122 up-regulation on GC-TFH cells, we analyzed CD122 expression in WT and Il6r−/− NP-specific GC-TFH cells from WT/Il6r−/− chimeras. We found that Il6r−/− NP-specific GC-TFH cells were CD122hi compared to WT NP-specific GC-TFH cells (Fig. 4F). To confirm the effect of IL-6 on CD122 expression, we analyzed CD25 (Fig. 4G) and CD122 (Fig. 4H) expression in vitro. As expected, CFSElo-CD4+ T cells expressed high levels of CD25 in control conditions, but progressively downregulated CD25 in the presence of increasing concentrations of anti-IL-2 Ab (Fig. 4G). The presence of rIL-6 did not significantly change the dynamics of CD25 expression (Fig. 4G). However, whereas CD25lo cells generated under anti-IL-2 Ab conditions expressed high levels of CD122, CD25lo cells generated in the anti-IL-2 + rIL-6 conditions were CD122lo (Fig. 4H). Correlating with the differences in CD122 expression, CD25loCD122lo cells from the anti-IL-2 + rIL-6 cultures expressed higher levels of Bcl-6 compared to CD25loCD122hi cells generated in the presence of anti-IL-2 Ab alone (Fig. 4I). These data indicated that intrinsic IL-6 signaling negatively regulates CD122 expression; thereby preventing CD122 up-regulation on TFH cells.
IL-21 and IL-6 overlap in their capacity to promote TFH cell development (23). Thus, we used our in vitro system to test the capacity of IL-21 to regulate Bcl-6 (Fig. 4J), CD122 (Fig. 1K), and CD25 (Fig. 1L) expression. Similar to the anti-IL-2+rIL-6 conditions, cells cultured in the presence of anti-IL-2+rIL-21 up-regulated Bcl-6 (Fig. 4J) and downregulated CD122 (Fig. 4K) compared to cells activated in the presence of anti-IL-2 Abs alone. No differences were detected in CD25 (Fig. 4L). Next, we studied the TFH cell response in WT and Il21r−/− mice infected with influenza (Fig. 4M–P). The frequency (Fig. 4M) and number (Fig. 4N) of NP-specific TFH cells, and the frequency of NP-specific TFH cells with a PD-1hi-GC-TFH cell phenotype (Fig. 4O) were similar in WT and Il21r−/− mice at all time points analyzed. We also found that WT and Il21r−/− NP-specific GC-TFH cells expressed equivalent levels of CD122 (Fig. 4P). As a control, CD122 expression in NP-specific TFH cells was increased in WT and Il21r−/− mice treated with anti-IL-6/R Abs (Fig. S4D). These data indicated that, whereas IL-21 could replace IL-6 signaling in vitro, it was dispensable for down-regulating CD122 and promoting influenza-specific GC-TFH responses in vivo. In contrast, IL-6 signaling was necessary and sufficient.
In the absence of IL-6, STAT5 deficiency rescues GC-TFH cells
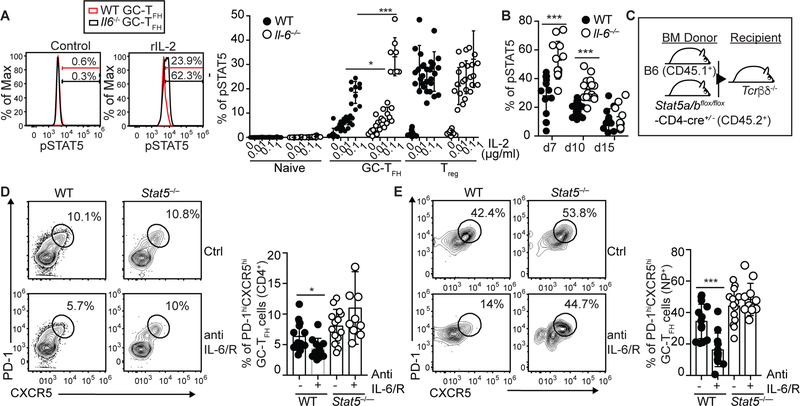
Given that IL-2-producing GC-TFH cells largely accumulated at the peak of the response and that GC-TFH cells express high levels of CD122, we hypothesized that the lack of GC-TFH cells observed in the Il6−/− mice was due to excessive IL-2/STAT5 signaling. To determine whether increased CD122 expression in Il6−/− TFH cells resulted in augmented responsiveness to IL-2, we stimulated cells from influenza-infected WT and Il6−/− mice with different concentrations of IL-2 and evaluated STAT5 phosphorylation in GC-TFH cells (Fig. 5A and B). We found that pSTAT5 was increased in Il6−/−compared to WT GC-TFH cells (Fig. 5A and B). Next, we generated WT/Stat5ab−/− mixed BM chimeras (Fig. 5C), infected them with influenza, treated them or not with anti-IL-6/R Abs, and enumerated WT and Stat5ab−/− GC-TFH cells on day 10 after infection (Fig. 5D and E). The frequency of WT GC-TFH cells was reduced by about 50% in the anti-IL-6/R relative to control-treated mice (Fig. 5D). In contrast, the frequency of Stat5ab−/− GC-TFH cells was similar in both groups (Fig. 5D). Similar results were obtained when we analyzed the NP-specific GC-TFH cell response (Fig. 5E). These results suggested that IL-6 signaling during the non-GC to GC-TFH cell transition phase prevented CD122 up-regulation, thereby limiting IL-2/Stat5 responsiveness and protecting GC-TFH cells from the deleterious effect of IL-2.
