Abstract
Due to the combined advantages of cellulose and nanoscale (diameter 20–60 nm), bacterial cellulose possesses a series of attractive features including its natural origin, moderate biosynthesis process, good biocompatibility, and cost-effectiveness. Moreover, bacterial cellulose nanofibers can be conveniently processed into three-dimensional (3D) intertwined structures and form stable paper devices after simple drying. These advantages make it suitable as the material for construction of organ-on-a-chip devices using matrix-assisted sacrificial 3D printing. We successfully fabricated various microchannel structures embedded in the bulk bacterial cellulose hydrogels and retained their integrity after the drying process. Interestingly, these paper-based devices containing hollow microchannels could be rehydrated and populated with relevant cells to form vascularized tissue models. As a proof-of-concept demonstration, we seeded human umbilical vein endothelial cells (HUVECs) into the microchannels to obtain the vasculature and inoculated the MCF-7 cells onto the surrounding matrix of the paper device to build a 3D paper-based vascularized breast tumor model. The results showed that the microchannels were perfusable, and both HUVECs and MCF-7 cells exhibited favorable proliferation behaviors. This study may provide a new strategy for constructing simple and low-cost in vitro tissue models, which may find potential applications in drug screening and personalized medicine.
Keywords: Bacterial cellulose, embedded 3D printing, sacrificial 3D printing, microphysiological systems, vascularization, breast cancer
Graphical Abstract
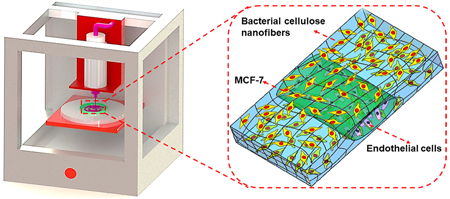
The three-dimensional (3D) printing technology circumvents many limitations associated with current biofabrication strategies through the combination of precisely controlled 3D structure,1,2 as well as accurately positioned biomaterials3,4 and cells.5 In particular, the biomimetic 3D structures enabled by 3D printing could allow for the generation of in vitro tissue models that reproduce the physiology or pathophysiology of their in vivo niches.6–10 The biomaterial matrixes provide multiple biological, structural, and mechanical supports in the regulation of cell behaviors and tissue morphogenesis,11–15 while cells derived from individuals or patients allow for personalization of these microphysiological systems.16,17
Conventionally, complex composite structures with hollow interiors reveal unique manufacturing challenges where internal tooling,18 commonly referred to as a core or mandrel, is required to define the microchannel internal features.19 Any configuration in which the mandrel is clamped within a composite component, requires sacrifice of tools or a more complex, foldable or inflatable method.20 At present, the sacrificial tooling technology uses materials including eutectic salts, ceramics, cast urethanes, and other similar materials.21–24 These technologies present challenges including (i) difficulty in handling due to fragile material properties; (ii) requirement for production with special tools; and (iii) limited design freedom attributing to production or the methods of removal.25
To this end, 3D printing again has emerged as a technique that allows for convenient creation of sacrificial molds with complex geometries for various applications including those in biomedicine.26 This process is based on the construction of a template using rapid prototyping technologies such as extrusion bioprinting.27–30 The deposited sacrificial molds are then used to form tissue-like constructs from various matrix materials containing complex, hollow microchannel structures after template removal. Nevertheless, many of the biomaterials and (bio)inks used in sacrificial 3D printing are relatively expensive limiting their utilization in the large-scale fabrication, for example, of tissue models for precision medicine applications. We therefore hypothesized that bacterial cellulose, an organic compound with the formula (C6H10O5)n biosynthesized by Gluconacetobacter xylinus (G. xylinus), has the potential to be used as the matrix material in sacrificial 3D printing, providing a cost-effective means to generate biomimetic tissue models. Indeed, bacterial cellulose has previously been reported for usage in tissue engineering of cartilage, skin, dental, nerve, and blood vessels, among others.31–36 It has also been manufactured in large quantities for various applications including medical implants for approximately 30 years.35 Many studies demonstrated that bacterial cellulose is a promising biomaterial due to its large specific surface area, abundance of surface hydroxyl groups,37 diverse and ascendant mechanical properties,38,39 cytocompatibility,40,41 and the fact that they are biodegradable and renewable.42,43
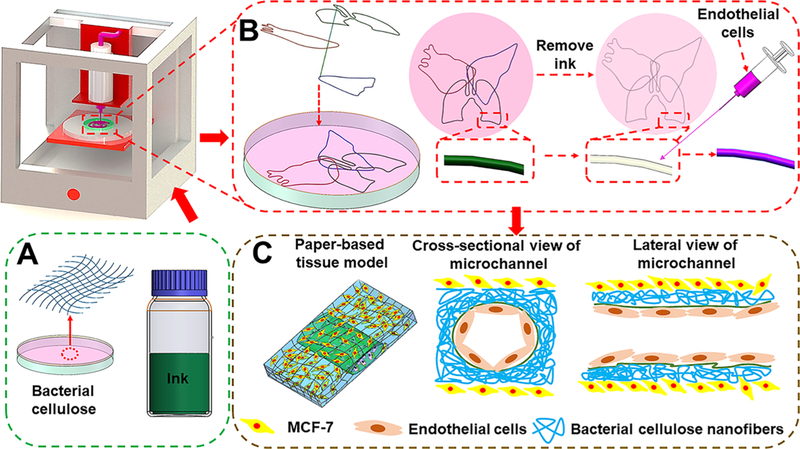
Here we report a new strategy of matrix-assisted sacrificial 3D printing for in vitro tissue modeling using a hydrophobic fugitive ink directly deposited within a bacterial cellulose hydrogel matrix, to form templates of arbitrary shapes (Figure 1). Upon drying to form a paper-like membrane, the fugitive ink could be selectively extracted, forming hollow perfusable microchannels. Such paper devices containing microchannels could be stored in the dry state possessing essentially infinite shelf life, and be rehydrated to fabricate relevant tissue models as needed, providing a simple and low-cost platform suitable for cell growth and amenable to large-scale application. The bacterial cellulose is not biodegradable.44,45 In addition, unlikewood cellulose, the bacterial cellulose hydrogel can be obtained without any hydrophilic modification of the cellulose material such as carboxymethylation46,47 or nanofibrillation.48,49 Meanwhile, the microchannels of the paper devices did not need any hydrophobic treatment to maintain their stability in culture medium, enabled by the superfine, nanoscale, but long bacterial cellulose fibers making the devices. These characteristics would in our case, facilitate the construction of in vitro tissue models for drug screening by allowing for extended culture periods. Indeed, hydrophobic treatment is required for paper devices constructed from shorter wood cellulose nanofiber-based matrixes to achieve water-resistance and structural stability when wetted.50 As a proof-of-concept study, MCF-7 cells were seeded onto the paper matrix of the device, while human umbilical vein endothelial cells (HUVECs) were used to populate the surface of the microchannels, creating a vascularized breast tumor model (Figure 1C).
