Abstract
Background
Our previous research revealed that membrane type 1-matrix metalloproteinase (MT1-MMP) is overexpressed and plays a crucial role in gastric cancer (GC) progression. Exosomes are important for GC carcinogenesis, and the exosomal contents are ideal biomarkers. However, the expression of exosomal MT1-MMP mRNA in serum and its potential significance in GC remains unclear.
Material/Methods
The expression of exosomal MT1-MMP mRNA in serum of patients with GC, chronic gastritis, or atypical hyperplasia, and healthy controls was detected using real-time quantitative RT-PCR. Serum CEA, CA19-9, and CA72-4 were also measured by electrochemiluminescence assay.
Results
Exosomes were isolated and identified in serum, and serum exosomal MT1-MMP mRNA was found to be higher in patients with GC compared with healthy controls and patients with chronic gastritis or atypical hyperplasia (all P<0.05). Serum exosomal MT1-MMP mRNA was significantly correlated with tumor diameter, differentiation, Borrmann type, invasion depth, lymphatic metastasis, distal metastasis, and TNM stage. The AUC of exosomal MT1-MMP mRNA was 0.788 (95% CI: 0.714–0.850) with 63.9% sensitivity and 87.1% specificity, and was higher than that of CEA (0.655 (95% CI: 0.573–0.730)). The combination of 2 markers gave an AUC of 0.821 (95% CI: 0.750–0.878), which was better than with the individual marker. The sensitivity, specificity, and positive and negative predictive values were 75.6%, 83.9%, 94.7%, and 47.3%, respectively. Moreover, the multiple logistic regression model showed that tumor diameter, differentiation, invasion depth, and exosomal MT1-MMP mRNA were the risk factors for lymphatic metastasis in GC.
Conclusions
Our results characterized serum exosomal MT1-MMP mRNA in GC, providing a foundation for discovering serum exosomes-targeted biomarkers for GC diagnosis.
MeSH Keywords: Biological Markers; Exosomes; Matrix Metalloproteinase 14; RNA, Messenger; Stomach Neoplasms
Background
Gastric cancer (GC) is the second leading cause of cancer-related deaths in both men and women, and an estimated 679 100 new cases are diagnosed in China each year [1]. Due to the absence of specific symptoms in early stage and the lack of early diagnostic markers, 80% of patients with GC are mostly asymptomatic and are often diagnosed in an advanced stage, missing the best opportunity for curative surgery [2]. The prognosis of GC varies remarkably according to tumor stages, with the 5-year survival rates ranging from greater than 90% for stage I to less than 5% for stage IV [3]. Thus, early detection of GC is critical to decrease the mortality rate and improve the prognosis of GC patients. Although gastroscopic screening greatly increased the diagnosis rate in early stage, the invasive nature and cost incurred have hampered its application. On the other hand, currently-used clinical serum tumor markers such as carcinoembryonic antibody (CEA), carbohydrate antibody 19-9 (CA19-9), and carbohydrate antibody 72-4 (CA72-4) have insufficient sensitivity and specificity for GC screening, which limits their clinical utility [4,5]. Therefore, it is urgent to find novel biomarkers with higher sensitivity and specificity to improve GC diagnosis.
Exosomes are small endosomal-derived vesicles (50–150 nm) characterized by a lipid bilayer, classic dish or cup morphology, and a buoyant density of 1.13–1.19 g/mL [6]. Its main protein markers are tetraspanins CD63 and CD9 and tumor susceptibility gene 101 (TSG101). Exosomes can be secreted by many different cell types; they facilitate cell-to-cell communication and participate in progression of various disease, including GC. During this process, exosomes are packed with RNAs, proteins, DNA, and lipids, which can reflect the pathological state of the parental tumor cells [7,8]. In particular, with the lipid bilayer structure protecting RNA from degradation, exosomes are stable in serum/plasma, and this makes exosomal RNAs a potential candidate as ideal non-invasive biomarkers of cancers [9,10]. Among them, non-coding RNA including microRNA (miRNA), long non-coding RNA (lncRNA), and circular RNA in serum/plasma have shown potential in GC diagnosis [9,11,12]. However, it remains uncertain whether exosomal mRNAs in serum can be used to effectively diagnose GC. Evidence shows mRNAs can be encapsulated into exosomes [13], and serum exosomal mRNAs are aberrance in digestive system cancer. Xu et al. [14] revealed that serum exosomal hnRNPH1 mRNA level in hepatocellular carcinoma patients is remarkably higher than in healthy controls, and is considered as a novel biomarker. Serum exosomal ECRG4 [15], WASF2, and ARF6 mRNAs [16] are also abnormally expressed in esophageal cancer and pancreatic cancer, which provides insights into the early diagnosis of digestive system cancer. The expression and clinical significance of serum exosomal mRNAs in GC has not yet been well described.
Membrane type 1-matrix metalloproteinase (MT1-MMP, MMP-14), a ‘master switch’ proteinase with a C-terminal sequence that acts as membrane-anchoring domain, is one of the critical factors during tumor procession [17]. In our previous study, MT1-MMP was found to be elevated in GC tissue, and enhances the invasion of GC cells [18]. Accumulating evidence also suggests that the increased expression of MT1-MMP mRNA is significantly correlated with TNM stage, metastasis, and the poor prognosis of GC [19–21]. However, these studies of MT1-MMP were primarily focused on tissue samples, although several findings addressing circulating levels of MT1-MMP have been reported in the literature. Kasurinen et al. found that MT1-MMP protein level of serum was elevated in 240 GC patients [22]. Furthermore, in a large-scale study of 810 GC patients, a high MT1-MMP mRNA expression in peripheral blood was shown to be correlated with metastasis [23]. Thus, circulating MT1-MMP might be a putative biomarker for GC. Nevertheless, it is well known that the stability of protein and mRNA in serum is easily affected by storage time and temperature [24–26]. These results have inspired us to try a more reliable, exosomal-based method. MT1-MMP has been found in exosomes derived from cancer cell lines and secreted into the extracellular space, which plays an important role in tumor progression [27,28]. However, to the best of our knowledge, data regarding exosomal MT1-MMP mRNA in serum of GC patients remains lacking so far.
Owing to the established importance of MT1-MMP in GC and previous studies reporting its existence in exosomes, we hypothesized that it might be a reliable biomarker for GC diagnosis. In the current study, the expression of serum exosomal MT1-MMP mRNA of GC patients in comparison with healthy controls was observed. Then, we evaluated its role in the prediction of GC, and analyzed its correlations to clinicopathological variables. Our study reveals that exosomal MT1-MMP mRNA in serum might be a reliable biomarker for GC diagnosis.
Material and Methods
Participant information
We enrolled 216 patients in Qilu Hospital of Shandong University from December 2016 to December 2017, including 33 in training phase and 183 in validation phase. Data on demographic and clinicopathological parameters were collected and recorded. GC was diagnosed according to the histopathological examination of tissue. Stage was according to the UICC/AJCC TNM staging system (2010). Exclusion criteria were: (1) gastric stromal tumor, (2) treatment with antibiotics or proton pump inhibitors (PPI) within 12 months, (3) history of immune system disease, (4) history of malignant tumor, (5) incomplete clinical information, (6) infectious disease, and (7) unwilling to participate in this study.
Sixteen patients (7 males and 9 females, age range 32–58 and 31–66 years, respectively) in training phase and 31 in validation phase (19 males and 12 females, age range 29–61 and 30–62 years, respectively) were enrolled, in addition to a group of healthy control subjects. Inclusion criteria for the healthy controls were: (1) normal physical examination results; (2) results in normal range for routine tests of blood, urine, and stools, erythrocyte sedimentation rate, liver function, renal function, electrolyte, blood glucose, and lipids; (3) electrocardiogram and imaging analysis including liver ultrasound and chest X-ray showed no abnormalities. For the healthy sample pool, we excluded candidates who had diabetes, irritable bowel syndrome, coeliac disease, or individuals who treating with antibiotics and/or probiotics within the last 12 months. This study was approved by the Medical Ethics Committee of Qilu Hospital of Shandong University (KYLL-2015-097). All participants signed informed consent.