Figure 5. Lack of GC-TFH cells in the absence of IL-6 signaling is STAT5 dependent.
(a) Cells obtained from the mLN of PR8-infected WT and Il6−/− mice were stimulated with the indicated amounts of rIL-2 for 15 minutes and STAT5 phosphorylation in PD-1hiBcl-6hiCXCR5hiCD4+B220– GC-TFH, CD4+B220–Bcl-6loCXCR5loCD25+Foxp3+ Treg and in PD-1loBcl-6loCXCR5loCD4+B220–CD25-Foxp3- naïve CD4+ T cells was determined by flow cytometry on day 10. Data are representative of three independent experiments. Data are shown as the mean ± SD (n=8–10 mice). (b) Kinetic of STAT5 phosphorylation in GC-TFH at different times after infection. Data were pooled from three independent experiments. Data are shown as the mean ± SD. (c-e) Irradiated Tcrb−/−Tcrd−/− mice were reconstituted with a 50:50 mix of BM from CD45.1+ C57BL/6 (WT) and CD45.2+ Stat5a/bfl/fl-Cd4cre/+ (Stat5−/−) donors (c). Two months later, chimeric mice were infected with PR8, treated or not with 250 μg of a mix of anti-IL-6 + anti-IL-6/R Abs on days 0, 2, 4, 6 and 8 after infection, and cells from the mLN were analyzed on day 10 post-infection. (d) Frequency of PD-1hiCXCR5hi GC-TFH cells within the WT and Stat5−/− CD4+ T cell compartments. (e) Frequency of PD-1hiCXCR5hi GC-TFH cells within the WT and Stat5−/− NP-specific CD4+ T cell compartments. Representative plots are shown. Data in the graph are shown as the mean ± SD. Data were pooled from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. P values were determined using a two-tailed Studentś t-test.
IL-6 prevents association of STAT5 to the Il2rb locus
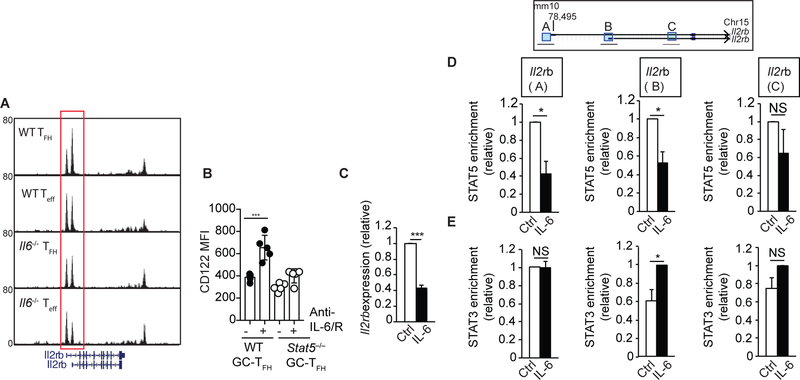
To better define the mechanisms by which IL-6 controls CD122 expression, we examined whether changes in CD122 expression in the absence of IL-6 correlated with changes in chromatin accessibility in the Il2rb locus in TFH and TEFF cells (Fig. 6A). We adoptively transferred naïve OTII (CD45.1+) cells into WT or Il6−/− recipient mice that were then infected with PR8-OTII one day later. Seven days after infection, donor-derived TFH and TEFF cells were sorted from WT or Il6−/− recipients and chromatin accessibility was examined via transposase-accessible chromatin-sequencing (ATAC-seq). We observed differences in chromatin accessibility near the 5’ end of the Il2rb locus between OTII-TFH and OTII-TEFF cells from WT recipients (Fig. 6A). Interestingly, the profile of the ATAC-seq peaks from OTII-TFH cells from Il6−/− recipient mice resembled the profile obtained in OTII-TEFF cells (Fig. 6A). As a control, no differences were detected in the Il2ra locus (Fig. S5A). These results indicated the presence of regulatory elements at the Il2rb locus that were responsive to IL-6 in TFH and TEFF cells.
