Abstract
Aging is associated with enhanced oxidative stress and increased susceptibility to numerous diseases. This relationship is particularly striking with respect to the incidence of fibrotic lung disease. To identify potential mechanisms underlying the association between aging and susceptibility to fibrotic lung disease we analyzed transcriptome data from 342 disease-free human lung samples as a function of donor age. Our analysis reveals that aging in lung is accompanied by modest yet progressive changes in genes modulating redox homeostasis, the TGF-beta 1 signaling axis, and the extracellular matrix (ECM), pointing to an aging lung functional network (ALFN). Further, the transcriptional changes we document are tissue-specific, with age-dependent gene expression patterns differing across organ systems. Our findings suggest that the age-associated increased incidence of fibrotic pulmonary disease occurs in the context of tissue-specific, age-dependent transcriptional changes. Understanding the relationship between age-associated gene expression and susceptibility to fibrotic pulmonary disease may allow for more accurate risk stratification and effective therapeutic interventions within this challenging clinical space.
Keywords: Oxidoreductase, Aging, Lung, Transforming growth factor beta, Oxidative stress, Pulmonary fibrosis, Extracellular matrix, Aging lung function network
1. Introduction
The free radical theory of aging proposes that biological aging results from cellular damage mediated by the chronic accumulation of oxidants (also referred to as reactive oxygen species (ROS)) [1–12]. While this assertion remains in dispute, there is growing evidence that oxidative stress does increase with age [13,14]. Further, aging is recognized as a major risk factor for disease states linked to chronic oxidative damage, many of which manifest in the lung including idiopathic pulmonary fibrosis (IPF), chronic obstructive pulmonary disease (COPD), and lung cancer [15–17]. Nevertheless, the molecular mechanisms mediating these associations remain incompletely understood [18].
Many diseases exhibit a positive correlation with age. The association between age and fibrotic lung disease is particularly strong, exemplified by the exponential increase in IPF incidence beginning with the fifth decade of life [19]. Fibrosis is defined by dysregulated wound healing and excessive deposition of extracellular matrix in response to injury. The process can impact several different organs including the lung, skin, kidney, heart and liver [20]. Perturbation of redox homeostasis and the increased oxidative stress that results are hypothesized to mediate the dysfunctional healing that defines fibrosis pathogenesis by disrupting interactions between epithelial cells, fibroblasts, and immune cells [17,21–24]. In IPF this relationship is illustrated by the reciprocal dysregulation of cellular oxidant and antioxidant systems, including upregulation of NADPH oxidases (e.g. NOX4), and downregulation of antioxidant pathways (e.g. NRF2 and glutathione biosynthesis) [16,25–32]. Whether these alterations are sufficient to explain the increased incidence of age-associated fibrotic lung disease, or represent effects of an established disease process, remains to be established.
Understanding the molecular events predisposing the aging lung to pulmonary disease is a crucial step allowing for the rational design of novel therapies. We reasoned that characterizing naturally occurring, age-dependent transcriptional changes in disease-free lung might provide insights into the mechanisms that underlie age-associated susceptibility to fibrotic pulmonary disease. Using RNAseq data from The Genotype-Tissue Expression (GTEx) project we evaluated global changes in the lung transcriptome as a function of donor age. Our analyses reveal coordinated, age-dependent regulation of genes encoding cellular oxidation/reduction effectors, the TGF-beta 1 signaling pathway, and components of the extracellular matrix in aging disease-free human lung. Importantly, these transcriptional alterations exhibit tissue-specificity and are not simply a function of aging cells. While modest in magnitude, the expression changes impact genes involved in a subset of specific biologic processes. Taken together, the analysis of the pulmonary transcriptome highlights candidate genes for consideration as fibrotic disease-risk biomarkers, as well as potential therapeutic targets. Further, we propose that these transcriptional changes reflect a lung microenvironment that, in response to pulmonary injury, is increasingly susceptible to developing fibrosis in an age-dependent manner.
2. Materials and methods
2.1. GTEx RNAseq data
RNAseq data used in our analyses were obtained from dbGaP (Accession phs000424.v7.p2, 06/2018) as part of the Genotype-Tissue Expression (GTEx) Project. GTEx is supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. This dataset includes information on 56,202 transcripts from 11,688 samples derived from 53 tissue types and 714 human donors. Our analyses utilize data from 342 human lung samples, 600 heart samples, 175 liver samples, and 387 sun-protected skin samples. Quantified RNA values are reported as transcripts per million reads (TPM).
2.2. Sample exclusion criteria
85 lung samples were removed from the GTEx dataset prior to analysis based on accompanying data indicating the donor suffered from a chronic interstitial lung disease.