Figure 1.
Schematics showing the procedure of fabricating a flexible microfluidic thin paper device by the matrix-assisted sacrificial 3D printing strategy. (A) Bacterial cellulose matrix and the ink of petroleum jelly-liquid paraffin. (B) Fabrication of a 3D paper-based construct containing multilayered microchannels. (C) Formation of a vascularized breast tumor model using the printed paper device.
Numerous studies have shown the capabilities of 3D printing for use in tissue and tissue model biofabrication.51,52 However, the relatively high costs of inks and matrix materials limit their large-scale production. To solve this problem, we present a strategy based on bacterial cellulose matrixes combined with 3D sacrificial printing, allowing us to conveniently generate embedded complex microchannel patterns at low cost. The schematic diagram in Figure 1B shows the fabrication process of the paper-based devices containing perfusable hollow microchannels for producing tissue models. Specifically, the hydrophobic petroleum jelly-liquid paraffin was chosen as the fugitive ink attributed to its temperature-responsiveness and immiscibility with the hydrophilic bacterial cellulose hydrogel matrix, facilitating its in-matrix 3D printing and subsequent removal from the matrix.
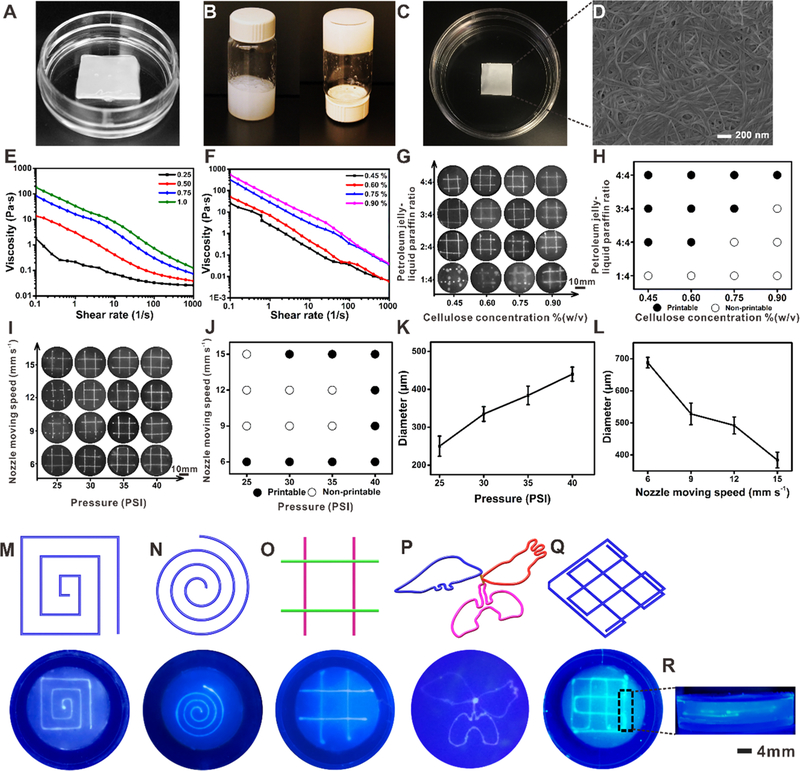
The bacterial cellulose membrane (Figure 2A) was obtained by culturing cost-effective G. xylinus through a multilayer fermentation method.53 The bacterial cellulose suspension (bacterial cellulose membranes were pulped with a mechanical homogenizer) possessed a hydrogel-like consistency when the concentration was higher than 0.9 w/v % due to the densely entangled matrix structure (Figure 2B). Then, the bacterial cellulose hydrogel was subject to air-drying to form a paper-like flexible membrane (Figure 2C and Figure S1). The scanning electron microscope (SEM) image showed that the bacterial cellulose membrane consisted of a network of intertwined superfine but long nanofibers, where the diameters of the nanofibers fell in the range of 20–60 nm and the pore size of the bacterial cellulose matrixes was 216.52 ± 77 nm (Figure 2D). The 3D network of the nanofibers and the porosity of bacterial cellulose matrixes (~38%, Figure S2A) could be helpful for the exchange of nutrients, oxygen to the cells, and wastes between the layers.54 This porosity could also enable the diffusion of cells during seeding and their subsequent migration and proliferation. The interconnected pores of the matrixes along with the hydrophilicity of the bacterial cellulose were proven to effectively hold medium with water retention values of more than 96% (Figure S2B), which would facilitate cell growth upon rehydration.
