Abstract
The UDP-glucuronosyltransferase (UGT) family of enzymes is important in the metabolic elimination of a variety of endogenous compounds such as bile acids, steroids, and fat-soluble vitamins, as well as exogenous compounds including many pharmaceuticals. The UGT2B subfamily is a major family of UGT enzymes expressed in human liver. The identification of novel mechanisms including post-transcriptional regulation by microRNA (miRNA) contributes to interindividual variability in UGT2B expression and is a crucial component in predicting patient drug response. In the present study, a high-resolution liquid chromatography–tandem mass spectrometry method was employed to measure UGT2B protein levels in a panel of human liver microsomal samples (n = 62). Concurrent in silico analysis identified eight candidate miRNAs as potential regulators of UGT2B enzymes. Comparison of UGT2B protein expression and candidate miRNA levels from human liver samples demonstrated a significant inverse correlation between UGT2B10 and UGT2B15 and one of these candidate miRNAs, miR-485-5p. A near-significant correlation was also observed between UGT2B7 and miR-485-5p expression. In vitro analysis using luciferase-containing vectors suggested an interaction of miR-485-5p within the UGT2B10 3′-untranslated region (UTR), and significant reduction in luciferase activity was also observed for a luciferase vector containing the UGT2B7 3′-UTR; however, none was observed for the UBT2B15 3′-UTR. UGT2B10 and UGT2B7 activities were probed using nicotine and 3′-azido-3′-deoxythymidine, respectively, and significant decreases in glucuronidation activity were observed for both substrates in HuH-7 and Hep3B cells upon overexpression of miR-485-5p mimic. This is the first study demonstrating a regulatory role of miR-485-5p for multiple UGT2B enzymes.
SIGNIFICANCE STATEMENT
The purpose of this study was to identify novel epigenetic miRNA regulators of the UGT2B drug-metabolizing enzymes in healthy human liver samples. Our results indicate that miRNA 485-5p is a novel regulator of UGT2B7 and UGT2B10, which play an important role in the metabolism of many commonly prescribed medications, carcinogens, and endogenous compounds. This study identified potential miRNA-UGT2B mRNA interactions using a novel proteomic approach, with in vitro experiments undertaken to validate these interactions.
Introduction
The UDP-glucuronosyltransferase (UGT) 2B enzyme subfamily is involved in the metabolic clearance of numerous endogenous compounds including steroid hormones and bile acids, as well as exogenous agents including a variety of drugs and carcinogens (Stingl et al., 2014). UGT2B enzymes are well-expressed in the liver (Ohno and Nakajin, 2009), comprising approximately one-half of the hepatic UGT expression as determined in previous proteomic studies (Fallon et al., 2013).
The hepatically expressed UGT2B isoforms include 2B4, 2B7, 2B10, 2B11, 2B15, and 2B17 (Mackenzie et al., 2005; Nakamura et al., 2008; Ohno and Nakajin, 2009; Jones and Lazarus, 2014). Members of the UGT2B subfamily are comprised of six independent exons, exhibiting lower sequence homologies than either the 1A or 2A enzymes, which exhibit an exon-sharing structure. UGT2Bs play an important role in drug metabolism and carcinogen detoxification. Therefore, identifying novel mechanisms involved in the regulation of UGT2B enzymes may provide insight into the mechanisms underlying overall metabolism in humans.
Recent studies have examined the role of microRNA (miRNA) in regulating UGT expression (Papageorgiou and Court, 2017; Wijayakumara et al., 2017; Tatsumi et al., 2018). miRNAs are short nucleotide fragments that bind in the 3′-untranslated region (UTR) of mRNA and reduce translation of those transcripts, either by disruption of the mRNA structure through protein interactions, which initiate deadenylation of the poly-A tail and uncapping of the methylated 5′-end (Fabian et al., 2011), or by sequestration of the mRNA transcripts (Liu et al., 2005). UGT1A gene expression has been shown to be regulated by miR-491-3p, with miR-491-3p expression levels inversely correlated with UGT1A3 and UGT1A6 mRNA levels in normal human liver, contributing to the observed interindividual variability in the expression of these enzymes (Dluzen et al., 2014). In addition, miR-376c exhibited an inverse expression pattern with androgen-metabolizing UGT2B15 and 2B17 in prostate cancer cells (Wijayakumara et al., 2015). The opposite relationship between miR-376c and UGT2B15/UGT2B17 expression was shown in healthy prostate tissue, suggesting that low expression of miR-376c may contribute to prostate cancer development in androgen-dependent tumors by dysregulation of androgen signaling (Wijayakumara et al., 2015; Margaillan et al., 2016). miR-216b-5p was also shown to play a role in UGT2B regulation, with miR-216b-5p regulating UGT2B4, 2B7, and 2B10 in cell lines (Dluzen et al., 2016). A recent study identified miR-3664-3p as a potential regulator of UGT2B7 expression and activity (Wijayakumara et al., 2017), with an inverse correlation observed between UGT2B7 and miR-3664-3p expression in a panel of human tissues. Potential regulation of UGT2B4 by miR-135a-5p and miR-410-3p was also observed in the same study, with UGT2B4 expression reduced after miR-135a-3p and miR-410-3p overexpression, a pattern supported by a significant inverse expression correlation observed between UGT2B4 and miR-135a-5p as well as miR-410-3p in normal human tissues. In a screening of the 2048 microRNAs listed in miRbase version 19, Papageorgiou and Court (2017) recently identified several miRNAs that altered UGT2B7 and UGT2B15 activity in vitro. Of these, only the expression of miR-455-5p was shown to be correlated with altered UGT2B15 expression or glucuronidation activity in a panel of human livers; no miRNAs were associated with altered UGT2B7 expression or activity in the same specimens.
In the present study, a proteomics approach was implemented to screen a panel of normal human liver specimens for miRNAs potentially important in regulating UGT2B gene expression, with potential miRNA candidates examined for functionality in vitro. Results from the present study indicate that miRNA miR-485-5p may be a functional regulator of hepatic UGT2B10 and UGT2B7 expression.
Materials and Methods
Chemicals and Reagents.
The pGL3-promoter luciferase and pRL-TK renilla plasmids as well as Dual-Luciferase Reporter Assay Kits were obtained from Promega (Madison, WI). All synthesized DNA oligonucleotides used for 3′-UTR amplification, site-directed mutagenesis, and polymerase chain reaction (PCR) analysis were obtained from Integrated DNA Technologies, Inc. (Coralville, IA) or Life Technologies (Carlsbad, CA). Lipofectamine 3000 transfection reagent and Pierce BCA Protein Assay Kits were obtained from Life Technologies. Dulbecco’s modified Eagle’s medium, penicillin/streptomycin (10,000 U/ml), and Opti-MEM Reduced Serum Media were purchased from Gibco (Carlsbad, CA), while FBS and the SpectraMax DuoLuc Reporter Assay were purchased from VWR (Radnor, PA). Human liver microsomes were purchased from XenoTech, LLC (Kansas City, KS). miRVana miR-485-5p mimic (#MC10837), and miRVana negative control mimic #1 (#4464058) were purchased from Ambion/Thermo Fisher Scientific (Waltham, MA). The qScript miRNA cDNA Synthesis Kit and PerfeCta SYBR Green SuperMix were purchased from Quanta Biosciences (Beverly, MA). Nicotine, uridine 5′-diphosphoglucuronic acid, alamethicin, trifluoroacetic acid, formic acid, and bovine serum albumin (BSA) were all purchased from Sigma-Aldrich (St. Louis, MO). 3′-Azido-3′-deoxythymidine (AZT), 3′-azido-3′-deoxythymidine-methyl-d3-β-D-glucuronide (sodium salt; d3-AZT-glucuronide), 3′-azido-3′-deoxythymidine β-D-glucuronide (sodium salt; AZT-glucuronide), nicotine-β-D-glucuronide, and nicotine-d3-β-D-glucuronide were purchased from Toronto Research Chemicals (Toronto, Canada). Urea and dithiothreitol were purchased from Fisher Scientific (Pittsburgh, PA). Iodoacetamide and ammonium bicarbonate were purchased from Acros Organics (Morris Plains, NJ), N-tosyl-L-phenylalanine chloromethyl ketone–treated trypsin was obtained from Worthington Biochemical Corporation (Freehold, NJ), and lysyl endopeptidase was purchased from Wako Chemicals (Richmond, VA). Water Oasis HLB columns were obtained from Waters Corporation (Milford, MA), while mass spectrometer–grade acetonitrile was purchased from Avantor (Center Valley, PA). Indexed retention time standard solutions were purchased from Biognosys AG (Cambridge, MA). All other chemicals were purchased from Fisher Scientific unless specified otherwise.
