Short abstract
Sensory neurons are chemically and functionally heterogeneous, and this heterogeneity has been examined extensively over the last several decades. These studies have employed a variety of different methodologies, including anatomical, electrophysiological, and molecular approaches. Recent studies using next-generation sequencing techniques have examined the transcriptome of single sensory neurons. Although these reports have provided a wealth of exciting new information on the heterogeneity of sensory neurons, correlation with functional types is lacking. Here, we employed retrograde tracing of cutaneous and muscle afferents to examine the variety of mRNA expression profiles of individual, target-specific sensory neurons. In addition, we used an ex vivo skin/nerve/dorsal root ganglion/spinal cord preparation to record and characterize the functional response properties of individual cutaneous sensory neurons that were then intracellularly labeled with fluorescent dyes, recovered from dissociated cultures, and analyzed for gene expression. We found that by using single-cell quantitative polymerase chain reaction techniques and a set of 28 genes, we can identify transcriptionally distinct groups. We have also used calcium imaging and single-cell quantitative polymerase chain reaction to determine the correlation between levels of mRNA expression and functional protein expression and how functional properties correlated with the different transcriptional groups. These studies show that although transcriptomics does map to functional types, within any one functional subgroup, there are highly variable patterns of gene expression. Thus, studies that rely on the expression pattern of one or a few genes as a stand in for physiological experiments, runs a high risk of data misinterpretation with respect to function.
Keywords: Dorsal root ganglion, primary afferents, nociceptors, muscle, mechanoreceptors
Introduction
Understanding complex diseases of the nervous system, like chronic pain, requires a detailed analysis of the molecular components of the cells that contribute to pain. Within the peripheral nervous system, primary afferents are the principal transducers of environmental stimuli and serve as the initial pathway by which noxious signals, in the form of action potentials, arrive in the central nervous system (CNS). How this information is interpreted and processed by cells within the CNS depends, in large part, on the functional properties of the sensory neuron responsible for transduction and transmission of this information. Primary afferents are a heterogeneous population of neurons comprised of cells with multiple neurochemically and functionally distinct identities that contribute to how they transduce environmental stimuli. There is a long history of attempts to link identified anatomic/neurochemical cellular characteristics to a cell’s functional properties, but progress in this domain has been hampered by this cellular complexity.
Concerted efforts have been made to determine unique sets of biomarkers that correspond to nociceptive and non-nociceptive neurons. Traditionally, this has been attempted by examining various proteins expressed in cells using immunohistochemical methods. Technological advances, including next-generation RNA sequencing, have expanded the definition of cellular identity, normalizing the use of transcriptional profiling and providing unparalleled insight into the transcriptional profiles within respective populations of nociceptive and non-nociceptive afferents. While these discoveries have provided a wealth of information, there still has not been a clear link made between transcriptional profile and functional properties.1,2 An additional complexity that has made this latter point challenging has been the limited use of transcriptional profiling in afferents that have been physiologically characterized within their native environments where their central and peripheral sites of termination remain intact. Examining afferent function and subsequent mRNA profile within the context of the peripheral tissue each cell innervates is critical because previous work has shown that the tissue innervated influences the way in which nociceptors respond to inflammation and injury.3,4
Here, we utilized a multifaceted approach that employed retrograde tracing of cutaneous and muscle afferents to examine the variety of expression profiles of individual, target-specified sensory neurons. We characterized the expression of 28 high priority candidate genes, chosen based on their use in previous characterizations, and their potential contributions to sensory transduction, using single-cell real-time quantitative polymerase chain reaction (qPCR). These results revealed significant diversity in patterns of expression between sensory neurons. In addition, we used our ex vivo skin/nerve/dorsal root ganglion (DRG)/spinal cord preparation to record and characterize the physiological response properties of individual cutaneous sensory neurons. Characterized neurons were collected and subjected to single-cell qPCR analysis of transcriptional profile. Using this approach, we were able to identify collections of genes and expression levels that correlated with physiological response properties of each neuron. Our results represent the first time that transcriptional profile has been correlated with neuronal responses to quantitative naturalistic stimulation and provide additional insight into the genes that contribute to response properties of nociceptive afferents.
Materials and methods
Animals
Experiments were conducted on adult (4–8 week) Swiss-Webster mice (Hilltop Farms or The Jackson Laboratory). Animals were group housed with a 12-h light–dark cycle and ad libitum access to food and water. All procedures were approved by the Institutional Animal Care and Use Committee at the University of Pittsburgh and used in accordance with Association for Assessment and Accreditation of Laboratory Animal Care-approved practices.
Retrograde labeling
Mice were anesthetized with isoflurane (2%) and an incision was made on the medial surface of the right thigh. The saphenous or femoral nerve was isolated with a minimum amount of soft tissue damage. Parafilm was placed behind the isolated nerve, and the chosen dye (1% Alexa 488-Isolectin-binding protein 4 (IB4), 1% Alexa 488-wheat germ agglutinin (WGA) or 0.5% cholera toxin subunit β (CTB)) was pressure injected into the appropriate nerve using a quartz pipette and a picospritzer. The wound was closed using silk sutures.
Single-cell survey design
Four animals per dye were back labeled 24 h before DRG collection. On the collection day, their L2 and L3 DRGs were removed and dissociated. Then, 8 to 10 back-labeled cells were randomly selected and picked up from each animal and stored at −80°C until processed further. Each cell’s RNA was then amplified and underwent qPCR until 6 to 8 viable samples (defined as having a glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cycle threshold (Ct) of less than 24) were obtained for each animal.
Cell dissociation and pickup
DRGs containing labeled cells were removed and dissociated as described previously.5 Briefly, DRGs were treated with papain (30 U) followed by collagenase CLS2 (10 U)/Dispase type II (7 U), centrifuged (1 min at 1000 r/min), triturated in Minimal Essential Medium, plated onto laminin-coated coverslips in 30 mm diameter dishes, and incubated at 37°C for 45 min. Dishes were removed and flooded with collection buffer (140 mM NaCl, 10 mM Glucose, 10 mM HEPES, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2). Single, labeled cells were identified using fluorescence microscopy, picked up using glass capillaries (World Precision Instruments) held by a 4-axis micromanipulator under bright-field optics, and transferred to tubes containing 3uL of lysis buffer (Epicentre, MessageBOOSTER kit). Cells were collected within 1 h of removal from the incubator and within 4 h of removal from the animals.
Single-cell amplification and qPCR
The RNA isolated from each cell was reverse transcribed and amplified using T7 linear amplification (Epicentre, MessageBOOSTER kit for cell lysate), run through RNA Cleaner & Concentrator-5 columns (Zymo Research), and analyzed using qPCR as described previously6 using optimized primers and SsoAdvanced SYBR Green Master Mix (Bio-Rad). Threshold cycle time (Ct) values were determined for each well.
Internal controls
In these studies, the expression levels are all reported relative to GAPDH level. To test for potential batch effects, we examined the variability of GAPDH expression in different batches of amplified cells. We routinely amplified in batches of 24 cells for efficient use of the premeasured reagents in the kits. We examined 11 amplification batches that contained virtually all of the cells reported here. We first ran PCR for GAPDH, and any cell with a Ct threshold (corrected for primer efficiency and dilution) of 24 or higher was excluded from further analysis. Of the 264 cells amplified in these 11 batches, 23 (9%) were excluded, while 241 (91%) were accepted for further qPCR analysis. The average Ct for each batch ranged from 16.6 to 20.3 with a mean of 18.3 and standard deviation of ±1.04. Thus, the average Ct for each batch was within 2 standard deviations of the mean suggesting that differences of GAPDH Cts in individual batches should not bias our results.
We also considered the relationship between relative level of GAPDH and three genes that are expressed in most if not all of the cells (i.e., Ret, NefH, Scn9a). These three genes are variably regulated in different populations, which should add variability to the comparison between them and GAPDH. A linear correlation between these three genes resulted in a slope = 1.12 and R2 = 0.53 for Ret; slope = 1.24, R2 = 0.63 for Scn9a, and slope = 1.93, R2 = 0.59 for NefH. Thus, the majority of the variability between these widely expressed genes and GAPDH are factors that these genes and GAPDH have in common, i.e., amplification and initial lysate amount.