Figure 2.
Formation of bacterial cellulose hydrogel and printing performance of the petroleum jelly-liquid paraffin inks inside bacterial cellulose hydrogel matrixes. (A) Photograph of the original bacterial cellulose membrane. (B) Photograph of the bacterial cellulose hydrogel matrix. (C, D) Photograph and SEM image, respectively, of the bacterial cellulose film formed by air-drying of a bacterial cellulose hydrogel matrix. (E) Viscosity profiles of the fugitive inks with different ratios of petroleum jelly to liquid paraffin as a function of shear rate. (F) Viscosity profiles of various concentrations of bacterial cellulose hydrogel matrixes as a function of shear rate. (G) and (H) Printing performance at various bacterial cellulose hydrogel concentrations and ink compositions. Filled circles: printable. Open circles: nonprintable. (I) and (J) Printability of the inks in relation to pressure and moving speed of the printhead. Filled circles: printable. Open circles: nonprintable. (K) Diameter of the extruded microfibers in relation to pressure at a constant printhead moving speed of 15 mm-s−1. (L) Diameter of the extruded microfibers in relation to printhead moving speed at a constant pressure of 35 psi. (M)–(O) Designs and photographs of continuously extruded single-layer patterns in bacterial cellulose hydrogel matrix. (P)–(R) Designs and photographs of multilayer patterns printed in bacterial cellulose hydrogel matrix.
During a 3D printing process as the nozzle moved inside the bacterial cellulose hydrogel matrix, the fugitive petroleum jelly-liquid paraffin ink was deposited as microfibers (Movie S1). Petroleum jelly was blended with liquid paraffin at different ratios to improve its rheological properties and enhance its printability at room temperature. The viscosity of the petroleum jelly-liquid paraffin-based ink gradually increased with elevated petroleum jelly ratio from 1:4 to 4:4 (petroleum jelly:liquid paraffin, Figure 2E). Since the rheological properties of the bacterial cellulose matrix was also a contributing factor, four different concentrations of bacterial cellulose matrixes were selected and their viscosities were measured. The viscosity of the bacterial cellulose matrix increased as the bacterial cellulose concentration increased within the shear rate range evaluated (Figure 2F). At the low viscosity of the ink at the ratio of 1:4, complete lines could not be formed in the bacterial cellulose matrix (when the latter also at low concentrations) (Figure 2G,H).46 On the contrary, the instability of the deposited ink was compensated by the high concentration of the bacterial cellulose matrix, and the areas of the microfibers were close to the defined dimensions. Therefore, the 0.6 v/v% of bacterial cellulose matrix and the petroleum jelly-liquid paraffin ink at the ratio of 3:4 were selected for all subsequent experiments.
We further examined the influence of nozzle moving speed and extrusion pressure on the printability. The map of printability presented that a balance between the nozzle moving speed and extrusion pressure achieved appropriate structural fidelity to obtain precisely printed patterns (Figure 2I,J). Within the printable conditions, the diameter of the deposited microfibers increased from 250 ± 26.6 (25 psi) to 440 ± 18.7 μm (40 psi) as a result of the increase of extrusion pressure from 25 to 40 psi, at a constant printhead moving speed of 15 mm-s−1 (Figure 2K). Meanwhile, the diameter of the deposited microfibers decreased from 688 ± 16.5 (6 mm s−1) to 384 ± 24.5 μm (15 mm s−1) when the nozzle moving speed was elevated from 6 to 15 mm-s−1, at a constant pressure of 35 psi (Figure 2L). The results indicated that increased diameter of lines was formed due to the larger amount of inks extruded under higher pressures, while the smaller size of lines was facilitated by the increasing nozzle moving speed.26 Therefore, 35 psi and 15 mm-s−1 were selected as the optimized extrusion pressure and nozzle moving speed, respectively.
To illustrate that the petroleum jelly-liquid paraffin ink could be continuously extruded within the bacterial cellulose matrix, we designed and printed two single-layered linear patterns that contained right-angle corners (Figure 2M) and that resembled a spiral arc (Figure 2N). It was clear that the structures were sufficiently stable during the printing processes without noticeable pinching of the ink and were smooth without localized ink accumulation, even at corners in the angular geometry when the nozzle moving directions were rapidly changed. Several multilayered patterns of the petroleum jelly-liquid paraffin ink were subsequently printed into the bacterial cellulose hydrogel matrixes (Figure 2O–Q), which showed well-separated ink microfibers in adjacent layers (Figure 2R), indicating that complex interconnected networks could be realized. These different patterns of the 3D-printed constructs maybe utilized to simulate various types of native tissues to meet the need of constructing different vascularized structures.
