Abstract
Dynamic turnover of cell-surface glycans is involved in a myriad of biological events, making this process an attractive target for in vivo molecular imaging. Metabolic glycan labeling coupled with ‘bioorthogonal chemistry’ has paved the way for visualizing glycans in living organisms. However, a two-step labeling sequence is required, which is prone to tissue penetration difficulties for the imaging probes. Here, by exploring the substrate promiscuity of endogenous glycosyltransferases, we developed a single-step fluorescent glycan labeling strategy by directly using fluorophore-tagged analogs of the nucleotide sugars. Injecting the fluorophore-tagged sialic acid and fucose into the yolk of zebrafish embryos at the one-cell stage enables systematic imaging of sialylation and fucosylation in live zebrafish embryos at distinct developmental stages. From these studies, we obtained insights into the role of sialylated and fucosylated glycans in zebrafish hematopoiesis.
Keywords: Glycan labeling, Fluorophore-tagged nucleotide sugar, sialylation, fucosylation, hematopoiesis
Graphical Abstract
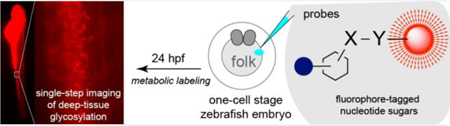
Single-step fluoresecent imaging of live zebrafish glycosylation via metabolic incorporation of fluorophore-tagged nucleotide sugars.
Introduction
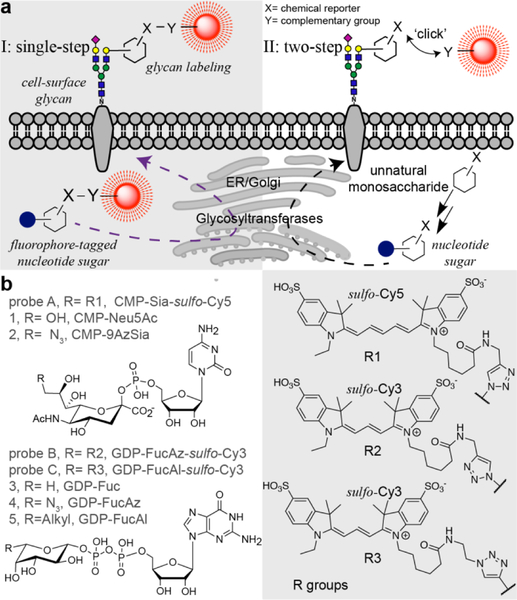
Metabolic oligosaccharide engineering (MOE) coupled with the bioorthogonal chemical reporter strategy has opened an avenue for labeling and visualizing glycans in living organisms.[1–5] In this approach, the cell’s glycan biosynthetic machinery is exploited to install a biorthogonal chemical group onto cell surface glycans, which is then covalently labeled in a secondary step with a complementary probe.[6] However, this strategy has a few intrinsic limitations, such as the biocompatibility of a chosen bioorthogonal reaction and poor deep-tissue penetration of the probes.[7]
Chemoenzymatic glycan editing, however, enables the direct incorporation of a fluorescently labeled monosaccharide without the requirement of a second-step covalent reaction. By using recombinant sialyltransferases (STs) and fucosyltransferases (FTs) with broad donor substrate scopes, sialic acid (Sia) and fucose (Fuc) conjugated with fluorescent dyes can be directly transferred to the cell surface from the corresponding nucleotide sugars.[8–11] Furthermore, recent findings have shown that fluorescently labeled trehalose and N-acetylglucosamine can be directly incorporated into the cell wall of M. tuberculosis[12] and intracellular O-GlcNAcylated proteins in cultured mammalian cells[13], respectively. Inspired by these findings, here, we sought to explore the feasibility of directly incorporating fluorophore-labeled monosaccharides into the cellular glycans of living organisms by exploiting the substrate promiscuity of endogenous glycosyltransferases (Fig. 1a).
Figure 1.
a), Comparison of the single-step fluorescent glycan visualization via metabolic incorporation of unnatural fluorophore-tagged nucleotide sugars versus the conventional, two-step approach. b), Nucleotide sugar analogs functionalized with a chemical reporter (alkyl and azido) or a fluorophore (Cy3 and Cy5) that were used in this study.
Results and Discussion
We assessed the incorporation of fluorophore-labeled monosaccharides in the live zebrafish embryo due to its optical transparency, external fertilization, and amenability to genetic and embryological manipulations.[14–16] First, we synthesized a fluorophore-labeled CMP-N-acetylneuraminic acid (CMP-Neu5NAc). Previous studies have shown that many sialyltransferases (STs) can tolerate large substituents at C9 of Neu5NAc.[17,18] Therefore, we synthesized CMP-Sia-sulfo-Cy5 (probe A) by coupling CMP-9AzSia[19] with Al-sulfo-Cy5 via ligand (BTTP) assisted copper-catalyzed azide-alkyl [3+2] cycloaddition (CuAAC)[20].
