Abstract
PURPOSE:
Defects in genes in the DNA-repair pathways significantly contribute to prostate cancer (PC) progression. We hypothesize that overexpression of DNA repair genes may also drive poorer outcomes in PC. The ribonucleotide reductase small subunit M2 (RRM2) is essential for DNA synthesis and DNA repair by producing dNTPs. It is frequently overexpressed in cancers, but there is very little known about its function in PC.
EXPERIMENTAL DESIGN:
The oncogenic activity of RRM2 in PC cells was assessed by inhibiting or overexpressing RRM2. The molecular mechanisms of RRM2 function were determined. The clinical significance of RRM2 overexpression was evaluated in 11 PC clinical cohorts. The efficacy of an RRM2 inhibitor (COH29) was assessed in vitro and in vivo. Last, the mechanism underlying the transcriptional activation of RRM2 in PC tissue and cells was determined.
RESULTS:
Knockdown of RRM2 inhibited its oncogenic function, while overexpression of RRM2 promoted epithelial mesenchymal transition in PC cells. The prognostic value of RRM2 RNA levels in PC was confirmed in 11 clinical cohorts. Integrating the transcriptomic and phospho-proteomic changes induced by RRM2 unraveled multiple oncogenic pathways downstream of RRM2. Targeting RRM2 with COH29 showed excellent efficacy. Thirteen putative RRM2-targeting transcription factors were bioinformatically identified, and FOXM1 was validated to transcriptionally activate RRM2 in PC.
CONCLUSIONS:
We propose that increased expression of RRM2 is a mechanism driving poor patient outcomes in PC and that its inhibition may be of significant therapeutic value.
Keywords: DNA damage, RRM2, FOXM1, COH29, prostate cancer
INTRODUCTION
Prostate cancer (PC) is the third leading cause of cancer death in American men. Advances in genomics allow the identification of putative drivers of cancer initiation and progression. Genomic instability results from DNA damage induced by genotoxic insults, dysfunction in pathways governing DNA repair, and abnormal cell-cycle control (1, 2). DNA-repair pathway defects are prevalent in advanced PC (3, 4). Defective DNA repair is correlated with an increased sensitivity to PARP inhibitors, platinum chemotherapy, or immunotherapy that can be exploited for clinical benefit (5, 6). Ironically, overexpression of DNA repair genes may also contribute to cancer progression: PC patients with BRCA1-expressing tumors more likely develop lethal cancer than those with BRCA1-non-expressing tumors (7).
The nucleotide metabolism enzyme, ribonucleotide reductase (RNR), is essential for DNA synthesis and DNA repair by producing dNTP. Abnormal dNTP levels lead to infidelity of DNA replication, causing an increase in genomic instability (8). RNR activity is coordinated with cell cycle progression to maintain the fine balance between dNTP production and DNA replication (9). During the cell cycle, levels of two RNR subunits, RRM1 and RRM2B, are constant but RRM2 levels fluctuate (10). As the rate-limiting RNR enzyme, RRM2 levels control the cell cycle-dependent activity of RNR. Overexpression of RRM2 is associated with poorer outcomes in multiple cancers (11, 12). In mice, overexpression of RRM2 induces the development of lung cancer. Defects in DNA mismatch repair synergistically promote RNR-induced carcinogenesis in lung neoplasms from RRM2 transgenic mice (13).
Although the oncogenic role of RRM2 has been linked to promotion of epithelial mesenchymal transition (EMT) and angiogenesis (14, 15), there has been no comprehensive molecular understanding of how RRM2 regulates key downstream biological processes and signaling to control tumor initiation and progression. In particular, very little is known about the function of RRM2 in PC. Here, we uncovered the oncogenic properties of RRM2 by integrating transcriptomic with phospho-proteomic analysis, and we determined the significant prognostic value of RRM2 in PC. Further, we elucidated the transcriptional regulation of RRM2 by FOXM1 in PC cells.
MATERIALS AND METHODS
Clinical cohort summary and characterization
Characteristics of PC patients in the Physicians’ Health Study (PHS) and Health Professionals Follow-up Study (HPFS) cohorts are shown in table S1 (16, 17). mRNA expression profiling from high-density tumor areas and adjacent normal prostate tissue was reported (18). Logistic regression was used to quantify the association of RRM2 expression (in quartiles) and lethal cancer (table S2). The analysis was adjusted for age at cancer diagnosis (linear), calendar year of diagnosis, body mass index, current smoking at diagnosis (binary), and Gleason grade. An additional eleven publicly-available PC cohorts are summarized in table S3.
Cell culture and PC tissue microarray
Normal human prostate cell lines (PWR-E1, PZ-HPV-7, RWPE-1) and human PC cell lines (LNCaP, 22Rv1, DU145, and PC-3) were purchased from ATCC (Manassas, VA). C4–2 cells were obtained from VitroMed (Burlington, NC). E006AA cells were provided by John T. Isaacs (The Johns Hopkins University School of Medicine, Baltimore, MD) and the LAPC-4 cell line was provided by Charles Sawyers (Memorial Sloan Kettering Cancer Center [MSK], New York, NY). Normal prostate cell lines were cultured in Keratinocyte Serum-Free Medium (K-SFM) (Kit Cat Number: 17005–042; Thermo Fisher Scientific). All other cell lines were maintained in 10% FBS supplemented with 2 mM of L-glutamine and antibiotic at 37°C in 5% CO2. Cells were authenticated by human short tandem repeat profiling at the MSK Integrated Genomics Operation Core Facility. The PC tissue microarray was purchased from US Biomax, Inc. (Cat. No. T195c).
Gene silencing and gene expression
SMARTpool siRNA were obtained from Dharmacon and transfected with RNAiMAX (Invitrogen). Cells were harvested 48 hours or 72 hours after transfection for protein and mRNA analysis. Lentiviral vectors encoding RRM2 were purchased from GeneCopoeia and transfected with psPAX2 packaging and pMD2.G envelope plasmid to HEK293FT cells for 2 days using Lipofectamine 3000 (Invitrogen). Thereafter, PC cells were infected with viral supernatants in the presence of 8 μg/ml polybrene. Stable cells were generated using puromycin selection. A list of siRNAs and constructs is provided in table S4. Efficiency of knockdown and overexpression was verified by qPCR and western blot.
RNA analysis, RNA-Seq and immunoblotting
Total RNA was extracted from cells and analyzed as previously described (19). TaqMan gene expression assays (Life Technology) were used for relative gene expression (table S5) by qRT-PCR. Transcript levels were normalized to levels of GAPDH transcript. RNA sequencing was performed by 50 million 2×50bp reads in the MSK Integrated Genomics Operation Core. RNA sequencing data were analysed by Partek Inc. (St. Louis, MO). The data are available from GEO (GSE117921-GSE117924). Proteins were extracted by RIPA buffer and protein concentration was determined by the Bradford method. Equal amounts of protein were loaded and resolved by SDS-PAGE, then transferred to polyvinylidene difluoride membranes for immunoblotting. All antibodies used are listed in table S4.
Phospho-kinase arrays (R&D Systems) were applied according to the manufacturer’s instructions. Briefly, protein was extracted from cells treated with siRNAs or inhibitors. Changes in kinase activity were visualized by chemiluminescence on autoradiography film and measured using ImageJ software.
Cell viability, cell cycle, apoptosis, and DNA damage analysis
Cells were treated with siRNAs or inhibitors. Cell viability was assessed using CellTiter-Glo luminescent cell viability assay (Promega). At 72 hours post-transfection, cell cycle and apoptosis were detected using the Muse cell cycle assay kit and the Muse Annexin V and dead cell kit (EMD Millipore). DNA damage was detected at 48 hours post-transfection of siRNAs or at 24 hours treatment of inhibitors in cells by using Muse multi-color DNA damage kit (EMD Millipore).