Cells and Cell Culture Conditions.
The human embryonic kidney 293 and human hepatocellular carcinoma Hep3B cell lines were purchased from American Type Culture Collection (Manassas, VA). The human hepatocellular carcinoma HuH-7 cell line was a kind gift from Dr. Jianming Hu (Penn State University, Hershey, PA). All cell lines were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS and 1% penicillin/streptomycin, and grown and maintained in 5% CO2 at 37°C.
Tissues.
Liver specimens and corresponding total RNA (n = 62) were obtained from the tissue bank at the H. Lee Moffitt Cancer Center Tissue Procurement Facility (Tampa, FL). All 62 subjects from whom liver specimens were obtained were Caucasian, with women comprising 44% (n = 27) of these subjects. All specimens were obtained during resection for hepatocellular carcinoma, were deemed normal after pathology assessment, and were isolated and frozen at −70°C within 3 hours post surgery. All protocols involving the collection and analysis of tissue specimens from these tissue banks were approved by their respective Institutional Review Boards and were in accordance with assurances filed with and approved by the United States Department of Health and Human Services.
miRNA Binding Site Predictions.
TargetScan is an in silico microRNA-mRNA binding prediction database that was used to generate a list of candidate microRNAs. The criteria for selection of candidate microRNAs included: 1) prediction to bind to individual UGT2B isoforms with a Context++ percentile above the 95th percentile within the TargetScan program, 2) prediction to bind to one or more of the other UGT2B isoforms, and 3) expression of candidate microRNA in human liver.
Real-Time Quantitative PCR.
Total liver RNA concentrations were determined using a Nanodrop (ND-1000) spectrophotometer (Thermo Scientific), diluted, and stored as aliquots in RNase/DNase-free water in a −80°C freezer. The cDNAs were synthesized using the qScript miRNA cDNA Synthesis Kit. Real-time PCR was performed with PerfeCta SYBR Green SuperMix, cDNA corresponding to 50 ng of RNA, 100 nM of a universal reverse primer (provided by the PerfeCta Kit), and 100 nM of the specific miRNA forward primer for either miR-135a-5p, miR-146a-3p, miR-196a-5p, miR-216a-5p, miR-216b-5p, miR-379-3p, miR-485-5p, miR-590-5p, miR-152-3p, or miR-23b-3p (Supplemental Table 1) as per the manufacturer’s protocol. Reactions were performed in 15 µl total volume in 384-well plates using the BioRad CFX 384 real-time PCR machine, with incubations performed at 95°C for 2 minutes, followed by 40 cycles at 95°C for 10 seconds, 60°C for 15 seconds and 70°C for 15 seconds. Each plate included a no DNA negative control, and all reactions were performed in quadruplicate. Expression analysis used the comparative CT method, adapted using the method detailed in Lamba et al. (2014) and Vandesompele et al. (2002). Briefly, the geometric mean of the expression of both miR152-3p and miR-23b-3p, which is the square root of the CT value for miR-152-3p multiplied by the CT value for miR-23b-3p, is used as a combined endogenous control for data normalization. miR152-3p and miR-23b-3p were used as controls since they were shown in previous studies to be more stable across human liver specimens than other controls (including U6 and RNU6B). The ΔCT value for the candidate microRNA was calculated by subtracting the geometric mean of the combined controls from the CT value of the candidate microRNA. The −ΔCt value was then used as the exponent to calculate the relative expression using a logarithm base 2, as is standard when using comparative analysis. The CT values were determined using the CFX Manager software (BioRad Laboratories).
All real-time values were normalized based on primer efficiencies. Briefly, each miRNA mimic was used as a template in the qScript miRNA cDNA synthesis reaction, as described previously for the tissue samples. The resulting cDNA was diluted over a 5-log range and each dilution was used as a template in quantitative PCRs identical to those described previously for the tissue samples. The CT values for each dilution were plotted against the log of the dilution factor, and the slope of the regression line was calculated. The efficiency of each primer was calculated as E, where E = 10−1/slope (Pfaffl, 2004). Relative expression values for each miRNA in each tissue sample were calculated by first adjusting all CT values for primer efficiency using log2(ECT), and then calculating relative expression using the standard 2ΔCt method described by Pfaffl (2004).
Proteomics Sample Preparation.
Human liver microsomal fractions were prepared from individual liver specimens as previously described (Coughtrie et al., 1986; Wiener et al., 2004). Protein concentration was measured using the Pierce BCA protein assay. Protein digestion was performed as previously described (Shi et al., 2018). Briefly, aliquots of 80 μg protein from liver microsomes were mixed with 0.2 μg of BSA internal standard. The protein mixture was precipitated with precooled acetone and washed with 80% ethanol. Protein reduction was performed by adding 4 mM dithiothreitol in 8 M urea/100 mM ammonium bicarbonate at 37°C for 45 minutes, after which 20 mM iodoacetamide solution in 8 M urea/100 mM ammonium bicarbonate was added for alkylation at room temperature for 30 minutes in the dark. The urea concentration was adjusted to 6 M with 100 mM ammonium bicarbonate solution prior to digestion. Lysyl endopeptidase was added for the first digestion step at a protein-to-enzyme ratio of 100:1 at 37°C for 6 hours. Samples were then diluted to 1.6 M urea with 50 mM ammonium bicarbonate, followed by a second digestion step with trypsin at a protein-to-enzyme ratio of 50:1 for overnight incubation at 37°C. Digestion was terminated by adding 1 μl of trifluoroacetic acid. Digested protein peptides were extracted and purified using Waters Oasis HLB columns according to the manufacturer’s instructions. Eluted peptides were dried in a SpeedVac SPD1010 concentrator and reconstituted in 80 μl of 3% acetonitrile containing 0.1% formic acid. The peptide samples were centrifuged and supplemented with synthetic indexed retention time standard solutions prior to liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis.
LC-MS/MS–Based Protein Quantification.
Peptide samples were analyzed on a TripleTOF 5600+ mass spectrometer (AB Sciex, Framingham, MA) coupled with an Eksigent 2D Plus LC system (Eksigent Technologies, Dublin, CA). Liquid chromatography separation was performed via a trap-elute configuration including a trapping column (ChromXP C18-CL, 120 Å, 5 μm, 0.3 mm cartridge; Eksigent Technologies) and an analytical column (ChromXP C18-CL, 120 Å, 150 × 0.3 mm, 5 μm; Eksigent Technologies). The mobile phase consisted of 0.1% formic acid in water (phase A) and 0.1% formic acid in acetonitrile (phase B). A total of 6 μg protein was injected for analysis. Peptides were trapped and cleaned on the trapping column with phase A delivered at a flow rate of 10 μl/min for 3 minutes before being separated on the analytical column with a gradient elution at a flow rate of 5 μl/min. The gradient time program was set as follows for column equilibration of phase B: 0–68 minutes at 3%–30%; 68–73 minutes at 30%–40%; 73–75 minutes at 40%–80%; 75–78 minutes at 80%; 78–79 minutes at 80%–3%; and, finally, 79–90 minutes at 3%. The TripleTOF instrument was operated in positive ion mode with ion spray voltage floating at 5500 V, ion source gas 1 at 28 psi, ion source gas 2 at 16 psi, curtain gas at 25 psi, and source temperature at 280°C.