To first validate that the Cy5-tagged CMP-Sia analog can be incorporated by sialyltransferases in live cells, we examined the feasibility of transferring Sia-sulfo-Cy5 onto the cell-surface of sialylation-defect Chinese Hamster Ovary (CHO) mutant Lec2, using three recombinant human STs (hST3Gal1, hST3Gal4, hST6Gal1).[21] ST6Gal1/2 and ST3Gal 1/2/3/4/5 are evolutionarily conserved in human and zebrafish genomes and STs in both species share high homology,[22–24] therefore it is reasonable to use the human homologs to examine the donor substrate scope. After incubation with STs and probe A, we detected intense Cy5 fluorescence on Lec2 cells, but not in the cells treated without STs (Fig. s1). Confirming that STs had the capability to incorporate probe A onto cell-surface glycans for the single-step glycan labeling.
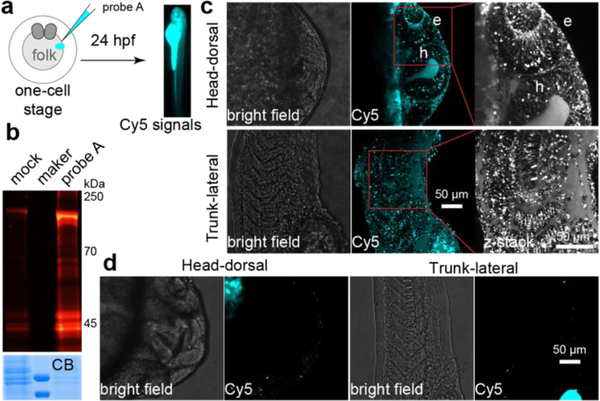
Next, we microinjected probe A into the yolk sack of zebrafish embryos at the one-cell stage,[25] which enables the injected nucleotide sugar to disperse and incorporate into all daughter cells during early zebrafish embryogenesis (Fig. 2a). The injected embryos were subsequently imaged using fluorescent microscope at distinct developmental stages. As shown in the supplemental Fig. s2, overwhelming fluorescent signals in the yolk due to the accumulation of probe A prevented the visualization of the incorporated Sia in other tissue structures during the early embryogenesis. To our delight, the incorporation of probe A in several distinct tissue structures became apparent starting at 14 hours’ post fertilization (hpf), and 28 hpf. Importantly, the probe-dependent Cy5 signal in live embryos is significantly reduced by coinjection of sialylation inhibitor 3FaxNeu5Ac[26] or natural sialylation precursor CMP-Neu5Ac with probe A (Fig. s3), suggesting specific labeling of cell-surface glycans in zebrafish.
Figure 2.
Metabolic incorporation of CMP-Sia-sulfo-Cy5 (probe A) onto sialylated glycan in live zebrafish embryos. a), Workflow of using probe A to fluorescently label sialylated glycan in zebrafish embryos. After a 24-hour incubation, the labeled glycoconjugates in live embryos were directly imaged. b), In-gel analysis of Cy5 fluorescence of labeled glycoproteins (top panel). The same counts of embryos were collected, deyolked, and the resulted lysis was loaded onto SDS-PAGE. The loading was evaluated by Coomassie blue staining (CB). c, d), The confocal images of embryos injected with probe A (c), or prequenched reaction mixture (d, mock control). Intensive Cy5 fluorescence was detected in the eye (e) and hindbrain (h) regions of the head, in the somatic tissue, muscle cells, and intro-hypochord of the trunk.
To confirm that probe A was metabolically incorporated into zebrafish glycoproteins, the lysates of the 24 hpf deyolked embryos were analyzed using an in-gel fluorescence assay. We detected strong A-dependent Cy5 signal in SDS-PAGE resolved proteins with a molecular weight ranging from 30–250 kD (Fig. 2b and s4), which was largely removed by PNGase F-assisted releasing of N-glycans or neuraminidase-directed hydrolyzing Sia from glycoproteins (Fig. s5). Together, these results strongly suggest that CMP-Sia-sulfo-Cy5 is metabolically incorporated in sialylated glycoproteins.
Once we confirmed the specific labeling of probe A into the cell-surface glycoproteins, we proceeded to perform systematic imaging of sialylation in zebrafish tissues at 24 hpf. We observed distinct Cy5 fluorescence in the head and trunk regions (Fig. 2c; movie s1 and s2). Specifically, in the eye and hindbrain regions of the brain, the hypochord in the trunk, and muscle cells in the somites exhibited strong Cy5 fluorescence. Importantly, in the control embryos that were injected with CMP-9AzSia and Al-sulfo-Cy5, only negligible background Cy5 signals was detected (Fig. 2d). These observations are consistent with what previously been reported by Bertozzi and coworkers,[17,18] in which BCNSia was used as the metabolic substrate and the detection was realized by the reaction with a fluorogenic tetrazine probe injected into the posterior caudal vein. In this approach, imaging can only be launched at 30 hpf, which is the earliest time that the caudal vein starts to form. Since the incorporation of unnatural analogs onto glycans could potentially interfere with their biological functions, we further analyzed the morphological defects of zebrafish embryos injected with different concentrations of probe A (2.5, 5 or 10 pmol). However, at 24 hpf we did not observe any apparent morphological defects in any of the injected embryos (Fig. s6), suggesting that probe A do not interfere with normal biological functions.