Samples collection
From all study participants, we collected 5 mL venous blood in vacutainer tubes (SSTTM II, BD-Belliver industrial Estate, Plymouth, UK) after fasting, and hemolysed blood specimens were discarded. Samples were centrifuged at 3000×g for 10 min after being kept at room temperature (20–25°C) for 1 h. Serum was stored at −80°C until analysis.
Isolation of exosomes from serum
The cell-free serum samples were thawed on ice, then centrifuged at 3000×g for 10 min, followed by filtering through a 0.22-μm filter (Millipore) to eliminate cell debris and other cellular organelles. To obtain the purified exosomes, we centrifuged serum at 50 000×g for 60 min to remove large microvesicles, following the method described in a previous study [29]. Then, exosomes were isolated using an exoEasy Maxi Kit (Cat: 76064, Qiagen, Germany) following the instructions. Briefly, 1 mL serum was mixed with 1 mL buffer XBP, and then added into the exoEasy spin column. After centrifuging at 500×g for 60 s, 10 mL buffer XWP was added. After centrifuging at 5000×g for 5 min, we added 400 μl Buffer XE to elute exosomes by centrifugation at 500×g for 5 min.
RNA extraction and quantitative real-time RT-PCR
Total RNA was extracted from serum exosomes with the exoRNeasy Serum/Plasma Midi Kit (Cat: 77044, Qiagen, Germany) according to the manufacturer’s instructions. Equal amounts of total RNA from each sample were then reverse-transcribed to cDNA using an All-in-one first-strand cDNA synthesis kit (cat. no. QP006, GeneCopoeia Company, Rockville, Maryland) according to the manufacturer’s instructions. For quantitative real-time RT-PCR (qRT-PCR) amplification, cDNA was amplified using CFX96 (BIO-RAD, USA) with All-in-one™ qPCR Mix (cat. no. QP001, GeneCopoeia Company). The primers of MT1-MMP were as follow: Forward: 5′-GGCGAGTATGCCACATACGA-3′, Reverse: 5′-GATGTCGGCCTGCTTCTCAT-3′. The size was 126 bp. GAPDH was purchased from GeneCopoeia Company (Rockville, Maryland). Amplification cycle conditions were set up as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 62°C for 20 s, and 72°C for 10 s. Melting curve analysis was used to confirm amplification specificity. Data were normalized to GAPDH, and relative expression levels were evaluated using the 2−ΔΔCt method. No-template and no-RT reactions were performed as negative controls.
Transmission electron microscopy (TEM)
Exosomes (20 μl) were fixed on 100-mesh carbon-coated, formvar-coated, nickel grids treated with poly-L-lysine for 30 min. After washing 3 times with PBS, samples were incubated with 1% glutaraldehyde for 5 min, and then were negatively stained using aqueous 4% uranyl acetate for 5 min. The excess stain was blotted off and the sample was air dried. Samples were then observed using a JEOL-1200EX transmission electron microscope.
Measurement of size distribution
The size distribution of exosomes in serum was detected on Flow NanoAnalyzer (NanoFCM Inc., Xiamen, China) according to the manufacturer’s instructions. Silica Nanospheres Cocktail (S16M-Exo, NanoFCM, Inc.) was used as the size standard to construct a calibration curve according to particle sizes and side scattering intensities.
Exosome protein quantification and Western blot analysis
Exosomes were suspended in RIPA lysis buffer on ice to solubilize the protein. Total protein concentration was determined with the BCA Protein Assay Kit (cat. no. P0012, Beyotime Biotechnology, China). Briefly, protein standards (0, 0.025, 0.05, 0.1, 0.2, 0.3, 0.4, and 0.5 mg/ml) were prepared with bovine serum albumin (BSA) diluted in RIPA buffer and PBS mixture (1: 1). Standards and samples were transferred to 96-well plates, and we then added 200 μl BCA working fluid. After incubating for 30 min at 37°C, the absorbance was read at 562 nm, and protein concentration was calculated according to the standard curve.
For Western blot analysis, an aliquot of total protein was separated by 10% sodium dodecyl sulfate polyacrylamide gels and was further electrotransferred onto PVDF membranes, followed by incubating with mouse anti-human CD63 antibody (Abcam, 1: 1000), rabbit anti-human TSG101 antibody (Abcam, 1: 1000), and rabbit anti-human CD9 antibody (Cell Signaling Technology, 1: 1000) overnight at 4°C. The specific horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit secondary antibodies were used to blot the target proteins. The signals of labeled proteins were detected by use of an enhanced chemiluminescence detection system.
Electrochemiluminescence assay for CEA, CA19-9, and CA72-4 concentrations in serum
Serum CEA, CA19-9, and CA72-4 were measured and evaluated using a Cobas E601 machine (Roche Diagnostics, Switzerland) following the manufacturer’s instructions.
Fecal occult blood tests
The gastrointestinal bleeding of all participants was detected using the Colloidal gold-based fecal occult blood diagnostic Kit (Chemtron Biotech Co. Shanghai, China).
Statistical analysis
The Kolmogorov-Smirnov test was used to determine the distribution of each group. Data were described by median and interquartile range. The level of exosomal MT1-MMP mRNA and CEA in serum among different groups, including clinicopathological variables, was evaluated by Mann-Whitney U or Kruskal-Wallis test. The relationship between exosomal MT1-MMP mRNA and CEA was determined by Pearson or Spearman’s correlation analysis. Receiver operating characteristic (ROC) analysis was used to evaluate the diagnostic value of these biomarkers in gastric cancer. The area under the curve (AUC), positive likelihood ratio (+LR), negative likelihood ratio (−LR), positive predictive value (+PV), and negative predictive value (−PV) were calculated. Youden index (sensitivity+specificity-1) was used to calculate cutoff values. For the risk factors of lymphatic metastasis, bivariate analysis was performed using the Mann-Whitney U or Fisher’s exact test. Variables with P<0.2 in the bivariate analysis were included into the multivariate regression models. Forward stepwise algorithms were used. In addition, odds ratio (OR) and 95% confidence interval (CI) were calculated. P<0.05 was considered statistically significant. SPSS V.25.0 software (Chicago, Illinois, USA) and MedCalc software (Version 8.0, Korea) were used in this study.
Results
Characteristics of cases
The cases in this study were divided into 2 stages: the training phase and the validation phase, as shown in the flow chart (Figure 1). Finally, 33 cases were enrolled in the training phase, including 16 healthy controls and 17 gastric cancer patients (10 males, age range 36–69 years and 7 females, age range 32–68 years). In the validation phase, we included 31 healthy controls, 31 chronic gastritis patients (18 males, age range 32–69 years, and 13 females, age range 33–66 years), 33 atypical hyperplasia patients (23 males, age range 33–73 years and 10 females, age range 38–73 years), and 119 gastric cancer patients (89 males, age range 25–82 years and 30 females, age ranged 23–74 years). There were 37, 29, 33, and 20 GC patients in stages I, II, III, and IV, respectively. The clinical pathological parameters of GC are summarized in Table 1.
Figure 1.
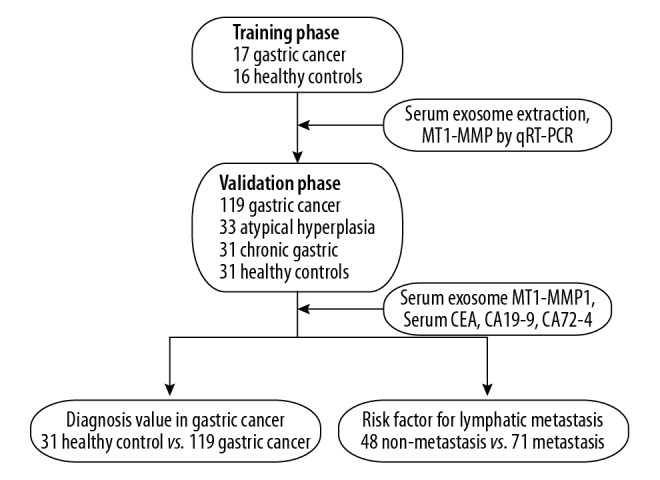
Study design and flow diagram.