Primer design and validation
Forward and reverse primer sequences were chosen for each gene within 500 bases of the 3′ poly A addition site. Primer sequences are listed in Table 1. Primer testing was done using cDNA generated using RNA from the whole DRG, as described in Jankowski et al.6 As described above for single cells,10 or 160 pg aliquots of the RNA was amplified as described previously for single cells. For each primer set, serial dilutions of these aliquots were used to calculate primer efficiencies over the range of RNA concentrations observed in single cells as described in Pfaffl.7 Expression of specific genes was corrected for these primer efficiencies,7 and expression level relative to GAPDH was determined. Primer specificity was also confirmed by demonstration of a single PCR product (single peak for melting curve) and a single band of the appropriate size on agarose gels.
Table 1.
PCR primer sequences used in this study.
| Gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| Asic1 | CAGGCCAGCTCTCCAATCTC | ACGTACACAGGTGATCTGCC |
| Asic2 | GCACCTGTGGAGGAAGTACG | CCCGCCCCAAACAAAAATCAG |
| Asic3 | GCAACACTCTGCTCCAGGAA | CGAGGTAACAGGTACGGTGG |
| Calca | TGACAGCATGGTTCTGGCTT | GTCCCCAGAAGAGCAAGAGG |
| GAPDH | ATGAATACGGCTACAGCAACAGG | CTCTTGCTCAGTGTCCTTGCTG |
| Gfra1 | GACTCGGAATCCAGCCTACG | TGTGCACTTGTCCTCTCGTG |
| Gfra2 | CTTGGGGAGAAGGGCTGTTG | AGGAGAAGAGAGAAGGGGCA |
| Gfra3 | GCTGGTGTCTTGACTGCTCT | GACCCAAGGGACTAGGGGAA |
| Mrgprd | CTGCTTCAGGCCAGCTCCTA | AGCATCTCTGTCACCTTGAGCA |
| Mrgpra3 | ATAGCCCTCTTCTGGTCCTAACT | AGGCTCTTCATCACGGCTCT |
| Nefh | TTGCCCAGTACACGCTCCTG | GAGTACACCCTGGCGTGGTT |
| Piezo2 | GACTGAGGCCACAGGGCT | GCAAGGAAAATGTCACACACTG |
| P2rx3 | GGTGCCTAAGCCTCTTCTGG | AGGGATGGCGCTGAGTAAAC |
| P2ry1 | GCCAGGACACTAACCCATCG | AACTGAAGGCCCACAAACCTC |
| P2ry2 | AGGGAGGGGTCCCTGGAATG | ACCTTTGCCCATTCCTGCCTA |
| Ret | CAACATGCCTACACGGTAAGTG | ATCTCAGGAACAGACAGACAATGG |
| Scn9a | AGAACTGTGTTTGGAGCCATCA | GGCTACTTACTCATTTTCTGGGAG |
| Scn10a | GGCACTGTCGCATAGGGGT | ACCCTCAGGTATTGTCCGGC |
| Scn11a | CGGAAGGCCTGAAGGACAGTT | CCATACACCCAAATCCGCAGC |
| Sst | GGTCTGCCAACTCGAACCCA | ATCGGGGGCCAGGAGTTAAG |
| Tac1 | CTGTGCGTCTCTCTCACGCT | CACGAAACAGGAAACATGCTGCT |
| Th | CACCATCCGGCGCTCCTTAG | GTCCAGCCACACACATGGGA |
| TrkA | GAGCCAAGTTTTGGTGCCAG | GATGCTGGCCATGAAGCAAG |
| TrkB | AAGATGTGTCCCTGGGCTTC | AAGTGAGTCACGAGCTGCC |
| TrkC | CTGCTCCCCATGGTTGTAGG | GGGAGGCTGGAAATGAGGTC |
| Trpa1 | TGAGCCACATGACAGAAGTCC | CTAAGCAGCAGCAACAACTGG |
| Trpm3.1 | TCAGAGTTTTTCAAGGGCTGGT | TGGCCTACCTGCAACTGATG |
| Trpv1 | GGCGAGACTGTCAACAAGATTGC | TCATCCACCCTGAAGCACCAC |
Skin-nerve-DRG ex vivo preparation
The ex vivo preparation has been described previously.8,9 Briefly, mice were anesthetized with a mixture of ketamine (90 mg/kg) and Xylazine (10 mg/kg) and perfused using 10 to 12°C oxygenated (95% O2/5% CO2) artificial cerebrospinal fluid (ACSF) (127 mM NaCl, 26 mM NaHCO3, 10 mM D-glucose, 2.4 mM CaCl2, 1.9 mM KCl, 1.3 mM MgSO4, 1.2 mM KH2PO4). The spinal column and right hindlimb were removed and placed in a circulating bath of the same oxygenated ACSF. The shaved hairy skin of the hindlimb with connected saphenous nerve, L1-L5 DRGs, and the associated spinal cord were isolated and transferred to a second recording chamber with circulating oxygenated ACSF, where they were heated to 31°C.
DRG neurons were impaled using quartz microelectrodes (>200 MOhm) containing 0.05% Alexa dye (555 or 488 nm) in 1 M potassium acetate. Electrical search stimuli were delivered using a suction electrode on the saphenous nerve. Mechanical receptive fields were located by probing with Von Frey filaments or glass rod. If no mechanical receptive field was found, thermal fields were detected by applying hot (52°C) or cold (0°C) saline (0.9%) to the skin.
Once the receptive field was located, peripheral conduction velocity was calculated using spike latency and the distance between stimulation and recording electrodes. Controlled mechanical stimuli (square waves) were presented using a force-modulating mechanical stimulator (Aurora Scientific) with a 1-mm diameter plastic foot. Thermal stimuli (rapid cooling to 4°C or a 12 s heat ramp from 31 to 52°C) were presented using a 3-mm Peltier element (Yale University Machine Shop). Cells were given 30 s to recover between stimuli. After electrophysiological characterization, cells were iontophoretically filled with Alexa dye. Only one cell per dye was injected per ganglion. Following the experiment, DRGs were removed, dissociated, and the labeled cells were collected as described above. Responses were analyzed offline using Spike2 software (Cambridge Electronic Design).
Automated hierarchical clustering
Back-labeled and functionally characterized cells were clustered using the unweighted pair group method with averaging (UPGMA) using the expression information obtained from single-cell PCR. Preprocessing for this data analysis consists of taking the ΔCt values and replacing the samples that failed to generate a value for a given gene with the detection limit for that gene. We then used MATLAB’s UPGMA implementation to agglomeratively cluster these neurons based on the Euclidean distance between their ΔCt values for each gene.
Calcium imaging of dissociated neurons
Back-labeled cutaneous neurons were dissociated from L2 and L3 DRGs as described above and incubated with 2 µM Fura2AM calcium indicator dye dissolved in Hank’s balanced salt solution (HBSS) for 30 to 60 min before recording. Coverslips were moved to an inverted fluorescence microscope (Olympus) where they were continuously superfused with HBSS using a tribarrel drug application device (Exfo Burleigh PCS-6000). A suitable field of view was located, and regions of interest (ROIs) were marked over cells identified with the white light transmittance. Experimentation and shutter control was automated (Lambda DG-4/Lambda 10-B, Sutter Instruments) and two application protocols were used: 1 s 50 µM αβmeATP, 1 s 50 mM K+ and 1 s 1 µM capsaicin, or 5 s 100 µM 2-methylthioADP, 1 s 50 mM K+ and 1 s 1 µM capsaicin. Following imaging, a 3-axis micromanipulator with borosilicate glass electrodes was used to pick up cells into 3 µL of lysis buffer (Epicentre, MessageBOOSTER), their ROI identifier was recorded, and the cells were stored at −80°C until amplified.
Statistical analysis
Comparisons for levels of gene expression were performed using t tests or Fisher’s exact test. Relationships between gene expression and Ca2+ influx were determined by Pearson’s correlation.
Results
We recovered and amplified mRNA from a total of 247 sensory neurons from 64 mice. Of these, 120 were cutaneous afferents, 81 were back-labeled from the saphenous nerve (12 mice), and 39 were identified and intracellularly labeled in our ex vivo preparation (36 mice). An additional 127 cells were muscle afferent fibers back labeled from the femoral nerve (16 mice).