2.3. Statistical analyses
Spearman correlation analysis was performed for each transcript as a function of donor age. Genes in disease-free lung whose expression significantly correlated with donor age (p-value < 0.05) were flagged and included in gene lists used in downstream analyses. Statistical analyses were completed using base R (v3.5.1) within RStudio (v1.1.453).
2.4. Functional network analysis (FNA)
To determine whether functionally related clusters exist among the genes significantly correlated with age by Spearman analysis, we queried our gene lists in the genome-scale integrated analysis of gene networks in human tissues (GIANT 2.0)33 using the “lung” network available for download at the URL: https://hb.flatironinstitute.org/download. The GIANT networks are tissue-specific gene-gene interaction networks that encode predicted functional interactions among genes based on gene co-expression, co-regulation by a transcription factor, and protein interactions [33,34]. To reduce network complexity, we set the edge weight minimum threshold to 0.5 to obtain the aging lung functional network (ALFN). The choice of 0.5 as a threshold was motivated by two factors. First, in comprehensive and systematic network comparisons, Huang et al. noted that, at the network level, prediction of disease genes scales with network size and edge density, so that including more interactions outweighs the downsides of false positive interactions [35]. Second, before integrating any data, the GIANT networks assume a baseline probability of 0.1 that two genes functionally interact. Thus, an edge weight of 0.5 in the network indicates that the integrated data predict a five-fold increase in confidence that the genes are functionally related. Therefore, the 0.5 threshold allowed for a network analysis of a reasonably dense, high confidence network, while removing a substantial amount of the noise. We then clustered the resulting network using the fast greedy community detection algorithm [36], implemented in the Gephi (v0.9.2) network visualization software [37]. This allowed the identification of clusters within the network having denser within-cluster connections compared to outsidecluster connections. Annotation of the resulting subnetworks was completed using the ‘gProfileR’ R package [38] resulting in the identification of functional enrichments.
2.5. Gene ontology (GO) analysis
Gene Ontology (GO) analysis was performed separately on gene lists containing all protein coding genes exhibiting positive correlation with age, or gene lists containing all protein coding genes demonstrating negative correlation with age, using the online classification software ToppGene [39] (https://toppgene.cchmc.org). Member genes within identified GO terms with significant age-dependent expression correlation were then visualized using base R (v3.5.1) and the ggplots2 package within RStudio (v1.1.453).
3. Results
3.1. Age-associated changes in the human lung transcriptome
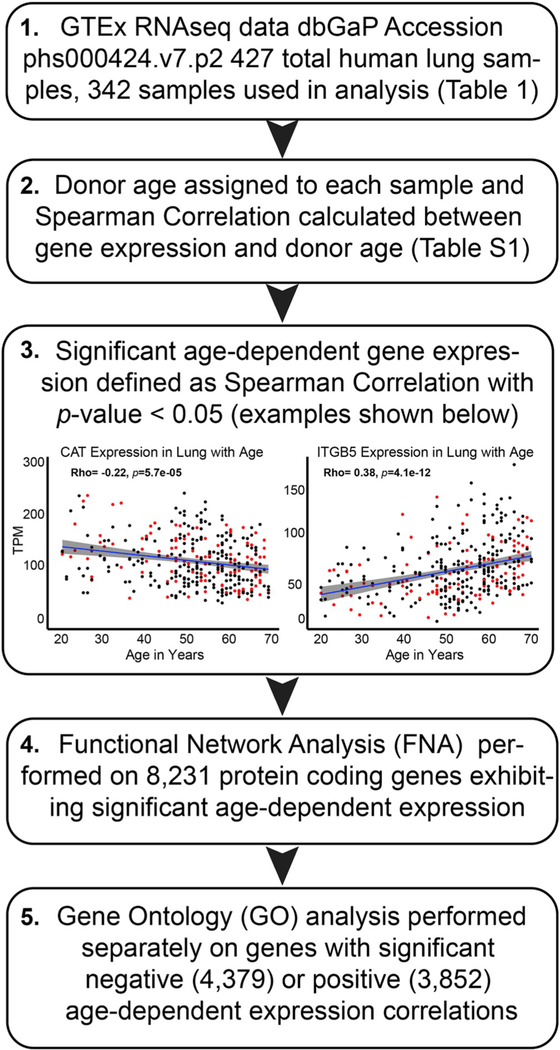
Using RNAseq data from the Genotype-Tissue Expression (GTEx) Project we evaluated transcriptional changes as a function of age in 342 disease-free human lung samples. The GTEx v7.p2 dataset contains 427 human lung samples, 85 of which were removed prior to analysis based on accompanying data indicating the donor suffered from a chronic interstitial lung disease (Table 1). Of the 342 samples included in our analysis, 236 were derived from male donors aged 21 to 70, while 106 were derived from female donors aged 21 to 70 (Table 1). Each sample analyzed included data for 56,202 RNA species, representing both protein coding and non-protein coding genes. Spearman correlation analysis was completed for each transcript as a function of donor age identifying 8,231 protein coding transcripts exhibiting significant age-dependent expression (p < 0.05; Fig. 1 and Table S1). Scatter plots for two representative genes (CAT and ITGB5) demonstrating significant age-dependent expression changes are included in Fig. 1 (panel 3). These examples visualize the patterns identified by our analysis pipeline resulting in the gene lists used in subsequent analyses.