In a typical procedure of sacrificial 3D printing, the microfibrous templates deposited are selectively removed to form hollow microchannels that are supported by the surrounding matri.50,55,56 We adopted a similar process to obtain hollow, perfusable microchannels in the bacterial cellulose matrix-based paper devices. Figure 3 shows the structure of a paper device containing multilayered microchannels using the sacrificial 3D printing technique. The first step extruded the fugitive petroleum jelly-liquid paraffin ink in separate layers within the bacterial cellulose hydrogel matrix. During the process of air drying, the bacterial cellulose hydrogel matrix flattened into a paper membrane that was still flexible, and no damages to the overall structure as well as the microchannels was observed (Figure 3A). Subsequently, the ink was removed from the microchannels of the dried paper film. The solid ink of petroleum jelly-liquid paraffin was liquefied by elevating the temperature to 70 °C, which rendered the ink easily removable from the dried matrix to form perfusable microchannels. Then, the membrane was washed with n-hexane and centrifuged to remove the ink from the microchannels. Moreover, the membrane was washed with ethanol and centrifuged triple times to remove n-hexane. Finally, the membrane was washed with distilled water three times and air-dried again at room temperature to form a clean paper device containing hollow, perfused microchannels (Figure 3B). The bacterial cellulose hydrogel matrix was demonstrated to have certain levels of self-healing ability possibly owing to the presence of hydroxyl groups in these molecules that allowed for the formation of intramolecular and intermolecular hydrogen bonds.57 In addition, during the drying process the capillary force would further facilitate the self-healing event.46 Indeed, our scratch experiment suggested that while the final cellulose matrix was slightly thinner in the region of scratch, the groove almost disappeared during the drying process (Figure S3).
Figure 3.
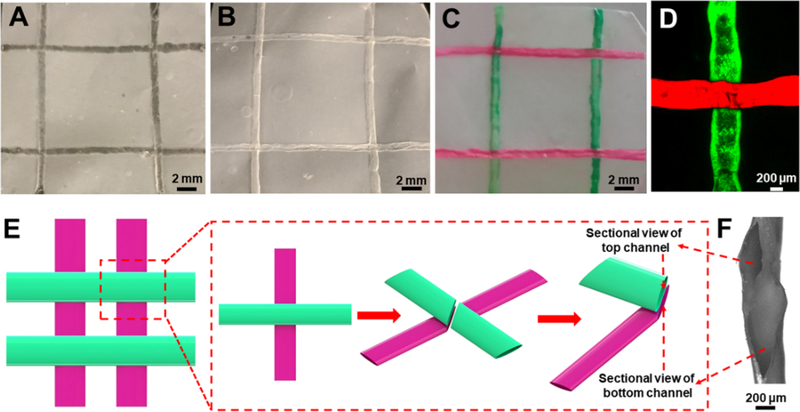
Paper-based devices containing hollow, perfusable, multilayered microchannels with sacrificial 3D printing. (A) Photograph of the paper device before removing the ink. (B) Photograph of the paper device after removing the ink. (C) Photograph of the paper device perfused with two different dye solutions in the embedded microchannels at two different layers. (D) Fluorescence micrograph showing the two microchannels perfused with different fluorescence microparticles at the cross-section. (E) Schematic diagram of the multilayered microchannels in the paper device. (F) Cross-sectional optical micrograph showing the two microchannels separated by a bacterial cellulose barrier in between.
To visualize the embedded microchannels in the paper device, two solutions containing differently colored dyes (Figure 3C, Movie S2) or fluorescent microparticles (Figure 3D) were injected. The solutions were successfully perfused into the two sets of crossing microchannels without mixing and no leakage of the liquid could be observed, indicating the 3D nature of the device possessing the microchannels at two depths. Indeed, a dense bacterial cellulose membrane was formed between the two crossing microchannels during the drying process, functioning as the barrier at the junction (Figure 3E,F). As such, with the proposed matrix-assisted sacrificial 3D printing method, it was possible to create 3D perfusable microchannels in paper devices. Importantly, the 3D nature of the initially deposited fugitive microfibers could be well-preserved after ink removal and drying, facilitating applications in tissue model biofabrication.
The ability to accurately screen anticancer pharmaceutical compounds can be potentially promoted through the use of biomimetic tumor models that better predict human responses, which plays a pivotal role in drug development.58 Microfluidic devices have been combined with tumor cells to create the vascularized tumor microenvironments59 allowing for kinetic examination of drug effects and cancer progression.60 Previously, we have also demonstrated the inclusion of sacrificially printed microchannels in gelatin-based hydrogel constructs to study diffusion-dependent drug toxicity.61
As a proof-of-concept study, we generated a vascularized breast tumor model by seeding green fluorescence protein (GFP)-HUVECs inside the microchannel, and MCF-7 cells were seeded onto the matrix of the paper device. It should be noted that, while the dried paper devices allow for extremely long shelf life and can be stored for extended periods of time, the devices can be rapidly rehydrated prior to tissue model fabrication, forming a critical advantage of our technology. In our case, we rehydrated the paper devices through autoclave, which at the same time served as the sterilization process. Autoclaving did not seem to induce any noticeable damage to our paper devices.