Soft agar assays, wound healing, and invasion assays
Soft agar assays were performed in 6-well tissue culture plates by placing cells (2–5 × 104/well) in 2 ml of 0.3% soft-agar above a 2-ml layer of 0.5% agar. After two weeks’ incubation, cells were stained with 1 mg/ml MTT in medium for 1 hour. Colonies were detected and counted using GelCount technology (Oxford Optronix Inc., Oxford, UK).
Cells were seeded in 6-well cell culture plates for 48 hours. A scratch was made with a pipette tip in a confluent area. Triplicates were prepared in each condition. Photographs of each scratch were taken at 24 hours after scratching.
Matrigel invasion assays were performed in Matrigel invasion chamber (Fisher Scientific) with cells on the top of chambers in serum-free media. 10% FBS in the lower chamber was used as chemo-attractant. After indicated times, cells in the bottom chamber were fixed in methanol and stained with crystal violet, photographed, and counted under phase-contrast microscopy.
Chromatin immunoprecipitations (ChIP)-Seq, ChIP-qPCR, and luciferase assays
Radical prostatectomy tissue was prepared and ChIP-Seq performed as described previously (20) with 6-ug antibodies to AR and H3K27Ac antibody. DNA sequencing libraries were prepared using the ThruPLEX-FD Prep Kit (Rubicon Genomics, Ann Arbor, MI). Libraries were sequenced using 50-base pair reads on the Illumina platform (Illumina, San Diego, CA) at Dana-Farber Cancer Institute. Related data analysis for ChIP-Seq and peak calling followed the same protocol as reported. The quantitation of signals on the 1-MB region of enhancer of RRM2 was performed from 4 normal prostate tissue samples and 4 tumor samples. The data are available from GEO (GSE118845).
ChIP-qPCR was performed using the ChIP-IT High Sensitivity Kit (Active Motif, #53040) with ChIP-grade H3K4me3, FOXM1 antibodies and native IgG, according to the manufacturer’s instructions. DNA was analyzed via qPCR. All ChIP experiments were completed with at least two biological replicates.
For luciferase reporter assays, siRNAs were transfected in cells for 24 hours and 500 ng of RRM2 promoter reporters (GeneCopoeia) containing 0kb or 3kb promoter sequence of human RRM2 were cotransfected with Lipofectamine 2000 (Invitrogen). At 24 hours post-transfection, cells were collected for luciferase assays according to Promega’s instructions.
dNTP pool assay
This assay was performed as previously described (21). Cells were lysed with 60% cold methanol and the dry pellet was resuspended in water. Two microliters of sample were used in the HIV-1 RT based primer extension assay. 5’ 32P-endlabeled primer (“P”; 5’-GTCCCTCTTCGGGCGCCA-3’) was individually annealed to one of four different templates (3’-CAGGGAGAAGCCCGCGGTN-5’). The template:primer complex was extended by HIV-1 RT to generate one additional nucleotide extension product (“P+1”) for one of four dNTPs contained in the dNTP samples extracted from the cells. In this assay, the molar amount of the P+1 product is equal to that of each dNTP contained in the extracted samples, which allowed us to calculate and compare the amounts of the cellular dNTPs for the different treatments. The dNTP amounts were normalized to 1 × 106 cells for the comparisons.
Xenograft studies
NOG-SCID mice were implanted subcutaneously with C4–2 cells. After palpable tumors developed (typically 100 mm3), six mice per group received either vehicle or COH29 (200 mg/kg) by oral gavage twice a day for 3 weeks. Tumors were measured twice a week using calipers. Tissues were collected for immunohistochemistry (IHC) staining and protein assays. Multiple proteins were assessed by IHC using antibodies of H2A.x (1:1000), MYC (1:100), RRM2 (1:2500), cleaved-caspase3 (1:300), prostate-specific antigen (PSA) (1:2000) (table S4). All animal care was in accordance with the guidelines of the Institutional Animal Care and Use Committee at MSK.
Bioinformatic analysis of clinical cohorts
Data for various clinical cohorts were obtained from cBioPortal for Cancer Genomics (22) and Oncomine (23). Heatmaps and volcano plots were generated using Rv3.4.3 (https://www.R-project.org). Pathway analysis was performed using GSEA (24) and ToppGene (25).
Statistical analysis
Results are reported as mean ± S.E. Comparisons between groups were performed using an unpaired two-sided Student’s t test or Wilcoxon test (p<0.05 was considered significant). Disease-free survival was examined for time since diagnosis. Cox proportional hazard regression was performed adjusting for clinical and demographic factors.
RESULTS
RRM2 functions as an oncogene in PC cells
RRM2 is highly expressed in most PC cell lines tested, regardless of androgen receptor (AR) status (fig. S1A). Four PC cell lines (LNCaP, C4–2, 22Rv1, and E006AA) have relatively high RRM2 protein levels, while PC-3 cells have lower RRM2 protein expression (fig. S1A). It has been reported that RRM2 is barely expressed in normal prostate tissue (26). However, in three widely used normal prostate cell lines, RRM2 was expressed at a high level similar to that in LNCaP cells (fig. S1B). We speculated that those immortalized prostate cell lines may not reflect the real gene profiling of normal prostate cells. We further detected RRM2 protein expression by IHC staining in a PC tissue microarray which included 9 cases of PC adenocarcinoma and 1 leiomyosarcoma, plus 2 tissue samples from normal prostates. Compared to the two normal prostates that had very low levels of RRM2, 5 of the 9 PC tissue samples showed high expression of RRM2 (fig. S1C), indicating the potential oncogenic role of RRM2 in PC.
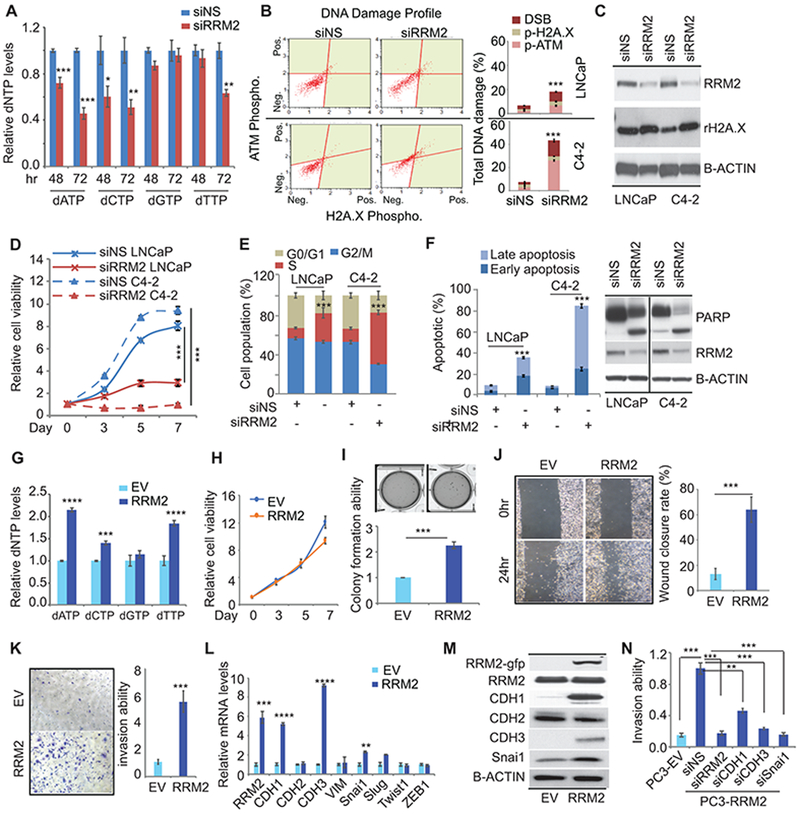
dNTP production (dATP, dCTP and dTTP) was significantly inhibited by knockdown of RRM2 (siRRM2) (Fig. 1A). In both LNCaP and C4–2 cells, siRRM2 induced DNA damage with significant activation of DNA damage markers, including upstream ATM and downstream H2A.X phosphorylation (Fig. 1B, 1C). Furthermore, siRRM2 led to cell growth inhibition, S phase arrest, and apoptosis (Fig. 1D, 1E, and 1F). Similar phenotypes induced by siRRM2 were also detected in 22Rv1 cells, which express AR-V7, and E006AA cells, which do not express AR (fig. S1D-S1H). Therefore, the tumor suppressive effects of RRM2 inhibition are not related to AR status and are not cell line specific.