Table 1.
Correlations between exosomal MT1-MMP mRNA, CEA, and clinicopathological variables [median (interquartile range)].
| Variables | N | MT1-MMP mRNA | U value | P value | CEA (ng/ml) | U value | P value | |
|---|---|---|---|---|---|---|---|---|
| Gender | Male | 89 | 5.045 (1.862, 8.786) | 1040 | 0.071 | 2.520 (1.725, 4.088) | 1029 | 0.062 |
| Female | 30 | 3.392 (0.229, 6.764) | 1.835 (1.250, 3.220) | |||||
| Age | ≤61 | 61 | 3.647 (1.161, 7.723) | 1540 | 0.223 | 2.310 (1.503, 3.525) | 1587 | 0.333 |
| >61 | 58 | 5.122 (1.820, 8.089) | 2.325 (1.740, 4.480) | |||||
| Cancer location | Antrum | 47 | 4.965 (0.558, 8.873) | 0.922* | 0.817 | 2.420 (1.603, 4.180) | 1.490* | 0.685 |
| Angulus | 23 | 5.037 (1.830, 7.160) | 2.240 (1.575, 3.900) | |||||
| Cardia | 28 | 3.952 (1.471, 7.228) | 2.165 (1.580, 3.170) | |||||
| Body | 21 | 5.045 (2.143, 8.256) | 2.910 (1.743, 4.440) | |||||
| Tumor diameter | ≤3 cm | 56 | 3.671 (0.230, 6.374) | 1184 | 0.002 | 2.210 (1.500, 3.115) | 1368.5 | 0.035 |
| >3 cm | 63 | 6.764 (2.387, 9.279) | 2.780 (1.665, 5.818) | |||||
| Differentiation | Well | 12 | 1.234 (0.096, 3.952) | 8.663* | 0.013 | 2.150 (1.945, 2.210) | 4.146* | 0.126 |
| Moderately | 25 | 5.199 (2.579, 9.291) | 2.860 (1.725, 5.778) | |||||
| Poorly | 82 | 5.066 (1.876, 8.482) | 2.305 (1.510, 3.930) | |||||
| Bormann | I | 19 | 1.748 (0.092, 5.516) | 11.037* | 0.012 | 2.070 (1.475, 2.403) | 5.693* | 0.128 |
| II | 57 | 4.448 (1.573, 7.058) | 2.370 (1.478, 4.088) | |||||
| III | 27 | 6.764 (3.371, 8.775) | 2.910 (1.913, 8.438) | |||||
| IV | 16 | 7.490 (2.776, 9.637) | 2.195 (1.630, 3.040) | |||||
| Invasion depth | T1 | 28 | 1.988 (0.074, 5.066) | 24.385* | <0.001 | 2.115 (1.500, 2.760) | 8.404* | 0.038 |
| T2 | 19 | 2.416 (1.322, 5.599) | 2.620 (1.840, 3.953) | |||||
| T3 | 23 | 5.199 (2.654, 7.588) | 2.060 (1.235, 3.218) | |||||
| T4 | 49 | 7.359 (3.196, 10.540) | 2.920 (1.718, 6.235) | |||||
| Lymphatic metastasis | No | 48 | 2.695 (0.138, 5.005) | 781 | <0.001 | 2.210 (1.535, 3.195) | 1370 | 0.070 |
| Yes | 71 | 7.028 (2.848, 9.391) | 2.620 (1.645, 6.765) | |||||
| Distal metastasis | No | 99 | 3.813 (1.207, 7.160) | 397 | <0.001 | 2.350 (1.595, 3.953) | 941.5 | 0.730 |
| Yes | 20 | 9.150 (6.476, 16.019) | 2.275 (1.585, 6.500) | |||||
| TNM Stage | I | 37 | 2.299 (0.095, 4.843) | 35.69* | <0.001 | 2.120 (1.473, 2.480) | 8.404* | 0.038 |
| II | 29 | 2.973 (1.802, 7.420) | 2.960 (1.785, 4.443) | |||||
| III | 33 | 6.983 (2.999, 9.578) | 2.780 (1.670, 8.343) | |||||
| IV | 20 | 9.150 (6.476,16.019) | 2.275 (1.585, 6.500) | |||||
| Hypertension | No | 89 | 5.088 (1.589, 8.521) | 1213 | 0.455 | 2.240 (1.515, 4.383) | 1268 | 0.682 |
| Yes | 30 | 4.032 (2.185, 7.219) | 2.330 (2.140, 3.170) | |||||
| Fecal occult blood | No | 84 | 4.625 (1.251, 7.801) | 1334 | 0.428 | 2.350 (1.670, 4.455) | 1277 | 0.260 |
| Yes | 35 | 4.965 (2.328, 8.844) | 2.300 (1.300, 3.283) | |||||
χ2 value.
Characterization of exosomes in serum
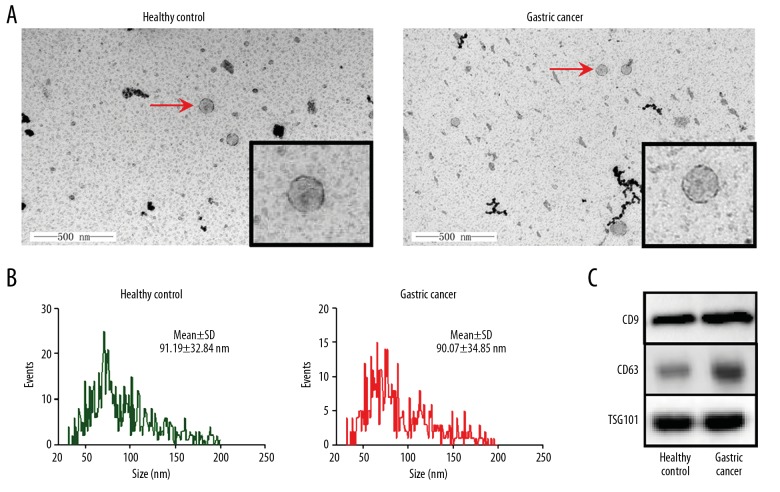
We isolated exosomes in serum of healthy controls and GC patients. Under transmission electron microscopy, exosomes showed the specific morphology with dish-like vesicles and double-lipid layer (Figure 2A). Flow NanoAnalyzer revealed that the distribution of exosomes from serum of GC patients was similar to that from healthy controls (Figure 2B), with diameters of 90.07±34.85 nm and 91.19±32.84 nm, respectively. The specific exosomes markers CD9, TSG101, and CD63 were also detected (Figure 2C).
Figure 2.
The characterization of exosomes in serum. Exosomes were isolated and characterized from human serum of healthy control and gastric cancer groups. (A) TEM micrographs. The magnification was 60 000×. (B) Size distribution of exosomes by Flow NanoAnalyzer. (C) Western blot analysis of exosomes markers CD9, CD63, and TSG101.