Validation of linear preamplification
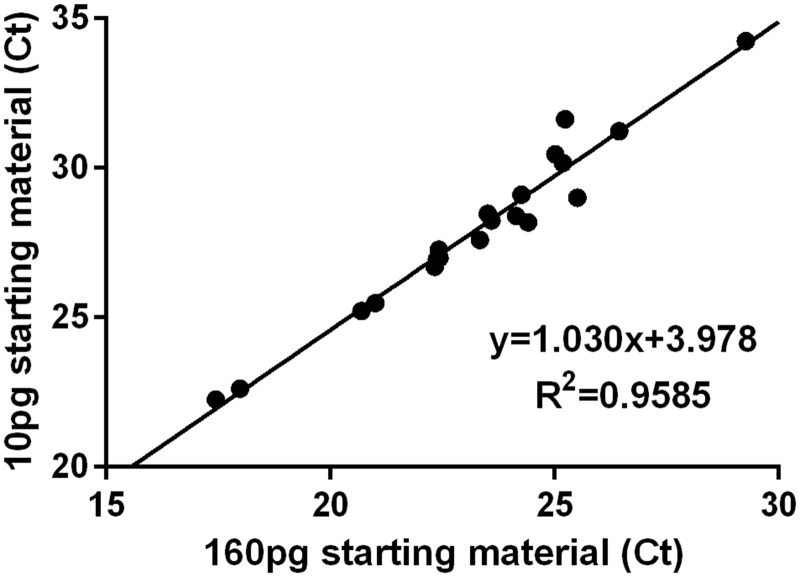
To verify that pre-amplification was independent of both the specific RNA sequence being amplified and the starting amount of the specific mRNA, we pre-amplified eight replicates of two different amounts of mRNA (10 pg and 160 pg) from Swiss-Webster lumbar whole DRG lysate. After pre-amplification, we pooled the replicate samples and proceeded through RNA column purification and qPCR, running 20 of our primer sets. The variable abundances of different mRNA templates in whole DRG lysate resulted in a wide distribution of Ct values, which allowed us to assess whether the amplification was dependent on the amount of starting transcript. As shown in Figure 1, the 16-fold difference between starting amounts of mRNA resulted in amplification not significantly different from linear (Runs test, p = 0.59) with a slope of 1.03 ± 0.05 indicating that the cDNA yield of amplification depended linearly on the starting mRNA amount. The 16-fold change in starting material would be predicted to impart a 4 Ct offset (24 = 16), which was also supported by the observed 3.98 ± 1.186 Ct offset. Thus, we concluded that the preamplification of each mRNA is linear to the extent of our ability to detect it.
Figure 1.
Preamplification linearity. 1 µl aliquots containing 10, 20, 40, 80, and 160 pg of whole DRG mRNA were individually amplified and the cycle thresholds obtained using qPCR.
Clustering cutaneous afferent types by single-cell qPCR
We identified 81 cutaneous neurons for single-cell qPCR by back-labeling the saphenous nerve with commonly used tracers (IB4 n = 14, WGA n = 56, and CTβ n = 11). Another 39 were collected and analyzed after being functionally characterized in our ex vivo skin-nerve-DRG preparation. This approach allowed the generation of a comprehensive distribution of a total of 120 cutaneous afferents, which were then clustered autonomously. To obtain these clusters, we employed the UPGMA using the expression information. For this analysis, we took the ΔCt values (relative to GAPDH). For those genes that failed to generate a measurable product the detection limit for that gene was used to calculate the ΔCt value. We then used MATLAB’s UPGMA implementation to agglomeratively cluster neurons based on the Euclidean distance between their ΔCt value for each gene.
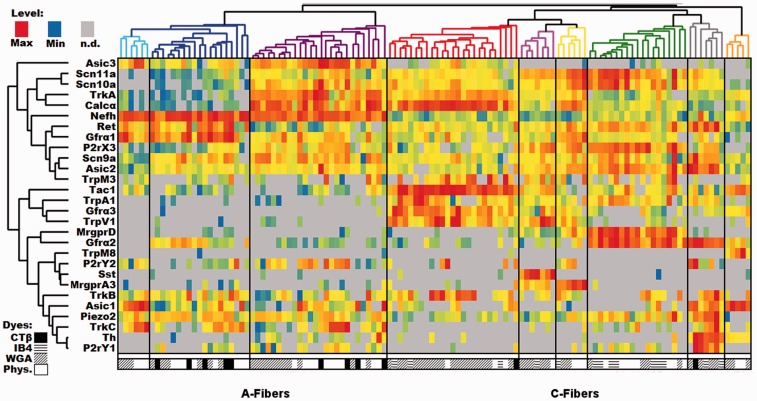
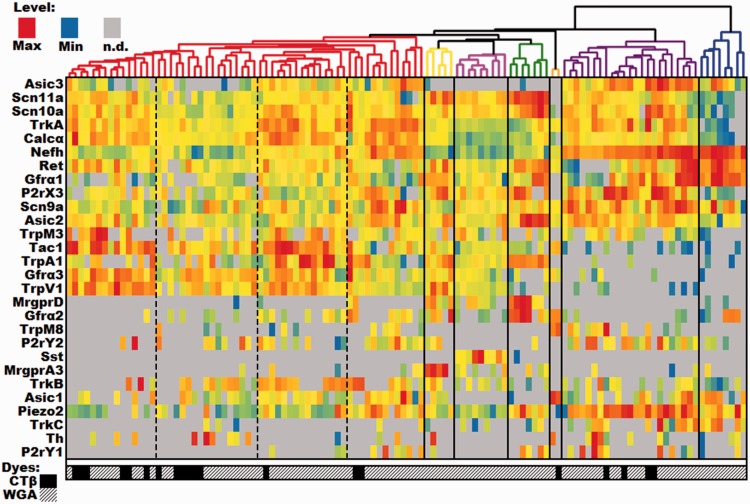
The distribution of ΔCt values for each gene was typically left-skewed. Therefore, to ensure that the upper mode of the distribution would be colored red, orange, or yellow, while the lower tail would be green or blue, heatmap temperatures for the histogram are scaled to expression, with red representing the maximum level, yellow representing 1/32 of the maximum, blue representing the minimum, and gray representing an undetectable level. Following this analysis, the individual clusters were characterized by their expression profiles and each different group assigned a name. The emergent groups we identified were most similar to the classification scheme put forth by Usoskin et al.1 and Zeisel et al.2
In our analysis, the root split in the cell dendrogram (Figure 2) divides the identified cutaneous neurons into two groups based on the relative level of expression of the myelinated neuron marker neurofilament heavy chain (Nefh), a part of the NF200 complex. The low-Nefh expression cluster can be further subdivided into six populations of putative and characterized unmyelinated fibers, each with an identifying gene marker. For example, the green color-coded population that expresses mitochondrial assembly protein (MAS)-related G-protein-coupled receptor, member D (Mrgprd), lacks classic biomarkers for peptidergic neurons (e.g. Calcα calcitonin gene-related peptide [CGRP] and Tac1 [substance P]). This group also contains some of the highest levels of P2rX3, Asic2, and Gfrα2 as would be expected from the extensive literature on this population.10–12
Figure 2.
DRG profiling of identified cutaneous afferents using single cell PCR. Single-cell qPCR results from identified cutaneous sensory neurons, with each column being a unique cell and rows representing genes. Heatmap temperatures for the histogram are scaled to expression, with red representing the maximum level, yellow representing 1/32 of the maximum level, blue representing the minimum level, and gray representing an undetectable level of transcript. Distributions were typically left-skewed, so scaling the heatmap in this way ensures the upper mode of the distribution will be colored red, orange, or yellow, while the lower tail is green or blue. Both cells and genes have been clustered using the same UPGMA algorithm. The symbols below each column indicate the source of each cell. WGA: wheat germ agglutinin; IB4: Isolectin-binding protein 4; CTB: cholera toxin subunit β; Phys: physiological characterized.
The pink-coded group of neurons is denoted by the high level of somatostatin (Sst) expression. Although the Sst group also expresses significant levels of Mas-related G-protein-coupled receptor, member A3 (Mrgpra3), it can be distinguished from the yellow-coded cluster of neurons with higher levels of Mrgpra3 expression, by its Sst expression and lower levels of Trka. Both clusters contain some level of Mrgpra3 and Trpv1 with low to undetectable levels of Gfrα2. The Sst population has been previously identified as being separate from the canonical “peptidergic” substance P (Tac1) population,13 but the uniqueness of these cells was not fully appreciated until recently.1,14 Similarly, Mrgpra3 was identified as a population unique from the Mrgprd population that is positive for TRPV1, P2X3, CGRP, and c-RET protein expression but not NF200 complex or substance P.15,16 Although on average there is 11 times more Mrgpra3 expressed in our A3 (yellow) subpopulation than in the Sst-containing group (t-test: p = 0.011), Mrgpra3 has been reported to co-localize with another marker for the Sst group, natriuretic peptide B (Nppb).17,18 Thus, there may be functional Mrgpra3 expression in both populations. The MrgprD, MrgprA3, and Sst groups are most likely to be the closest analogs for the population that was previously identified based on staining with the Isolectin B4.