Figure 6. IL-6 signaling limits CD122 expression by inhibiting STAT5 association to the Il2rb locus.
(a) OTII (CD45.1+) cells were adoptively transferred into WT and Il6−/− recipient mice. One day later, recipient mice were infected with PR8-OTII. Seven days after infection, donor-derived CD45.1+CXCR5hiSLAMloCD4+CD19- TFH and CD45.1+CXCR5loSLAMhiCD4+CD19- Teff cells were sorted from the mLN of WT and Il6−/− recipient mice and ATAC-seq was performed. USCS genome browser tracks displaying chromatin accessibility peaks at the Il2rb locus are shown. Results are representative of two biological replicates from two independent experiments. (b) WT/Stat5−/− mixed BM chimeras were infected with PR8, treated or not with 250 μg of a mix of anti-IL-6+anti-IL-6/R Abs on days 0, 2, 4, 6 and 8 post-infection, and expression of CD122 in WT and Stat5−/− PD-1hiCXCR5hi NP-specific GC-TFH cells was determined on day 10 after infection. Data in the graph are shown as the mean ± SD (n=3–5 mice per group). Data are representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. P values were determined using a two-tailed Studentś t-test. (c-e) CD4+ T cells from the spleen of naïve B6 mice were activated in vitro with plate-bound anti-CD3/CD28 Abs in the presence of rIL-6 or control PBS. (c) The expression of Il2rb was determined by RT-PCR on day 3. Data are normalized to Rps18 and represented as fold change in expression relative to the control condition. Data are shown as the mean ± SD (n=5). (d-e) CD4+ T cells activated in the presence of IL-6 or control PBS were crosslinked with paraformaldehyde at 72h. Chromatin samples were immunoprecipitated with anti-STAT5 (d) or anti-STAT3 (e) and the indicated regions of the Il2rb locus were monitored by qPCR. Data are shown as the mean ± SEM (n=3). Samples were normalized to the total input followed by subtraction of the isotype control to account for unspecific antibody binding and represented as relative fold change enrichment. Data are shown as the mean ± SEM (n=3). Data are representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. P values were determined using a two-tailed Studentś t-test
CD122 expression is positively regulated in response to IL-2/STAT5 signaling (19, 20). Corresponding with this idea, using our WT/Stat5ab−/− chimeras we found that increased CD122 expression observed after IL-6 blockade was STAT5 dependent (Fig. 6B). Given that STAT3 competes with STAT5 for DNA binding to some of its target genes (29), we considered the possibility that IL-6 signaling prevented CD122 expression by interfering with the ability of STAT5 to associate to the Il2rb locus. Chromatin immunoprecipitation sequencing studies (ChIP-seq) have been performed previously to study binding of STAT3 and STAT5 to the Il17a locus (29). We reanalyzed these data and identified three STAT5 binding sites, one located at the promoter region (Region A), and two down-stream of the transcription start site (Regions B and C). STAT3 ChIP-seq peaks were found at the same sites, thus suggesting that STAT3 and STAT5 bound to the same regions of the Il2rb locus (Fig. S5). Next, we activated CD4+ T cells in the presence of rIL-6 or control PBS and used ChIP-qRT-PCR to study whether IL-6 signaling affected STAT5 binding to the Il2rb locus (Fig. 6C–E). As expected, we observed reduced Il2rb expression in the IL-6 treated cells (Fig. 6C). We also found that STAT5 association to Regions A and B of the Il2rb locus was significantly diminished in the presence of IL-6 (Fig. 6D), which correlated with reduced gene expression (Fig. 6C). In contrast, STAT3 association at Region B was significantly enriched in IL-6 relative to control-stimulated cells (Fig. 6E). Collectively, our results indicated that IL-6/STAT3 signaling prevented association of STAT5 with the Il2rb locus, thereby preventing CD122 up-regulation and the subsequent initiation of a deleterious-feedback loop of TCR/IL-2/STAT5 signaling that results in increased responsiveness to IL-2.