Next, we expanded this design to directly probe another type of glycosylation known as fucosylation. We prepared a Cy3-tagged GDP-Fuc analog by conjugating the GDP-FucAz[27] with Al-sulfo-Cy3 (Fig. s7b). However, embryos injected with GDP-FucAz-sulfo-Cy3 (probe B) exhibited gross morphological defects including bent tails and twisted notochord. Furthermore, defects increased in severity as concentration of probe B increased (Fig. s8). The same phenotype was also observed in the GDP-FucAz injected group, and in embryos injected with a mixture of GDP-FucAz and Al-sulfo-Cy3. As an alternative, we chose to use the GDP-6-alkylfucose (GDP-FucAl) that has better biocompatibility to prepare a Cy3-tagged analog (GDP-Fuc-sulfo-Cy3, Fig. s7a).[28] To our delight, embryos treated with GDP-Fuc-sulfo-Cy3 (probe C) exhibited no apparent morphological defects compared to the natural GDP-Fuc treated groups.
To image fucosylated glycans in live zebrafish embryos, we injected the sulfo-Cy3-bearing probe B and probe C into the yolk of one-cell stage embryos and performed confocal imaging of the embryos at 24 hpf (movie s3 and s4). Embryos injected with both probes showed intense Cy3 fluorescence in the brain and trunk regions without any observable differences in labeling patterns. By contrast, control group embryos showed no specific labeling except for dye trapped within the yolk region. Importantly, C-dependent Cy3-labeling was suppressed when probe C was coinjected with fucosylation inhibitor 2-fluoro-fucose[26] or the natural donor GDP-Fuc (Fig. s9). In addition, the Cy3-associated fluorescence in embryo-lysate was significantly abolished after PNGase F treatment (Fig. s10). These results strongly suggest that GDP-Fuc-sulfo-Cy3 is metabolically incorporated into zebrafish glycans. Similar to what we observed for recombinant human STs, recombinant human FTs were also capable of adding Cy3-tagged Fuc onto Lec2 glycans by using probe C as a substrate, including hFuT6, hFuT7 and hFuT9. In labeled-Lec2 cells, we imaged robust Cy3 labeling compared to the cells treated without hFuTs (Fig. s11).
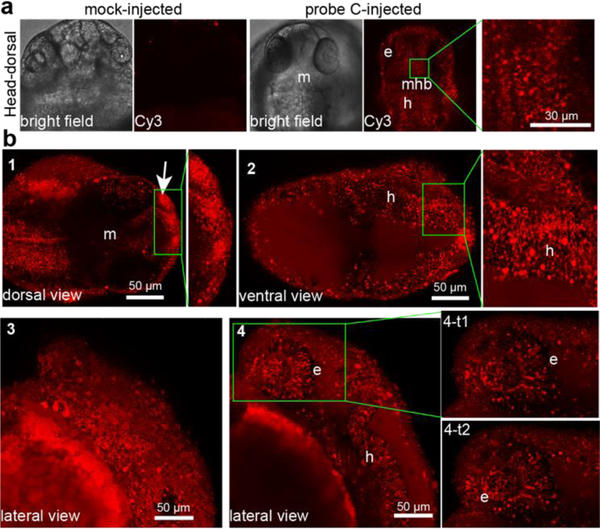
Due to the toxicity of probe B, systematic imaging of fucosylation was conducted with probe C, even though probe C showed weaker incorporation. Confocal images of live zebrafish embryos injected with probe C at 24 hpf revealed a highly-localized distribution of labeled fucosides in the head (lateral, ventral and dorsal views) and trunk regions (lateral view) (Fig. 3 and s 11; movie s5 and s6). Abundant fucosylation was specifically detected within the eye, olfactory placode, midbrain, hindbrain, hypochord, dorsal fin, blood vessels and the caudal hematopoietic tissue (cht). The incorporation appeared more intense in neural structures such as the midbrain and the hindbrain. Interestingly, the midbrain-hindbrain boundary (mhb) did not show any incorporation of the unnatural fucose. Moreover, this strategy also enabled time-lapse tracking of fucosylated glycans in live zebrafish embryos. For example, Cy3-labeled fucoside was found to be abundantly expressed in the retina and the optical cup of eyes, and dynamically moving along the ganglion cells of acetylated microtubules, which are regarded as stable, long-lived microtubules (Fig. 3b 4-t1 and 4-t2; movie s7).
Figure 3.