Figure 1: The oncogenic roles of RRM2 in prostate cancer.
(A) dNTP production in small interfering RNA (siRNA)-transfected cells. dNTP was detected at both 48 hours and 72 hours post-transfection of siRNAs in C4-2 cells. siNS, nonspecific siRNA. (B) siRRM2-induced DNA damage. DNA damage marker activation was monitored in LNCaP and C4-2 cells using Muse multi-color DNA damage kit. (C) Activation of H2A.X was confirmed by immunoblotting. (D) and (E) Analysis of cell proliferation (D) and cell cycle (E) in transfected cells. (F) Apoptosis detected by Annexin V assays and immunoblots. (G) dNTP production in empty vector (EV)/RRM2-overexpressing PC-3 cells (PC3-EV; PC3-RRM2). (H) Cell proliferation in stable cells. (I) soft agar assays of stable PC-3 cells. The colony numbers were normalized to those in the control cells. (J) Wound healing assays after the scratch done for 24 hours. (K) Invasion assays after cells were plated for 48 hours. (L) and (M) EMT marker expression detected by qPCR (L) and immunoblots (M) in both EV- and RRM2-expressing PC-3 cells. (N) Invasion assays after multiple siRNAs were transfected in PC3-RRM2 cells. Figure 1 values represent the mean ± S.E. of three independent experiments. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001 vs control groups treated with empty vector (EV) or with nonspecific (siNS) siRNA.
To further understand the oncogenic functions of RRM2, we established RRM2-expressing stable LNCaP and PC-3 cell lines. Overexpression of RRM2 significantly increased dNTP production while knockdown of RRM2 decreased dNTP (Fig. 1G). Surprisingly, increased RRM2 expression did not affect 2D cell growth (Fig. 1H), but promoted anchorage-independent 3D colony formation in RRM2-overexpressing PC-3 cells (PC3-RRM2) (Fig. 1I). LNCaP-RRM2 cells showed both increasing 2D cell growth and 3D colony formation (fig. S2A, S2B). PC3-RRM2 cells had a 6-fold higher wound healing rate and level of invasiveness than control PC-3 cells (Fig. 1J, 1K), indicating that RRM2 induced EMT. Mesenchymal markers including N-cadherin (CDH2) and vimentin (VIM) were not affected by increased RRM2 expression, while the EMT markers (SNAI1 and SLUG) were up-regulated in PC3-RRM2 cells (Fig. 1L). Intriguingly, P-cadherin (CDH3), a cell-to-cell adhesion molecule, was up-regulated 9-fold in PC3-RRM2 cells, while E-cadherin (CDH1) expression was increased about 5-fold (Fig. 1L, 1M). CDH3 has been observed in the development and progression of multiple cancers including PC (27). CDH3 and CDH1 co-regulate collective migration in breast cancer (28). Therefore, RRM2 may promote collective migration by upregulating key regulators such as CDH1, CDH3, and SNAI1. This hypothesis was supported by showing that knockdown of RRM2 and each of these EMT regulators significantly attenuated RRM2-induced invasiveness in PC3-RRM2 cells (Fig. 1N, fig. S2F). In LNCaP-RRM2 cells, 2.4-fold increased invasiveness was detected, coupled with a 5-fold upregulation of CDH2 and CDH3 (fig. S2C-S2E), indicating RRM2 may activate specific mediators of EMT or collective migration markers during different disease states. Taken together, this provides strong evidence that RRM2 has oncogenic properties in vitro.
RRM2 overexpression is prognostically significant in PC patients
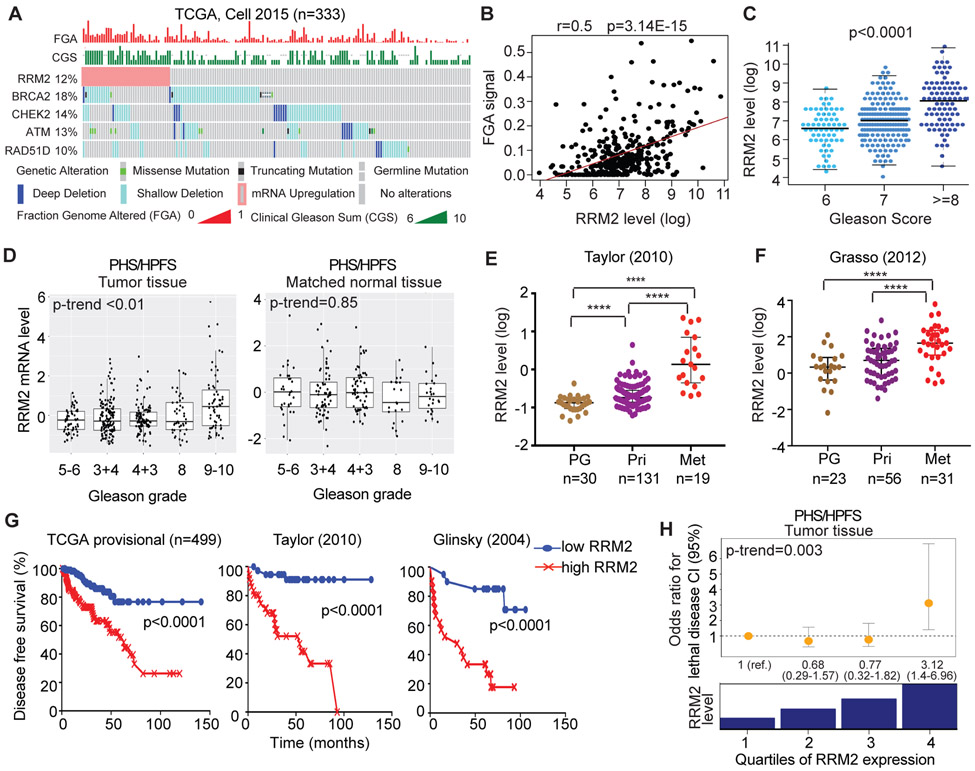
Overexpression of RRM2 leads to increased genomic instability (29), contributing to cancer progression and resistance to anti-cancer treatments (30). The copy number variation measured by fraction of the genome altered (FGA) has been shown to be associated with Gleason score and the development of metastases in PC patients (3, 31). Here, for the first time, the positive correlation between RRM2 level and overall FGA was observed in both the Cancer Genome Atlas (TCGA) cohort (3) (r=0.5, p=3.14E-15) (Fig. 2A, 2B) and Taylor cohort (32) (r=0.5, p=1.7E-07) (fig. S3C). In the two advanced PC cohorts (SU2C/PCF and Kumar cohorts) (4, 33), weaker but significant correlations were seen (fig. S3E, S3F). Correspondingly, a significant positive correlation between RRM2 level and Gleason score was observed in TCGA (Fig. 2C) and Taylor cohort (fig. S3D). Consistent with other studies (3), alteration of DNA repair genes (BRCA2, CHEK2, ATM, and RAD51D) in the TCGA cohort was significantly correlated with high FGA (Fig. 2A and fig. S3A). Higher RRM2 levels were significantly associated with heterozygous loss of CHEK2 and ATM (fig. S3B), suggesting their contribution to the higher FGA and Gleason score in PC.
Figure 2: Overexpression of RRM2 positively associates with poorer clinical outcomes in multiple prostate cancer clinical cohorts.