DISCUSSION
Here we show that fully differentiated TFH cells secrete large amounts of IL-2, a potent inhibitor of TFH cells (12–14). Despite producing IL-2, however, TFH cells were resistant to IL-2 signaling. Our data demonstrated that maintaining IL-2 hyporesponsiveness in TFH cells during the non-GC to GC-TFH cell transition was fundamental for the generation of influenza-specific GC-TFH cells. Importantly, IL-6 did not limit IL-2 responsiveness by a CD25-dependent mechanism, but instead prevented CD122 expression. Mechanistically, IL-6 signaling inhibited CD122 up-regulation in response to TCR stimulation by preventing STAT5 association to the Il2rb locus, which precluded the initiation of a positive-feedback loop of TCR/IL-2/STAT5 signaling that would result in increased responsiveness to IL-2. Collectively, our results demonstrate that IL-6 signaling fine-tunes the threshold of IL-2 responsiveness of TFH cells, allowing them to receive TCR stimulation and produce IL-2 without succumbing to its deleterious effects.
Our data suggest a model by which the requirement for IL-6 depends on the relative availability of IL-2, hence clarifying previous conflicting studies regarding the role of IL-6 in controlling TFH cell responses. In this model, in the absence of IL-2, or when the environmental levels of IL-2 are below the minimum necessary threshold for TFH cell suppression, IL-6 signaling is dispensable. In contrast, when the IL-2 availability exceeds the threshold for TFH cell suppression, intrinsic IL-6 signaling is required for preventing IL-2 mediated suppression of the TFH cell response. The environmental availability of IL-2 is the result of a tightly regulated balance between IL-2-producing and IL-2-consuming cells (15, 16, 31). This balance, however, varies throughout the infection depending on timing and anatomical location. For example, early after infection CD25+Treg cells (15, 31) and CD25+DCs (16) consume T cell-derived IL-2, thereby limiting IL-2 availability and favoring TFH cell development during the DC phase. As the immune response to progresses, however, TFH cells migrate into the B cell follicles where CD25-expressing cells are scarce. As such, Tregs in B cell follicles characteristically express low levels of CD25 (34, 37, 38) and CD25+DCs preferentially localize outside the B cell follicles and quickly deregulate CD25 after activation (16). Thus, high density of IL-2-producing TFH cells combined with the scarcity of “IL-2 consumers” generates a relatively high-IL-2 environment during the non-GC to GC-TFH transition phase. Under such conditions, IL-6 is required for preventing excessive IL-2 signaling and generating GC-TFH cells.
The capacity of TFH cells to produce IL-2 is not entirely surprising since sustained TCR stimulation, which is required for the maintenance of GC-TFH cells, potently induces IL-2 production (4, 5, 10, 11, 39). Moreover, while Blimp-1 represses Il2 transcription as part of a negative feedback loop that hinders IL-2 production after T cell activation (40, 41), Bcl-6 positively regulates IL-2 secretion (18). Thus, given that TFH cells characteristically express high levels of Bcl-6, low levels of Blimp-1, and establish prolonged cognate interactions with APCs, it is likely that they will produce large amounts of IL-2. One caveat to our study is that, due to the lack of Tfh-conditional knock out mice, we did not examine the effect of IL-2 produced by Tfh cells. Thus, further investigations will be required to define the role played by IL-2-secreting TFH cells in GC dynamics. In summary, our study indicates that TFH cell homeostasis is controlled by a tightly regulated balance between the relative levels of IL-2 and IL-6, rather than by the absolute expression of IL-2 or IL-6 alone, thus offering a new perspective for how TFH cell responses are regulated.
STUDY DESIGN
The goal of this study was to determine the role of IL-6 signaling in the TFH cell response to influenza. Flow cytometry was used to characterize TFH cells in WT and Il-6−/− mice at different times after infection. The main conclusions were validated using mixed bone marrow chimeras and molecular studies. All mice were infected with subtlethal doses of influenza virus. Mice did not receive supportive care, but body condition was monitored daily. A sample size of 4–5 mice/group/time-point was used for the in vivo experiments. This number was sufficient to generate enough cells for analysis and to detect statistically significant differences between groups while minimizing the use of laboratory animals. The investigators were not blinded when performing the experiments. Control and experimental groups were age and sex-matched. Males and females were used in these experiments.