Systematic visualization of fucosylation in live zebrafish by metabolic incorporation of GDP-FucAl-sulfo-Cy3 (probe C) onto fucosylated glycans. a), confocal images of embryos injected with probe C or prequenched reaction mixture (mock control). In probe C-injected embryos, strong Cy3 fluorescence was detected in the eye (e), midbrain (m) and hindbrain (h) regions, while only negligible background fluorescence was detected in the midbrain-hindbrain boundary (mhb) as shown in the high magnification of the midbrain inset of head-dorsal view. b), Dorsal view (1 and 2) of a head in different depth, the developing mouth showed intensive fucosylation (arrow). Lateral views of head surface (3) and in deep-tissue at different time points (4, 4-t1 and 4-t2).
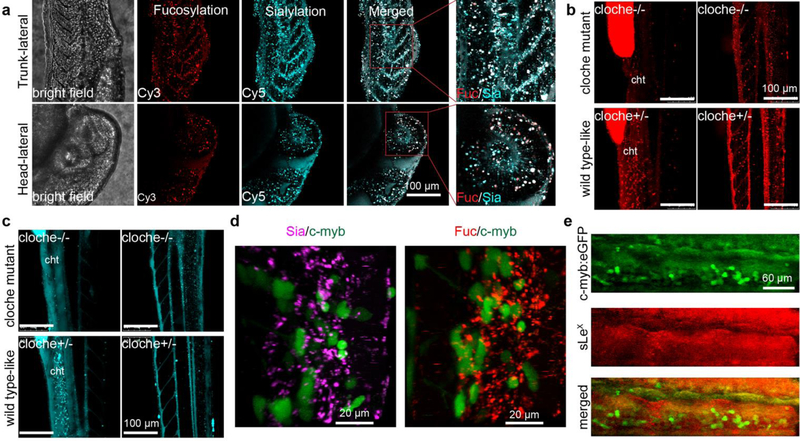
The ability to simultaneously visualize distinct biomolecules in vivo is of essential importance for understanding normal and disease processes. We then evaluated the feasibility to visualize sialylated and fucosylated glycans in parallel. To this end, we co-injected probe A and probe C at a 1:1 molar ratio into the yolk of zebrafish embryos and conducted confocal imaging at different time points (Fig. 4a and s12). As shown in Fig. 4a, sialylated glycans have broader distributions than fucosylated counterparts at 24 hpf embryos. The blood vessel is the region showing the strongest co-localization of both glycans. However, these two types of glycans also exhibit many distinct distribution patterns. For example, sialylation was abundantly detected in the notochord and myotome region, while fucosylation was generally localized in the vascular network. In addition, the labeled fucoside decayed at a much faster rate than the labeled sialosides; by 72 hpf, the Cy3-associated signal disappeared almost completely in the brain; whereas abundant Cy5 signal was still detectable at this time (Fig. s13).
Figure 4.
Simultaneous visualization of the deep-tissue fucosylation (Cy3 in red channel) and sialylation (Cy5 in cyan channel). a), The lateral view of zebrafish embryos co-injected with probe A and probe C at 24 hpf. b, c), The trunk (lateral view of cloche mutant (cloche −/−) or wildtype-like siblings (cloche+/−) at 48 hpf was visualized at different depth. The fluorescence in yolk was an endogenic control for the success of microinjections. d), In the cht region of Tg(c-Myb:eGFP) embryos, HSCs (green channel) do not co-localized with the labeled fucosides and sialosides. e), In Tg(c-Myb:eGFP) embryos, HSCs do not co-localize with cells expressing sLeX (red) in the cht region.
Next, we directly compared the labeling patterns of the single-step glycan labeling to the two-step bioorthogonal chemical reporter strategy previously well-established. We co-injected probe C and GDP-FucAz into the same embryo, and the metabolically incorporated FucAz was fluorescently labeled with Al-sulfo-Cy5 by BTTPS-assisted CuAAC reaction at 24 hpf. In deep tissues such as the cht region, we only observed the Cy3 signal, but not that of Cy5. Likewise, only a very weak Cy5 signal was detected in the trunk and head regions, which is likely due to the poor permeability of labeling reagents into the deep-tissue of live zebrafish embryos. The observation that a much stronger Cy5 signal was detected at the tip of the tail compared to the Cy3 signal is likely caused by the same issue (Fig. s14). Similar results were also observed in probe C and GDP-FucAl coinjected zebrafish embryos (Fig. s15).
To exclude the possibility that the observed labeling patterns were generated by unincorporated probes circling in the blood or diffusing into the tissue matrix, a hematopoietic mutant line known as ‘Cloche’[29] was used to repeat our labeling experiments. In this mutant (cloche−/−), no blood circling system including cht, is formed due to the failure to specify all blood cell types in the developing embryos. We injected embryos from cloche mutant heterozygous (cloche+/−) parents with probe A or C. A distinct labeling pattern was detected in the trunk and head regions of wildtype-like sibling embryos (cloche+/−) that has normal hemato-vascular cell types, as we had previously observed. In the cloche−/− embryos that are easily distinguished by the lack of blood flow after 26 hpf, the loss of the labeling was observed in tissues such as cht due to the loss of vascular development (Fig. 4b, c, movie s8–s11). Based on these observations, we reasoned that the background fluorescence generated by the free, unincorporated probes in the bloodstream is negligible.