The red-coded Tachkykinin-1-containing (Tac1) cluster is likely a large component of the canonical unmyelinated cutaneous peptidergic neurons, which also express high levels of Calcα, TrkA, Gfrα3, and Trpv1.19–22 Histological methods of identifying peptidergic afferents tend to rely on the products of these five transcripts, which also map to functional differences,9 indicating that at least for this population, our unbiased methods correspond to previous functional and anatomical classification schemes.
The gray-coded tyrosine hydroxylase-containing (TH) cluster included other known markers of this population such as Gfrα2, Ret, P2ry1, Piezo2, Trkc, and Asic1.1,23 The relatively low prevalence of this population is not surprising given that only 10% to 15% of the mouse DRG neurons express TH. This cutaneous population is poorly labeled by IB424 and identified C-low threshold mechanoreceptive fibers that have been shown to express TH, do not bind IB4 or express CGRP.23,25
The final group within the low-Nefh expressing cluster is the orange-coded group which is primarily distinguished by the expression of transient receptor potential cation channel, member M8 (Trpm8). In addition to Trpm8, these cells express relatively high levels of Asic1, Gfrα3, and Tac1. Notably, these fibers express a complement of sodium channels, which is unique among unmyelinated sensory neurons. They lack detectable levels of Scn11a (Nav1.9) and have low to undetectable levels of Scn10a (Nav1.8), both of which are typically associated with small afferents and are expressed at high levels in cells expressing low levels of Nefh.
The high-Nefh expressing cluster is also clearly split into classical peptidergic and non-peptidergic groups. The purple-coded Calcα group has high levels of Calcα, Trka, and Asic3. The non-peptidergic myelinated fibers are divided into two groups. The dark blue-coded cluster has high levels of Gfrα1, Ret, and piezo2, whereas the light blue-coded cluster expressed high levels of Asic1, Trkc, and Asic3. This latter group appears to closely match the NF3 group reported by Usoskin et al.1
Relationship of Ca2+ response to relative transcript levels
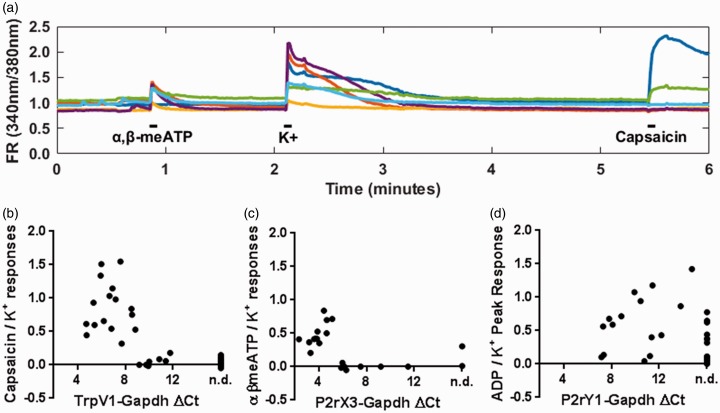
Results from the single cutaneous neuron qPCR analysis reveal a wide range of transcript expression levels between cutaneous afferents, with several thousand-fold differences between maximum and minimum levels. In addition, small but measurable transcript levels are occasionally expressed in neuron types not thought to contain the associated protein (e.g. Calcα in the non-peptidergic MrgprD population). These observations suggest the possibility that our technique may be detecting transcripts that are not translated to protein. To address this issue and examine the relationship between function and transcript levels, we used calcium imaging of 39 back-labeled cutaneous sensory neurons obtained from five mice and single-cell qPCR to compare agonist responses in cultured neurons to the associated receptors’ transcript levels for individual neurons (Figure 3). For these experiments, we chose Trpv1, P2rx3/P2rx2, and P2ry1 as target transcripts because their respective receptors possess specific agonists, and evidence suggests they are differentially expressed across the DRG cell subpopulations. For example, P2X3 and P2X2 are reported to be expressed in nonpeptidergic C-fibers;26,27 Trpv1 should be expressed largely in the peptidergic population28 and P2ry1 should be expressed in both large and small cells.29,30
Figure 3.
Calcium imaging reveals apparent thresholds of function for two ionotropic channels. (a) Example fluorescence ratio (FR) traces in response to 50 µM α,β-meATP, 1 M KCl, and 1 µM Capsaicin (100 mM ADP not shown). Ratio of agonist to KCl responses versus relative receptor transcript expression for (b) TrpV1, (c) P2rX3, and (d) P2rY1. GAPDH: glyceraldehyde 3-phosphate dehydrogenase.
In these experiments, we examined transcript levels in cutaneous back-labeled cells that exhibited calcium transients evoked by α,β-meATP, capsaicin, or ADP (Figure 3(a)). Nonresponsive cutaneous neurons, collected from the same cultures, were analyzed in parallel. Capsaicin responders and nonresponders were clearly divided by their relative Trpv1 transcript level (Figure 3(b)). Neurons responsive to capsaicin always expressed detectable Trpv1 transcript (16/16), while detection in non-responders was rare. Interestingly, several noncapsaicin-sensitive cells were found to express low levels of TrpV1 (7 of 23). Further analysis of this result showed a clear threshold relationship between transcript levels and whether a cell would respond to capsaicin. Specifically, neurons that did not respond to capsaicin had TrpV1 ΔCt levels over 9 (signifying lower levels of transcript expression), while every cell below 9 ΔCt responded to capsaicin (Figure 3(b)).
Applying this threshold to findings from our single-cell survey results demonstrated that in our Tac1, red-coded peptidergic group, where TrpV1 expression levels ranged from ΔCt 4.57 to 10.39, (mean ΔCt = 6.5), 15 of 25 cells should express functional levels. Likewise, for the Mrgpra3 group (ΔCt 6.03–7.54, mean ΔCt = 7.01), three of six would be predicted to have functional TRPV1, whereas for the Sst group (ΔCt 4.58–7.05, mean ΔCt = 5.98), five of seven would be predicted to contain functional TRPV1. Finally, in the MrgprD group, no cells (0/19 predicted functional) expressed Trpv1 at functional levels. These results agree with previously published immunohistochemistry, which primarily show Trpv1 expression in CGRP+/NF200− cells.26,28,31 Among responders, however, there is no statistical correlation between ΔCt and absolute or relative response amplitude.
Similarly, P2rx3 levels were related to cellular response to α,β-meATP (Figure 3(c)). Only 1 of 9 neurons with a P2rx3 ΔCt over 6 exhibited a significant α,β-meATP response, while all 11 neurons with a ΔCt below 6 was responsive to α,β-meATP (Figure 3(c)). The responsive cell without P2rX3 transcript had the highest recorded expression level of P2rx2 (ΔCt = 6.70), another ATP receptor. P2rx2 was only observed in three responsive neurons (ΔCt = 6.70, 10.82, 12.35) and the other two expressing P2rx2 were not responsive to α,β-meATP. Applying a 6 ΔCt threshold to findings from our single-cell survey indicates that, within different groups, 16 of 19 Mrgprd, five of six Mrgpra3 and six of seven Sst cells likely expressed functional levels of P2rx3. In the Tac1 group, only 2 of 22 cells expressed sufficient levels of P2rx3. Interestingly, 8 of 22 of myelinated cells expressed levels of P2rx3 that would be predicted to be functional. This is not entirely unexpected, given that large or myelinated sensory neurons have been observed expressing P2rx3 via in situ hybridization,32 immunohistochemistry,33 and function.34,35
Unlike the two ionotropic channels, relative expression levels of the metabotropic receptor P2ry1 are poorly related to 2-methylthioADP responsiveness (Figure 3(d)). Only 11 of 18 ADP responders expressed detectable levels of P2ry1 (Average ΔCt 10.54), while 5 of 13 ADP nonresponders also expressed P2ry1 (Average ΔCt 10.00). Neither the percent of cells with detectable expression (Fisher’s exact test: p = 0.29) nor the expression levels (t-test: p = 0.70) were significantly different between the samples.