Table 1. Age and sex distributions in the GTEx human lung RNAseq data.
Of the 427 lung samples 85 were flagged for removal prior to analysis as the donor suffered from an interstitial lung disease (ILD). Age and sex distributions shown below were calculated based on the 342 samples analyzed.
| Lung samples in dataset |
# Samples with ILD |
# Samples used in analysis |
# Male samples |
Male age range |
Male median age |
Male mean age |
# Female samples |
Female age range |
Female median age |
Female mean age |
|---|---|---|---|---|---|---|---|---|---|---|
| 427 | 85 | 342 | 236 | 21–70 | 54 | 52.45 | 106 | 21–70 | 54 | 51.85 |
Fig. 1. Workflow used to analyze age-dependent gene expression data from disease-free human lung.
Exclusion criteria, along with donor age/sex distributions, are shown in Table 1. Spearman correlations between donor age and the 56,202 transcripts analyzed are included in Table S1. Representative genes exhibiting the significant age-dependent expression required for inclusion in downstream analyses are included in panel 3 (red dots indicate female donors; black dots indicate male donors).
3.2. Functional network analysis identifies processes likely altered in aging human lung
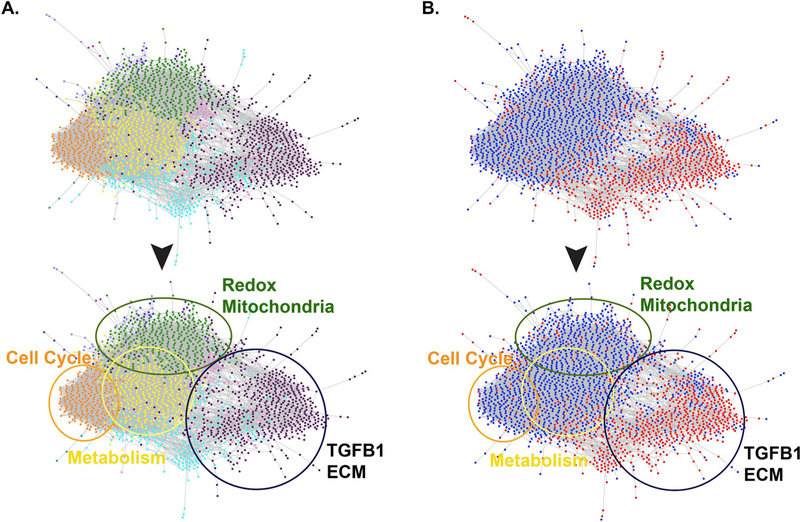
In order to identify the biologic processes potentially affected by age-associated transcriptional changes in the lung, we interrogated the list of 8,231 genes exhibiting significant age-dependent expression correlation through a functional network analysis workflow based upon the genome-scale integrated analysis of gene networks in human tissues (GIANT 2.0) [33]. The GIANT networks are tissue-specific, gene-gene interaction networks that encode predicted functional interactions among genes based on published gene co-expression, co-regulation by a transcription factor, and protein-protein interactions. Each putative interaction has an associated probability, with high probabilities corresponding to highly likely interactions. By setting the edge weight minimum threshold to 0.5, our analysis focused on strongly predicted interactions [35]. This effective filtering trimmed our list of 8,231 genes down to a highly interconnected aging lung functional network (ALFN) composed of 2,073 genes. We then clustered the ALFN using the fast greedy community detection algorithm [36], implemented in the Gephi network visualization software [37], to identify gene subgroups within the network having denser within-cluster connections compared to outside-cluster connections. These distinct clusters effectively correspond to functionally related subsets of genes within the network and are illustrated by shared dot color (Fig. 2A). In order to annotate these clusters we used the R package ‘gProfileR’ [38]. This resulted in the identification of four major subnetworks, descriptions of which were manually curated based upon formal enrichment analysis and visualized using color-coordinated circles (Fig. 2A, Table S2). These sub-networks are described in Fig. 2 as Redox/Mitochondria (green circle), Cell Cycle (orange circle), Metabolism (yellow circle), and TGFB1/ECM (black circle).