MATERIALS AND METHODS
Mice.
C57BL/6 (B6), B6.SJL-Ptprca Pepcb/BoyJ (B6.CD45.1), B6.129S2-IL6tm1Kopf/J (Il-6−/−), B6.129P2-Tcrβtm1MomTcrδtm1Mom (Tcrb−/−Tcrd−/−), B6.129S4-Il2ratm1Dw/J (Cd25−/−), B6.Cg-Tg(Lck-cre)3779Nik/J, B6;SJL-Il6ratm1.1Drew/J (Il6rafl/fl), C57BL/6-Tg(TcraTcrb)425Cbn/J (OTII), B6.129S6-Foxp3tm1DTR (FoxP3-DTR), and B6N.129-Il21rtm1Kopf/J mice were originally obtained from Jackson Laboratories. Il21-mCherry-Il2-emGFP dual-reporter transgenic mice were obtained from W. J. Leonard (NHLBI). Il6ra fl/fl mice were crossed to B6.Cg-Tg(Lck-cre)3779Nik/J mice to generate B6. Il6ra fl/fl 1fl/fl-Lckcre/+mice (Il6r−/−). BM from Stat5a/bfl/fl-Cd4cre/+ mice was obtained from Dr. John J. O’Shea (National Institutes of Health) All mice were bred in the University of Alabama at Birmingham (UAB) animal facility. All experimental procedures involving animals were approved by the UAB Institutional Animal Care and Use Committee and were performed according to guidelines outlined by the National Research Council.
Infections, BM chimeras and in vivo treatments.
Infections were performed intranasally (i.n) with 6,500 VFU of A/PR8/34 (PR8) influenza virus in 100 μl of PBS or with 500 VFU of PR8-OTII influenza virus in 100 μl of PBS. BM chimeras were generated by irradiating the indicated recipient mice with 950 Rads from an X-ray source delivered in two equal doses administered 4–5 hours apart. Following the second dose of irradiation, mice were intravenously injected with 5 × 106 total BM cells and were allowed to reconstitute for 8–10 weeks before influenza infection. In indicated experiments, experimental animals received an intraperitoneal injection of 50 μg/kg of DT (Sigma) at the indicated time points. In some experiments, mice were intraperitoneally administered recombinant IL-2 (National Cancer Institute) at the indicated time points and doses. In some experiments mice were treated with 250 μg of a mix of anti-IL-6/R (15A7) and anti-IL-6 (MP5–20F3) neutralizing antibodies, both obtained from BioXcell.
Cell purification and adoptive transfer.
CD45.1+CD45.2- and CD45.1+CD45.2+ OTII cells were purified from the spleens of naïve CD45.1+CD45.2- and CD45.1+CD45.2+ OTII mice by positive selection with anti-CD4 beads (Miltenyi Biotec). Purified OTII cells (1× 106) were transferred i.v into either B6 or Il6−/− recipient mice. One day later, the recipient mice were infected with PR8-OTII influenza virus. Three days after infection, CD4+ T cells from the mLN of WT and Il6−/− recipient mice were positively selected with anti-CD4 beads (Miltenyi Biotec). Purified CD4+ T cells were mixed so that the mixture contained a 1:1 ratio of CD45.1+CD45.2- (WT-primed) and CD45.1+CD45.2+ (Il6−/−-primed) OTII cells. Cell numbers were then normalized to the concentration of OTII cells and 2×103 cells of the 1:1 mixture of WT-primed and Il6−/−-primed OTII cells were adoptively transferred into WT and Il6−/− mice that were previously infected with PR8-OTII influenza virus 5 days earlier.
In vitro studies.
Naïve CD4+ cells were purified from spleens of B6 mice using positive selection with anti-CD4 MACS beads (Miltenyi Biotec). Purified cells were activated with anti-CD3 (clone 145–2C11, 2.5μg/mL) and anti-CD28 (clone 37.51, 2.5μg/mL) antibodies in the presence of the indicated concentration of anti-IL-2 neutralizing antibodies (JES6–1A12 and S4B6–1, BioXcell) with or without rIL6 (R&D) at indicated concentrations. Cells were cultured for 48 h at 37 °C in 125 μl in round- bottomed 96-well plates in RPMI-1640 supplemented with sodium pyruvate, HEPES (pH 7.2–7.6 range), nonessential amino acids, penicillin, streptomycin, 2-mercaptoethanol and 10% heat-inactivated FCS (all from Gibco).