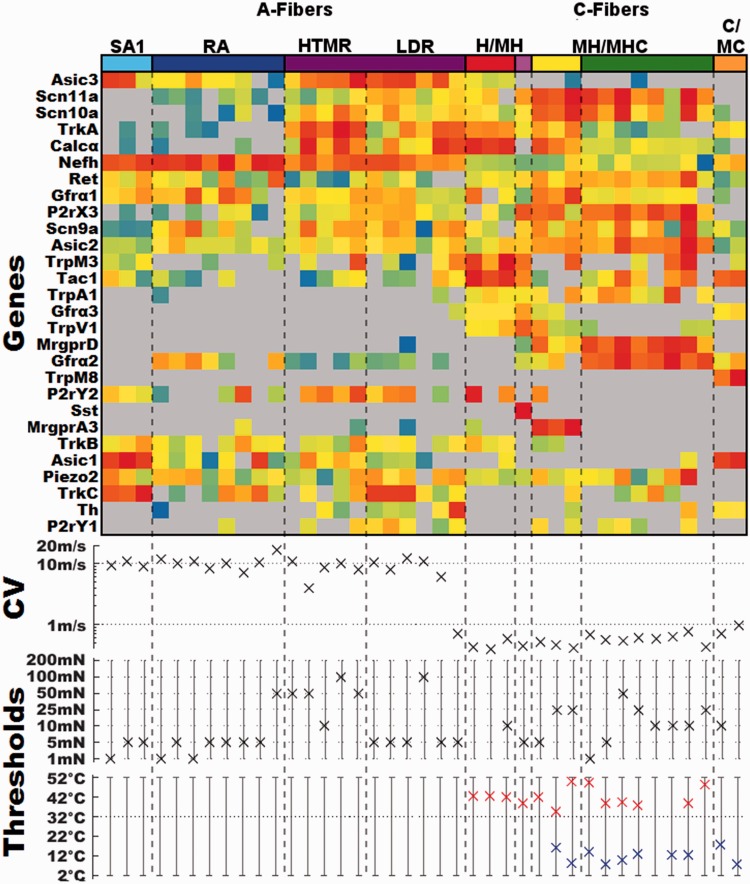
Distribution of functionally characterized cutaneous afferents within expression derived groups
Of the 120 identified cutaneous afferents depicted in (Figure 2), 39 were collected following characterization of their functional properties (Figure 4). Of these, 39 cells were represented in all but one (TH) of the nine groups. We have separated out these fibers and report their expression profiles, conduction velocities, and functional properties, including threshold data for different stimulus modalities (Figure 4). This comparison shows that different types of characterized fibers consistently mapped to single-cell clusters. For example, three characterized slowly adapting Type I mechanoreceptors (SAI) mapped together in the light blue-coded cluster. Likewise, myelinated rapidly adapting (RA) fibers all mapped to the dark blue-coded cluster. The purple-coded cluster contains all the characterized myelinated nociceptors with high mechanical thresholds (HTMRs). This cluster also contains fibers that have low initial mechanical thresholds. Many of the latter fibers had initial thresholds of 5 mN, the same as the majority of well-defined low threshold mechanoreceptors. This group of fibers exhibits a rapidly adapting response to low-threshold stimulation, which becomes slowly adapting as the intensities increase. These fibers are also capable of encoding the intensity of the mechanical stimulus over a large range of intensities. In order to reflect the functional properties of these cells, we are classifying them as having a large dynamic range (LDR) and have separated the two functional subdivisions of the purple-coded group by a dashed line. In addition, there was one characterized C-fiber that mapped to this group. It expressed very high levels of TH but differed from the TH group by the lack of Gfrα2 and Ret expression and high levels of Calcα and Trka.
Figure 4.
Expression profiles and the associated functional properties of characterized cutaneous neurons. Top panel: transcriptionally defined group color code and expression profiles of individual characterized cells Middle panel: conduction velocity for each cell. Bottom panel: mechanical, heat, and cold thresholds for each cell. Functional type abbreviations; SAI: slowly adapting Type I mechanoreceptor; RA: myelinated rapidly adapting mechanoreceptor; HTMR: myelinated high threshold mechanoreceptor; LDR: myelinated large dynamic range; H/MH: unmyelinated fibers responsive to heat (H) or mechanical and heat (MH); MH/MHC: unmyelinated fibers responsive to mechanical and heat (MH) or responsive to mechanical, heat and cold (MHC); M: unmyelinated fibers responding only to mechanical; C/MC: unmyelinated fibers responding to cold (C) or mechanical and cold (MC).
Within the C-fiber groups we found that the red-coded Tac1 group contained both mechanically insensitive, heat-sensitive C-fibers (CH), and those that responded to both mechanical and heat (CMH). The three clusters corresponding to the classical nonpeptidergic or IB4 binding population, Sst (purple), MrgprA3 (yellow), and MrgprD (green) contained cells that were classified as C-mechanical (CM) fibers (no response to heat or cold), CMH (mechanical and heat, but not cold), and CMHC (C-mechanical, heat, and cold). Furthermore, when comparing the response of the heat-sensitive cells, those in the Tac1/Trpv1 group had higher peak instantaneous firing rates (range 50.8–56.4 Hz) compared to other heat-sensitive fibers (range = 1.65–14.43 Hz) in response to heat stimulation as has been reported previously when comparing the CH population to C-polymodal (responsive to more than one modality) population.9
The two characterized cells in the orange-coded Trpm8 group were both cold sensitive and displayed higher peak instantaneous firing rates in response to cold (29.4 and 158 Hz) relative to the other eight cold-sensitive fibers (range = 0.65–5.14 Hz) which mapped to the MrgprD and MrgprA3 clusters. This agrees with our recent finding that Trpm8 expressing cold-sensitive fibers have similarly high instantaneous frequencies.36–38 This unique property may be due in part to the cluster’s lack of the tetrodotoxin (TTX)-insensitive Scn10a and Scn11a (Nav1.8 and 1.9, respectively).
Clustering muscle afferent types by single-cell qPCR
To compare results from cutaneous afferent profiling, we collected 113 identified muscle afferents following back labeling from the femoral nerve using WGA (n = 93) and CTβ (n = 20). We used the same clustering procedure to autonomously place the individual cells into seven defined groups (Figure 5). In the construction of the heat map for muscle afferents, we used the same ordering of genes to allow for easier comparison with the cutaneous neurons. This cluster analysis produced fewer groups than determined for cutaneous afferents. The expression patterns of 28 genes analyzed in individual muscle afferents showed a greater heterogeneity within some of these muscle afferent clusters when compared to their cutaneous counterparts. This is most obvious in the large red-coded peptidergic group that is denoted by the expression of high levels of Tac1 and/or Calcα. This group appears to contain several subgroups. Most obvious is the subgroup expressing relatively high levels of Nefh, suggesting that they may be thinly myelinated fibers, possibly Group III. This group also appears to be distinct from the other clusters by its moderate expression of ASIC3 but low levels of TRPV1. This group is also distinguished by high expression of Calcα with little or no expression of Tac1.
Figure 5.
DRG profiling of identified muscle afferents using single-cell PCR. Single-cell qPCR results from identified muscle sensory neurons, with each column being a unique cell and rows representing genes. Heatmap temperatures for the histogram are scaled to expression, with red representing the maximum level, yellow representing 1/32 of the maximum level, blue representing the minimum level, and gray representing an undetectable level of transcript. Distributions were typically left-skewed, so scaling the heatmap in this way ensures the upper mode of the distribution will be colored red, orange, or yellow, while the lower tail is green or blue. The genes have been clustered using the UPGMA algorithm. The genes are listed in the same order as the cutaneous neurons in Figure 2 for ease of comparison. The symbols below each column indicate the source of each cell. Dashed lines denote putative subgroups within the red cluster. WGA: wheat germ agglutinin; CTB: cholera toxin subunit β.