To generate the reference spectral library, information-dependent acquisition (IDA) was performed on three pooled human liver microsomal samples. The IDA method was set up with a 250-millisecond time-of-flight (TOF) mass spectrometry (MS) scan from 400 to 1250 Da, followed by an MS/MS scan in high-sensitivity mode from 100 to 1500 Da (50-millisecond accumulation time, 10 ppm mass tolerance, charge state from +2 to +5, rolling collision energy, and dynamic accumulation) of the top 30 precursor ions from the TOF-MS scan. The IDA data from the pooled liver microsomes were searched by MaxQuant (version 1.5.3.30; Max Planck Institute of Biochemistry, Martinsried, Germany). The human proteome fasta file with 20,237 protein entries downloaded from Uniport on March 2, 2017, was used as the reference sequences for the search. Trypsin/P was used as the protease. Peptide length was between 7 and 25 residues with up to two missed cleavage sites allowed. Carbamidomethyl was set as a fixed modification. A false discovery rate of 0.01 was used as the cutoff for both peptide and protein identification.
All liver microsomal samples were analyzed using a sequential window acquisition of all theoretical (SWATH) mass spectra method comprised of a 250-millisecond TOF-MS scan from 400 to 1250 Da, followed by MS/MS scans from 100 to 1500 Da performed on all precursors in a cyclic manner. The isolation scheme was set as SWATH (100 v/w). The accumulation time was 50 milliseconds per isolation window, resulting in a total cycle time of 2.8 seconds. The spectral alignment and target data extraction of the SWATH mass spectra data were performed on Skyline-daily (version 3.7.1.11271; University of Washington, Seattle, WA) with the reference spectral library generated from the IDA searches. The MS1 and MS/MS filtering were both set as TOF mass analyzer with resolution powers of 30,000 and 15,000, respectively. The retention time prediction was based on the autocalculated regression implemented in the indexed retention time calculator. Proper peak selection was checked manually with the automated assistance of Skyline-daily. The surrogate peptides used for the quantification of UGT2B4, 2B7, 2B10, 2B15, 2B17, and BSA are listed in Supplemental Table 2. These peptides were selected based on uniqueness and chromatographic performance. The peak areas of the top three-to-five fragment ions were summed and normalized to the internal standard BSA for quantification.
Construction of Reporter Plasmids.
The 3′-UTRs of UGT2B7, 2B10, and 2B15 were inserted into the XbaI restriction enzyme site lying 3′ of and adjacent to the luciferase gene within the pGL3-promotor vector (Promega) to simulate the sequence position in the native UGT2B mRNA transcript. To include the entire 3′-UTR sequence, primers were designed to amplify the full 3′-UTRs of UGT2B10 (1138 nucleotides) and UGT2B15 (467 nucleotides), flanked by the TCTAGA XbaI restriction sequence plus an additional four nucleotides to ensure the restriction site nucleotides were not lost during PCR amplification (Supplemental Table 1). The luciferase-containing plasmid used for analysis of the 3′-UTR of UGT2B7 (251 nucleotides) was created as described in a previous study (Dluzen et al., 2016).
To identify the miR-485-5p–binding site within the 3′-UTR of UGT2B10, four nucleotides within the miRNA recognition element (MRE) (see Fig. 1) identified by TargetScan were removed by site-directed mutagenesis, following the manufacturer’s protocols. To identify the miR-485-5p–binding site within the 3′-UTR of UGT2B7, two sites were similarly mutated by deleting four nucleotides in two potential MREs within the 3′-UTR of UGT2B7 (Fig. 1). The first site was selected based on its similarity to the predicted binding site of UGT2B10, with the exception of two uracils in the potential UGT2B7 MRE in place of two cytosines in the UGT2B10 MRE. The second site was selected due to the presence of five nucleotides with a Watson-Crick binding pair and a guanine-uracil (GU) wobble pair in the second nucleotide of the miRNA seed sequence. These potential binding sites are located 35–55 nucleotides (termed site 1) and 230–251 nucleotides (termed site 2) downstream from the UGT2B7 translational stop codon. Verification of the manually identified MREs within the 3′-UTRs of UGT2B7 and UGT2B10 was accomplished by removal of those predicted binding sites. Using the pGL3-UGT2B7 3′-UTR and the pGL3-UGT2B10 3′-UTR plasmids as templates, four nucleotides in each potential MRE were deleted using the mutagenesis primers listed in Supplemental Table 1. All sequences used in this study were confirmed by Sanger sequencing analysis carried out by Genewiz (Plainfield, NJ).
Fig. 1.
Prediction of miR-485-5p binding to UGT2B10 and UGT2B7 3′UTRs. The miRNA prediction algorithm TargetScan database was used to identify miRNA binding candidates for five UGT2B mRNAs: UGT2B4, 2B7, 2B10, 2B15, and 2B17. miR-485-5p was predicted to bind to the 3′-UTR of UGT2B10; two proposed binding sites for miR-485-5p to the 3′-UTR of UGT2B7 are shown. Numbers listed correspond to the MRE 3′ locations of the translational stop codon for the corresponding UGT.
Luciferase Assays.
Human embryonic kidney 293 cells were seeded onto 12-well plates at 50,000 cells per well. After 24 hours, cells were cotransfected with 380 ng of UGT2B 3′-UTR–containing pGL3 luciferase plasmid, 20 ng of the pRL-TK control vector, and either 100 nM scrambled miRNA control or 100 nM miR-485-5p mimic. For each well, 2 µl of Lipofectamine 3000 transfection reagent was added to the plasmids and miRNA mimics in Opti-MEM and incubated for 15 minutes according to the manufacturer’s protocol. Whole cell homogenates were lysed 24 hours post transfection using the passive lysis buffer provided with the Dual-Luciferase Reporter Assay Kit. Firefly and Renilla luciferase activity was measured separately with a luminometer upon addition of Luciferase Assay Reagent II and Stop & Glo Reagent, respectively (Bio-tek Synergy HT, Winooski, VT). All luciferase assay experiments were performed in triplicate, with each experimental and control group representing the results of triplicate treatments.
Glucuronidation Assays.
HuH-7 or Hep3B cells were transfected with either 100 nM scrambled miRNA controls or miR-485-5p mimic using 8 μl of Lipofectamine 3000 in 10 cm dishes, according to the manufacturer’s protocol. Cells were collected 48 hours post transfection. Cell pellets underwent five freeze-thaw cycles and were homogenized by passing them through a 27 gauge needle 10 times prior to storage at −80°C. Protein concentration was determined using the BCA Protein Assay Kit (Pierce Chemical, Rockford, IL) and measured using the Synergy Neo and Gen5 Data Analysis Software (BioTek).