(A)-(C) The correlation of gene alteration with the fraction of genome altered (FGA) and Gleason grades in TCGA cohort was visualized in (A). The statistical quantitation was shown in (B) and (C). (D) The correlation of RRM2 level with Gleason grade in tumor and matched normal tissues in the PHS/HPFS cohorts. (E) and (F) The correlation of RRM2 expression with tumor progression in Taylor and Grasso cohorts. RRM2 levels were analyzed in prostate gland (PG), primary (Pri), and metastasis (Met) tissue samples. (G) The association of RRM2 expression and the disease-free survival in the TCGA, Taylor, and Glinsky cohorts. (H) The correlation of RRM2 with the risk of lethal prostate cancer over long-term follow-up, independent from clinical characteristics and Gleason grade, in the combined HPFS and PHS prostate cancer cohorts. The odd ratios for lethal disease were adjusted for Gleason score. ****, p<0.0001 vs comparator groups.
Besides publicly available clinical cohorts, we further validated the prognostic significance of overexpression of RRM2 within the HPFS (n=254) and the PHS (n=150), two cohorts with long-term follow-up for fatal outcomes. Significantly higher levels of RRM2 were observed with increasing Gleason score (p-trend <0.01) (Fig. 2D, left panel) in tumor, but not in matched normal tissue (p-trend=0.85) (Fig. 2D, right panel). In both Taylor and Grasso cohorts (32, 34), the levels of RRM2 were significantly higher in metastatic tissue than in primary and normal tissue (Fig. 2E, 2F). In the TCGA, Taylor, and Glinsky cohorts (3, 32, 35), RRM2 levels in the highest quartile were significantly associated with an increased rate of biochemical recurrence (Fig. 2G).
Importantly, in HPFS/PHS cohorts, high RRM2 expression was significantly associated with increased risk of lethal disease (table S2). When combining the HPFS and PHS cohorts, RRM2 RNA levels in the highest quartile, compared to the lowest quartile, were associated with a 4.0 times higher risk of lethal disease (95% CI, 2.1 to 7.7) (table S2). When adjusting for patient and tumor characteristics including centrally reviewed Gleason grade, this association remained strong (odds ratio, 3.1, 95% CI, 1.4 to 7.0; table S2 and Fig. 2H). There was no association with lethal PC for RRM2 in matched normal tissues (fig. S3G). Altogether, these results suggest that increased expression of RRM2 is associated with adverse biology and poor outcomes.
Transcriptomic changes induced by regulation of RRM2 unraveled the clinically relevant RRM2 signature in PC
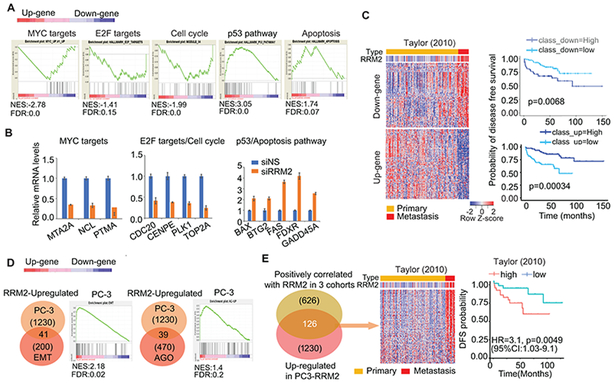
Although the oncogenic role of RRM2 has been reported in several cancer types, an understanding of the comprehensive molecular mechanisms by which it functions is lacking. We detected global transcriptomic changes induced by siRRM2 (fig. S4A). The gene ontology (GO) analysis revealed that inhibition of RRM2 repressed genes enriched in some key biological processes (e.g., cell proliferation, cell death, protein metabolic process, and cell cycle) and oncogenic pathways (e.g., MYC pathway, PLK pathway, WNT signaling, Aurora B signaling, and FOXM1 pathway) (fig. S4B). The gene set enrichment analysis (GSEA) further defined the highly enriched pathways (MYC, E2F, Cell cycle, p53 signaling, apoptosis) (Fig. 3A). Targets of MYC, Cell cycle, and E2F pathways were significantly inhibited in siRRM2-treated cells (Fig. 3B). Five common genes in the p53 and apoptosis gene set pathways were validated to be upregulated by siRRM2 treatment (Fig. 3B).
Figure 3: Transcriptomic analysis unraveled the molecular mechanism of RRM2 function.
(A) Gene set enrichment analysis (GSEA) of transcriptomic changes. The histograms showed the distribution of select top GSEA molecular signatures: MYC targets (MYC_up_V1_up gene set); E2F targets (hallmark_E2F_targets gene set), cell cycle (Module_54 gene set), p53 pathway (hallmark_p53_pathway), and apoptosis (hallmark apoptosis). Up-gene (up-regulated genes); Down-gene (down-regulated genes). (B) Multiple targets of pathways were validated by quantitative reverse transcription PCR (qRT-PCR). (C) siRRM2-regulated gene profiling and the correlation of these genes with disease-free survival (DFS) in Taylor cohort. (D) Significant enrichment in EMT and Angiogenesis gene sets in RRM2-overexpressing PC-3 cells. (E) RRM2-regulated 126-gene profiling and correlation with the clinical outcome in clinical cohorts. 126 genes were revealed by overlapping 1230 up-regulated genes in PC3-RRM2 cells with 627 common genes positively correlated with RRM2 overexpression in three prostate cancer cohorts (TCGA, Kumar, and SU2C/PCF). The correlation of these genes with disease-free survival was analyzed in the Taylor cohort.
To evaluate the clinical relevance of inhibition of RRM2, we first looked at siRRM2-down-regulated genes that were positively correlated with RRM2 in the Taylor cohort (named as “Down-gene”), as well as up-regulated genes which negatively correlated with RRM2 expression level (named as “Up-gene”) (Fig. 3C). The heat maps showed that the Down-gene set was activated in metastatic patients with high RRM2 expression (Fig. 3C, left panel). These genes were significantly correlated with disease-free survival (Fig. 3C, right panel). The findings were validated in five other PC clinical cohorts, supporting the clinical relevance of the genes regulated by inhibition of RRM2 (fig. S4C).
To fully understand the oncogenic role of RRM2 in PC, we evaluated the transcriptomic changes in PC-3-RRM2 cells (fig. S4D). GSEA analysis showed significant enrichment in EMT and angiogenesis (AGO) gene sets (Fig. 3D) (36, 37), which strongly supported the phenotypes induced by RRM2 overexpression (Fig. 1I-1N). The up-regulated gene sets induced by RRM2 overexpression may contribute to its oncogenic properties in PC. Following integration of RRM2-induced genes in cell lines and genes, which are positively correlated with RRM2 up-regulation in three clinical cohorts, we identified the key 126 RRM2-regulated genes relevant to clinical outcome (Fig. 3E). These 126 genes were up-regulated in metastatic patients in two additional clinical cohorts (fig. S4E).
Integration of two data sets from siRRM2 RNA-Seq in C4–2 cells and RRM2-expressing RNA-Seq in PC-3 cells provided an in-depth understanding of the RRM2-regulated molecular mechanisms. Fifty-eight common genes were significantly enriched in oncogenic pathways (FOXM1, PLK1, MYC, and mTORC1 signaling) and in glycolysis and cell cycle–related hallmark gene sets (fig. S4F).