Three other potential subgroups within the red-coded group that appear to be comprised of unmyelinated Group IV muscle afferents. Two of these groups that appear to be peptidergic subgroups have much in common with their cutaneous counterparts, co-expressing high levels of Trpv1, Trpa1, and Gfrα3. The dissimilarities driving the separation of these two subgroups in the analysis appears to be differences in TrkB transcript levels and possibly ASIC3 expression. The third group of putative group IV fibers expresses lower or undetectable levels of Tac1, Calcα, Asic3, and Trpa1. Interestingly, many of these cells were obtained by CTB back labeling that is typically used to target myelinated, somatic afferents. However, it is also effective at labeling visceral afferents.31,39
There were also several muscle afferent groups that exhibited unexpected similarities with cutaneous C-fibers groups. Most notably, we observed MrgprD (green) and MrgprA3 (yellow) expressing cells that were thought to be restricted to the cutaneous population. The relative expression of all 28 genes analyzed here was consistent across cutaneous and muscle afferents within these groups. Sst expressing neurons (magenta) was also present in both populations. However, in this case, the co-expression of other genes differed somewhat from the cutaneous group, most notably due to the lack of Mrgpra3 expression. In addition, the orange TrpM8 group appears to be similar to the cutaneous counterpart including the lack of the TTX-insensitive sodium channels Scn10a and Scn11a (Nav1.8 and 1.9, respectively) and the expression of Tac1, Asic1, and Gfrα2. There were many other Trpm8-expressing muscle afferents that mapped to different groups including putative myelinated fibers in the magenta group. This suggests that unlike the cutaneous population, Trpm8-expressing muscle afferents represent more heterogeneous groups of afferent fibers.
One of the cutaneous C-fiber groups missing from muscle group IV fibers was the TH group. Interestingly, while the percentage of cells expressing high levels of Th was similar between the cutaneous and other populations of muscle afferent, the co-expression of other sampled genes differed in the TH-expressing muscle fibers causing them to be split into several of the other clusters.
The three remaining groups appear to be representative of myelinated fibers due to the high levels of Nefh expression. The blue-coded group has expression profiles indicative of proprioceptors including high levels of Nefh expression and little or no Scn11a or Scn10a. These cells also uniformly expressed high levels of Ret and Gfrα1. For the most part, the purple-coded group has similar expression levels for many of the same genes including high levels of Nefh, Asic2, Asic3, and Piezo2. This suggests that this group represents some type of myelinated mechanoreceptors and possibly some that are chemoreceptors given the expression of Asic3, P2rx3, P2ry1, and P2ry2 in many of these afferents.
Discussion
Clustering of identified single cutaneous sensory neurons
Here, we have shown the ability to use a linear amplification protocol and qPCR methodology to assign cells to functional transcriptional groups using the relative expression of 28 genes. Our automated clustering algorithm isolated nine clusters of cutaneous sensory neurons. While previous RNASeq studies of single DRG neurons have used afferents with unspecified target tissues,1,2,40,41 we have identified eight transcriptionally defined classes of cutaneous afferents that closely resemble previously identified groups.1,40,41 However, a recent deep sequencing study2 has reported several additional groupings of DRG neurons. The clusters we have identified here will be discussed with reference to this study. For comparison, we have mapped the seven to eight highest expressed genes in our individual clusters to the expression data available at http://mousebrain.org/.
Using this approach, our SA1 group maps to the PSNF2 group reported by Zeisel et al.,2 whereas the LTMR-RA group corresponds to their PSNF1 group. Our LDR group matches the PSNF3 group and the myelinated HTMR clusters the PSPEP1 group. Among the C-fibers, our Tac1 cluster is analogous to their PSPEP3 group. The MrgprA3 cluster matches the PSNP4 group, whereas our Sst cluster closely corresponds to their PSNP6 and the TH cluster is representative of their PSNP1 group. While they identified three TRPM8 groups, our cutaneous cluster most closely resembles their PSPEP8 group. Finally, our MrgprD cluster closely matched both the PSNP2 and PSNP3 groupings.
In a very recent study by Zheng et al.,42 they employed a series of genetic mouse models to label specific subsets of cutaneous sensory neurons. Overall, the results presented here are in good agreement with their findings. For example, they report a high level of expression of Mrgprd in their nonpeptidergic C-fiber group and a high level of P2ry1 expression in their population of C-LTMRs which is analogous to our TH cluster. However, there are some small differences as they also report high levels of Trpm8 expression in these cells, where we find a lack of Trpm8 in our cutaneous TH cluster. However, we do see moderate expression of TH in our Trpm8. This distinction is most likely due to differences in clustering strategies.
Relationship between relative expression level and function
We have been able to demonstrate a strong relationship between the relative level of gene expression and function for two receptor/ion channels in individual cutaneous sensory neurons. Although we did not detect a statistical correlation between gene expression and magnitude of Ca2+ transients, a well-defined level of gene expression occurred where Ca2+ influx was observed. We also found no relationship between the relative level of P2ry1 in cells that responded to ADP. In fact, ADP application evoked significant calcium transients in eight cells that did not contain measurable levels of P2ry1, suggesting that P2ry1 mRNA may not be maintained at a constant level and is transcribed on demand or post-transcriptionally modified. Alternatively, there may be another ADP receptor.
Functionally characterized cutaneous sensory neurons
Segregation of functionally characterized cutaneous sensory neurons in appropriate individual clusters adds further validation to the results presented here. Within the A-fiber population, the Aß SAI and RA fibers exhibit narrow somal action potentials and separate into two distinct groups. The remaining myelinated fibers map to the purple-coded cluster in which all cells exhibited broad somal action potentials. These cells contain myelinated peptidergic high threshold mechanoreceptors (HTMRs) and afferents functionally characterized as fast conducting myelinated fibers with low initial mechanical thresholds that also encode stimulus intensities into the noxious range (LDRs). Of the six neurons in the LDR group, three were found to express high levels of Trkc. These afferents may be analogous to TrkC-positive fibers that form circumferential endings around hair follicles, as they respond to graded mechanical stimuli43 (see Figure S5 in Bai et al.).
The other two myelinated fibers in this group express relatively high levels of Trka and Calcα. Recent reports show that two types of fibers make circumferential endings around hair follicles; one that contains TrkC and the other CGRP. Interestingly, both subtypes responded most vigorously to hair tugging.43,44 While Bai et al.43 considered the TrkC expressing group to be low-threshold mechanoreceptors based on their response to light stroking of the hairy skin, Ghitani et al.44 describe the Calcα-expressing neurons as nociceptors due to the vigorous response to tugging on the hairs.
For the C-fiber clusters, the Tac1 cluster contained 2 CH fibers and 1 CMH fiber. The Sst, MrgprA3, and MrgprD clusters were similar in that all the cells were mechanically sensitive containing CM, CMH, C-mechanical and cold, and CMHC fibers. This matches previous studies characterizing cutaneous fibers expressing mrgprd and mrgpra3.16,45 Finally, the TrpM8 cluster contains cold-sensitive cells. Fibers in his group have several characteristics that separate them from other C-fibers. For example, cooling-sensitive TrpM8 fibers express little or no Nav 1.8 or 1.9, suggesting that this population has a complement of voltage-gated sodium channels that is more similar to myelinated fibers. The lack of two major TTX-resistant sodium channels also raises the possibility that these cells are the TTX-sensitive small cold fiber population.46 In addition, this difference in their voltage-gated ion channel complement could potentially underlie the high instantaneous frequency that these fibers display in response to cooling.36
Another intriguing finding in the TrpM8 group is the expression of Tac1. It is present without significant expression of other classical markers of peptidergic fibers such as Calcα, Gfrα3, or Trka.47,48 Earlier genetic studies on the distribution of TrpM8 were inconsistent, with one group showing significant overlap with CGRP and substance P,49 while the other reported no significant overlap with either CGRP or substance P.48 Interestingly, the recent study by Zeisel et al.2 indicates Tac1 is co-expressed with TRPM8 in all three groups of TRPM8 fibers. Intriguingly, the presence of Tac1 in these fibers could provide insight into the mechanism of cold allodynia following injury. Release of substance P in lamina I could positively modulate activity in NK1R-positive lamina I projection neurons. This finding also suggests the possibility that these cooling fibers could play a role in the thermal grill illusion. For example, substance P release from these terminals in lamina I could increase sensitivity of adjacent NK1R containing neurons to relatively innocuous thermal inputs.
Clustering of single identified muscle sensory neurons
Studies by Usoskin et al.1 and Chiu et al.40 have provided information on the molecular profiles of putative muscle afferent populations. However, these profiles were defined based on their expression of parvalbumin, a marker of proprioceptor afferents. No studies to date have defined the expression patterns of group III and IV fibers. Although our results for identified muscle afferents do not clearly define these conduction velocity groups, our data do provide meaningful information on putative myelinated and unmyelinated muscle afferent subpopulations.
Many of the high Nefh expressing muscle afferents also express high levels of Asic3. This channel is a known proton sensing channel but has also been linked to modulation of mechanosensation,50 nociceptive processing,51–54 and modulation of cardiovascular reflexes.55 Nefh neurons also appear to contain the mechanosensitive receptor, piezo2,56 which may contribute to a mechanosensory function of these particular myelinated afferents. Whether these are of the proprioceptor or group III mechanoreceptor subtype remains to be elucidated.