ATAC-seq analysis.
OTII (CD45.1+) cells were adoptively transferred into WT (C57Bl/6) and Il6−/− recipient mice. One day later, recipient mice were infected with PR8-OTII. Seven days after infection, 30,000 donor-derived CD45.1+CXCR5hiSLAMloCD4+CD19- TFH and CD45.1+CXCR5loSLAMhi CD4+CD19- TEFF cells were sorted from the mLN of WT and Il6−/− recipient mice using a FACSAria (BD Biosciences) and ATAC-seq was performed as previously described (42). Sorted TFH and TEFF cells were lysed in ice-cold lysis buffer (10mM TrisCl pH 7.4, 10mM NaCl, 3mM MgCl2, 0.1% Igepal). Transposase reactions were performed using Tn5 transposase (Illumina) and were incubated for 30 minutes at 37°C. DNA MinElut Kit (Qiagen) was used for DNA purification. Libraries were amplified using Nextara primers with NEBNext High-Fidelity 2X PCR Master Mix (New England Biolegends), and reactions were purified with the PCR Purification Kit (Qiagen). Libraries were sequenced on a 1×50 bp paired end run on a HiSeq2500 instrument in a rapid run mode at the University of Alabama at Birmingham Heflin Center for Genomic Science.
The raw fastq data files were trimmed to remove primer adapters using Trim Galore! version 0.4.1. Trimmed sequences were aligned to mm9 reference genome from UCSC using Bowtie2 version 2.2.9 with the option ‘-X. Aligned reads were then sorted followed by removal of duplicates using Picard version 2.6.0 SortSam and MarkDuplicates, respectively (Picard: http://broadinstitute.github.io/picard). Peak calling was then performed using Model-based Analysis of ChIP-seq (MACS2) callpeak version 2.1.1.20160309 using these options: ‘-B’, ‘-q 0.05’, ‘--call-summits’, ‘—nomodel’, ‘--nolambda’, and --keep-dup all’. For visualization of the peaks in the UCSC Genome Browser (http://genome.ucsc.edu/), alignment files were normalized using deepTools version 2.3.3 bamCoverage using the options: ‘--normalizeTo1× 2150570000’, and ‘--ignore For Normalization chrX chrM’.
Statistical analysis.
GraphPad Prism software (Version 7) was used for data analysis. The statistical significance of differences in mean values was determined using a two-tailed Student’s t test. P values of less than 0.05 were considered statistically significant.
Supplementary Material
Fig. S1. TFH and GC B cell responses in the absence of IL-6.
Fig. S2. IL-2 production by TFH cells.
Fig. S3. IL-6 signaling fine-tunes IL-2 responsiveness of TFH cells.
Fig. S4. IL-6 signaling prevents CD122 expression.
Fig. S5. STAT3 and STAT5 binding to the Il2rb locus
ACKNOWLEDGEMENTS.
The authors would like to thank W. J. Leonard (US National Institutes of Health) for providing the Il21-mCherry-Il2-emGFP dual reporter transgenic mice and T.S. Simpler, R. Burnham and U. Mudunuru for animal husbandry.
FUNDING.
This work was supported by the University of Alabama at Birmingham (UAB) and National Institutes of Health grants 1R01 AI110480 to A.B.-T, R01 AI116584 to B.L, R01AI061061 to A.W, R56AI127800 and R01AI134972 to K.J.O, and NIAMS Intramural Research Program funds to J.J.O. The X-RAD 320 unit was purchased using a Research Facility Improvement Grant, 1 G20RR022807–01, from the National Center for Research Resources, National Institutes of Health. Support for the UAB flow cytometry core was provided by grants P30 AR048311 and P30 AI027767.
Footnotes
COMPETING INTERESTS
The authors declare that they have no competing interests.
DATA AVAILABILITY.
The data that support the findings of this study are available from the corresponding author upon request. ATAC-seq data are available from GEO under accession code GSE124588. Non-commercially available mice and reagents generated in this study will be available upon request.