RRM2 inhibitor COH29 induced molecular mechanisms of tumor suppression
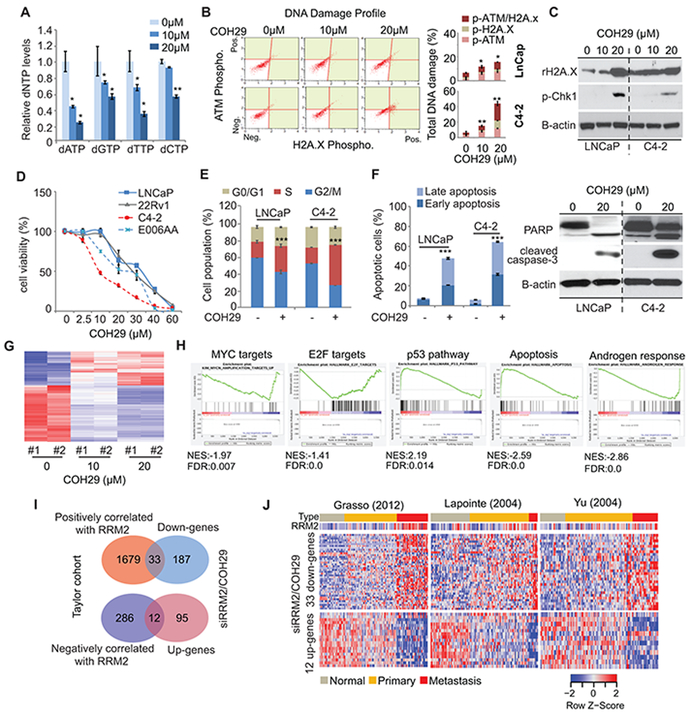
Given the potential oncogenic role of RRM2, multiple RNR inhibitors have been developed for clinical application. The recent novel RNR inhibitor COH29 was designed to target a ligand pocket of RRM2 and showed low side effects (38, 39). COH29 is currently in a phase I trial to treat multiple solid tumors. Here, we assessed the effects of COH29 on PC cells. COH29 inhibited dNTP production due to inhibition of RRM2 activity in multiple cancer cell lines. Similarly, dose-dependent inhibition of dNTP production by COH29 was detected in C4–2 cells (Fig. 4A). Similar to siRRM2, COH29 led to DNA damage (Fig. 4B, 4C); strong cell growth inhibition in multiple PC cell lines (Fig. 4D); S-phase arrest (Fig. 4E, fig. S5A); and apoptosis (Fig. 4F, fig. S5B).
Figure 4: The tumor suppressive role of COH29 (RRM2 inhibitor).
(A) dNTP production in COH29-treated C4-2 cells. Two doses of COH29 were used in C4-2 cells, and dNTP production was detected after 24 hours of treatment. (B) and (C) Activation of DNA damage markers. (D) Cell proliferation assay. (E) Cell cycle analysis after 48 hours of COH29 treatment. (F) COH29-induced apoptosis. (G) The global mRNA changes induced by COH29 in C4-2 cells, after 48 hours of treatment. (H) GSEA analysis of mRNA profiling in 20 μM of COH29-treated cells. The histograms showed the distribution of select top GSEA molecular signatures. Up-gene (COH29-induced up-regulated genes); Down-gene (COH29-induced down-regulated genes). NES, normalized enrichment score; FDR, false discovery rate. (I) and (J) Identification of targeted genes by inhibition of RRM2. Genes affected by inhibition of RRM2 in cells were overlapped with genes correlated with RRM2 overexpression in Taylor cohort to reveal 33 down-regulated genes and 12 up-regulated genes (I). These gene panels were validated in three additional prostate cancer cohorts (J). The Figure 4 values represent the mean ± S.E. of three independent experiments. *, p<0.05; **, p<0.01; ***, p<0.001 vs control groups.
The COH29-induced global transcriptomic changes are dose-dependent (Fig. 4G). More than 98% of the altered gene expression in high (20 μM) and low (10 μM) doses of COH29 overlapped (fig. S5C, S5D); therefore, only the RNA-Seq dataset of high-dose COH29 was applied for the sequential GO analysis. GSEA analysis of the mRNA changes induced by COH29 exhibited similar gene set enrichment as was seen with siRRM2 treatment, including MYC targets, E2F targets, the p53 pathway, and the apoptosis pathway (Fig. 4H). Additionally, COH29 inhibited the androgen response gene set (Fig. 4H). Compared to the siRNA approach, COH29 induced wider transcriptomic changes (fig. S5E). We applied GO analysis by using genes whose expression were similarly altered (219 down-regulated genes; 106 up-regulated genes) with COH29 and siRRM2 treatment. The top five inhibited pathways were PLK, MYC activation, FOXM1, Aurora B, and telomerase pathways, while p53 downstream pathway and MYC repressive pathway were activated with both approaches to inhibit RRM2 activity (fig. S5F). The major inhibited biological processes included cell cycle, RNA processing, response to DNA damage, and chromosome organization–related processes (fig. S5G). Intriguingly, besides apoptosis, DNA damage, and cell proliferation, the processes for signal transduction, regulation of kinase activity, and inhibition of phosphorylation were activated by inhibition of RRM2 (fig. S5G).
There are 33 genes that were inhibited by siRRM2/COH29 and positively correlated with RRM2 up-regulation in the Taylor cohort; 12 genes were up-regulated with inhibition of RRM2, but negatively correlated with RRM2 up-regulation (Fig. 4I). This RRM2-regulated gene profile was validated in three clinical cohorts (Fig. 4J). The 33 down-regulated genes were activated in metastatic samples, while the 12 up-regulated genes were down-regulated in metastatic samples (Fig. 4J). RRM2 showed oncogenic activity in colorectal and breast cancer progression (13, 14). Therefore, we applied the 33 down-regulated genes in the Gaedcke colorectal cohort and Curtis breast cancer cohort (40, 41). The expression of the 33 genes are significantly up-regulated in aggressive tumor in both cohorts (fig. S5H, S5I). These results suggested that this gene panel can be targeted by inhibition of RRM2 in multiple cancer types in which RRM2 drives aggressiveness.
Phospho-proteomic changes are induced by modulating RRM2 expression in PC cells
The GO analysis of genes upon inhibition of RRM2 revealed that signal transduction and kinase activity processes were affected (fig. S5G). Therefore, we applied the phospho-kinase array in siRRM2- and COH29-treated C4–2 cells (Fig. 5A, fig. S6A). The array detected the phosphorylation of 43 human kinases, which are involved in several key oncogenic signaling processes. Both treatments significantly inhibited AKT/mTOR signaling, leading to the downstream effectors inhibition. Besides AKT/mTOR signaling, both treatments inhibited multiple components in STAT signaling and SFK signaling (Fig. 5A, 5B, fig. S6A, S6B). The inhibition of these signaling pathways induced by COH29 was dose dependent (Fig. 5A, 5B). These results were confirmed by western blotting (Fig. 5C). The activation of DNA damage markers was detected when the activity of these kinases was inhibited. Together, multiple oncogenic signaling pathways were repressed by inhibiting RRM2.
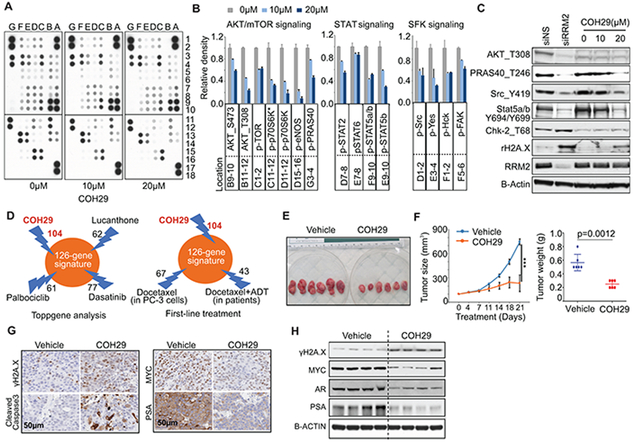
Figure 5: Phospho-proteomic changes induced by inhibition of RRM2 and COH29 efficacy in vivo.