Expression of exosomal MT1-MMP mRNA in serum was increased in gastric cancer
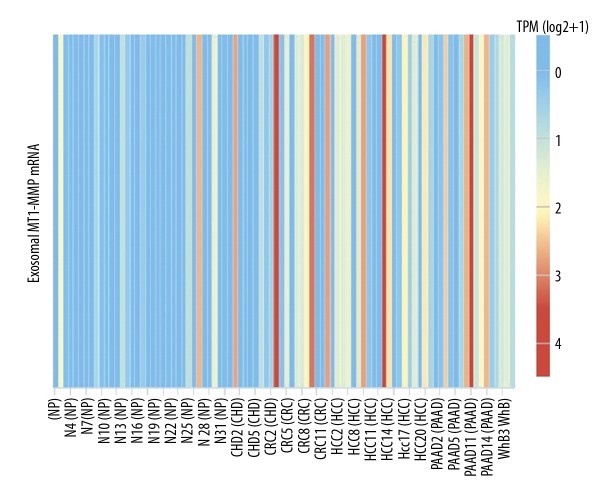
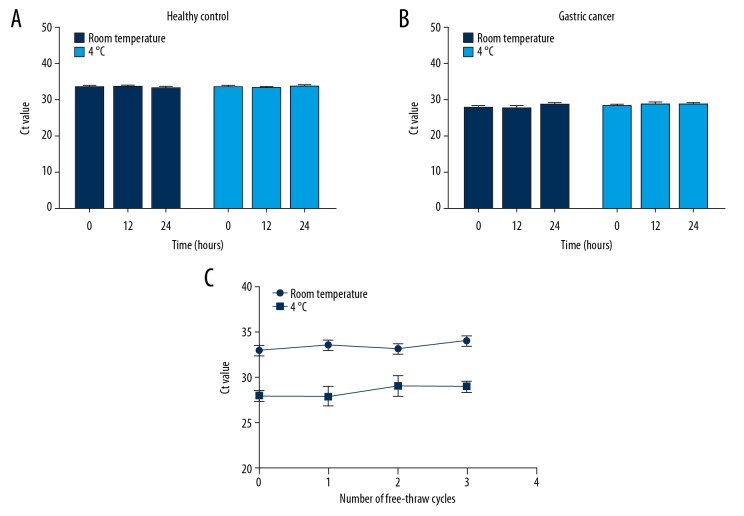
The online database exoRBase (http://www.exorbase.org/), a repository of mRNA derived from RNA-seq data analyses of human blood exosomes [30], was used to observe the expression of exosomal MT1-MMP mRNA in some diseases. The results showed that MT1-MMP mRNA was upregulated in colorectal cancer, hepatocellular carcinoma, and pancreatic adenocarcinoma (Figure 3). However, its expression and trend were not validated using qRT-PCR, and there are no available data on exosomal MT1-MMP mRNA in GC. To observe the stability of exosomal MT1-MMP mRNA in serum, we divided 1-mL aliquots of serum and stored them either at room temperature or 4°C for 12 h to 24 h (Figure 4A, 4B). Aliquots stored at −80°C were freeze-thawed repeatedly for 1 to 3 cycles (Figure 4C). Our findings revealed that none of the above treatments had a significant effect on the expression of exosomal MT1-MMP mRNA in healthy controls and GC patients, which confirms that exosomal MT1-MMP mRNA is stable in serum.
Figure 3.
The expression of exosomal MT1-MMP mRNA in blood based on the online database exoRBase. The results based on RNA-seq data analyses showed that exosomal MT1-MMP mRNA in blood was upregulated in CRC, HCC, and PAAD (http://www.exorbase.org/). NP – normal person; CHD – coronary heart disease; CRC – colorectal cancer; HCC – hepatocellular carcinoma; PAAD – pancreatic adenocarcinoma; WHB – whole blood.
Figure 4.
The stability of exosomal MT1-MMP mRNA in serum in harsh environments. The serum of healthy control and gastric cancer groups were collected and divided into 1-mL aliquots. (A, B) Serum samples were stored for 12 h or 24 h at room temperature or at 4°C. The fresh serum 2 h after collection was used for control. (C) Serum samples were freeze-thawed for 1 to 3 cycles at −80°C. Exosomes were isolated from the above-treated serum. MT1-MMP was detected by qRT-PCR and Ct value was calculated. Results are presented as means±SD of 3 independent experiments.
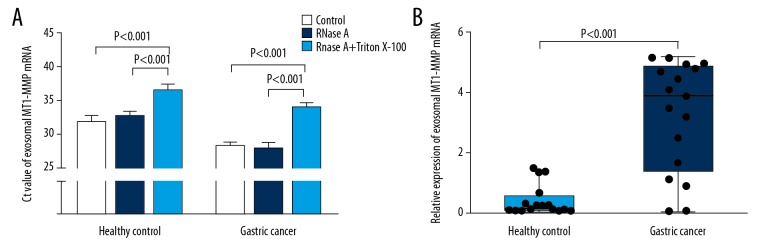
We further confirmed that MT1-MMP mRNA was indeed present in exosomes. The expression of MT1-MMP mRNA from exosomes was little changed after treatment with RNase A. However, MT1-MMP mRNA in exosomes was significantly decreased when exosomes were penetrated with Triton X-100, followed by RNase A (Figure 5A). These results suggest that exosomes derived from serum contained MT1-MMP mRNA. We next verified the expression of MT1-MMP mRNA in serum exosomes of all cases by qRT-PCR. The concentration of RNA extracted from serum exosomes was not significantly different among healthy controls and patients with chronic gastritis, atypical hyperplasia, or gastric cancer. The concentrations of RNA were (13.64±3.53 ng/μl), (15.09±5.00 ng/μl), (16.09±5.44 ng/μl), and (15.36±4.85 ng/μl), respectively. After normalization relative to the level of GAPDH, the results in Supplementary Figure 5B showed that serum exosomal MT1-MMP mRNA in GC patients (3.900 (1.390–4.865)) was more highly expressed than in healthy controls (0.180 (0.095–0.588), P<0.001).
Figure 5.
The expression of exosomal MT1-MMP mRNA in serum of healthy control and gastric cancer groups in the training phase. (A) Exosomes were isolated from serum of healthy control and gastric cancer groups. They were divided into 3 equal portions, and were then untreated (control) or treated with RNase A or RNase A plus 0.1% Triton X-100 for 45 min at 37°C. qRT-PCR was performed to detect MT1-MMP expression. Results are presented as means±SD of 3 independent experiments. (B) The levels of exosomal MT1-MMP mRNA were compared in serum of healthy control (n=16) and gastric cancer groups (n=17).
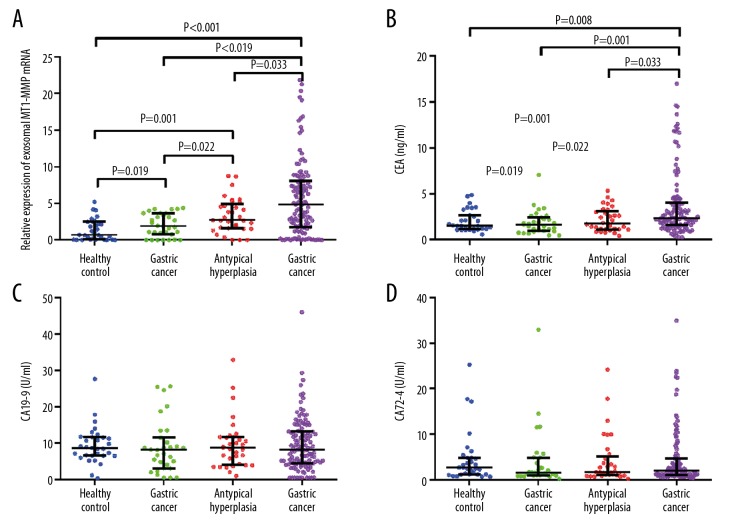
Then, we increased the sample size, and the results from Figure 6A demonstrated that exosomal levels of MT1-MMP mRNA in patients with GC (4.819 (1.748–8.023)) were much higher than in healthy controls (0.694 (0.066–2.464)) and in patients with chronic gastritis (1.886 (0.779–3.631)) or atypical hyperplasia (2.725 (1.566-4.878)) (all P<0.05). Meanwhile, the level of exosomal MT1-MMP mRNA in atypical hyperplasia was higher than that of chronic gastritis patients and healthy controls (all P<0.05). The exosomal MT1-MMP mRNA in chronic gastritis patients was higher than in healthy controls (P<0.05). We also assessed the expression of CEA, CA19-9, and CA72-4 in serum. The results showed that CEA in GC patients (2.31 (1.59–4.00) ng/ml) was higher than in healthy controls (1.52 (1.17–2.66) ng/ml) and in patients with chronic gastritis (1.65 (0.98–2.42) ng/ml) or atypical hyperplasia (1.77 (1.13–3.13) ng/ml) (all P<0.05). However, there was no difference in patients with chronic gastritis or atypical hyperplasia and healthy controls (Figure 6B, all P>0.05). Moreover, the expressions of CA19-9 and CA72-4 were not different among the 4 groups (Figure 6C, 6D, all P>0.05). Thus, we then evaluated the clinical value of CEA and serum exosomal MT1-MMP mRNA in GC patients based on the above results.