Of the low Nefh, putative unmyelinated muscle afferents, several groups could be defined. The non-peptidergic (low Tac1/Calcα), putative group IV afferents were Asic3 negative but can contain TrpV1 and P2rX3. Both of these latter receptors have been shown to have roles in cardiovascular reflex regulation57,58 and muscle pain59,60 and appear to be contained within a population of muscle afferents with responsiveness to muscle metabolites such as lactate, ATP, and protons.53,61,62 These cells have been found to be of the putative metabonociceptor and innocuous metaboreceptor subtypes.62 While Asic3 expression has been linked to each of these processes,51–55 its expression does not necessarily overlap with TRPV1 or P2Xr3.62 Our current data do show a population of unmyelinated muscle afferents that are peptidergic and express Trpv1, Asic3, and P2rx3. It is possible that these cells fall within the nociceptor muscle afferent subtype and may respond to multiple stimulus modalities.62 P2ry1 did not appear to have robust expression in many muscle afferent subtypes, but this G-protein-coupled receptor has been shown to regulate a variety of muscle afferent responses, including modulation of nociception, mechanical, and chemical responses and cardiovascular reflexes.63
The results presented here from single-cell qPCR on identified and functionally characterized cells demonstrates the utility of this approach to unlock the potential of the vast amount of information contained in the present and future next-gen RNAseq studies.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Funding for this work was provided by NINDS NS023725 (HRK) NIAMS AR069951 (KMA).
References
- 1.Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 2015; 18: 145–153. [DOI] [PubMed] [Google Scholar]
- 2.Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, Codeluppi S, Furlan A, Lee K, Skene N, Harris KD, Hjerling-Leffler J, Arenas E, Ernfors P, Marklund U, Linnarsson S. Molecular architecture of the mouse nervous system. Cell 2018; 174: 999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gold MS, Flake NM. Inflammation-mediated hyperexcitability of sensory neurons. Neurosignals 2005; 14: 147–157. [DOI] [PubMed] [Google Scholar]
- 4.Malin S, Molliver D, Christianson JA, Schwartz ES, Cornuet P, Albers KM, Davis BM. TRPV1 and TRPA1 function and modulation are target tissue dependent. J Neurosci 2011; 31: 10516–10528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malin SA, Davis BM, Molliver DC. Production of dissociated sensory neuron cultures and considerations for their use in studying neuronal function and plasticity. Nat Protoc 2007; 2: 152–160. [DOI] [PubMed] [Google Scholar]
- 6.Jankowski MP, Lawson JJ, McIlwrath SL, Rau KK, Anderson CE, Albers KM, Koerber HR. Sensitization of cutaneous nociceptors after nerve transection and regeneration: possible role of target-derived neurotrophic factor signaling. J Neurosci 2009; 29: 1636–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 2001; 29: 45e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McIlwrath SL, Lawson JJ, Anderson CE, Albers KM, Koerber HR. Overexpression of neurotrophin-3 enhances the mechanical response properties of slowly adapting type 1 afferents and myelinated nociceptors. Eur J Neurosci 2007 2007; 26: 1801–1812. [DOI] [PubMed] [Google Scholar]
- 9.Lawson JJ, McIlwrath SL, Woodbury CJ, Davis BM, Koerber HR. TRPV1 unlike TRPV2 is restricted to a subset of mechanically insensitive cutaneous nociceptors responding to heat. Pain 2008; 9: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong X, Han S-k, Zylka MJ, Simon MI, Anderson DJ. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell 2001; 106: 619–632. [DOI] [PubMed] [Google Scholar]
- 11.Rau KK, McIlwrath SL, Wang H, Lawson JJ, Jankowski MP, Zylka MJ, Anderson DJ, Koerber HR. Mrgprd enhances excitability in specific populations of cutaneous murine polymodal nociceptors. J Neurosci 2009; 29: 8612–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zylka MJ, Rice FL, Anderson DJ. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron 2005; 45: 17–25. [DOI] [PubMed] [Google Scholar]
- 13.Hökfelt T, Elde R, Johansson O, Luft R, Nilsson G, Arimura A. Immunohistochemical evidence for separate populations of somatostatin-containing and substance P-containing primary afferent neurons in the rat. Neurosci 1976. 1976; 1: 131–136. [DOI] [PubMed] [Google Scholar]
- 14.Stantcheva KK, Iovino L, Dhandapani R, Martinez C, Castaldi L, Nocchi L, Perlas E, Portulano C, Pesaresi M, Shirlekar KS, Castro Reis F, Paparountas T, Bilbao D, Heppenstall PA. A subpopulation of itch-sensing neurons marked by Ret and somatostatin expression. EMBO Rep 2016; 17: 585–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Tang Z, Surdenikova L, Kim S, Patel KN, Kim A, Ru F, Guan Y, Weng H-J, Geng Y, Undem BJ, Kollarik M, Chen Z-F, Anderson DJ, Dong X. Sensory neuron-specific GPCR Mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell 2009; 139: 1353–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han L, Ma C, Liu Q, Weng H-J, Cui Y, Tang Z, Kim Y, Nie H, Qu L, Patel KN, Li Z, McNeil B, He S, Guan Y, Xiao B, LaMotte RH, Dong X. A subpopulation of nociceptors specifically linked to itch. Nat Neurosci 2013; 16: 174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishra SK, Hoon MA. The cells and circuitry for itch responses in mice. Science 2013; 340: 968–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Polgár E, Solinski HJ, Mishra SK, Tseng P-Y, Iwagaki N, Boyle KA, Dickie AC, Kriegbaum MC, Wildner H, Zeilhofer HU, Watanabe M, Riddell JS, Todd AJ, Hoon MA. Circuit dissection of the role of somatostatin in itch and pain. Nat Neurosci 2018; 21: 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skofitsch G, Jacobowitz DM. Calcitonin gene-related peptide coexists with substance P in capsaicin sensitive neurons and sensory ganglia of the rat. Peptides 1985; 6: 747–754. [DOI] [PubMed] [Google Scholar]
- 20.Hokfelt T, Kellerth J, Nilsson G, Pernow B. Substance p: localization in the central nervous system and in some primary sensory neurons. Science 1975; 190: 889–890. [DOI] [PubMed] [Google Scholar]
- 21.Tominaga M, Caterina MJ, Malmberg AB, Rosen TA, Gilbert H, Skinner K, Raumann BE, Basbaum AI, Julius D. The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron 1998; 21: 531–543. [DOI] [PubMed] [Google Scholar]
- 22.Bennett DLH, Boucher TJ, Michael GJ, Popat RJ, Malcangio M, Averill SA, Poulsen KT, Priestley JV, Shelton DL, McMahon SB. Artemin has potent neurotrophic actions on injured C-fibres. J Peripher Nerv Syst 2006; 11: 330–345. [DOI] [PubMed] [Google Scholar]
- 23.Li L, Rutlin M, Abraira VE, Cassidy C, Kus L, Gong S, Jankowski MP, Luo W, Heintz N, Koerber HR, Woodbury CJ, Ginty DD. The functional organization of cutaneous low-threshold mechanosensory neurons. Cell 2011; 147: 1615–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brumovsky P, Villar MJ, Hökfelt T. Tyrosine hydroxylase is expressed in a subpopulation of small dorsal root ganglion neurons in the adult mouse. Exp Neurol 2006; 200: 153–165. [DOI] [PubMed] [Google Scholar]
- 25.Albers KM, Woodbury CJ, Ritter AM, Davis BM, Koerber HR. Glial cell-line-derived neurotrophic factor expression in skin alters the mechanical sensitivity of cutaneous nociceptors. J Neurosci 2006; 26: 2981–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo A, Vulchanova L, Wang J, Li X, Elde R. Immunocytochemical localization of the vanilloid receptor 1 (VR1): relationship to neuropeptides, the P2X3 purinoceptor and IB4 binding sites. Eur J Neurosci 1999; 11: 946–958. [DOI] [PubMed] [Google Scholar]
- 27.Petruska JC, Cooper BY, Gu JG, Rau KK, Johnson RD. Distribution of P2X1, P2X2, and P2X3 receptor subunits in rat primary afferents: Relation to population markers and specific cell types. J Chem Neuroanat 2000; 20: 141–162. [DOI] [PubMed] [Google Scholar]