The cht region is a transient vascular network formed by the ramification of the caudal vein plexus, provides a necessary niche microenvironment for supporting the development of nascent hematopoietic stem cells (HSCs).[30] The cht is of critical importance for the maturation of these blood cell and immune cell types. To explore the possible biological functions of glycans within this compartment, we imaged probe A and probe C-injected zebrafish in the background of hematopoietic marker Tg(cMyb:eGFP)[31] whose HSCs produced by definitive hematopoiesis can be tracked by the expression of eGFP (Fig. 4d, movie s12, and s13). Definitive hematopoiesis produces multi-potent blood cell types that give rise to multiple lineages through cellular intermediates, and supports the production of all blood cells throughout adulthood. Within the cht region, we observed that the fluorescently labeled sialosides and fucoside were present in cells that are in direct contact with the eGFP-expressing HSCs, but not within the HSCs themselves. Previous studies have reported that sialyl-Lewis X (sLeX, Siaα2–3Galβ1–4(Fucα1–3)-GlcNAc) plays critical roles in mediating leukocyte adhesion and lymphocyte homing,[32,33] we immunostained sLeX epitopes in Tg(c-Myb:eGFP) embryos with an anti-CLA antibody (Fig. 4d). Despite the high fluorescence background, the intensive staining within the cht region was predominantly found in the extracellular matrix, rather than co-localizing with HSCs. This observation strongly suggests that the interactions mediated by sialylated and fucosylated glycans such as sLeX are involved in the hematovascular cell biology in zebrafish.
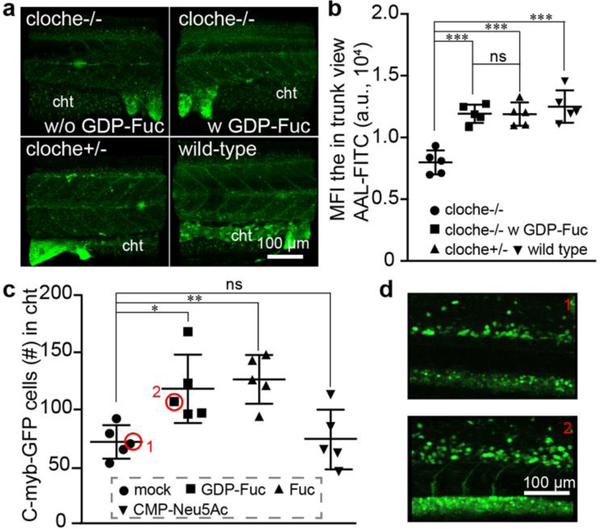
During these imaging studies, we made an intriguing observation: compared to their sibling embryos (cloche+/−), the incorporation of GDP-Fuc based Probe C was significantly decreased in the mutant embryos (cloche−/−); whereas the incorporation of CMP-Sia-based probe A was essentially unchanged (Fig. 4b and 4c). Interestingly, compared to the untreated or the probe A injected groups approximately 5-fold more cloche−/− mutant embryos were still alive at 48 hpf in the probe C-microinjected group (Fig. s16a). These observations prompted us to examine the expression of fucoside biosynthesis enzymes in the cloche mutants. We performed RT-qPCR analysis of gene expression of key components of the GDP-Fuc de novo biosynthesis and salvage pathways (Fig. s17a), and found that the mRNA levels of fucose kinase (FUK) and GDP-Fuc transporter (SLC35C1) were significantly downregulated in the cloche mutants (cloche−/−) (Fig. s17b). Consistent with those changes, the global fucosylation in cloche−/− embryos was significantly down-regulated compared to that of the wildtype embryos or their siblings (cloche+/−) as assessed by fluorescein-conjugated Aleuria Aurantia Lectin (AAL-FITC) staining (AAL is a lectin specific for α1–3- and α1–6-linked fucose) (Fig. 5a). By administering GDP-Fuc into embryos of the cloche mutants (cloche−/−) at the one-cell stage, the global fucosylation level can be largely rescued (Fig. 5b).
Figure 5.
Impairing the global decrease of fucosylation in cloche mutant (cloche−/−) by the administration of the precursor monosaccharide into the yolk of embryos at one-cell stage. a), The 3D confocal images of the fucosylation in the trunk of different embryos that were stained by AAL-FITC. The wild-type, cloche mutant (cloche−/−), wildtype-like sibling (cloche+/−), and GDP-Fuc-injected cloche mutant was collected at 48 hpf and stained with AAL-FITC after fixation and permeabilization. b), The fluorescent intensity in the trunk view of embryos was quantified and presented by mean fluorescence intensity (MFI) via ImageJ. c, d), Confocal imaging-assisted counting of HSCs cells in the cht. Tg(c-Myb:eGFP) embryos at 1–2 cell stage were injected with Fuc, GDP-Fuc, CMP-Neu5Ac, or mock loading buffer, and the HSCs were quantified at 48 hpf. The * represents p<0.05, the ** represents p<0.01, the *** represents p<0.001 and the ns represents not significant in the T-test.