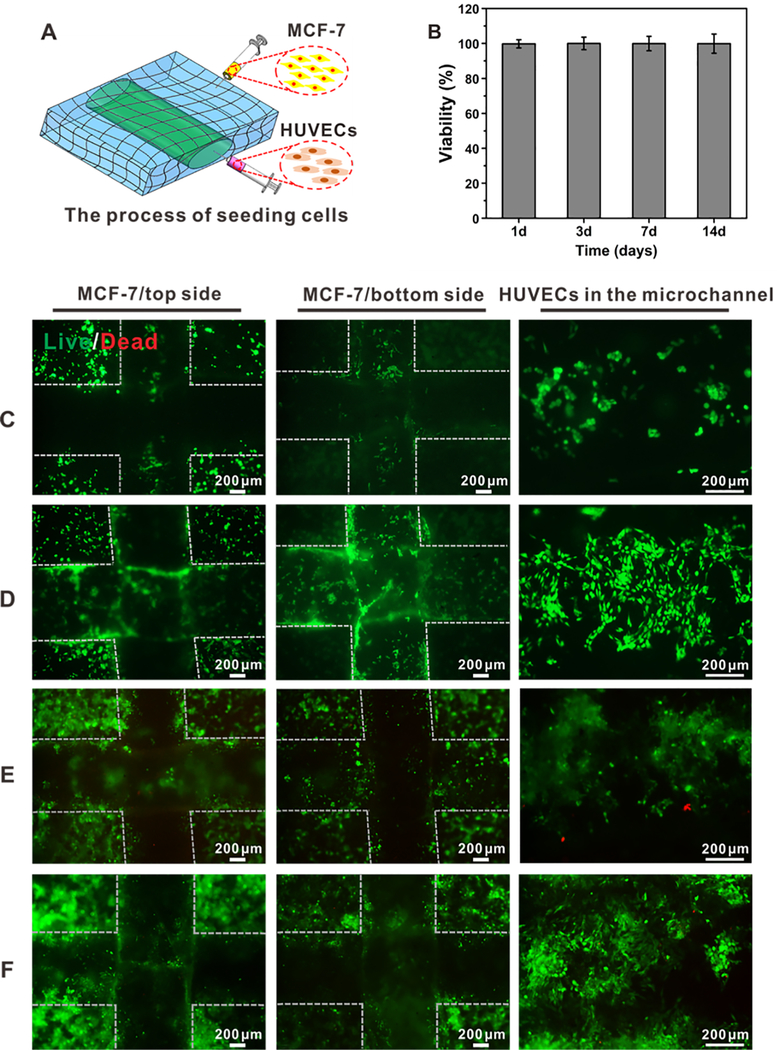
To create a uniform monolayer of endothelial cells attached to the inner surface of the microchannels, the device was manually flipped at 30 min after a suspension of GFP-HUVECs was injected into the microchannel, and maintained still for another 30 min, following which MCF-7 cells were seeded onto surfaces on both sides of the paper device (Figure 4A). The fluorescence micrographs indicated that the HUVECs and MCF-7 cells were homogeneously distributed on the microchannels and within the matrixes of the 3D paper-based devices, and the constructs could maintain their intact structures. Live/dead staining further revealed that the viabilities of HUVECs and MCF-7 cells in the constructs were both high (Figure 4C–F). The percentages of viable cells within the microchannels and in the matrixes exceeded 90% at 1, 3, 7, and 14 days of culture (Figure 4B). Furthermore, cell proliferation assessment was further performed (Figure S4). These results indicated that the ink could be completely removed with n-hexane, and there was perhaps no residue that would affect the behaviors of the two types of cells. In addition, it denoted the good cytocompatibility of the bacterial cellulose matrix, consistent with previous reports.62,63 As such, it was suggested that the porous network of bacterial cellulose nanofibers enabled cells to attach and proliferate, potentially allowing us to expand to cultures of various cell types for tissue model construction.64
Figure 4.
Cell viability in the paper-based vascularized breast tumor model. (A) Schematic showing the seeding procedure of the two cell types, HUVECs in the microchannel and MCF-7 cells on the paper matrix. (B) Quantitative analysis of viable cells at 1, 3, 7, and 14 days of culture. (C)–(F) Fluorescence micrographs showing live/dead staining at 1, 3, 7, and 14 days of culture, respectively. The dotted lines demarcate the borders of the microchannels.
During the 14 days of culture, endothelial cells and tumor cells proliferated, which covered the interior surface of the microchannel and were uniformly grown on the matrix, respectively. The projection and reconstructed confocal microscopy images of the MCF-7 cells in the paper matrix are shown in Figure 5A,B. However, the MCF-7 cells did not seem to migrate into the paper matrix but mostly proliferated along its surfaces (Figure S5), possibly due to the presence of dense, intertwined cellulose nanofibers (Figure 2D). At 14 days, the HUVECs also proliferated and became confluent to form an intact endothelium in the microchannel (Figure 5C,D). The super-imposed confocal Z-project reconstruction image in Figure 5E clearly indicated the intact endothelium on the surface of the microchannel and the densely populated MCF-7 cells in the paper matrix. Favorable spreading and proliferation of both cell types indicated the good potential of these paper-based devices for engineering complex tissue models with built-in vascular networks. In particular, the vascular endothelial growth factor produced by the tumor cells could possibly have caused rapid proliferation of the endothelial cells.65
Figure 5.