Glucuronidation assays included HuH-7 or Hep3B cell homogenate (50 µg), substrate, 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, and 4 mM uridine 5′-diphosphoglucuronic acid, and were performed at 37°C. For UGT2B7 activity assessment, cell homogenate was incubated with 5 mM AZT for 60 minutes; for UGT2B10, cell homogenate was incubated with 500 µM nicotine for 2 hours (Chen et al., 2007; Walsky et al., 2012). Incubation times were confirmed to be within the linear phase as determined in time-course experiments. Glucuronidation reactions were spiked with either d3-AZT-glucuronide for UGT2B7 or d3-nicotine glucuronide for UGT2B10, and terminated by adding an equal volume of cold 100% acetonitrile. Reactions were centrifuged for 10 minutes at 16,000g and supernatant was collected for analysis on LC-MS/MS. Human liver microsomes were used as a positive control, with d3-labeled standards spiked into each sample to confirm peak identity. As a negative control, assays containing human liver microsomes were performed without UDP-glucuronic acid. Glucuronide metabolites were analyzed on a Sciex QTRAP 6500 LC-MS/MS System, using the specific conditions listed subsequently. Quantification was based on multiple independent assays utilizing internal standards and validated standard curves of the glucuronide metabolites, using Sciex MultiQuant 2.1 software. A minimum of four experiments was performed for each assay.
AZT-glucuronide formation was analyzed using a 1.7 µm ACQUITY UPLC BEH C18 analytical column (2.1 mm × 50 mm; Waters) in series with a 0.2 µm Waters assay frit filter (2.1 mm). Gradient elution was performed at a flow rate of 0.4 ml/min, starting with 95% buffer A (5 mM ammonium acetate 0.1% formic acid) and 5% buffer B (100% acetonitrile) for 30 seconds, a subsequent linear gradient to 50% buffer B over 2.5 minutes, and then 95% buffer B maintained over the next 2 minutes for re-equilibration. AZT-glucuronide and d3-AZT-glucuronide were monitored using multiple reactions monitoring with mass-to-charge ratio (m/z) transitions of m/z 442.000 →125.100 for AZT-glucuronide and m/z 445.000 → 128.100 for d3-AZT-glucuronide. Identification of AZT-glucuronide was confirmed using an AZT-glucuronide standard.
Nicotine glucuronides were analyzed using a Waters ACQUITY UPLC BEH HILIC 1.7 µm analytic column (2.1 × 100 mm). Gradient conditions were performed at a 0.4-ml/min flow rate, starting with 80% buffer B (5 mM NH4Ac in 90% acetonitrile) for 1 minute and 50 seconds, followed by a steep gradient to 100% buffer A (5 mM NH4Ac in 60% acetonitrile) for 3 minutes, and then back to 80% buffer B for the remainder of the 10-minute run. Nicotine glucuronide was determined using multiple reactions monitoring, with mass transitions of m/z 339.1 → 163.1 for nicotine glucuronide and m/z 163.1 → 106.0 for d3-nicotine glucuronide.
Statistical Analysis.
The entire set (n = 62) of liver specimens available to the investigators was used for the liver proteomics analysis; an initial power calculation was not performed a priori. Statistical analysis was performed using Graphpad Prism 5 software (La Jolla, CA). For all studies examining the effects of overexpressed miR-485-5p miRNA mimic, the one-tailed Student’s t test was performed. The correlation between UGT2B protein levels and miRNA expression was described by the Pearson correlation coefficient with a one-tailed P value. Error bars presented in Figs. 3 and 4 are S.D.
Fig. 3.
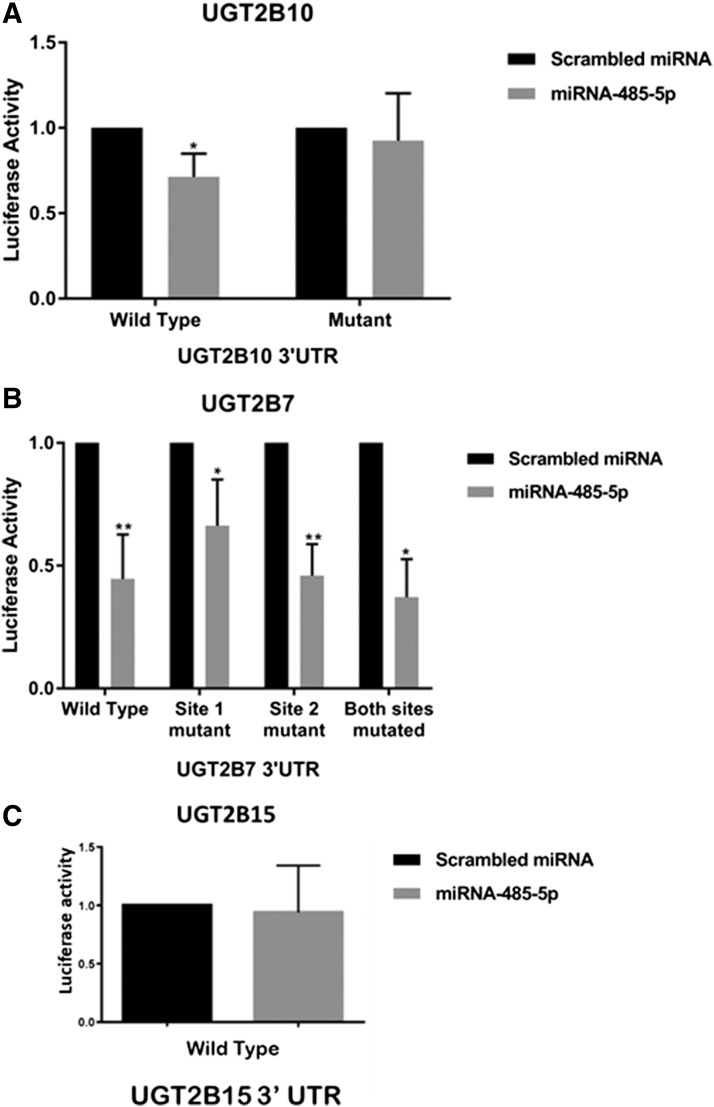
UGT2B 3′-UTR luciferase activity in the presence of miR-485-5p mimics. (A) The pGL3-UGT2B10 3′-UTR luciferase vector (Wild Type) contains 1086 nucleotides of the UGT2B10 3′-UTR including the MRE for miR-485-5p. The pGL3-UGT2B10 3′-UTR MRE deletion vector (Mutant) contains a four-nucleotide deletion within the potential miR-485-5p MRE seed sequence. The two columns on the left-hand side were performed using transfections of the luciferase vector containing the wild-type UGT2B10 3′-UTR; the two columns on the right-hand side were performed using the luciferase vector containing the UGT2B10 3′-UTR with its seed sequence deletion mutation. (B) The pGL3-UGT2B7 3′-UTR luciferase vector contains 251 nucleotides of the UGT2B7 3′-UTR including the MRE for each of the two predicted binding sites for miR-485-5p. The pGL3-UGT2B7 3′-UTR MRE deletion vectors each contain a four-nucleotide deletion within the potential miR-485-5p MRE seed sequence. The two columns on the left-hand side were performed using transfections of the luciferase vector containing the wild-type UGT2B7 3′-UTR; the next six columns on the right-hand side were performed using the luciferase vector containing the UGT2B7 3′-UTR with seed sequence deletion mutations at site 1, site 2, or both sites together, as indicated in the graph. (C) The pGL3-UGT2B15 3′-UTR luciferase vector contains 467 nucleotides of the UGT2B15 3′-UTR. Luciferase reporter vectors were cotransfected into human embryonic kidney 293 cells with the pRL-TK renilla control vector along with 100 nM of miR-485-5p mimic or scrambled miRNA control. Columns represent the mean ± S.D. of at least three independent experiments and are normalized to the scrambled miRNA-transfected control. *P < 0.05; **P < 0.01, all compared with the referent scrambled control miRNA cotransfected with the luciferase vector containing the corresponding wild-type UGT2B 3′-UTR.
Fig. 4.