Figure 6.
The level of serum exosomal MT1-MMP mRNA in healthy control, gastric cancer, chronic gastritis, and atypical hyperplasia groups. Serum of healthy control, gastric cancer, chronic gastritis, and atypical hyperplasia groups were collected. (A) Exosomes were isolated and MT1-MMP mRNA was detected by qRT-PCR. (B–D) CEA, CA19-9, and CA72-4 in serum were also measured.
Correlation between serum exosomal MT1-MMP mRNA and clinicopathological features in gastric cancer
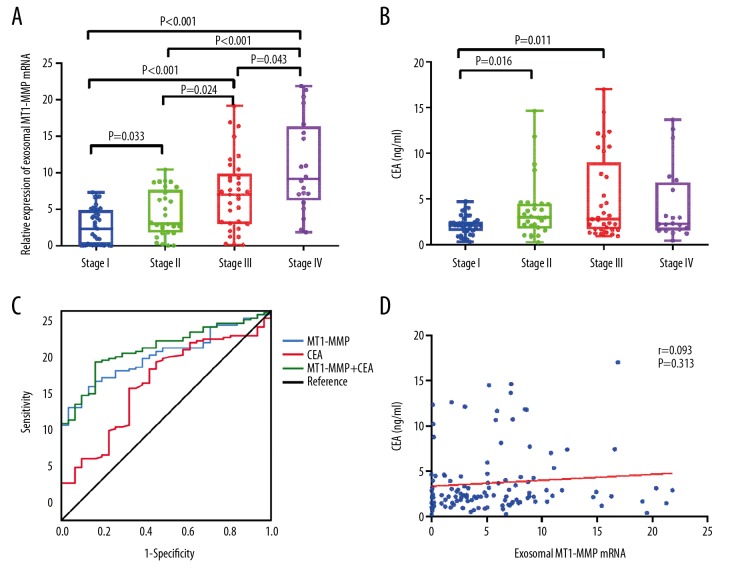
Table 1 summarizes that the correlation of exosomal MT1-MMP mRNA and CEA with clinicopathological variables, including sex, age, and lymphatic metastasis. Higher expression of exosomal MT1-MMP mRNA was statistically correlated with tumor diameter (P=0.002), differentiation (P=0.013), Borrmann type (P=0.012), invasion depth (P<0.001), lymphatic metastasis (P<0.001), distal metastasis (P<0.001), and TNM stage (P<0.001). There was no significant relationship between exosomal MT1-MMP mRNA and sex, age, cancer location, hypertension, or fecal occult blood (all P>0.05). CEA was associated with tumor diameter (P=0.035), invasion depth (P=0.038), and TNM stage (P=0.038). A significant correlation was observed between exosomal MT1-MMP mRNA and TNM stage (r=0.549, P<0.001), suggesting that upregulation of MT1-MMP mRNA is correlated with clinical progression of GC. Figure 7A shows that MT1-MMP mRNA in stage IV (9.149 (6.200–16.320)) was much higher than stage I (2.299 (0.090–4.883)), II (2.973 (1.784–7.621)), and III (6.983 (2.975–9.869)) (all P<0.05). The expression of MT1-MMP mRNA in stage III was higher than that in stage II and stage I (all P<0.05), while the level in stage II was higher than that in stage I (P<0.05). Our findings reveal that exosomal MT1-MMP mRNA might be a reliable biomarker for GC stage. In addition, Figure 7B shows that CEA of stage III (2.78 (1.66–8.97) ng/ml) and stage II (2.96 (1.72–4.46) ng/ml) were higher than that of stage I (2.12 (1.46–2.54) ng/ml) (all P<0.05).
Figure 7.
The diagnostic value of serum exosomal MT1-MMP mRNA in gastric cancer. The expression of exosomal MT1-MMP mRNA (A) and CEA (B) in serum were analyzed among GC patients with different TNM stages. MT1-MMP mRNA were normalized to GAPDH. (C) ROC analysis for the diagnosis of GC using serum exosomal MT1-MMP mRNA and CEA. (D) The correlation between exosomal MT1-MMP mRNA and CEA in GC was analyzed.
Predictive value of exosomal MT1-MMP mRNA in serum of gastric cancer
Because exosomal MT1-MMP mRNA and CEA were both related with GC procession, we further assessed the predictive value of these 2 biomarkers. ROC analysis was performed to compare exosomal MT1-MMP mRNA and CEA, and the AUC was calculated. As shown in Figure 7C, the AUC of exosomal MT1-MMP mRNA (0.788 (95% CI: 0.714–0.850)) was higher than that of CEA (0.655 (95% CI: 0.573–0.730)), achieving a high classification power for GC patients and healthy controls. The corresponding cutoff value, sensitivity, specificity, +PV, and −PV of MT1-MMP and CEA were 2.89 and 2.10 ng/ml, 63.9% and 63.0%, 87.1% and 67.7%, 95.0% and 88.2%, and 38.6% and 32.3%, respectively (Table 2). This indicated exosomal MT1-MMP mRNA might be a more reliable marker than CEA for GC patients. Furthermore, we attempted to combine MT1-MMP mRNA and CEA for GC diagnosis. ROC analysis showed that the combination of the 2 markers gave an AUC of 0.821 (95% CI: 0.750–0.878), which was better than each alone, presenting a significant increase of classification power for GC. The sensitivity, specificity, +PV, and −PV were 75.6%, 83.9%, 94.7%, and 47.3%, respectively. In addition, there was no correlation between exosomal MT1-MMP mRNA and CEA (Figure 7D, r=0.093, P>0.05).
Table 2.
The predictive value of CEA and exosomal MT1-MMP mRNA in gastric cancer.
| Cut-off | Sens. (95% CI) | Spec. (95% CI) | +LR (%) | −LR (%) | +PV (%) | −PV (%) | AUC (95% CI) | |
|---|---|---|---|---|---|---|---|---|
| MT1-MMP | 2.89 | 63.9 (54.6–72.5) | 87.1 (70.1–96.3) | 4.95 | 0.41 | 95 | 38.6 | 0.788 (0.714–0.850) |
| CEA | 2.10 | 63.0 (53.7–71.7) | 67.7 (48.6–83.3) | 1.95 | 0.55 | 88.2 | 32.3 | 0.655 (0.573–0.730) |
| MT1-MMP+CEA | 75.6 (66.9–83.0) | 83.9 (66.3–94.5) | 4.69 | 0.29 | 94.7 | 47.3 | 0.821 (0.750–0.878) |
Sens. – sensitivity; Spec. – specificity; +LR – positive likelihood ratio; −LR – negative likelihood ratio; +PV – positive predictive value; −PV – negative predictive value.
Risk factors for lymphatic metastasis of gastric cancer
Lymphatic metastasis is common in GC progression, although the causes of lymphatic spread remain unclear [31]. Exosomes can mediate tumor metastasis and have been recognized as potential biomarkers [32,33]. We also found that exosomal MT1-MMP mRNA was correlated with lymphatic metastasis. Therefore, we further explored the risk factors of lymphatic metastasis in GC patients. Our study sample included 48 lymphatic non-metastasis patients and 71 metastasis patients. In the univariate logistic regression, several basic factors were considered, including sex, age, and cancer location and are summarized in Table 3. The results showed that tumor diameter, differentiation, Bormann type, invasion depth, and MT1-MMP mRNA were closely correlated with lymphatic metastasis (all P<0.05) and might be risk factors.
Table 3.