(A)-(B) Phospho-kinase array analysis after 24-hour COH29 treatment in C4-2 cells. The whole-cell lysates were collected for human phospho-kinase array analysis. Each membrane contains kinase-specific antibodies (number indicated). Relative phosphorylation of spots was quantified by Image J software and the value of vehicle (0 μM) was set up as “1” (B). (C) Validation of phospho-kinase array by immunoblots. For siRNA-treated groups, cell lysates were collected after transfection for 48 hours. The representative blots for each condition are shown and the values represent the mean ± S.E. of two independent experiments. (D) Druggable RRM2 signatures. Top three drugs from ToppGene analysis and COH29 are shown (left panel). Numbers of genes down-regulated by docetaxel (in PC-3 cells) and docetaxel + ADT (in patients) are shown (right panel). (E)-(F) Antitumor effects of COH29 in vivo. Tumor volumes and weights were measured following oral administration of COH29 (200 mg/kg) in established C4-2 xenograft tumors (n=6 per group). Values are means ± S.E. ***P<0.001 versus vehicle mice. (G)-(H) Regulation of key genes by COH29 in vivo. Multiple genes regulated by COH29 were assessed in xenograft tumors by immunohistochemistry staining (G) or immunoblotting (H).
RRM2 inhibitor COH29 has antitumor effect in vivo
Since the 126 genes induced by RRM2 are highly correlated with clinical outcome, targeting them may provide therapeutic benefit (Fig. 3E). We applied ToppGene analysis by using the 126-gene signature (25). The top 3 drugs that target the signature are lucanthone, palbociclib, and dasatinib. The numbers of genes for each drug potentially targeting the signature are shown (Fig. 5D). We also evaluated whether docetaxel as the first-line treatment of PC affects these genes by using two profiling datasets of docetaxel treatment in PC-3 cells (GSE83654) and docetaxel plus androgen deprivation therapy (ADT) in patients (42). Of 126 genes, 67 (from docetaxel-treated PC-3 cells) and 43 genes (from docetaxel + ADT-treated patients) were down-regulated (Fig. 5D, right panel). Compared to the other drugs mentioned, COH29 inhibited the highest number of genes in the signature (104 of 126 genes) (Fig. 5D), due to its RRM2-specific inhibition that was previously reported (39).
We further assessed the antitumor activity of COH29 in vivo. After 3 weeks of treatment, COH29-treated tumor bearing mice had significantly lower tumor volume (231 mm3) and tumor weight (0.27 g) than the vehicle group (744 mm3 and 0.57 g) (Fig. 5E, 5F), without significant difference in body weight of the mice (fig. S6C). The COH29-treated xenografts showed increased H2A.X phosphorylation and cleaved caspase-3, and decreased MYC and PSA (Fig. 5G, 5H). We further detected significant inhibition of AR mRNA and protein expression in COH29-treated tumors (Fig. 5H and fig. S6D). These results are consistent with the repression of androgen response genes by COH29 in vitro (Fig. 4H). Interestingly, we did not observe inhibition of AR by siRRM2 treatment (data not shown), indicating COH29-inhibited AR may not be directly associated with inhibition of RRM2. Together, COH29 has antitumor effects in vivo in a human PC xenograft model.
RRM2 is transcriptionally activated by FOXM1 in PC
Several studies have focused on transcriptional activation and delayed degradation of RRM2 in cancers (29, 43-45). Amplification and mutation of RRM2 is rare in PC. Transcriptional regulation may largely contribute to the overexpression of RRM2 in PC. We applied H3K27Ac ChIP-Seq in prostate normal and tumor tissues. Significantly higher H3K27Ac binding signal was detected at the enhancer of RRM2 (1Mb region) in PC (n=4, p<0.05), compared to normal prostate tissue (n=4) (Fig. 6A), indicating more activated RRM2 transcription in PC than in normal tissue.
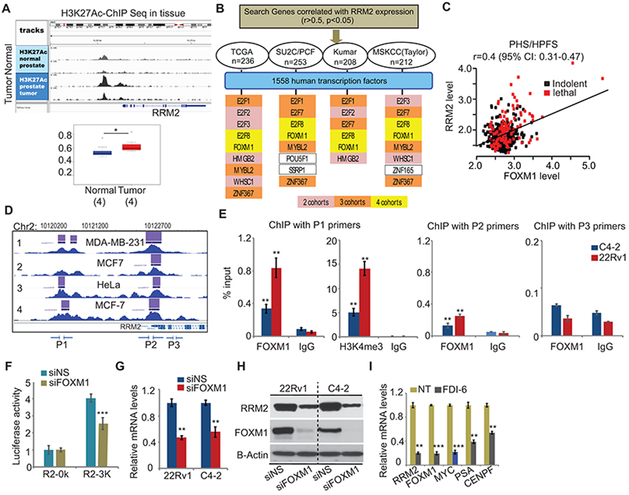
Figure 6: Transcriptional activation of RRM2.
(A) H3K27Ac ChIP-Seq in tissues. The binding signal on RRM2 enhancer (1Mb region) was quantified. (B) The strategy to identify RRM2-targeting transcription factors (TFs). Human TFs were selected in the genes positively correlated with RRM2 expression in prostate cancer cohorts. Yellow/orange/pink bars indicate that TFs appear in four/three/two cohorts. (C) The correlation of FOXM1 and RRM2 in PHS/HPFS cohorts. (D) FOXM1 binding on RRM2 promoter in cancer cells. The overview of multiple ChIP-Seq datasets were extracted from Cistrome Data browser. P1-P3 primers were designed for FOXM1 ChIP-PCR. (E) FOXM1 or H3K4me3-ChIP-PCR on RRM2 promoter. (F) RRM2 promoter activity regulated by FOXM1. The reporters without (R2-0K) or with (R2-3K) 3kb RRM2 promoter sequence were transfected in siRNA-treated 22Rv1 cells. (G) and (H) Inhibition of RRM2 expression by siFOXM1 in 22Rv1 and C4-2 cells. (I) Inhibition of FOXM1 targets by FDI-6 (20 μM) in 22Rv1 cells. The Figure 6 values represent the mean ± S.E. of three independent experiments. *, p<0.05; **, p<0.01; ***, p<0.001 vs control groups.
A few transcription factors (TFs) have been identified that target RRM2 in different cancers (e.g., E2F1, E2F3 and MYCN) (43-45), but there are no studies of transcriptional regulation of RRM2 in PC cells. Here, we developed a strategy that combines PC clinical cohorts and TF databases to seek potential TFs targeting RRM2. We searched genes that positively correlated with RRM2 expression (r>0.5, p<0.05) in four widely used clinical cohorts (Fig. 6B). Among all the candidates, 13 genes are TFs. Interestingly, E2F1 and E2F3 have been demonstrated to target RRM2 in colorectal cancer and KB cells (43, 44).
A correlation matrix from the TCGA (primary tumor samples) and SU2C/PCF (metastatic samples) cohorts not only showed the relationships between RRM2 and all potential RRM2-targeting TFs but also showed the correlations among the TFs (fig. S7A). It revealed different correlation patterns for the TCGA and SU2C/PCF cohorts, indicating that disease state may contribute to the specificity of RRM2-targeting TFs in PC. The subset of RRM2-targeting TFs which correlate with significant clinical outcome should be more meaningful and could facilitate the development of future therapeutic strategies. Among 13 TFs, both FOXM1 and E2F8 were shared in all the cohorts (Fig. 6B). FOXM1 has been reported to be one of the major drivers in PC (46). Besides those four cohorts (Fig. 6B), the positive correlation between FOXM1 and RRM2 was also observed in PHS/HPFS cohorts (Fig. 6C). Similar to RRM2, FOXM1 level is highly correlated with disease-free survival in the Taylor cohort (fig. S7B). For both FOXM1 and RRM2, higher levels are associated with lethal disease (p<0.0001) (fig. S7C).