Fig. 2. Functional Network Analysis performed on transcriptome data from disease-free human lung reveals biologic processes whose effector genes exhibit age-dependent expression.
A. Of the 8,231 genes found to demonstrate significant expression correlation with age, 2,073 met criteria for inter-connectedness. These genes self-assembled into the aging lung functional network (ALFN), which is further divided into 4 major subnetworks: Redox/Mitochondria (green circle), Cell Cycle (orange circle), Metabolism (yellow circle), TGFB1/ECM (black circle). Characterization of the major subnetworks was based upon manual review of associated GO analysis (Table S2). B. Network overlaid with Spearman Rho values (blue dots indicate genes exhibiting significant negative correlation with age; red dots indicate significant positive correlation with age). Redox/Mitochondria, Cell Cycle, and Metabolism subnetworks are enriched with genes negatively correlated with age in disease-free lung (blue dots), while the TGFB1/ECM subnetwork is enriched with genes positively correlated with age (red dots).
Importantly, by using this approach the analysis was blind to whether expression of specific member genes within the network were positively or negatively correlated with age, only that the correlation was significant. To address the question of directional correlation between significant gene expression changes and age, we next overlaid the associated Spearman Rho values for each gene onto the network map (Fig. 2B). A positive rho value (red dot) indicates a gene whose expression increases with age, while a negative value (blue dot) represents a gene whose expression decreases with age in disease-free human lung. Intriguingly, a striking separation emerged from within the self-assembled network, revealing that genes exhibiting enhanced expression with age (red dots) enriched in the subnetwork dominated by ECM and TGF-beta 1 pathway members (Fig. 2A&B). Conversely, genes exhibiting reduced expression with age (blue dots) were clustered within the subnetworks characterized by redox effectors, mitochondrial function, metabolism and cell cycle regulation (Fig. 2A&B). The inferred implication of this dichotomy is that the functionally-related processes represented by the subnetworks may themselves be coordinately regulated by the transcriptional alterations we report. In this way, the transcriptional data derived from aging disease-free lung can be viewed as a surrogate for the associated biologic function. Ultimately these data suggest that the biologic pathways governing the TGFB1/ECM axis become more active with age, while those underlying metabolism and redox homeostasis exhibit reduced functionality with age.
3.3. Expression of oxidoreductase-related genes negatively correlates with age in human lung
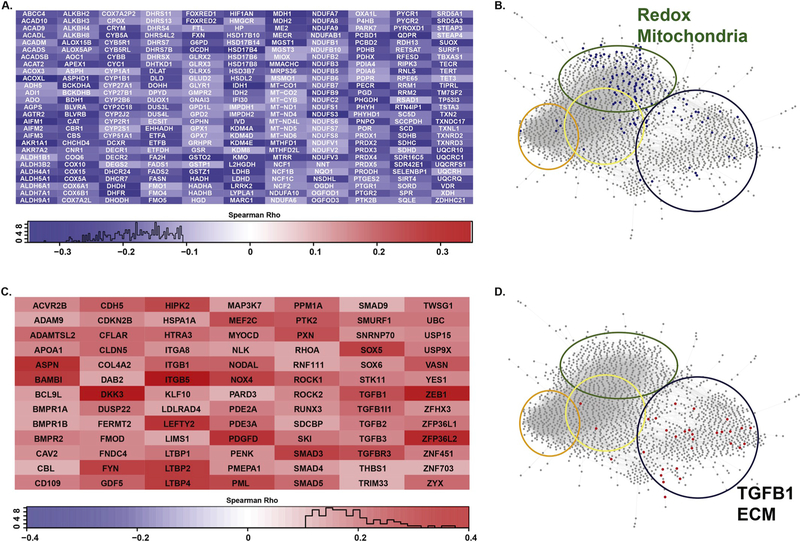
Given the bifurcation observed within the self-assembled functional network between genes positively or negatively correlated with age, we next conducted formal gene ontology analysis on the complete lists of significant positively and negatively regulated genes, using ToppGene [39]. Of the 8,231 protein coding transcripts identified as significantly correlating with age, 4,379 exhibited a significant negative correlation (Spearman rho <0; p < 0.05), while 3,852 a significant positive age-dependent correlation (Spearman rho > 0; p < 0.05) (Table 2, Table S3). GO analysis performed on these gene lists identified terms overlapping with the self-assembled subnetworks observed for the ALFN shown in Fig. 2. Specifically, GO terms for oxidation/reduction activity and mitochondrial function were notably enriched in the results for genes displaying a significant negative correlation with age (Table 2, Table S4). Similarly, genes exhibiting a significant positive correlation with age resulted in GO terms describing ECM remodeling and TGF-beta 1 signaling (Table 2, Table S5). When the intersection of representative GO term member genes are overlaid on the self-assembled functional networks, the terms segregated into their respective functionally derived