All vertebrates, including zebrafish, has two waves of hematopoiesis[34,35] The earlier of these is known as the primitive wave and the later one as the definitive wave. The primitive wave is responsible for the production of red blood cell types that can facilitate tissue oxygenation as the embryo undergoes rapid growth; whereas definitive or adult hematopoiesis provides the organism with long-term HSCs that are capable of unlimited self-renewal and generating all mature hematopoietic lineages. The cloche mutant (cloche−/−) exhibits serious defects in both waves, but interestingly the absence of vascular cells is partially rescued by 48 hpf.[29]
To try and decipher the role that fucosylation might play during early zebrafish hematopoietic development, we injected Fuc or GDP-Fuc into transgenic zebrafish lines harboring reporters of these two stages of hematopoiesis and followed their development. Although no convincing findings were made using reporter lines of the primitive wave, in the transgenic zebrafish line Tg(cMyb:eGFP) whose definitive blood progenitors can be directly traced by eGFP-expression, Fuc and GDP-Fuc injection distinctively increased the population of eGFP-labeled HSCs in the cht niche region between 48–72 hpf. These changes were not detectable in mock and CMP-Neu5Ac administrated groups (Fig. 5c and 5d). Finally, we also identified significantly larger number of live cloche−/− mutant embryos 48 hpf after the administration of Fuc or GDP-Fuc compared to CMP-Neu5Ac-treated or mock-treated groups (Fig. s16b).
Conclusion
Previous studies by Bertozzi and us demonstrated that unnatural UDP-GalNAc bearing an azide tag and GDP-fucose bearing an azide or alkyne tag can be used by endogenous ppGalNActransferases and fucosyltransferases, respectively, and incorporated into cellular glycoconjugates in zebrafish embryos[27,36] Bioorthogonal chemistry can then be employed for their visualization as early as at the two-cell stage. Via this approach, rapid glycan migration to the cleavage furrow of mitotic cells was observed despite poor tissue penetration of fluorescent probes used for their detection.
In the current study, we have demonstrated that fluorophore-tagged CMP-sialic acid and GDP-fucose (probes A-C) injected into the yolk of zebrafish embryos are incorporated onto cellular glycoconjugates, enabling the direct deep-tissue visualization of sialylated and fucosylated glycans. Using this single-step approach, time lapse, high-resolution, and sensitive confocal imaging of these biologically important glycans in zebrafish embryogenesis is realized. Importantly, this approach enables direct visualization of the dynamic turnover of glycans in live embryos. For example, we observed that in the head region, fucosylatd glycans have a quicker turnover rate than sialylated glycans.[24,37,38]
Studies by Stanley, Haltiwanger, and Taniguchi have shown that knockout of FUT8, POFUT1 or POFUT2 is lethal to mice.[39–42] In addition, congenital mutations of the Golgi localized GDP-fucose transporter SLC35C1 cause leukocyte adhesion deficiency type II, whose manifestation is severe developmental and immune deficiencies.[43] In line with these previous observations, we discovered that cloche mutation leads to down-regulation of fuk and slc35c1, and accordingly reduced cell-surface fucosylation, which can be partially compensated by exogenously introduced Fuc or GDP-Fuc. Interestingly, not only did Fuc or GDP-Fuc injection expand the HSC-population in the cht niche, but this treatment also increased the survival of the cloche mutant significantly. Our findings combined with previous reports[44–47] underscore the critical role of fucosylation in hematopoiesis, especially in the definitive wave, though the exact molecular mechanism for our observed phenotypes is still waiting to be further explored.
Experimental Section
Maintenance of zebrafish embryos:
Adult wild-type AB zebrafish were kept at 28 °C on a 14-h light/10-h dark cycle under standard laboratory conditions. Fertilized embryos were obtained from natural spawning and were maintained in embryo medium (150 mM NaCl, 0.5 mM KCl, 1.0 mM CaCl2, 0.37 mM KH2PO4, 0.05 mM Na2HPO4, 2.0 mM MgSO4, 0.71 mM NaHCO3 in deionized water, pH 7.4). At about 24 hpf, embryos were dechorionated with pronase (1 mg/mL in embryo medium) for releasing larvae from the embryos, rinsed three times with medium, and transferred into embryo medium containing PTU (0.2 mM) to prevent pigment formation. Zebrafish experiments were approved by the UC-San Diego Animal Care and Use Committee.