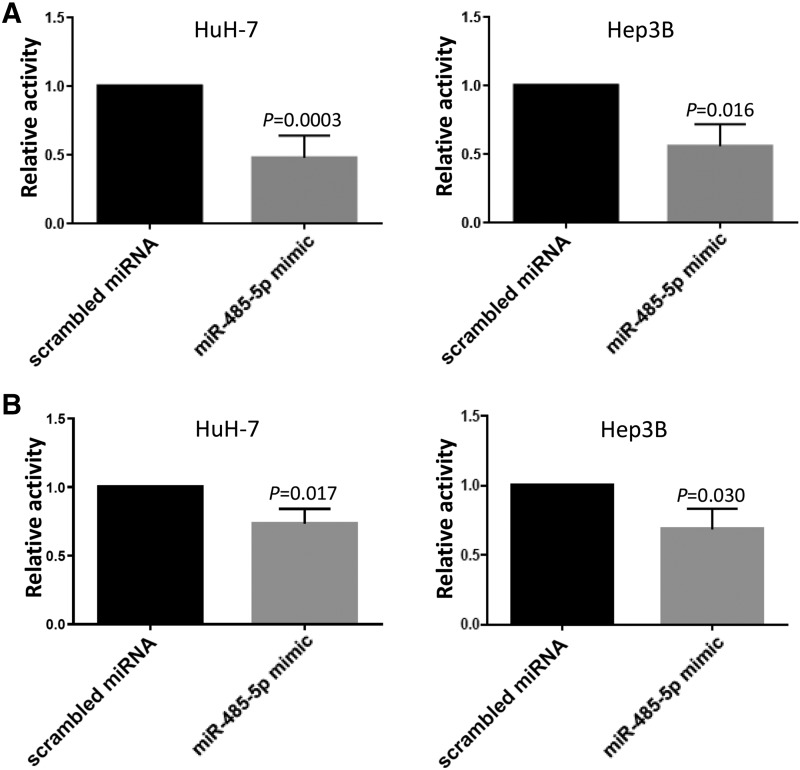
Glucuronidation activity in the presence of miR-485-5p mimic. (A) UGT2B10 glucuronidation activity against nicotine in HuH-7 cells transfected with 100 nM miR-485-5p mimic or 100 nM of scrambled miRNA control in HuH-7 and Hep3B cells. Columns represent the mean ± S.E. of four independent experiments. (B) UGT2B7 glucuronidation activity against AZT in HuH-7 cells transfected with 100 nM scrambled or miR-485-5p mimic. Columns represent the mean ± S.D. of six independent experiments.
Results
The miRNAs predicted to bind to UGT2B mRNA were identified using TargetScan. Candidate miRNAs were selected if the TargetScan context score was in the 95th percentile for at least one of the UGT2Bs and the miRNA was predicted to bind to at least one other 2B member. While miR-485-5p was only predicted to bind to a single UGT2B (UGT2B10), it was included in the current study because only miR135a-5p exhibited a higher score profile (including context++ score and conserved branch length). The final miRNA candidates (with their predicted targeted UGT2Bs in parenthesis) were miR-379-5p (UGT2B4 and UGT2B10), miR-135a-5p (UGT2B4, 2B10, 2B15, and 2B17), miR-590-5p (UGT2B4, 2B15, and 2B17), miR-216a-5p (UGT2B7 and 2B10), miR-216b-5p (UGT2B4, 2B7, and 2B10), miR-146a-3p (UGT2B7 and 2B10), miR-485-5p (UGT2B10), and miR-196a-5p (UGT2B4 and 2B7).
The expression of candidate miRNAs was examined by real-time PCR using total RNA from a panel of 62 individual normal human liver specimens. All candidate miRNAs amplified successfully, with miR-590-5p exhibiting the highest expression, followed by miR-379-5p and miR-216b-5p (Table 1). While miR-485-5p was the least expressed miRNA, it was expressed with a CT value of less than 29 for all 62 liver specimens examined.
TABLE 1.
Quantitative PCR data for miRNA in normal human liver specimensa
| MicroRNA | Mean | S.D. | Maximum Value | Minimum Value |
|---|---|---|---|---|
| miR-379-5p | 0.6540 | 0.3377 | 1.7136 | 0.2418 |
| miR-135a-5p | 0.0290 | 0.0400 | 0.2792 | 0.0007 |
| miR-590-5p | 1.1699 | 0.7811 | 4.4453 | 0.2338 |
| miR-216a-5p | 0.0716 | 0.0505 | 0.2085 | 0.0127 |
| miR-216b-5p | 0.0215 | 0.0197 | 0.0934 | 0.0004 |
| miR-146a-3p | 0.0089 | 0.0061 | 0.0268 | 0.0022 |
| miR-196a-5p | 0.2286 | 0.8009 | 6.3672 | 0.0140 |
| miR-485-5p | 0.0031 | 0.0020 | 0.0111 | 0.0009 |
The miRNA expression levels were normalized using the method detailed in Lamba et al. (2014).
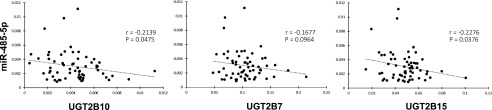
The relative abundance of each of the UGT2B enzymes within the same panel of human liver specimens was determined by LC-MS/MS SWATH mass spectra analysis. Pearson correlation analysis was performed to test if miRNA levels were negatively associated with individual UGT protein levels. A significant inverse correlation was observed between the expression of both UGT2B10 (P = 0.047, r = −0.214) and UGT2B15 (P = 0.038, r = −0.228) and the expression of miR-485-5p in the 62 human liver specimens (Fig. 2). A trend toward an inverse correlation was also observed for miR-485-5p and the expression of UGT2B7 (P = 0.096, r = −0.167). For both UGT2B4 and UGT2B17, there were no significant associations with the expression of any of the tested miRNA candidates in the 62 human liver specimens examined in this study. For UGT2B17, five samples were removed from the data set since they were previously shown to be from individuals homozygous for a known UGT2B17 whole-gene deletion polymorphism (Gallagher et al., 2007), and therefore exhibited no UGT2B17 expression regardless of miRNA levels. None of the other miRNAs tested exhibited a significant correlation (i.e., with a P < 0.05) between the expression of any of the UGT2B enzymes and any of the other seven miRNAs tested.
Fig. 2.
Expression of UGT2B7, 2B10, and 2B15 vs. miR-485-5p in human liver samples. Expression levels of miR-485-5p were quantified using quantitative reverse-transcription PCR and normalized to the geometric mean of miR-152-3p and miR-23b-3p in the same samples. The Y-axis represents the natural log transformed values for miR-485-5p. The X-axis represents the ratio of UGT2B7, 2B10, or 2B15 to the BSA internal standard, transformed to the natural log value.
To test if overexpressed miR-485-5p negatively regulated the expression of UGT2B enzymes, the 3′-UTRs of UGT2B7, 2B10, and 2B15 were cloned downstream from the luciferase gene within the pGL-3 reporter vector. Each vector was cotransfected with pRL-TK Renilla transfection control plasmid and either 100 nM scrambled miRNA control or miR-485-5p mimic. A significant (P = 0.022) 29% decrease in luciferase activity was observed after cotransfection of 100 nM miR-485-5p mimic with the wild-type UGT2B10 3′-UTR–containing pGL-3 vector in human embryonic kidney 293 cells (Fig. 3A). When comparing cells transfected with the UGT2B10 3′-UTR seed deletion mutant plasmid, there was a near-identical level of luciferase activity observed for cells cotransfected with miR-485-5p compared with cells cotransfected with the scrambled mimic control miRNA. A significant (P = 0.0060) 55% decrease in luciferase activity was observed for cells cotransfected with 100 nM of miR-485-5p mimic together with the pGL-3 vector containing the UGT2B7 3′-UTR (Fig. 3B). No effect on luciferase activity was observed for cells cotransfected with 100 nM of miR-485-5p mimic and the pGL-3 vector containing the 3′-UTR of UGT2B15 (Fig. 3C).