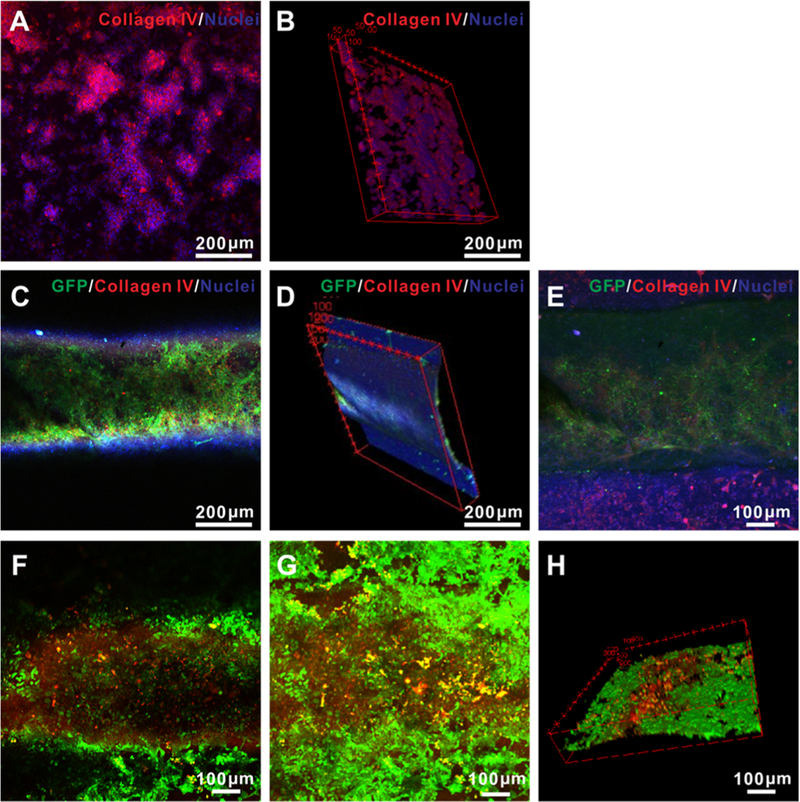
(A)–(E) Confocal microscopy images of (A) projection and (B) cross-sectional views showing the matrix of the 3D paper-based device populated with MCF-7 cells immunostained for collagen IV (red) and nuclei (blue), after 14 days of culture. (C) Projection and (D) cross-sectional views of the microchannel region populated with GFP-HUVECs in the 3D paper-based device, immunostained for collagen IV (red) and nuclei (blue) after 14 days of culture. (E) Projection view of the entire volume showing copresence of both MCF-7 cells in the matrix and HUVECs in the microchannel. (F)–(H) Confocal microscopy images of (F) single-layer view, (G) projection view, and (H) 3D views of live/dead staining of both MCF-7 cells in the matrix and HUVECs in the microchannel treated with tamoxifen at 10 μM for 48 h.
To evaluate the drug response of the vascularized breast tumor constructs, 10 μM of tamoxifen was injected from the endothelialized microchannels, and the paper devices were cultured for another 48 h. The spatial distribution of the live and dead cells in our vascularized breast tumor model was further revealed in superimposed confocal images, revealing cytotoxicity (Figure 5F–H). While still preliminary, these results have indicated the potential of our printed paper-based devices in constructing in vitro tissue models for applications in drug screening and possibly personalized medicine.
In summary, we have reported a strategy to rapidly fabricate biomimetic tissue models based on bacterial cellulose matrix using sacrificial 3D printing. The optimal balance of rheological properties between the bacterial cellulose matrix and the fugitive ink enabled stable 3D printing, while dehydration of the bacterial cellulose matrix and removal of the ink led to generation of well-separated microchannels in the 3D volume inside an ultrafine paper film. Using this strategy, microchannel structures with different degrees of complexity can be generated in bacterial cellulose paper films designed to suit for construction of different tissue models. More interestingly, these dried paper devices may be stored for extended periods of time, and upon rehydration, the 3D volumes are restored to facilitate cell growth. Since bacterial cellulose is nonbiodegradable and the matrix does not disperse in the medium, the devices should in principle last relatively long during in vitro cultures. In addition, all the materials including the matrix (bacterial cellulose) and the ink (petroleum jelly liquid paraffin) were cost-effective. Our calculations revealed that a single device was less than 4 cents in cost, as shown in Table S1 and Figure S6.We anticipate that our unique paper-based devices containing hollow, perfusable microchannels enabled by sacrificial 3D printing, which feature long shelf lives, highly porous structure, high water-holding capacity, and good biocompatibility, will likely provide a new platform for constructing low-cost tissue models potentially at larger scales. In fact, 3D printing was selected as the manufacturing method due to its automation process, which will further bring down the fabrication cost to a very low value in the longer-term through high-throughput robotic dispensing processes, further making the fabrication potentially scalable.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (K99CA201603, R00CA201603, R21EB025270, R21EB026175, R01EB028143), the Brigham Research Institute, and the New England Anti-Vivisection Society. X.C. acknowledges the support from the National Natural Science Foundation of China (81503025).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nano-lett.9b00583.
Materials and methods, photograph of the flexibility of bacterial cellulose film, porosity and water retention of the paper devices, photographs showing self-healing ability of the bacterial cellulose hydrogel matrix, quantification of proliferation of the cells, confocal microscopy images, dimensions of a typical paper device used to generat the tumor models, table of material cost analysis (PDF)
3D printing process of the petroleum jelly-liquid paraffin ink inside the bacterial cellulose hydrogel matrix (MP4) Injection of colored dyes to visualize the embedded microchannels in the paper device (MP4)
The authors declare no competing financial interest.