Confocal imaging of embryos:
For imaging of probe A-C or other control cargo injected embryos, the 1–2 cell stage zebrafish embryos were collected, injected with probes or control cargo into the yolk, and maintained in embryo medium at 28 °C. After dechorionation with pronase at 24 hpf, the zebrafish embryos were transferred into medium containing 0.2 mM PTU. Then, those zebrafish embryos were anesthetized in embryo medium containing 0.2 mM PTU and 2.4 mM tricaine before mounted using 1% low-melting agarose in a glass-bottomed dish and subjected to confocal microscope imaging directly. During the imaging, the embryos were maintained in embryo medium containing 0.2 mM PTU. The confocal images were collected on a Leica TSC SP5 confocal microscope equipped with a Resonant Scanner using Leica Application Suite Advanced Fluorescence 2.7.3.9723. The embryos were imaged under a water immersion objective (HCX IRAPO L N.A.= 0.95). FITC was excited with an Argon 488 nm laser at 20% power, Cy3 was excited with a HeNe 543 nm laser and Cy5 was excited with a HeNe 633 nm laser with power fixed at 20%. The images were 512 × 512 pixels in frame with 32 line averages. The z-step was 0.5 μm, and ten of the images were stacked to generate confocal images in ImageJ (version 1.50i). For the generation of AVI files, the images were taken with z-step of 0.5 μm, stacked and converted into AVI files in ImageJ. The generation of 3D images was performed using IMARIS8 (version 8.1.2). For the metabolic labeling of fucosylation with GDP-FucAz in live embryos, we injected the mixture of GDP-FucAz and probe C into the same yolk. The embryos were collected at 24 hpf, and labeled with BTTPS-assisted CuAAC reaction. In brief, the live embryos were washed with E3 medium and incubated in CuAAC E3 medium containing 100 μM Al-sulfo-Cy5, 50 μM Cu2+ (BTTPS:Cu2+=2:1 premixture) and 2.5 mM ascorbate sodium. After 30 minutes incubation, the BCS was added to quench this reaction, embryos were washed three times with E3 and imaged directly with confocal microscope. The Cy3 and Cy5 fluorescence were directly imaged using the same confocal settings depicted above.
Supplementary Material
Acknowledgements
Financial support was from the NIH (R01GM113046, R01GM111938, and R01GM093282 to P.W., P41GM103390, P01GM107012, and R01GM130915 to K.W.M., R01DK074482 to D.T.).
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- [1].Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR, Science 2008, 320, 664–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Laughlin ST, Bertozzi CR, Proc. Natl. Acad. Sci. U.S.A 2009, 106, 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Laughlin ST, Bertozzi CR, ACS Chem. Biol 2009, 4, 1068–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Xie R, Dong L, Huang R, Hong S, Lei R, Chen X, Angew. Chem. Int. Ed. Engl 2014, 53, 14082–14086. [DOI] [PubMed] [Google Scholar]
- [5].Kang K, Joo S, Choi JY, Geum S, Hong SP, Lee SY, Kim YH, Kim SM, Yoon MH, Nam Y, et al. , Proc. Natl. Acad. Sci. U.S.A 2015, 112, E241–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Prescher JA, Bertozzi CR, Nat. Chem. Biol 2005, 1, 13–21. [DOI] [PubMed] [Google Scholar]
- [7].Bertozzi CR, Acc. Chem. Res 2011, 44, 651–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Capicciotti CJ, Zong C, Sheikh MO, Sun T, Wells L, Boons GJ, J. Am. Chem. Soc 2017, 139, 13342–13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sun T, Yu SH, Zhao P, Meng L, Moremen KW, Wells L, Steet R, Boons G-J, J. Am. Chem. Soc 2016, 138, 11575–11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Li J, Chen M, Liu Z, Zhang L, Felding BH, Moremen KW, Lauvau G, Abadier M, Ley K, Wu P, ACS Cent. Sci 2018, 4, 1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hong S, Shi Y, Wu NC, Grande G, Douthit L, Wang H, Zhou W, Sharpless KB, Wilson IA, Xie J, et al. , Nat. Commun 2019, 10, 1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Backus KM, Boshoff HI, Barry CS, Boutureira O, Patel MK, D’Hooge F, Lee SS, Via LE, Tahlan K, Barry CE, et al. , Nat. Chem. Biol 2011, 7, 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tan HY, Eskandari R, Shen D, Zhu Y, Liu TW, Willems LI, Alteen MG, Madden Z, Vocadlo DJ, J. Am. Chem. Soc 2018, 140, 15300–15308. [DOI] [PubMed] [Google Scholar]
- [14].Zon LI, Peterson RT, Nat. Rev. Drug Discov 2005, 4, 35–44. [DOI] [PubMed] [Google Scholar]
- [15].Lieschke GJ, Currie PD, Nat. Rev. Genet 2007, 8, 353–367. [DOI] [PubMed] [Google Scholar]