SUPPLEMENTARY MATERIALS
Supplementary material and methods
REFERENCES
- 1.Qi H, T follicular helper cells in space-time. Nature reviews. Immunology 16, 612–625 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Vinuesa CG, Linterman MA, Yu D, MacLennan IC, Follicular Helper T Cells. Annual review of immunology 34, 335–368 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, Lao C, Crotty S, ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity 34, 932–946 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goenka R, Barnett LG, Silver JS, O’Neill PJ, Hunter CA, Cancro MP, Laufer TM, Cutting edge: dendritic cell-restricted antigen presentation initiates the follicular helper T cell program but cannot complete ultimate effector differentiation. J Immunol 187, 1091–1095 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deenick EK, Chan A, Ma CS, Gatto D, Schwartzberg PL, Brink R, Tangye SG, Follicular helper T cell differentiation requires continuous antigen presentation that is independent of unique B cell signaling. Immunity 33, 241–253 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballesteros-Tato A, Randall TD, Priming of T follicular helper cells by dendritic cells. Immunology and cell biology 92, 22–27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumjohann D, Okada T, Ansel KM, Cutting Edge: Distinct waves of BCL6 expression during T follicular helper cell development. J Immunol 187, 2089–2092 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Xu H, Li X, Liu D, Li J, Zhang X, Chen X, Hou S, Peng L, Xu C, Liu W, Zhang L, Qi H, Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature 496, 523–527 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Shi J, Hou S, Fang Q, Liu X, Liu X, Qi H, PD-1 Controls Follicular T Helper Cell Positioning and Function. Immunity 49, 264–274 e264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumjohann D, Preite S, Reboldi A, Ronchi F, Ansel KM, Lanzavecchia A, Sallusto F, Persistent antigen and germinal center B cells sustain T follicular helper cell responses and phenotype. Immunity 38, 596–605 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Kerfoot SM, Yaari G, Patel JR, Johnson KL, Gonzalez DG, Kleinstein SH, Haberman AM, Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity 34, 947–960 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nurieva RI, Podd A, Chen Y, Alekseev AM, Yu M, Qi X, Huang H, Wen R, Wang J, Li HS, Watowich SS, Qi H, Dong C, Wang D, STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. The Journal of biological chemistry 287, 11234–11239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S, STAT5 is a potent negative regulator of TFH cell differentiation. The Journal of experimental medicine 209, 243–250 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD, Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity 36, 847–856 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leon B, Bradley JE, Lund FE, Randall TD, Ballesteros-Tato A, FoxP3+ regulatory T cells promote influenza-specific Tfh responses by controlling IL-2 availability. Nature communications 5, 3495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Lu E, Yi T, Cyster JG, EBI2 augments Tfh cell fate by promoting interaction with IL-2-quenching dendritic cells. Nature 533, 110–114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, Ellyard JI, Parish IA, Ma CS, Li QJ, Parish CR, Mackay CR, Vinuesa CG, The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31, 457–468 (2009). [DOI] [PubMed] [Google Scholar]
- 18.DiToro D, Winstead CJ, Pham D, Witte S, Andargachew R, Singer JR, Wilson CG, Zindl CL, Luther RJ, Silberger DJ, Weaver BT, Kolawole EM, Martinez RJ, Turner H, Hatton RD, Moon JJ, Way SS, Evavold BD, Weaver CT, Differential IL-2 expression defines developmental fates of follicular versus nonfollicular helper T cells. Science 361, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malek TR, The biology of interleukin-2. Annual review of immunology 26, 453–479 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Au-Yeung BB, Smith GA, Mueller JL, Heyn CS, Jaszczak RG, Weiss A, Zikherman J, IL-2 Modulates the TCR Signaling Threshold for CD8 but Not CD4 T Cell Proliferation on a Single-Cell Level. J Immunol 198, 2445–2456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Yan X, Zhong B, Nurieva RI, Wang A, Wang X, Martin-Orozco N, Wang Y, Chang SH, Esplugues E, Flavell RA, Tian Q, Dong C, Bcl6 expression specifies the T follicular helper cell program in vivo. The Journal of experimental medicine 209, 1841–1852, S1841–1824 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, Wang YH, Watowich SS, Jetten AM, Tian Q, Dong C, Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity 29, 138–149 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eto D, Lao C, DiToro D, Barnett B, Escobar TC, Kageyama R, Yusuf I, Crotty S, IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PloS