To assess the transcription control of RRM2 by FOXM1, we first identified FOXM1 binding region to the RRM2 promoter from published FOXM1 ChIP-Seq. In breast cancer cells and HeLa cells, there were two major FOXM1 binding peaks in the promoter of RRM2 (Fig. 6D). Although no FOXM1 ChIP-Seq data in PC cells is available, ChIP-Seq of transcriptional activation marks (including H3K4me3, H3K27Ac, and POLR2A) in multiple PC cell lines provided supporting evidence that the FOXM1-binding regions of RRM2 promoter are activated (fig. S7D). Two pairs of primers (P1 and P2) were designed to evaluate FOXM1 binding in PC cells, based on the ChIP-Seq data from other cells. A negative control primer pair (P3) located in non-FOXM1 binding region was used (Fig. 6D). ChIP-qPCR in C4–2 and 22Rv1 cells showed stronger FOXM1 binding signals in the distal P1 region of the promoter than in the proximal P2 region, supported by significant H3K4me3 binding in the same region (Fig. 6E). More significant FOXM1 binding in 22Rv1 than in C4–2 could be due to higher expression of FOXM1 in 22Rv1 (Fig. 6H). As a positive control, we assessed FOXM1 binding at the CENPF (identified FOXM1 target) promoter in the same ChIP-qPCR. Strong FOXM1 binding at the CENPF promoter was detected in both cell lines (fig. S7E, S7F).
The activity of the RRM2 promoter reporter was reduced 40% in siFOXM1-treated 22Rv1 cells (Fig. 6F), due to 80% reduction of FOXM1 expression by siFOXM1 (fig. S7G). Consistently, siFOXM1 led to approximately 50% reduction of RRM2 mRNA and protein expression (Fig. 6G, 6H). Besides siRNAs, a small molecule inhibitor of FOXM1 (FDI-6) repressed multiple FOXM1 targets, including RRM2, MYC, PSA, and CENPF (Fig. 6I). Altogether, FOXM1 can transcriptionally activate RRM2 expression by directly binding to the promoter.
DISCUSSION
The well-established function of RRM2 is to maintain the balance of dNTP pools for DNA synthesis and DNA repair. However, there has been no comprehensive mechanistic investigation that defines the downstream biological consequences and signaling pathways of RRM2 in cancer, specifically in PC. This study showed for the first time the oncogenic properties of RRM2 in PC. We unraveled and validated the significant prognostic value of RRM2 in 11 PC clinical cohorts. Integrative analysis of transcriptomic and phospho-proteomic changes regulated by RRM2 revealed oncogenic mechanisms of RRM2 in PC.
Abnormal RRM2 degradation induced imbalance of the dNTP pool and genome instability (29). In our analysis, we found that overexpressed RRM2 is highly correlated with deletion of CHEK2 or ATM, which may lead to higher FGA and Gleason score (fig. S3B). Overexpression of RRM2 could be required to compensate for the DNA repair deficiency induced by deletion of DNA repair genes (e.g., CHEK2 or ATM). However, the correlation of RRM2 with FGA and clinical Gleason sum (CGS) is not completely dependent on the deletion of both genes, since some tumors showed high RRM2 expression without deletion of CHEK2 and ATM (Fig. 2A).
Similar to what was reported in breast cancer (14), overexpression of RRM2 in PC cells promoted 3D colony formation and invasion. Unexpectedly, RRM2 activated atypical EMT progression by up-regulating E-cadherin (CDH1) and P-cadherin (CDH3). High CDH1 expressed PC-3 cells showed more aggressive phenotypes (47). Similarly, moderate CDH1 levels present in 4T1 breast cancer cells could promote cancer metastasis by increasing the collective migration (48). The essential role of CDH3 in collective migration could promote tumorigenesis in prostate, colon, ovary, and breast cancers (49). Co-expression of CDH1 and CDH3 leads to the aggressive biology behavior in breast cancer cells due to interruption of the interaction between CDH1 and catenins (48). Therefore, up-regulation of CDH1 and CDH3 by RRM2 overexpression could be one of the driving forces of the more aggressive phenotype in PC-3-RRM2 cells. Furthermore, for the first time, we demonstrated that several transcription factor networks (FOXM1, MYC, APC/C/CDH1, E2F targets) and oncogenic pathways (PLK1, mTORC1, Aurora B, Rho GTPases signaling) are regulated by RRM2. All these TFs and signaling pathways have been reported to contribute to PC progression.
Besides exploring RRM2 function, we uncovered the regulatory mechanisms of RRM2 overexpression in PC. The fact that more transcriptional activation of RRM2 was observed in tumor tissue than in normal prostate tissue led us to develop a bioinformatic strategy to identify the RRM2-targeting TFs. Multiple ChIP-Seq in PC cells showed that the FOXM1 binding region of RRM2 promoter is highly transcriptionally activated. Thus, the cooperation of FOXM1 and other TFs regulating RRM2 is a potential avenue for future study. FOXM1 was reported as a master regulator of the aggressive PCS1 subtype tumors (50). In our study, we identified FOXM1 as the key regulator of RRM2, and inhibition of RRM2 effectively repressed the FOXM1 pathway (Fig. S4F, S5F), suggesting the feedback loop between FOXM1 and RRM2. We believe that interruption of this regulatory feedback loop may specifically target aggressive subtype tumors.
In summary, our study integrated the data from cell lines with clinical cohorts to reveal that overexpression of RRM2 is associated with poor outcomes and is potentially a driver of aggressive PC. Use of an RRM2-targeted inhibitor does inhibit RRM2 oncogenic activity in vivo. Furthermore, we defined the underlying molecular mechanisms and determined the transcriptional activation of RRM2 by FOXM1.
Supplementary Material
Translational Relevance.
Despite the recognized oncogenic activity of ribonucleotide reductase small subunit M2 (RRM2) in multiple cancers, the lack of knowledge of its function in prostate cancer limits the therapeutic application of its inhibitors. Here, we defined the mechanisms of the oncogenic function of RRM2 and unraveled its prognostic value in prostate cancer. Moreover, we identified FOXM1 as the driver of RRM2 overexpression in prostate cancer. Importantly, we elucidated the effectiveness and molecular mechanisms of an RRM2 inhibitor, COH29. Overall, our study not only reveals RRM2 as a prognostic marker for advanced prostate cancer, but also provides support for prostate cancer clinical trials targeting RRM2.
Acknowledgments
Financial support: This research was funded in part through the NIH/NCI Cancer Center Support Grant to Memorial Sloan Kettering Cancer Center (P30 CA008748) and dNTP assays were funded in part through NIH/NIGMS R01 GM104198 to B. Kim and NIH/NIAID R01 AI136581 to B. Kim.
Disclosure of potential conflicts of interest: This research was funded in part through the NIH/NCI Cancer Center Support Grant to Memorial Sloan Kettering (P30 CA008748). As of February 18, 2019, P.W. Kantoff reports the following disclosures for the last 36-month period: he has investment interest in Context Therapeutics LLC, DRGT, Placon, Seer Biosciences, and Tarveda Therapeutics; he is a company board member for Context Therapeutics LLC; he is a consultant/scientific advisory board member for BIND Biosciences, Inc., Bavarian Nordic Immunotherapeutics, DRGT, GE Healthcare, Janssen, Metamark, New England Research Institutes, Inc., OncoCellMDX, Progenity, Sanofi, Seer Biosciences, Tarveda Therapeutics, and Thermo Fisher; and he serves on data safety monitoring boards for Genentech/Roche and Merck. J. Armenia is currently an employee of AstraZeneca, but was not when he performed this work. All other authors declare no potential conflicts of interest.