- [16].White R, Rose K, Zon L, Nat. Rev. Cancer 2013, 13, 624–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ramya TNC, Weerapana E, Liao L, Zeng Y, Tateno H, Liao L, Yates JR, Cravatt BF, Paulson JC, Mol. Cell Proteomics 2010, 9, 1339–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Agarwal P, Beahm BJ, Shieh P, Bertozzi CR, Angew. Chem. Int. Ed. Engl 2015, 54, 11504–11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Han S, Collins BE, Bengtson P, Paulson JC, Nat. Chem. Biol 2005, 1, 93–97. [DOI] [PubMed] [Google Scholar]
- [20].Wang W, Hong S, Tran A, Jiang H, Triano R, Liu Y, Chen X, Wu P, Chem. Asian J 2011, 6, 2796–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].North SJ, Huang HH, Sundaram S, Jang-Lee J, Etienne AT, Trollope A, Chalabi S, Dell A, Stanley P, Haslam SM, J. Biol. Chem 2010, 285, 5759–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Petit D, Teppa E, Mir AM, Vicogne D, Thisse C, Thisse B, Filloux C, Harduin-Lepers A, Mol. Biol. Evol 2015, 32, 906–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Teppa RE, Petit D, Plechakova O, Cogez V, Harduin-Lepers A, Int. J. Mol. Sci 2016, 17, 1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chang LY, Teppa E, Noel M, Gilormini PA, Decloquement M, Lion C, Biot C, Mir AM, Cogez V, Delannoy P, et al. , Int. J. Mol. Sci 2019, 20, 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kimmel CB, Law RD, Dev. Biol 1985, 108, 78–85. [DOI] [PubMed] [Google Scholar]
- [26].Rillahan CD, Antonopoulos A, Lefort CT, Sonon R, Azadi P, Ley K, Dell A, Haslam SM, Paulson JC, Nat. Chem. Biol 2012, 8, 661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dehnert KW, Beahm BJ, Huynh TT, Baskin JM, Laughlin ST, Wang W, Wu P, Amacher SL, Bertozzi CR, ACS Chem. Biol 2011, 6, 547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].del Amo DS, Wang W, Jiang H, Besanceney C, Yan AC, Levy M, Liu Y, Marlow FL, Wu P, J. Am. Chem. Soc 2010, 132, 16893–16899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Reischauer S, Stone OA, Villasenor A, Chi N, Jin SW, Martin M, Lee MT, Fukuda N, Marass M, Witty A, et al. , Nature 2016, 535, 294–298. [DOI] [PubMed] [Google Scholar]
- [30].Gore AV, Pillay LM, Venero Galanternik M, Weinstein BM, Wiley Interdiscip. Rev. Dev. Biol 2018, 7, e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DYR, Traver D, Nature 2010, 464, 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Foxall C, Watson SR, Dowbenko D, Fennie C, Lasky LA, Kiso M, Hasegawa A, Asa D, Brandley BK, J. Cell Biol 1992, 117, 895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Springer TA, Cell 1994, 76, 301–314. [DOI] [PubMed] [Google Scholar]
- [34].Davidson AJ, Zon LI, Oncogene 2004, 23, 7233–7246. [DOI] [PubMed] [Google Scholar]
- [35].Clements WK, Traver D, Nat. Rev. Immunol 2013, 13, 336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Baskin JM, Dehnert KW, Laughlin ST, Amacher SL, Bertozzi CR, Proc. Natl. Acad. Sci. U.S.A 2010, 107, 10360–10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bardor M, Nguyen DH, Diaz S, Varki A, J. Biol. Chem 2005, 280, 4228–4237. [DOI] [PubMed] [Google Scholar]
- [38].Gilormini P-A, Lion C, Vicogne D, Guérardel Y, Foulquier F, Biot C, J. Inherit. Metab. Dis 2018, 41, 515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shi S, Stanley P, Proc. Natl. Acad. Sci. U.S.A 2003, 100, 5234–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Luo Y, Koles K, Vorndam W, Haltiwanger RS, Panin VM, J. Biol. Chem 2006, 281, 9393–9399. [DOI] [PubMed] [Google Scholar]
- [41].Wang X, Inoue S, Gu J, Miyoshi E, Noda K, Li W, Mizuno-Horikawa Y, Nakano M, Asahi M, Takahashi M, et al. , Proc. Natl. Acad. Sci. U.S.A 2005, 102, 15791–15796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Schneider M, Al-Shareffi E, Haltiwanger RS, Glycobiology 2017, 27, 601–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hellbusch CC, Sperandio M, Frommhold D, Yakubenia S, Wild MK, Popovici D, Vestweber D, Gröne H-J, von Figura K, Lübke T, et al. , J. Biol. Chem 2007, 282, 10762–10772. [DOI] [PubMed] [Google Scholar]
- [44].Zhou L, Li LW, Yan Q, Petryniak B, Man Y, Su C, Shim J, Chervin S, Lowe JB, Blood 2008, 112, 308–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Xia L, McDaniel JM, Yago T, Doeden A, McEver RP, Blood 2004, 104, 3091–3096. [DOI] [PubMed] [Google Scholar]
- [46].Bigas A, Robert-Moreno A, Espinosa L, Int. J. Dev. Biol 2010, 54, 1175–1188. [DOI] [PubMed] [Google Scholar]
- [47].Myers J, Huang Y, Wei L, Yan Q, Huang A, Zhou L, Transfusion 2010, 50, 2660–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.