In silico analysis of 3′-UTR sequences by TargetScan only predicted an MRE site in UGT2B10. However, proteomic and luciferase assay data suggested that miR-485-5p also potentially interacts with the 3′-UTR of UGT2B7. To identify the mRNA sequence that miR-485-5p may be binding to in the UGT2B7 3′-UTR sequence, two potential miR-485-5p MREs were manually identified within the UGT2B7 3′-UTR, and each was mutated individually to remove four nucleotides from their seed sequence. For site 1, nucleotides corresponding to 45–48 downstream from the stop codon were deleted, while nucleotides corresponding to 245–248 downstream from the stop codon were deleted for site 2 (see Fig. 1). A third plasmid was also created with both sites mutated. As shown in Fig. 3B, similar levels of inhibition of luciferase activity were observed in cells transfected with any of the three UGT2B7 3′-UTR–mutated luciferase plasmids when cells were cotransfected with miR-485-5p mimic versus scrambled mimic. These data suggest that neither of the manually predicted sites were acting as MREs for miR-485-5p within the UGT2B7 3′-UTR.
In previous studies, several UGT2B enzymes including UGT2B10 and 2B7 were shown to be expressed in both the HuH-7 and Hep3B cell lines (Dluzen et al., 2016). The expression of these UGTs in these cell lines was confirmed in the present study, with UGT2B7 demonstrating between 5- and 20-fold higher levels of expression than UGT2B10 in the two lines (data not shown). These cell lines also exhibit endogenous expression of miR-485-5p (Zhang et al., 2018), making them excellent cell line models to examine the potential regulation of 2B enzymes by miR-485-5p. The expression of miR-485-5p in both cell lines was confirmed in the present study, with it being expressed at levels that were ∼1.8-fold higher in Hep3B cells compared with HuH-7 cells (data not shown). HuH-7 and Hep3B cells were transfected with miR-485-5p mimic and in vitro glucuronidation assays were performed using homogenate from miRNA-transfected cells and either the UGT2B10-specific substrate, nicotine, or the UGT2B7-specific substrate, AZT. Significant decreases in nicotine glucuronide formation of 52% and 44% were observed in HuH-7 (P = 0.0003) and Hep3B (P = 0.016) cell homogenates, respectively, after overexpression of 100 nM miR-485-5p mimic in assays containing 500 µM nicotine (Fig. 4A). Similarly, significant decreases of 27% (P = 0.017) and 31% (P = 0.030) in AZT-glucuronide formation were observed in HuH-7 and Hep3B, respectively, after transfection with 100 nM miR-485-5p (Fig. 4B).
Discussion
The exploration of potential miRNA regulators of metabolizing enzymes may provide insight into the variability of an individual’s ability to detoxify UGT2B-specific substrates. Using a proteomic approach, the current study examined the expression of five hepatic UGT2B enzymes and eight miRNAs predicted in silico to bind to UGT2B 3′-UTR sequences in 62 normal human liver samples. While UGT2B10 and 2B15 were the only UGT2B enzymes to exhibit a statistically significant inverse expression relationship with one of the miRNA candidates examined in this study (miR-485-5p), UGT2B7 showed a similar but nonsignificant inverse relationship with the same miRNA, indicating the potential for downregulation of multiple UGT2B family members by miR-485-5p. An effect by miR-485-5p on the expression of both UGT2B10 and 2B7 was confirmed in assays with a luciferase vector containing the 3′-UTR of either UGT2B10 or UGT2B7; a similar effect was not observed for UGT2B15. This effect was further validated in two different cell lines, with overexpression of the miR-485-5p mimic resulting in significantly decreased UGT2B10 and UGT2B7 activities against known specific substrates in HuH7 and Hep3B cell homogenates. These data strongly suggest that miR-485-5p plays an important role in regulating the expression of these two UGT enzymes. In addition, the current study measured UGT2B protein levels in a panel of liver specimens by SWATH-MS analysis, with expression levels of candidate miRNA quantified by quantitative PCR in the same specimens. The ability to measure total protein levels in biologic samples using this technique can provide a highly sensitive and reproducible tool (Collins et al., 2017) to be applied to the study of miRNA regulation in specific tissues.
UGT2B10 is a key enzyme involved in the N-glucuronidation of many drugs that contain an amine group (Lu et al., 2017), including the antipsychotics loxapine, clozapine, and chlorpromazine, as well as the tricyclic amine drugs amitriptyline and imipramine (Kato et al., 2013). A diminished ability to detoxify these drugs could lead to liver toxicity in individuals with low expression of UGT2B10. In addition, UGT2B10 is the major UGT responsible for the clearance of nicotine and its major metabolite, cotinine (Chen et al., 2007), and is important in the metabolism and detoxification of tobacco-specific nitrosamines present in tobacco and smokeless tobacco products (Chen et al., 2008). Therefore, downregulation of UGT2B10 by miRNA including miR-485-5p could potentially lead to higher levels of exposure to nicotine or tobacco-specific nitrosamines, which could affect nicotine addiction and susceptibility to the carcinogenic effects of tobacco-specific nitrosamines.
In silico analysis predicted a miR-485-5p/UGT2B10 3′-UTR MRE located at 280–300 base pairs 3′ of the translation stop codon, and deletion analysis of this MRE site demonstrated a specific effect by overexpression of the miR-485-5p mimic. The UGT2B7 3′-UTR contains no predicted miR-485-5p MRE within its 3′-UTR based on an in silico screening using TargetScan; however, the predicted MRE in the UGT2B10 3′-UTR is similar to a region in the UGT2B7 3′-UTR located 35 nucleotides downstream from the UGT2B7 translational stop codon (site 1) (see Fig. 1). This proposed UGT2B7 MRE contains two uracils in place of two cytosines within the UGT2B10 MRE, and one less binding pair at the 3′-UTR end of this potential UGT2B7 MRE. The GU wobble in RNA may account for predictions that include guanine and uracil pairs in place of Watson and Crick binding partners. Another possible site (site 2) for binding within the 3′-UTR of UGT2B7 is located 230 nucleotides downstream from its translational stop codon. This site shares only six nucleotides with the predicted UGT2B10 MRE with one GU pair wobble pair and an additional A in the flanking 3′-end of the UGT2B7 mRNA sequence. Interestingly, deletion of these sites within the UGT2B7 3′-UTR of the pGL3 vector resulted in no diminished effect by miR-485-5p mimic overexpression on luciferase activity, suggesting that neither of these sites acts as a functional MRE within the UGT2B7 3′-UTR for miR-485-5p.
The other members of the UGT2B family in this study did not show any significant associations between hepatic UGT2B protein and expression of the miRNAs examined. Although there were no predicted binding sites within the 3′-UTR of UGT2B15 for miR-485-5p, luciferase assays were performed for this gene due to a significant inverse expression relationship observed between miR-485-5p and UGT2B15 expression in the panel of liver specimens examined in this study. In addition, the UGT2B15 3′-UTR sequence contains a possible MRE located 132 nucleotides downstream from the UGT2B15 stop codon, where the eight nucleotide binding pairs on the 3′ end of the miRNA include two GU wobble pairs. However, unlike that observed for UGT2B10 and 2B7, no effect on luciferase activity was observed for in vitro assays using a luciferase vector containing the UGT2B15 3′-UTR.