REFERENCES
- (1).Gao Q; He Y; Fu JZ; Liu A; Ma L Biomaterials 2015, 61, 203–215. [DOI] [PubMed] [Google Scholar]
- (2).Kingsley DM; Dias AD; Chrisey DB; Corr DT Biofabrication 2013, 5,No. 045006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Ma X; Yu C; Wang P; Xu W; Wan X; Lai CSE; Liu J; Koroleva-Maharajh A; Chen S Biomaterials 2018, 185, 310–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Kant RJ; Coulombe KL K. Acta Biomater. 2018, 69, 42–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Gao T; Gillispie GJ; Copus JS; Pr AK; Seol YJ; Atala A; Yoo JJ; Lee SJ Biofabrication 2018, 10, No. 034106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Prendergast ME; Montoya G; Pereira T; Lewicki J; Solorzano R; Atala A Microphysiological Systems 2018, 2 (2), 1. [Google Scholar]
- (7).Wang Z; Lee SJ; Cheng HJ; Yoo JJ; Atala A Acta Biomater. 2018, 70, 48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Koch L; Deiwick A; Franke A; Schwanke K; Haverich A; Zweigerdt R; Chichkov B Biofabrication 2018, 10, No. 035005. [DOI] [PubMed] [Google Scholar]
- (9).Moroni L; Burdick JA; Highley C; Lee SJ; Morimoto Y; Takeuchi S; Yoo JJ Nat. Rev. Mater. 2018, 3, 21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Zhang YS; Arneri A; Bersini S; Shin SR; Zhu K; Goli-Malekabadi Z; Aleman J; Colosi C; Busignani F; Dell’Erba V; Bishop C; Shupe T; Demarchi D; Moretti M; Rasponi M; Dokmeci MR; Atala A; Khademhosseini A Biomaterials 2016, 110, 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Mongera A; Rowghanian P; Gustafson HJ; Shelton E; Kealhofer DA; Carn EK; Serwane F; Lucio AA; Giammona J; Campas O Nature 2018, 561, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Atallah P; Schirmer L; Tsurkan M; Putra Limasale YD; Zimmermann R; Werner C; Freudenberg U Biomaterials 2018,181, 227–239. [DOI] [PubMed] [Google Scholar]
- (13).Liu Z; Tang M; Zhao J; Chai R; Kang J Adv. Mater. 2018, 30, No. 1705388. [DOI] [PubMed] [Google Scholar]
- (14).Jang J; Park JY; Gao G; Cho DW Biomaterials 2018, 156, 88–106. [DOI] [PubMed] [Google Scholar]
- (15).Mieremet A; Rietveld M; van Dijk R; Bouwstra JA; El Ghalbzouri A Tissue Eng. Part A 2018, 24, 873–881. [DOI] [PubMed] [Google Scholar]
- (16).Ahadian S; Civitarese R; Bannerman D; Mohammadi MH; Lu R; Wang E; Davenport-Huyer L; Lai B; Zhang B; Zhao Y; Mandla S; Korolj A; Radisic M Adv. Healthcare Mater. 2018, 7, 1700506. [DOI] [PubMed] [Google Scholar]
- (17).Shih YV; Varghese S Biomaterials 2019, 198, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Qu J; Ye F; Chen D; Feng Y; Yao Q; Liu H; Xie J; Yang J Adv. Colloid Interface Sci. 2016, 230, 29–53. [DOI] [PubMed] [Google Scholar]
- (19).Postiglione G; Alberini M; Leigh S; Levi M; Turri S ACS Appl. Mater. Interfaces 2017, 9, 14371–14378. [DOI] [PubMed] [Google Scholar]
- (20).Gu L; Yu G; Li CW Anal. Chim. Acta 2018, 997, 24–34. [DOI] [PubMed] [Google Scholar]
- (21).Foster EJ; Zahed N; Tallon C Small 2018, 14, 1870215. [DOI] [PubMed] [Google Scholar]
- (22).Wu G; Jacobi von Wangelin A Chem. Sci. 2018, 9, 1795–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Sapoznik E; Niu G; Zhou Y; Prim PM; Criswell TL; Soker S PLoS One 2018, 13, No. e0192654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Prasopthum A; Shakesheff KM; Yang J Biofabrication 2018, 10, No. 025002. [DOI] [PubMed] [Google Scholar]
- (25).Cerchiari A; Garbe JC; Todhunter ME; Jee NY; Pinney JR; LaBarge MA; Desai TA; Gartner ZJ Tissue Eng. Part C 2015, 21, 541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhang YS; Davoudi F; Walch P; Manbachi a.; Luo X; Dell’Erba V; Miri AK; Albadawi H; Arneri A; Li X; Wang X; Dokmeci MR; Khademhosseini A; Oklu R Lab Chip 2016, 16, 4097–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Daly AC; Pitacco P; Nulty J; Cunniffe GM; Kelly DJ Biomaterials 2018, 162, 34–46. [DOI] [PubMed] [Google Scholar]
- (28).Sochol RD; Sweet E; Glick CC; Venkatesh S; Avetisyan A; Ekman KF; Raulinaitis A; Tsai A; Wienkers A; Korner K; Hanson K; Long A; Hightower BJ; Slatton G; Burnett DC; Massey TL; Iwai K; Lee LP; Pister KS; Lin L Lab Chip 2016, 16, 668–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Femmer T; Kuehne AJ; Wessling M Lab Chip 2014, 14, 2610–2613. [DOI] [PubMed] [Google Scholar]
- (30).Shaw-Stewart J; Lippert T; Nagel M; Nuesch F; Wokaun A ACS Appl. Mater. Interfaces 2011, 3, 309–316. [DOI] [PubMed] [Google Scholar]
- (31).O’Donnell N; Okkelman IA; Timashev P; Gromovykh TI; Papkovsky DB; Dmitriev RI Acta Biomater. 2018, 80, 85. [DOI] [PubMed] [Google Scholar]
- (32).Lv X; Feng C; Liu Y; Peng X; Chen S; Xiao D; Wang H; Li Z; Xu Y; Lu M Theranostics 2018, 8, 3153–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Silva MA; Leite YKC; de Carvalho CES; Feitosa MLT; Alves MMM; Carvalho FAA; Neto BCV; Miglino MA; Jozala AF; de Carvalho MAM PeerJ 2018, 6, No. e4656. [Google Scholar]