one 6, e17739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harker JA, Lewis GM, Mack L, Zuniga EI, Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science 334, 825–829 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poholek AC, Hansen K, Hernandez SG, Eto D, Chandele A, Weinstein JS, Dong X, Odegard JM, Kaech SM, Dent AL, Crotty S, Craft J, In vivo regulation of Bcl6 and T follicular helper cell development. J Immunol 185, 313–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas PG, Brown SA, Yue W, So J, Webby RJ, Doherty PC, An unexpected antibody response to an engineered influenza virus modifies CD8+ T cell responses. Proceedings of the National Academy of Sciences of the United States of America 103, 2764–2769 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fonseca VR, Ribeiro F, Graca L, T follicular regulatory (Tfr) cells: Dissecting the complexity of Tfr-cell compartments. Immunological reviews 288, 112–127 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Olson MR, Verdan FF, Hufford MM, Dent AL, Kaplan MH, STAT3 Impairs STAT5 Activation in the Development of IL-9-Secreting T Cells. J Immunol 196, 3297–3304 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun HW, Wei L, Vahedi G, Kanno Y, O’Shea JJ, Laurence A, Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nature immunology 12, 247–254 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Yu CR, Kim HP, Liao W, Telford WG, Egwuagu CE, Leonard WJ, Key role for IL-21 in experimental autoimmune uveitis. Proceedings of the National Academy of Sciences of the United States of America 108, 9542–9547 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandiyan P, Zheng L, Ishihara S, Reed J, Lenardo MJ, CD4+CD25+Foxp3+ regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4+ T cells. Nature immunology 8, 1353–1362 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Pandiyan P, Conti HR, Zheng L, Peterson AC, Mathern DR, Hernandez-Santos N, Edgerton M, Gaffen SL, Lenardo MJ, CD4(+)CD25(+)Foxp3(+) regulatory T cells promote Th17 cells in vitro and enhance host resistance in mouse Candida albicans Th17 cell infection model. Immunity 34, 422–434 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Haines CJ, Gutcher I, Hochweller K, Blumenschein WM, McClanahan T, Hammerling G, Li MO, Cua DJ, McGeachy MJ, Foxp3(+) regulatory T cells promote T helper 17 cell development in vivo through regulation of interleukin-2. Immunity 34, 409–421 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Botta D, Fuller MJ, Marquez-Lago TT, Bachus H, Bradley JE, Weinmann AS, Zajac AJ, Randall TD, Lund FE, Leon B, Ballesteros-Tato A, Dynamic regulation of T follicular regulatory cell responses by interleukin 2 during influenza infection. Nature immunology 18, 1249–1260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ray JP, Marshall HD, Laidlaw BJ, Staron MM, Kaech SM, Craft J, Transcription factor STAT3 and type I interferons are corepressive insulators for differentiation of follicular helper and T helper 1 cells. Immunity 40, 367–377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi YS, Eto D, Yang JA, Lao C, Crotty S, Cutting edge: STAT1 is required for IL-6-mediated Bcl6 induction for early follicular helper cell differentiation. J Immunol 190, 3049–3053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wing JB, Kitagawa Y, Locci M, Hume H, Tay C, Morita T, Kidani Y, Matsuda K, Inoue T, Kurosaki T, Crotty S, Coban C, Ohkura N, Sakaguchi S, A distinct subpopulation of CD25(−) T-follicular regulatory cells localizes in the germinal centers. Proceedings of the National Academy of Sciences of the United States of America 114, E6400-E6409 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ritvo PG, Churlaud G, Quiniou V, Florez L, Brimaud F, Fourcade G, Mariotti-Ferrandiz E, Klatzmann D, Tfr cells lack IL-2Ralpha but express decoy IL-1R2 and IL-1Ra and suppress the IL-1-dependent activation of Tfh cells. Sci Immunol 2, (2017). [DOI] [PubMed] [Google Scholar]
- 39.Malek TR, Castro I, Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity 33, 153–165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martins GA, Cimmino L, Liao J, Magnusdottir E, Calame K, Blimp-1 directly represses Il2 and the Il2 activator Fos, attenuating T cell proliferation and survival. The Journal of experimental medicine 205, 1959–1965 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong D, Malek TR, Cytokine-dependent Blimp-1 expression in activated T cells inhibits IL-2 production. Journal of immunology 178, 242–252 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ, ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. TFH and GC B cell responses in the absence of IL-6.
Fig. S2. IL-2 production by TFH cells.
Fig. S3. IL-6 signaling fine-tunes IL-2 responsiveness of TFH cells.
Fig. S4. IL-6 signaling prevents CD122 expression.
Fig. S5. STAT3 and STAT5 binding to the Il2rb locus