Descriptive statistics and bivariate analysis of the associations between predictor variables and lymphatic metastasis.
| Variables | N | Non-metastasis (%) | Netastasis (%) | χ2 | P value | |
|---|---|---|---|---|---|---|
| Gender | Male | 89 | 37 (41.6) | 52 (58.4) | 0.224 | 0.636 |
| Female | 30 | 11 (36.7) | 19 (63.3) | |||
| Age | ≤61 | 61 | 23 (37.7) | 38 (62.3) | 0.360 | 0.548 |
| >61 | 58 | 25 (43.1) | 33 (56.9) | |||
| Cancer location | Antrum | 47 | 19 (40.4) | 28 (59.6) | 3.797 | 0.284 |
| Angulus | 23 | 12 (52.2) | 11 (47.8) | |||
| Cardia | 28 | 12 (42.9) | 16 (57.1) | |||
| Body | 21 | 5 (23.8) | 16 (76.2) | |||
| Tumor Diameter | ≤3 cm | 56 | 38 (67.9) | 18 (32.1) | 33.290 | <0.001* |
| >3 cm | 63 | 10 (15.9) | 53 (84.1) | |||
| Differentiation | Well | 12 | 12 (100.0) | 0 (100.0) | 26.515 | <0.001* |
| Moderately | 25 | 14 (56.0) | 11 (44.0) | |||
| Poorly | 82 | 22 (26.8) | 60 (73.2) | |||
| Bormann | I | 19 | 14 (73.7) | 5 (26.3) | 18.084 | <0.001* |
| II | 57 | 25 (43.9) | 32 (56.1) | |||
| III | 27 | 8 (29.6) | 19 (70.4) | |||
| IV | 16 | 1 (6.3) | 15 (93.7) | |||
| Invasion depth | T1 | 28 | 27 (96.4) | 1 (3.6) | 53.934 | <0.001* |
| T2 | 19 | 9 (47.4) | 10 (52.6) | |||
| T3 | 23 | 3 (13.0) | 20 (87.0) | |||
| T4 | 49 | 9 (18.4) | 40 (81.6) | |||
| Hypertension | No | 89 | 34 (38.2) | 55 (61.8) | 0.668 | 0.414 |
| Yes | 30 | 14 (46.7) | 16 (53.3) | |||
| Fecal Occult Blood | No | 84 | 34 (40.5) | 50 (59.5) | 0.002 | 0.962 |
| Yes | 35 | 14 (40.0) | 21 (60.0) | |||
| MT1-MMP mRNA | 119 | 2.695 (0.138, 5.005)** | 7.028 (2.848, 9.391)** | −5.000*** | <0.001* | |
| CEA (ng/ml) | 119 | 2.210 (1.535, 3.195)** | 2.620 (1.645, 6.765)** | −1.809*** | 0.070* | |
Included into the multiple logistic regression models (P<0.2);
median (interquartile range);
calculated using Mann-Whitney U test.
Baseline variables that were considered clinically relevant or that showed a univariate relationship with lymphatic metastasis were entered into a multiple logistic regression model. Variables for inclusion were carefully chosen, given the number of events available, to ensure parsimony of the final model. Candidate variables with a p value <0.2 on the univariate analysis were included in the multivariable model. In the multiple logistic regression model, tumor diameter (OR=4.172, 95% CI: 1.168–1.900, P=0.028), differentiation (OR=6.224, 95% CI: 1.843–21.019, P=0.003), invasion depth (OR=1.771, 95% CI: 1.012–3.101, P=0.045), and MT1-MMP mRNA (OR=1.29, 95% CI: 1.086–1.533, P=0.004) were associated with lymphatic metastasis and were independent risk factors (Table 4).
Table 4.
Risk factors for lymphatic metastasis in multiple logistic regression models.
| Variables | b | P | OR | 95% CI |
|---|---|---|---|---|
| Tumor diameter | 1.428 | 0.028 | 4.172 | 1.168–14.900 |
| Differentiation | 1.828 | 0.003 | 6.224 | 1.843–21.019 |
| Invasion depth | 0.572 | 0.045 | 1.771 | 1.012–3.101 |
| MT1-MMP mRNA | 0.255 | 0.004 | 1.290 | 1.086–1.533 |
OR – odds ratio; 95% CI – 95% confidence intervals.
Discussion
Gastric cancer is a highly malignant tumor, and early diagnosis can significantly improve the survival rate of patients [3]. However, the current non-invasive and invasive methods used for GC diagnosis still have some limitations, including tissue damage and low sensitivity [4,33]. In this study, we isolated exosomes, and observed exosomal MT1-MMP mRNA was stable in serum of healthy controls and GC patients. Exosomal MT1-MMP mRNA in serum was significantly higher in patients with GC, and was correlated with TNM stage and lymphatic metastasis. Further analysis showed that the diagnostic value of exosomal MT1-MMP mRNA was better than that of CEA in GC, with a higher sensitivity and specificity, and it was shown to be an independent risk factor for lymphatic metastasis of GC. Our findings suggest that exosomal MT1-MMP mRNA in serum might be a potential biomarker for GC diagnosis.
CEA, CA19-9, and CA72-4 are the most frequently used biomarkers in clinical practice for GC. However, its low sensitivity and specificity prevent it from improving early diagnosis of GC, and these serum biomarkers are not be recommended for preoperative evaluation and staging of GC by the National Comprehensive Cancer Network guidelines [4,5]. This was also supported by our findings that CA19-9 and CA72-4 were not significantly changed in patients with GC, chronic gastritis, atypical hyperplasia, and healthy controls. However, several studies have reported that CA19-9 and CA72-4 are elevated in patients with GC [34,35]. A possible explanation of this disagreement is that race, sample size, and technique in our study may be different from those of other studies. Consistent with previous studies, our results also show that CEA is increased in GC and is related with TNM stage [34,35]. Interestingly, CEA was not different in chronic gastritis, atypical hyperplasia, and healthy controls. This indicates that CEA is not a good early diagnostic marker and might not participate in the early progress of GC. It is remarkable that after a systematic review based on previous publications, Shimada et al. [5] concluded that CEA, CA19-9, and CA72-4 are useful for detecting recurrence and distant metastasis, predicting patient survival, and monitoring after surgery. The clinical significance of these 3 markers were not evaluated in our study because we focused on early diagnosis and discovering new biomarkers for GC.
Tumorigenesis involves genetic and epigenetic alterations, and the molecules involved in this process may be potential biomarkers, and it was recently suggested that the extracellular vesicles secreted by cells where exosomes are located are of research interest. Exosomes were first described, in the 1980s, as vesicles released by reticulocytes and were initially considered to be mere waste receptacles [36]. Recent studies show that exosomes can be secreted by cancer cells into body fluids, including blood, and are positively associated with tumor progression [37]. In 2009, Qu et al. [38] first reported that GC cell-derived exosomes promoted GC cell proliferation by activation of the PI3K/Akt and MAPK/ERK pathways. The following studies revealed that the concentration of exosomes is significantly elevated in the serum of GC patients [39,40]. This is also confirmed by the current study in which we successfully isolated and characterized exosomes in serum of GC patients. The growing interest in defining the clinical relevance of exosomes in GC has led to the identification of either organ- or disease-specific exosomal contents. Exosomal mRNAs have been receiving increased research attention, and serum levels of some exosomal mRNAs, such as hnRNPH1 and ECRG4 mRNA, are remarkably higher in some cancers [14,15]. These studies support the idea that exosomal mRNAs in serum are a source of new biomarkers for cancer. However, their expression and clinical value in GC remain to be investigated.
MT1-MMP can degrade or cleave a variety of substrates, including collagens, other MMPs (e.g., pro-MMP-2) and cell surface proteins (e.g., ICAM-1 and CD44), which play a central role in remodeling of the extracellular matrix [28,41]. We have previously reported that MT1-MMP is overexpressed in GC tissue and is involved in GC invasion, indicating that it might be a therapeutic target or biomarker [18]. Previous studies showed that MT1-MMP is contained in exosomes and is released into the extracellular space [27,28]. However, there have been no reports on MT1-MMP mRNA in exosomes. In this study, we identify the presence of MT1-MMP mRNA in serum exosomes of GC for the first time. Due to limitations imposed by clinical practice, specimens are often stored prior to testing, and this can cause degradation of some serum biomarkers. No changes of exosomal MT1-MMP mRNA were found in serum samples kept at room temperature or 4°C for 24 h and subjected to 3 freeze-thaw cycles. RNA is labile and ribonuclease is known to be present in serum, which results in a variation of RNA with time and temperature [26]. A possible explanation for this is that exosomes are stable in serum [9,10], and the lipid bilayer structure protects MT1-MMP mRNA from degradation. Although there was no further evidence, our result might support this explanation in some extent.