- (34).Rebelo AR; Archer AJ; Chen X; Liu C; Yang G; Liu Y Sci. Technol Adv. Mater. 2018, 19, 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Hou Y; Wang X; Yang J; Zhu R; Zhang Z; Li YJ Biomed. Mater. Res. Part A 2018, 106, 1288–1298. [DOI] [PubMed] [Google Scholar]
- (36).Jiang P; Ran J; Yan P; Zheng L; Shen X; Tong HJ Biomater. Sci. Polym. Ed. 2018, 29, 107–124. [DOI] [PubMed] [Google Scholar]
- (37).Chen S; Teng Q Materials (Basel) 2017, 10, E846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Teodoro KBR; Sanfelice RC; Mattoso LHC; Correa DS J. Nanosci. Nanotechnol 2018, 18, 4876–4883. [DOI] [PubMed] [Google Scholar]
- (39).Yu J; Huang TR; Lim ZH; Luo R; Pasula RR; Liao LD; Lim S; Chen CH Adv. Healthcare Mater. 2016, 5, 2983–2992. [DOI] [PubMed] [Google Scholar]
- (40).Martinez Avila H; Feldmann EM; Pleumeekers MM; Nimeskern L; Kuo W; de Jong WC; Schwarz S; Muller R; Hendriks J; Rotter N; van Osch GJ; Stok KS; Gatenholm P Biomaterials 2015, 44, 122–33. [DOI] [PubMed] [Google Scholar]
- (41).Arias SL; Shetty A; Devorkin J; Allain JP Acta Biomater. 2018, 77, 172–181. [DOI] [PubMed] [Google Scholar]
- (42).Luo H; Zhang Y; Li G; Tu J; Yang Z; Xiong G; Wang Z; Huang Y; Wan YJ Biomater. Appl. 2017, 32, 265–275. [DOI] [PubMed] [Google Scholar]
- (43).Luo H; Li W; Ao H; Li G; Tu J; Xiong G; Zhu Y; Wan Y Mater. Sci. Eng., C 2017, 76, 94–101. [DOI] [PubMed] [Google Scholar]
- (44).Walker LP; Wilson DB Bioresour. Technol. 1991, 36, 3–14. [Google Scholar]
- (45).Li J; Wan Y; Li L; Liang H; Wang J Mater. Sci. Eng., C 2009, 29 (5), 1635–1642. [Google Scholar]
- (46).Shin S; Hyun J ACS Appl. Mater. Interfaces 2017, 9, 26438–26446. [DOI] [PubMed] [Google Scholar]
- (47).Rashad A; Mustafa K; Heggset EB; Syverud K Biomacromolecules 2017, 18, 1238–1248. [DOI] [PubMed] [Google Scholar]
- (48).Palaganas NB; Mangadlao JD; de Leon ACC; Palaganas JO; Pangilinan KD; Lee YJ; Advincula RC ACS Appl. Mater. Interfaces 2017, 9, 34314–34324. [DOI] [PubMed] [Google Scholar]
- (49).Torres-Rendon JG; Kopf M; Gehlen D; Blaeser A; Fischer H; Laporte LD; Walther A Biomacromolecules 2016, 17, 905–913. [DOI] [PubMed] [Google Scholar]
- (50).Miri AK; Khalilpour A; Cecen B; Maharjan S; Shin SR; Khademhosseini A Biomaterials 2019, 198, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Skardal A; Devarasetty M; Kang HW; Seol YJ; Forsythe SD; Bishop C; Shupe T; Soker S; Atala AJ Visualized Exp. 2016, No. e53606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Daly AC; Cunniffe GM; Sathy BN; Jeon O; Alsberg E; Kelly DJ Adv. Healthcare Mater. 2016, 5, 2353–2362. [DOI] [PubMed] [Google Scholar]
- (53).Picheth GF; Pirich CL; Sierakowski MR; Woehl MA; Sakakibara CN; de Souza CF; Martin AA; da Silva R; de Freitas RA Int. J. Biol. Macromol 2017, 104, 97–106. [DOI] [PubMed] [Google Scholar]
- (54).Jeong SI; Lee SE; Yang H; Jin Y-H; Park C-S; Park YS Mol Cell. Toxicol. 2010, 6, 370–377. [Google Scholar]
- (55).Kolesky DB; Truby R; Gladman AS; Busbee TA; Homan KA; Lewis JA Adv. Mater. 2014, 26, 3124–3130. [DOI] [PubMed] [Google Scholar]
- (56).Kolesky DB; Homan KA; Skylar-Scott MA; Lewis JA Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 3179–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Habibi Y; Lucia LA; Rojas OJ Chem. Rev. 2010, 110, 3479–3500. [DOI] [PubMed] [Google Scholar]
- (58).Wang HF; Ran R; Liu Y; Hui Y; Zeng B; Chen D; Weitz DA; Zhao CX ACS Nano 2018, 12, 11600–11609. [DOI] [PubMed] [Google Scholar]
- (59).Chang TC; Mikheev AM; Huynh W; Monnat RJ; Rostomily RC; Folch A Lab Chip 2014, 14, 4540–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Tsai HF; Trubelja A; Shen AQ; Bao GJR Soc, Interface 2017, 14, 20170137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Massa S; Sakr MA; Seo J; Bandaru P; Arneri A; Bersini S; Zare-Eelanjegh E; Jalilian E; Cha BH; Antona S; Enrico A; Gao Y; Hassan S; Acevedo JP; Dokmeci MR; Zhang YS; Khademhosseini A; Shin SR Biomicrofluidics 2017, 11, No. 044109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Shi Z; Li Y; Chen X; Han H; Yang G Nanoscale 2014, 6, 970–977. [DOI] [PubMed] [Google Scholar]
- (63).Backdahl H; Helenius G; Bodin A; Nannmark U; Johansson BR; Risberg B; Gatenholm P Biomaterials 2006, 27, 2141–2149. [DOI] [PubMed] [Google Scholar]
- (64).Yin N; Stilwell MD; Santos TMA; Wang H; Weibel DB Acta Biomater. 2015, 12, 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Alvarez-García A; González A; Alonso-González C; Martínez-Campa C; Cos SJ Pineal Res. 2013, 54, 373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.