Several of the miRNAs selected as candidates for this study were predicted to bind to multiple UGT2B isoforms. While enzymes within the UGT1A and UGT2A (excluding UGT2A3) subfamilies share the same 3′-UTR, the UGT2B genes are independent of each other, exhibiting no exon sharing. However, there is relatively high homology between 3′-UTRs for several of the UGT2B genes. While the 3′-UTR for UGT2B7 is only roughly one-third the length of UGT2B10, this sequence exhibits over 80% homology to comparable sequences within UGT2B10 as determined using the ExPASy Bioinformatics Resource Portal SIM alignment tool (https://www.expasy.org). However, only miR-485-5p demonstrated a significant association with UGT2B expression, and only for UGT2B10 and 2B7. Previous studies identified miR-491-3p as a candidate to bind to the 3′-UTR of all UGT1A isoforms, which contain the same 3′-UTR (Dluzen et al., 2014). miR-491-3p was shown to regulate the expression of UGT1A1, 1A3, and 1A6, but not UGT1A4 or 1A9, and significant inverse associations were found in human liver specimens between miR-491-3p and the expression of both UGT1A3 and 1A6 but not UGT1A1. It was suggested that differences in secondary structure could be responsible for this differential regulation (Dluzen et al., 2014). A similar mechanism could also be playing a role in the differential regulation of UGT2B enzymes.
miR-485-5p has been shown to be involved in the regulation of a number of other cellular pathways. The long noncoding RNA, NEAT1, was found to reduce the levels of miR-485-5p expression, and a corresponding increase in STAT3 expression was observed in both HuH-7 and Hep3B cells (Zhang et al., 2018). When STAT3 is repressed, hepatocellular carcinoma (HCC) cell migration and invasion is reduced and low levels of miR-485-5p are associated with increased HCC cancer metastasis (Zhang et al., 2018). miR-485-5p has also been linked to liver cancer by targeting the extracellular matrix metalloproteinase inducer. Reduced miR-485-5p expression and associated increases in extracellular matrix metalloproteinase inducer expression were correlated with increased HCC tumor progression and metastasis (Sun et al., 2015). A similar association between low miR-485-5p expression and HCC tumor progression and metastasis was observed in studies by Guo et al. (2015), with stanniocalcin 2 identified as a potential miR-485-5p target. Other studies have implicated miR-485-5p regulation of several tumor suppressor genes, with high miR-485-5p expression associated with greater tumor suppression and better patient prognosis (Kang et al., 2015; Lou et al., 2016; Huang et al., 2018). The results of the present study demonstrate an effect by miR-485-5p on UGT2B10 and UGT2B7 expression and suggest that variations in miR-485-5p expression could also play an important role in liver toxicity in patients treated with drugs metabolized by these two enzymes, including epirubicin, zidovudine, and codeine for UGT2B7 (Innocenti et al., 2001; Court et al., 2003), and loxapine, amitriptyline, and chlorpromazine for UGT2B10 (Lu et al., 2017).
Previous in vitro studies had implicated miR-216b-5p in the regulation of several UGT2B enzymes (Dluzen et al., 2016). While the expression of miR-216b-5p was shown to be expressed in pooled liver RNA, an analysis of miR-216b versus UGT2B expression was not performed in human tissues in that study. Interestingly, a significant inverse relationship between the expression of miR-216b-5p and UGT2B enzymes was not observed in the 62 liver specimens examined in the present study. Potentially, differences in the expression of miR-216b-5p in hepatic tissue versus cell lines could help explain this discrepancy. miR-216b-5p was shown to be expressed up to 900-fold lower in liver than in some other human tissues including pancreas (Dluzen et al., 2016). Interestingly, overexpression of UGT2B4, 2B7, and 2B15 reduced lipid accumulation in the Panc-1 pancreatic cell line (Dates et al., 2015). Excess lipid levels in cancer cells are known to contribute to proliferation in pancreatic cells (Beloribi-Djefaflia et al., 2016), suggesting that it is possible that miR-216b-5p is playing a far more critical role in regulating the expression of UGT2Bs in the pancreas than in the liver.
In summary, the present study suggests that miR-485-5p exhibits a regulatory role for UGT2B10 and UGT2B7. This regulation could impact liver toxicity in response to doses considered safe for a number of the top over-the-counter and prescription drugs.
Acknowledgments
The authors thank the Mass Spectrometry Core facility at Washington State University Spokane for their help with LC-MS/MS.
Abbreviations
- AZT
3′-azido-3′-deoxythymidine
- BSA
bovine serum albumin
- GU
guanine-uracil
- HCC
hepatocellular carcinoma
- IDA
information-dependent acquisition
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- miRNA
microRNA
- MRE
miRNA recognition element
- MS
mass spectrometry
- m/z
mass-to-charge ratio
- PCR
polymerase chain reaction
- SWATH
sequential window acquisition of all theoretical
- TOF
time of flight
- UGT
UDP-glucuronosyltransferase
- UTR
untranslated region
Authorship Contributions
Participated in research design: Sutliff, Watson, Chen, Zhu, Lazarus.
Conducted experiments: Sutliff, Shi, Hunt.
Contributed new reagents or analytic tools: Sutliff, Shi, Watson, Hunt, Chen, Zhu, Lazarus.
Performed data analysis: Sutliff, Shi, Watson, Hunt, Chen, Zhu, Lazarus.
Wrote or contributed to the writing of the manuscript: Sutliff, Shi, Watson, Chen, Zhu, Lazarus.
Footnotes
Funding for these studies was provided by the National Institutes of Health National Institutes of Environmental Health Sciences [Grant R01-ES025460] (to P.L.); the National Institutes of Health National Heart, Lung, and Blood Institute [Grant R01-HL126969] (to H.-J.Z.); and the Health Sciences and Services Authority of Spokane County [Grant WSU002292] (to the Washington State University College of Pharmacy and Pharmaceutical Sciences).