- 28.Michael GJ, Priestley JV. Differential expression of the mRNA for the vanilloid receptor subtype 1 in cells of the adult rat dorsal root and nodose ganglia and its downregulation by axotomy. J Neurosci 1999; 19: 1844–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura F, Strittmatter SM. P2Y1 purinergic receptors in sensory neurons: contribution to touch-induced impulse generation. Proc Nat Acad Sci 1996; 93: 10465–10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burnstock G. Physiology and pathophysiology of purinergic neurotransmission. Physiol Rev 2007; 87: 659–797. [DOI] [PubMed] [Google Scholar]
- 31.Christianson JA, McIlwrath SL, Koerber HR, Davis BM. Transient receptor potential vanilloid 1-immunopositive neurons in the mouse are more prevalent within colon afferents compared to skin and muscle afferents. Neurosci 2006; 140: 247–257. [DOI] [PubMed] [Google Scholar]
- 32.Chen C-C, Akopian AN, Sivilottit L, Colquhoun D, Burnstock G, Wood JN. A P2X purinoceptor expressed by a subset of sensory neurons. Nature 1995; 377: 428–431. [DOI] [PubMed] [Google Scholar]
- 33.Bradbury EJ, Burnstock G, McMahon SB. The expression of P2X3 purinoreceptors in sensory neurons: effects of axotomy and glial-derived neurotrophic factor. Mol Cell Neurosci 1998; 12: 256–268. [DOI] [PubMed] [Google Scholar]
- 34.Dowd E, McQueen DS, Chessell IP, Humphrey PPA. P2X receptor-mediated excitation of nociceptive afferents in the normal and arthritic rat knee joint. Br J Pharmacol 1998; 125: 341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamilton SG, McMahon SB, Lewin GR. Selective activation of nociceptors by P2X receptor agonists in normal and inflamed rat skin. J Physiol 2001; 534: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jankowski MP, Rau KK, Koerber HR. Cutaneous TRPM8-expressing sensory afferents are a small population of neurons with unique firing properties. Physiol Rep 2017; 5: e13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmermann K, Hein A, Hager U, Kaczmarek JS, Turnquist BP, Clapham DE, Reeh PW. Phenotyping sensory nerve endings in vitro in the mouse. Nat Protoc 2009; 4: 174–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campero M, Baumann TK, Bostock H, Ochoa JL. Human cutaneous C fibres activated by cooling, heating and menthol. J Physiol 2009; 587: 5633–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fasanella KE, Christianson JA, Chanthaphavong RS, Davis BM. Distribution and neurochemical identification of pancreatic afferents in the mouse. J Comp Neurol 2008; 509: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiu IM, Barrett LB, Williams EK, Strochlic DE, Lee S, Weyer AD, Lou S, Bryman GS, Roberson DP, Ghasemlou N, Piccoli C, Ahat E, Wang V, Cobos EJ, Stucky CL, Ma Q, Liberles SD, Woolf CJ. Transcriptional profiling at whole population and single cell levels reveals somatosensory neuron molecular diversity. eLife 2014; 3: e04660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li CL, Li KC, Wu D, et al. Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res2016; 26(1): 83: 102. [DOI] [PMC free article] [PubMed]
- 42.Zheng Y, Liu P, Bai L, Trimmer JS, Bean BP, Ginty DD. Deep sequencing of somatosensory neurons reveals molecular determinants of Intrisicphysiological properties. Neuron 2019; 103: 598–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bai L, Lehnert BP, Liu J, Neubarth NL, Dickendesher TL, Nwe PH, Cassidy C, Woodbury CJ, Ginty DD. Genetic identification of an expansive mechanoreceptor sensitive to skin stroking. Cell 2015; 163: 1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghitani N, Barik A, Szczot M, Thompson JH, Li C, Le Pichon CE, Krashes MJ, Chesler AT. Specialized mechanosensory nociceptors mediating rapid response to hair pull. Neuron 2017; 95: 944–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rau KK, McIlwrath SL, Wang H, et al. Mrgprd enhances excitability in specific populations of cutaneous murine polymodal nociceptors. J Neurosci 2009; 29: 8612–8619. [DOI] [PMC free article] [PubMed]
- 46.Sarria I, Ling J, Gu JG. Thermal sensitivity of voltage-gated Na+ channels and A-type K+ channels contributes to somatosensory neuron excitability at cooling temperatures. J Neurochem 2012; 122: 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang T, Jing X, DeBerry JJ, Schwartz ES, Molliver DC, Albers KM, Davis BM. Neurturin overexpression in skin enhances expression of TRPM8 in cutaneous sensory neurons and leads to behavioral sensitivity to cool and menthol. J Neurosci 2013; 33: 2060–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dhaka A, Earley TJ, Watson J, Patapoutian A. Visualizing cold spots: TRPM8-expressing sensory neurons and their projections. J Neurosci 2008; 28: 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takashima Y, Daniels RL, Knowlton W, Teng J, Liman ER, McKemy DD. Diversity in the neural circuitry of cold sensing revealed by genetic axonal labeling of transient receptor potential melastatin 8 neurons. J Neurosci 2007; 27: 14147–14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sluka KA, Price MP, Breese NM, Stucky CL, Wemmie JA, Welsh MJ. Chronic hyperalgesia induced by repeated acid injections in muscle is abolished by the loss of ASIC3, but not ASIC1. Pain 2003; 106: 229–239. [DOI] [PubMed] [Google Scholar]
- 51.Chen W-N, Lee C-H, Lin S-H, Wong C-W, Sun W-H, Wood JN, Chen C-C. Roles of ASIC3, TRPV1 and Nav1.8 in the transition from acute to chronic pain in a mouse model of fribromyalgia. Mol Pain 2014; 10: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walder RY, Rasmussen LA, Rainier JD, Light AR, Wemmie JA, Sluka KA. ASIC1 and ASIC3 play different roles in the development of hyperalgesia after inflammatory muscle injury. J Pain 2010; 11: 210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ross JL, Queme LF, Cohen ER, Green KJ, Lu P, Shank AT, An S, Hudgins RC, Jankowski MP. Muscle IL1ß drives ischemic myalgia via ASIC3-Mediated sensory neuron sensitization. J Neurosci 2016; 36: 6857–6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naves LA, McCleskey EW. An acid-sensing ion channel that detects ischemic pain. Braz J Med Biol Res 2005; 38: 1561–1569. [DOI] [PubMed] [Google Scholar]
- 55.Tsuchimochi H, Yamauchi K, McCord JL, Kaufman MP. Blockade of acid-sensing ion channels attenuates the augmented exercise pressor reflex in rats with chronic femoral artery occlusion. J Physiol 2011; 589: 6173–6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coste B, Mathur J, Schmidt M, Earley TJ, Ranade S, Petrus MJ, Dubin AE, Patapoutian A. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 2010; 330: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith SA, Leal AK, Williams MA, Murphy MN, Mitchell JH, Garry MG. The TRPV1 receptor is a mediator of the exercise pressor reflex in rats. J Physiol 2010; 588: 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCord JL, Tsuchimochi H, Kaufman MP. P2X2/3 and P2X3 receptors contribute to the metaboreceptor component of the exercise pressor reflex. J Appl Physiol 1985; 109: 14116–11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saloman JL, Chung MK, Ro JY. P2X3 and TRPV1 functionally interact and mediate sensitization of trigeminal sensory neurons. Neurosci 2013; 232: 226–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shinoda M, Ogino A, Ozaki N, Urano H, Hironaka K, Yasui M, Sugiura Y. Involvement of TRPV1 in nociceptor behavior in a rat model of cancer pain. J Pain 2008; 9: 687–699. [DOI] [PubMed] [Google Scholar]
- 61.Light AR, Hughen RW, Zhang J, Rainier J, Liu Z, Lee J. Dorsal root ganglion neurons innervating skeletal muscle respond to physiological combinations of protons, ATP, and lactate mediated by ASIC3, P2X and TRPV1. J Neurophysiol 2008; 100: 1184–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jankowski MP, Rau KK, Ekmann KM, Anderson CE, Koerber HR. Comprehensive phenotyping of group III and IV muscle afferents in mouse. J Neurophysiol 2013; 109: 2374–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Queme LF, Ross JL, Lu P, Hudgins RC, Jankowski MP. Dual modulation of noociceptor and cardiovascular reflexes during peripheral ischemia through P2Y1 receptor-dependent sensitization of muscle afferents. J Neurosci 2016; 36: 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]