GC development is a complex process, involving a progression from normal condition to chronic gastritis to atypical hyperplasia to gastric cancer. Our findings demonstrated that the level of serum exosomal MT1-MMP mRNA in 4 stages is GC>atypical hyperplasia>chronic gastritis>healthy control, probably because during the progression of GC, the abnormal cells release exosomes containing MT1-MMP mRNA, which can be targeted to recipient cells. Once attached to the target cell, exosomes can deliver MT1-MMP mRNA into its cytosol via several means, such as receptor-ligand interaction, internalization by endocytosis and/or phagocytosis, or fusion with the cell membrane [7]. Therefore, MT1-MMP plays a role promoting GC progression, and this signal can be detected in serum. This indicates that exosomal MT1-MMP mRNA has the potential to be utilized as GC biomarkers. Our results also showed levels increase with TNM stages and are correlated with clinicopathological variables of GC, including differentiation and lymphatic metastasis. Thus, we evaluated the diagnostic value of serum exosomal MT1-MMP mRNA, and found that it has a good diagnostic value, with 63.9% sensitivity and 87.1% specificity, which are better than with CEA. Our results show that exosomal MT1-MMP mRNA might be a better biomarker. To obtain better accuracy for GC detection, we combined these 2 markers in logistic regression model, resulting in a better AUC than with either alone, suggesting that the combination of these 2 markers has a better value for GC diagnosis. Interesting, although +PV was not different, the −PV was increased from 38.6% to 47.3%, indicating an improved ability to detect true-negative patients. Taken together, the results show that serum exosomal MT1-MMP mRNA can help distinguish patients with GC from healthy controls, which may improve early diagnosis and lead to earlier therapy for GC.
Exosomes have been confirmed to participate in the critical steps in cancer progression, and have many potential applications in both diagnostics and therapeutics. This has drawn the interest of researchers. Currently, there are several methods to isolate exosomes from body fluid or cell supernatant. Ultracentrifugation is considered the criterion standard [42], but it is time-consuming and requires capital investment and an experienced operator. More importantly, different pre-processing protocols are used in different labs, and this makes comparison of results between studies virtually impossible. Standardized methods for isolating exosomes are lacking. Many laboratories have attempted to develop a simple, accurate, and reliable strategy for isolating exosomes, similar to ultracentrifugation. Fan et al. [43] constructed a novel method by integrating the rapid magnetic exosome-enrichment platform and the Ru(bpy)32+-polymer amplified electrochemiluminescence (ECL) strategy. In addition, some commercial kits have also been developed based on size-based filters, antibody-based capture, polymer-based precipitation reagents, and spin column-based method [44,45]. These methods have various advantages and disadvantages. In the current study, the exoEasy Maxi Kit was used to isolate exosomes in serum, and this kit has been used in other studies [45,46]. In our study, to obtain the purified exosomes, we first centrifuged serum at 50 000×g for 60 min to remove large microvesicles, using a method previously described [29]. Then, we used the exoEasy Maxi Kit. We compared exosomes isolated from the kit and those from ultracentrifugation using TEM, NTA, and Western blot analysis, showing that the kit can capture the intact exosomes population of expected size and number, which is consistent with previous studies [45]. This indicates that the exoEasy Maxi Kit is suitable for routine extraction of exosomes.
Lymphatic metastasis is common in GC progression, and results in poor prognosis [31]. We previously found that MT1-MMP is related to lymphatic metastasis [18], as well as serum exosomal MT1-MMP mRNA. Because patients with lymphatic metastasis require different treatments and have different disease outcomes, we further explored its risk factors. The results demonstrate that tumor diameter, differentiation, and invasion depth are independent risk factors, and this agrees with previous findings [47,48]. Moreover, the increased exosomal MT1-MMP mRNA indicates higher risk for lymphatic metastasis of GC, and is an independent risk factor. Dong et al. [19] reported that MT1-MMP mRNA is overexpressed in GC tissue and is significantly associated with lymphatic metastasis. Thus, we speculate that exosomal MT1-MMP mRNA-expressing cells may have much greater potential to generate lymphatic metastasis. Blockage of MT1-MMP might be an effective therapeutic target. However, the current study provides no evidence regarding the predictive value and underlying mechanism of exosomal MT1-MMP in lymphatic metastasis. Although the role of MT1-MMP in metastasis has been discussed, there has been no previously published report on exosomal MT1-MMP in GC [21,49]. Further study is required to determine its clinical predictive value and to elucidate the underlying mechanism in lymphatic metastasis.
Conclusions
In this study, we illustrated that serum exosomal MT1-MMP mRNA is significantly increased in GC patients, and is correlated with TNM stage and lymphatic metastasis. Serum exosomal MT1-MMP mRNA might be an effective biomarker for GC diagnosis. However, the limitations of our study included its retrospective nature and its relatively small number of patients. Multicenter studies with larger sample sizes are needed to confirm our findings, and could help gain deeper insights into the role of exosomal MT1-MMP. Moreover, GC cells can secrete exosomes; therefore, whether changes in serum exosomal MT1-MMP mRNA are caused by tumor cell-derived exosomes requires further investigation.
Acknowledgements
We thank Dr. Dong Sun for technical help.
Footnotes
Conflict of interest
None.
Source of support: This work was supported by the National Natural Science Foundation of China (81572070, 31500741), the Natural Science Foundation of Shandong Province (BS2014SW022, ZR2016HM37), and the Key Research and Development Project of Shandong Province (2016GSF201124, 2015GSF118152)
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. Cancer J Clin. 2016;66(2):115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Zong L, Abe M, Seto Y, et al. The challenge of screening for early gastric cancer in China. Lancet. 2016;388(10060):2606. doi: 10.1016/S0140-6736(16)32226-7. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Yu JC, Kang WM, et al. Treatment strategy for early gastric cancer. Surg Oncol. 2012;21(2):119–23. doi: 10.1016/j.suronc.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Pasechnikov V, Chukov S, Fedorov E, et al. Gastric cancer: Prevention, screening and early diagnosis. World J Gastroenterol. 2014;20(38):13842–62. doi: 10.3748/wjg.v20.i38.13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimada H, Noie T, Ohashi M, et al. Clinical significance of serum tumor markers for gastric cancer: A systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer. 2014;17(1):26–33. doi: 10.1007/s10120-013-0259-5. [DOI] [PubMed] [Google Scholar]
- 6.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581–93. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 7.Tkach M, Thery C. Communication by extracellular vesicles: Where we are and where we need to go. Cell. 2016;164(6):1226–32. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 8.Fu M, Gu J, Jiang P, et al. Exosomes in gastric cancer: Roles, mechanisms, and applications. Mol Cancer. 2019;18(1):41. doi: 10.1186/s12943-019-1001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Shao Y, Zhang X, et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015;36(3):2007–12. doi: 10.1007/s13277-014-2807-y. [DOI] [PubMed] [Google Scholar]