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Beloribi-Djefaflia S, Vasseur S, Guillaumond F. (2016) Lipid metabolic reprogramming in cancer cells. Oncogenesis 5:e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Blevins-Primeau AS, Dellinger RW, Muscat JE, Lazarus P. (2007) Glucuronidation of nicotine and cotinine by UGT2B10: loss of function by the UGT2B10 codon 67 (Asp>Tyr) polymorphism. Cancer Res 67:9024–9029. [DOI] [PubMed] [Google Scholar]
- Chen G, Dellinger RW, Sun D, Spratt TE, Lazarus P. (2008) Glucuronidation of tobacco-specific nitrosamines by UGT2B10. Drug Metab Dispos 36:824–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins BC, Hunter CL, Liu Y, Schilling B, Rosenberger G, Bader SL, Chan DW, Gibson BW, Gingras AC, Held JM, et al. (2017) Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat Commun 8:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughtrie MW, Burchell B, Bend JR. (1986) A general assay for UDPglucuronosyltransferase activity using polar amino-cyano stationary phase HPLC and UDP[U-14C]glucuronic acid. Anal Biochem 159:198–205. [DOI] [PubMed] [Google Scholar]
- Court MH, Krishnaswamy S, Hao Q, Duan SX, Patten CJ, Von Moltke LL, Greenblatt DJ. (2003) Evaluation of 3′-azido-3′-deoxythymidine, morphine, and codeine as probe substrates for UDP-glucuronosyltransferase 2B7 (UGT2B7) in human liver microsomes: specificity and influence of the UGT2B7*2 polymorphism. Drug Metab Dispos 31:1125–1133. [DOI] [PubMed] [Google Scholar]
- Dates CR, Fahmi T, Pyrek SJ, Yao-Borengasser A, Borowa-Mazgaj B, Bratton SM, Kadlubar SA, Mackenzie PI, Haun RS, Radominska-Pandya A. (2015) Human UDP-glucuronosyltransferases: effects of altered expression in breast and pancreatic cancer cell lines. Cancer Biol Ther 16:714–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dluzen DF, Sun D, Salzberg AC, Jones N, Bushey RT, Robertson GP, Lazarus P. (2014) Regulation of UDP-glucuronosyltransferase 1A1 expression and activity by microRNA 491-3p. J Pharmacol Exp Ther 348:465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dluzen DF, Sutliff AK, Chen G, Watson CJ, Ishmael FT, Lazarus P. (2016) Regulation of UGT2B expression and activity by miR-216b-5p in liver cancer cell lines. J Pharmacol Exp Ther 359:182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, et al. (2011) miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol 18:1211–1217. [DOI] [PubMed] [Google Scholar]
- Fallon JK, Neubert H, Hyland R, Goosen TC, Smith PC. (2013) Targeted quantitative proteomics for the analysis of 14 UGT1As and -2Bs in human liver using NanoUPLC-MS/MS with selected reaction monitoring. J Proteome Res 12:4402–4413. [DOI] [PubMed] [Google Scholar]
- Gallagher CJ, Muscat JE, Hicks AN, Zheng Y, Dyer AM, Chase GA, Richie J, Lazarus P. (2007) The UDP-glucuronosyltransferase 2B17 gene deletion polymorphism: sex-specific association with urinary 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol glucuronidation phenotype and risk for lung cancer. Cancer Epidemiol Biomarkers Prev 16:823–828. [DOI] [PubMed] [Google Scholar]
- Guo GX, Li QY, Ma WL, Shi ZH, Ren XQ. (2015) MicroRNA-485-5p suppresses cell proliferation and invasion in hepatocellular carcinoma by targeting stanniocalcin 2. Int J Clin Exp Pathol 8:12292–12299. [PMC free article] [PubMed] [Google Scholar]
- Huang RS, Zheng YL, Li C, Ding C, Xu C, Zhao J. (2018) MicroRNA-485-5p suppresses growth and metastasis in non-small cell lung cancer cells by targeting IGF2BP2. Life Sci 199:104–111. [DOI] [PubMed] [Google Scholar]
- Innocenti F, Iyer L, Ramírez J, Green MD, Ratain MJ. (2001) Epirubicin glucuronidation is catalyzed by human UDP-glucuronosyltransferase 2B7. Drug Metab Dispos 29:686–692. [PubMed] [Google Scholar]
- Jones NR, Lazarus P. (2014) UGT2B gene expression analysis in multiple tobacco carcinogen-targeted tissues. Drug Metab Dispos 42:529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Ren MP, Zhao L, Li CP, Deng MM. (2015) miR-485-5p acts as a negative regulator in gastric cancer progression by targeting flotillin-1. Am J Transl Res 7:2212–2222. [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Izukawa T, Oda S, Fukami T, Finel M, Yokoi T, Nakajima M. (2013) Human UDP-glucuronosyltransferase (UGT) 2B10 in drug N-glucuronidation: substrate screening and comparison with UGT1A3 and UGT1A4. Drug Metab Dispos 41:1389–1397. [DOI] [PubMed] [Google Scholar]
- Lamba V, Ghodke-Puranik Y, Guan W, Lamba JK. (2014) Identification of suitable reference genes for hepatic microRNA quantitation. BMC Res Notes 7:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. (2005) MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 7:719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou C, Xiao M, Cheng S, Lu X, Jia S, Ren Y, Li Z. (2016) MiR-485-3p and miR-485-5p suppress breast cancer cell metastasis by inhibiting PGC-1α expression. Cell Death Dis 7:e2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Xie Q, Wu B. (2017) N-glucuronidation catalyzed by UGT1A4 and UGT2B10 in human liver microsomes: assay optimization and substrate identification. J Pharm Biomed Anal 145:692–703. [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, Miners JO, Owens IS, Nebert DW. (2005) Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics 15:677–685. [DOI] [PubMed] [Google Scholar]
- Margaillan G, Lévesque É, Guillemette C. (2016) Epigenetic regulation of steroid inactivating UDP-glucuronosyltransferases by microRNAs in prostate cancer. J Steroid Biochem Mol Biol 155:85–93. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Nakajima M, Yamanaka H, Fujiwara R, Yokoi T. (2008) Expression of UGT1A and UGT2B mRNA in human normal tissues and various cell lines. Drug Metab Dispos 36:1461–1464. [DOI] [PubMed] [Google Scholar]
- Ohno S, Nakajin S. (2009) Determination of mRNA expression of human UDP-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction. Drug Metab Dispos 37:32–40. [DOI] [PubMed] [Google Scholar]
- Papageorgiou I, Court MH. (2017) Identification and validation of the microRNA response elements in the 3′-untranslated region of the UDP glucuronosyltransferase (UGT) 2B7 and 2B15 genes by a functional genomics approach. Biochem Pharmacol 146:199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. (2004) Quantification strategies in real-time PCR, in A-Z of Quantitative PCR (Bustin SA. ed) pp 87–112, CA International University Line, La Jolla, CA. [Google Scholar]
- Shi J, Wang X, Lyu L, Jiang H, Zhu HJ. (2018) Comparison of protein expression between human livers and the hepatic cell lines HepG2, Hep3B, and Huh7 using SWATH and MRM-HR proteomics: focusing on drug-metabolizing enzymes. Drug Metab Pharmacokinet 33:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl JC, Bartels H, Viviani R, Lehmann ML, Brockmöller J. (2014) Relevance of UDP-glucuronosyltransferase polymorphisms for drug dosing: a quantitative systematic review. Pharmacol Ther 141:92–116. [DOI] [PubMed] [Google Scholar]
- Sun X, Liu Y, Li M, Wang M, Wang Y. (2015) Involvement of miR-485-5p in hepatocellular carcinoma progression targeting EMMPRIN. Biomed Pharmacother 72:58–65. [DOI] [PubMed] [Google Scholar]
- Tatsumi N, Tokumitsu S, Nakano M, Fukami T, Nakajima M. (2018) miR-141-3p commonly regulates human UGT1A isoforms via different mechanisms. Drug Metab Pharmacokinet 33:203–210. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsky RL, Bauman JN, Bourcier K, Giddens G, Lapham K, Negahban A, Ryder TF, Obach RS, Hyland R, Goosen TC. (2012) Optimized assays for human UDP-glucuronosyltransferase (UGT) activities: altered alamethicin concentration and utility to screen for UGT inhibitors. Drug Metab Dispos 40:1051–1065. [DOI] [PubMed] [Google Scholar]
- Wiener D, Fang JL, Dossett N, Lazarus P. (2004) Correlation between UDP-glucuronosyltransferase genotypes and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone glucuronidation phenotype in human liver microsomes. Cancer Res 64:1190–1196. [DOI] [PubMed] [Google Scholar]
- Wijayakumara DD, Hu DG, Meech R, McKinnon RA, Mackenzie PI. (2015) Regulation of human UGT2B15 and UGT2B17 by miR-376c in prostate cancer cell lines. J Pharmacol Exp Ther 354:417–425. [DOI] [PubMed] [Google Scholar]
- Wijayakumara DD, Mackenzie PI, McKinnon RA, Hu DG, Meech R. (2017) Regulation of UDP-glucuronosyltransferases UGT2B4 and UGT2B7 by MicroRNAs in liver cancer cells. J Pharmacol Exp Ther 361:386–397. [DOI] [PubMed] [Google Scholar]
- Zhang XN, Zhou J, Lu XJ. (2018) The long noncoding RNA NEAT1 contributes to hepatocellular carcinoma development by sponging miR-485 and enhancing the expression of the STAT3. J Cell Physiol 233:6733–6741. [DOI] [PubMed] [Google Scholar]