REFERENCES
- 1.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, et al. Punctuated evolution of prostate cancer genomes. Cell 2013;153:666–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature 2011;470:214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 2015;163:1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beer TM, Kwon ED, Drake CG, Fizazi K, Logothetis C, Gravis G, et al. Randomized, double-blind, phase III trial of ipilimumab versus placebo in asymptomatic or minimally symptomatic patients with metastatic chemotherapy-naive castration-resistant prostate cancer. J Clin Oncol 2017;35:40–7. [DOI] [PubMed] [Google Scholar]
- 6.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917–21. [DOI] [PubMed] [Google Scholar]
- 7.Fiorentino M, Judson G, Penney K, Flavin R, Stark J, Fiore C, et al. Immunohistochemical expression of BRCA1 and lethal prostate cancer. Cancer Res 2010;70:3136–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar D, Abdulovic AL, Viberg J, Nilsson AK, Kunkel TA, Chabes A. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res 2011;39:1360–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aye Y, Li M, Long MJ, Weiss RS. Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene 2015;34:2011–21. [DOI] [PubMed] [Google Scholar]
- 10.Chabes A, Thelander L. Controlled protein degradation regulates ribonucleotide reductase activity in proliferating mammalian cells during the normal cell cycle and in response to DNA damage and replication blocks. J Biol Chem 2000;275:17747–53. [DOI] [PubMed] [Google Scholar]
- 11.Grade M, Hummon AB, Camps J, Emons G, Spitzner M, Gaedcke J, et al. A genomic strategy for the functional validation of colorectal cancer genes identifies potential therapeutic targets. Int J Cancer 2011;128:1069–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kretschmer C, Sterner-Kock A, Siedentopf F, Schoenegg W, Schlag PM, Kemmner W. Identification of early molecular markers for breast cancer. Mol Cancer 2011;10:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu X, Page JL, Surtees JA, Liu H, Lagedrost S, Lu Y, et al. Broad overexpression of ribonucleotide reductase genes in mice specifically induces lung neoplasms. Cancer Res 2008;68:2652–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah KN, Wilson EA, Malla R, Elford HL, Faridi JS. Targeting ribonucleotide reductase M2 and NF-kappaB activation with didox to circumvent tamoxifen resistance in breast cancer. Mol Cancer Ther 2015;14:2411–21. [DOI] [PubMed] [Google Scholar]
- 15.Zhang K, Hu S, Wu J, Chen L, Lu J, Wang X, et al. Overexpression of RRM2 decreases thrombspondin-1 and increases VEGF production in human cancer cells in vitro and in vivo: implication of RRM2 in angiogenesis. Mol Cancer 2009;8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giovannucci E, Liu Y, Platz EA, Stampfer MJ, Willett WC. Risk factors for prostate cancer incidence and progression in the Health Professionals Follow-Up Study. Int J Cancer 2007;121:1571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steering Committee of the Physicians’ Health Study Research Group. Final report on the aspirin component of the ongoing Physicians’ Health Study. N Engl J Med 1989;321:129–35. [DOI] [PubMed] [Google Scholar]
- 18.Penney KL, Sinnott JA, Tyekucheva S, Gerke T, Shui IM, Kraft P, et al. Association of prostate cancer risk variants with gene expression in normal and tumor tissue. Cancer Epidemiol Biomarkers Prev 2015;24:255–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Xie RL, Croce CM, Stein JL, Lian JB, van Wijnen AJ, et al. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A 2011;108:9863–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet 2015;47:1346–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollenbaugh JA, Tao S, Lenzi GM, Ryu S, Kim DH, Diaz-Griffero F, et al. dNTP pool modulation dynamics by SAMHD1 protein in monocyte-derived macrophages. Retrovirology 2014;11:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 2009;37:W305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Liu X, Wang YH, Yeh SD, Chen CL, Nelson RA, et al. The prognostic value of ribonucleotide reductase small subunit M2 in predicting recurrence for prostate cancers. Urologic oncology 2014;32:51.e9–19. [DOI] [PubMed] [Google Scholar]
- 27.Patel IS, Madan P, Getsios S, Bertrand MA, MacCalman CD. Cadherin switching in ovarian cancer progression. Int J Cancer 2003;106:172–7. [DOI] [PubMed] [Google Scholar]
- 28.Ribeiro AS, Sousa B, Carreto L, Mendes N, Nobre AR, Ricardo S, et al. P-cadherin functional role is dependent on E-cadherin cellular context: a proof of concept using the breast cancer model. J Pathol 2013;229:705–18. [DOI] [PubMed] [Google Scholar]
- 29.D’Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell 2012;149:1023–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature 2013;501:355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hieronymus H, Schultz N, Gopalan A, Carver BS, Chang MT, Xiao Y, et al. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci U S A 2014;111:11139–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer cell 2010;18:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 2016;22:369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487:239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest 2004;113:913–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Groger CJ, Grubinger M, Waldhor T, Vierlinger K, Mikulits W. Meta-analysis of gene expression signatures defining the epithelial to mesenchymal transition during cancer progression. PLoS One 2012;7:e51136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masiero M, Simoes FC, Han HD, Snell C, Peterkin T, Bridges E, et al. A core human primary tumor angiogenesis signature identifies the endothelial orphan receptor ELTD1 as a key regulator of angiogenesis. Cancer cell 2013;24:229–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen MC, Zhou B, Zhang K, Yuan YC, Un F, Hu S, et al. The novel ribonucleotide reductase inhibitor COH29 inhibits DNA repair in vitro. Mol Pharmacol 2015;87:996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou B, Su L, Hu S, Hu W, Yip ML, Wu J, et al. A small-molecule blocking ribonucleotide reductase holoenzyme formation inhibits cancer cell growth and overcomes drug resistance. Cancer Res 2013;73:6484–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaedcke J, Grade M, Jung K, Camps J, Jo P, Emons G, et al. Mutated KRAS results in overexpression of DUSP4, a MAP-kinase phosphatase, and SMYD3, a histone methyltransferase, in rectal carcinomas. Genes Chromosomes Cancer 2010;49:1024–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajan P, Stockley J, Sudbery IM, Fleming JT, Hedley A, Kalna G, et al. Identification of a candidate prognostic gene signature by transcriptome analysis of matched pre- and post-treatment prostatic biopsies from patients with advanced prostate cancer. BMC Cancer 2014;14:977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang Z, Gong C, Liu H, Zhang X, Mei L, Song M, et al. E2F1 promote the aggressiveness of human colorectal cancer by activating the ribonucleotide reductase small subunit M2. Biochem Biophys Res Commun 2015;464:407–15. [DOI] [PubMed] [Google Scholar]
- 44.Gong C, Liu H, Song R, Zhong T, Lou M, Wang T, et al. ATR-CHK1-E2F3 signaling transactivates human ribonucleotide reductase small subunit M2 for DNA repair induced by the chemical carcinogen MNNG. Biochim Biophys Acta 2016;1859:612–26. [DOI] [PubMed] [Google Scholar]
- 45.Valentijn LJ, Koster J, Haneveld F, Aissa RA, van Sluis P, Broekmans ME, et al. Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc Natl Acad Sci U S A 2012;109:19190–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aytes A, Mitrofanova A, Lefebvre C, Alvarez MJ, Castillo-Martin M, Zheng T, et al. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer cell 2014;25:638–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Celia-Terrassa T, Meca-Cortes O, Mateo F, Martinez de Paz A, Rubio N, Arnal-Estape A, et al. Epithelial-mesenchymal transition can suppress major attributes of human epithelial tumor-initiating cells. J Clin Invest 2012;122:1849–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elisha Y, Kalchenko V, Kuznetsov Y, Geiger B. Dual role of E-cadherin in the regulation of invasive collective migration of mammary carcinoma cells. Sci Rep 2018;8:4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vieira AF, Paredes J. P-cadherin and the journey to cancer metastasis. Mol Cancer 2015;14:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ketola K, Munuganti RSN, Davies A, Nip KM, Bishop JL, Zoubeidi A. Targeting prostate cancer subtype 1 by Forkhead box M1 pathway inhibition. Clin Cancer Res 2017;23:6923–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.