- 10.Jalalian SH, Ramezani M, Jalalian SA, et al. Exosomes, new biomarkers in early cancer detection. Anal Biochem. 2019;571:1–13. doi: 10.1016/j.ab.2019.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Tang W, Fu K, Sun H, et al. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. 2018;17(1):137. doi: 10.1186/s12943-018-0888-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang N, Wang L, Yang Y, et al. A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem Biophys Res Commun. 2017;493(3):1322–28. doi: 10.1016/j.bbrc.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–76. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu H, Dong X, Chen Y, et al. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin Chem Lab Med. 2018;56(3):479–84. doi: 10.1515/cclm-2017-0327. [DOI] [PubMed] [Google Scholar]
- 15.Mao L, Li X, Gong S, et al. Serum exosomes contain ECRG4 mRNA that suppresses tumor growth via inhibition of genes involved in inflammation, cell proliferation, and angiogenesis. Cancer Gene Ther. 2018;25(9–10):248–59. doi: 10.1038/s41417-018-0032-3. [DOI] [PubMed] [Google Scholar]
- 16.Kitagawa T, Taniuchi K, Tsuboi M, et al. Circulating pancreatic cancer exosomal RNAs for detection of pancreatic cancer. Mol Oncol. 2019;13(2):212–27. doi: 10.1002/1878-0261.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castro-Castro A, Marchesin V, Monteiro P, et al. Cellular and molecular mechanisms of MT1-MMP-dependent cancer cell invasion. Annu Rev Cell Dev Biol. 2016;32:555–76. doi: 10.1146/annurev-cellbio-111315-125227. [DOI] [PubMed] [Google Scholar]
- 18.Dong Z, Xu X, Du L, et al. Leptin-mediated regulation of MT1-MMP localization is KIF1B dependent and enhances gastric cancer cell invasion. Carcinogenesis. 2013;34(5):974–83. doi: 10.1093/carcin/bgt028. [DOI] [PubMed] [Google Scholar]
- 19.Dong Y, Chen G, Gao M, et al. Increased expression of MMP14 correlates with the poor prognosis of Chinese patients with gastric cancer. Gene. 2015;563(1):29–34. doi: 10.1016/j.gene.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Naseh G, Mohammadifard M, Mohammadifard M. Upregulation of cyclin-dependent kinase 7 and matrix metalloproteinase-14 expression contribute to metastatic properties of gastric cancer. IUBMB Life. 2016;68(10):799–805. doi: 10.1002/iub.1543. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Li S, Deng L, et al. Decreased MT1-MMP in gastric cancer suppressed cell migration and invasion via regulating MMPs and EMT. Tumour Biol. 2015;36(9):6883–89. doi: 10.1007/s13277-015-3381-7. [DOI] [PubMed] [Google Scholar]
- 22.Kasurinen A, Tervahartiala T, Laitinen A, et al. High serum MMP-14 predicts worse survival in gastric cancer. PLoS One. 2018;13(12):e0208800. doi: 10.1371/journal.pone.0208800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mimori K, Fukagawa T, Kosaka Y, et al. A large-scale study of MT1-MMP as a marker for isolated tumor cells in peripheral blood and bone marrow in gastric cancer cases. Ann Surg Oncol. 2008;15(10):2934–42. doi: 10.1245/s10434-008-9916-z. [DOI] [PubMed] [Google Scholar]
- 24.Cuhadar S, Koseoglu M, Atay A, et al. The effect of storage time and freeze-thaw cycles on the stability of serum samples. Biochem Med (Zagreb) 2013;23(1):70–77. doi: 10.11613/BM.2013.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddi KK, Holland JF. Elevated serum ribonuclease in patients with pancreatic cancer. Proc Natl Acad Sci USA. 1976;73(7):2308–10. doi: 10.1073/pnas.73.7.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsui NB, Ng EK, Lo YM. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem. 2002;48(10):1647–53. [PubMed] [Google Scholar]
- 27.Hakulinen J, Sankkila L, Sugiyama N, et al. Secretion of active membrane type 1 matrix metalloproteinase (MMP-14) into extracellular space in microvesicular exosomes. J Cell Biochem. 2008;105(5):1211–18. doi: 10.1002/jcb.21923. [DOI] [PubMed] [Google Scholar]
- 28.Sanderson RD, Bandari SK, Vlodavsky I. Proteases and glycosidases on the surface of exosomes: Newly discovered mechanisms for extracellular remodeling. Matrix Biol. 2019;75–76:160–69. doi: 10.1016/j.matbio.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W, Yu ZL, Wu M, et al. Magnetic and folate functionalization enables rapid isolation and enhanced tumor-targeting of cell-derived microvesicles. ACS Nano. 2017;11(1):277–90. doi: 10.1021/acsnano.6b05630. [DOI] [PubMed] [Google Scholar]
- 30.Li S, Li Y, Chen B, et al. exoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46(D1):D106–12. doi: 10.1093/nar/gkx891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng JY, Liang H. Clinical significance of lymph node metastasis in gastric cancer. World J Gastroenterol. 2014;20(14):3967–75. doi: 10.3748/wjg.v20.i14.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013;32(3–4):623–42. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuoka T, Yashiro M. Biomarkers of gastric cancer: Current topics and future perspective. World J Gastroenterol. 2018;24(26):2818–32. doi: 10.3748/wjg.v24.i26.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim DH, Oh SJ, Oh CA, et al. The relationships between perioperative CEA, CA 19-9, and CA 72-4 and recurrence in gastric cancer patients after curative radical gastrectomy. J Surg Oncol. 2011;104(6):585–91. doi: 10.1002/jso.21919. [DOI] [PubMed] [Google Scholar]
- 35.Ning S, Wei W, Li J, et al. Clinical significance and diagnostic capacity of serum TK1, CEA, CA 19-9 and CA 72-4 levels in gastric and colorectal cancer patients. J Cancer. 2018;9(3):494–501. doi: 10.7150/jca.21562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnstone RM, Adam M, Hammond JR, et al. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262(19):9412–20. [PubMed] [Google Scholar]
- 37.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 38.Qu JL, Qu XJ, Zhao MF, et al. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig Liver Dis. 2009;41(12):875–80. doi: 10.1016/j.dld.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 39.Yang H, Fu H, Wang B, et al. Exosomal miR-423-5p targets SUFU to promote cancer growth and metastasis and serves as a novel marker for gastric cancer. Mol Carcinog. 2018;57(9):1223–36. doi: 10.1002/mc.22838. [DOI] [PubMed] [Google Scholar]
- 40.Fu H, Yang H, Zhang X, et al. Exosomal TRIM3 is a novel marker and therapy target for gastric cancer. J Exp Clin Cancer Res. 2018;37(1):162. doi: 10.1186/s13046-018-0825-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itoh Y. Membrane-type matrix metalloproteinases: Their functions and regulations. Matrix Biol. 2015;44–46:207–23. doi: 10.1016/j.matbio.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Momen-Heravi F, Balaj L, Alian S, et al. Impact of biofluid viscosity on size and sedimentation efficiency of the isolated microvesicles. Front Physiol. 2012;3:162. doi: 10.3389/fphys.2012.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan Z, Yu J, Lin J, et al. Exosome-specific tumor diagnosis via biomedical analysis of exosome-containing microRNA biomarkers. Analyst. 2019;144(19):5856–65. doi: 10.1039/c9an00777f. [DOI] [PubMed] [Google Scholar]
- 44.Zeringer E, Li M, Barta T, et al. Methods for the extraction and RNA profiling of exosomes. World J Methodol. 2013;3(1):11–18. doi: 10.5662/wjm.v3.i1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Enderle D, Spiel A, Coticchia CM, et al. Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column-based method. PLoS One. 2015;10(8):e0136133. doi: 10.1371/journal.pone.0136133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delic D, Eisele C, Schmid R, et al. Urinary exosomal miRNA signature in type II diabetic nephropathy patients. PLoS One. 2016;11(3):e0150154. doi: 10.1371/journal.pone.0150154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim DY, Joo JK, Ryu SY, et al. Factors related to lymph node metastasis and surgical strategy used to treat early gastric carcinoma. World J Gastroenterol. 2004;10(5):737–40. doi: 10.3748/wjg.v10.i5.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren G, Cai R, Zhang WJ, et al. Prediction of risk factors for lymph node metastasis in early gastric cancer. World J Gastroenterol. 2013;19(20):3096–107. doi: 10.3748/wjg.v19.i20.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng L, Jiao W, Mei H, et al. miRNA-337-3p inhibits gastric cancer progression through repressing myeloid zinc finger 1-facilitated expression of matrix metalloproteinase 14. Oncotarget. 2016;7(26):40314–28. doi: 10.18632/oncotarget.9739. [DOI] [PMC free article] [PubMed] [Google Scholar]