clusters (Fig. 3B&D). Due to the filtering parameters used to generate the ALFN, many genes demonstrating significant age-dependent expression were removed (8,231 down to 2,073). For example, the GO term entitled “oxidoreductase activity” (GO:0016491), is strongly enriched for genes downregulated with age (p = 3.92 × 10 −22; Table 2), with 297 member genes exhibiting significant negative Spearman correlations (Fig. 3A, Table S4). However, only a third of these genes passed our functional network filter, which required an edge weight value of at least 0.5. Similarly, for the GO term entitled “response to transforming growth factor beta” (GO:0071559), many genes with significant positive age correlations (Table 2, Fig. 3C) were also filtered from the functional network analysis. This observation may reflect an inherent weakness or incompleteness in the functional network annotations. Despite this loss of granularity, the genes represented by these GO terms clustered within their predicted functional nodes (Fig. 3B&D), supporting the inference that corresponding biologic processes may be altered with age in disease-free lung.
Table 2. GO analysis highlighting terms that intersect the Functional Network Analysis.
Included results reflect representative GO terms identified by running all genes exhibiting significant negative or positive age-dependent correlation in disease-free human lung and overlap with the ALFN analysis shown in Fig. 2. An exhaustive list of enriched terms is included in Table S3.
| 4,379 genes negatively correlated with age | 3,852 genes positively correlated with age | ||||
|---|---|---|---|---|---|
| ID | Name | q-value FDR B &H |
ID | Name | q-value FDR B &H |
| GO:0016491 | oxidoreductase activity | 3.92E-22 | GO:0022610 | biological adhesion | 7.97E-32 |
| GO:0016651 | oxidoreductase activity, acting on NAD(P)H | 1.43E-11 | GO:0007155 | cell adhesion | 3.77E-31 |
| GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors | 1.23E-09 | GO:0030334 | regulation of cell migration | 5.72E-20 |
| GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor | 1.28E-06 | GO:2000145 | regulation of cell motility | 7.19E-20 |
| GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor | 2.59E-05 | GO:0040012 | regulation of locomotion | 1.27E-19 |
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | 2.93E-05 | GO:0071559 | response to transforming growth factor beta | 5.70E-11 |
| GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 1.47E-04 | GO:0071560 | cellular response to transforming growth factor beta stimulus | 4.19E-10 |
| GO:0008379 | thioredoxin peroxidase activity | 4.71E-02 | GO:0007179 | transforming growth factor beta receptor signaling pathway | 1.61E-08 |
| GO:0055114 | oxidation-reduction process | 6.68E-25 | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 4.63E-03 |
| GO:1990204 | oxidoreductase complex | 1.18E-13 | GO:0042542 | response to hydrogen peroxide | 6.39E-03 |
| GO:0005777 | peroxisome | 1.18E-07 | GO:0030054 | cell junction | 4.35E-15 |
| GO:0044439 | peroxisomal part | 4.66E-05 | GO:0031012 | extracellular matrix | 8.22E-13 |
Fig. 3. Member genes from GO terms significantly enriched for age-dependent expression in disease-free human lung.
A. 297 genes from GO:0016491 (Oxidoreductase Activity; 297/749, Bonferroni corrected p-value = 7.84e-22) exhibit significant negative correlation with age in disease-free lung. Genes are listed in alphabetical order. Inclusion indicates Spearman correlation with age reaches p-value < 0.05. Intensity of box color corresponds to Spearman Rho values whose distributions are reported as histograms below each heatmap. B. Member genes from GO:0016491 that overlap with the ALFN analysis and overlaid on the network map (blue dots). Colored circles correspond to the 4 major subnetworks defined in Fig. 2. C. 91 genes from GO:0071559 (Response to TGFB1; 91/234, Bonferroni corrected p-value = 5.70e-11) exhibit significant positive correlation with age in disease-free lung. D. Member genes from GO:0071559 that overlap with the ALFN analysis and overlaid on the network map (red dots). Colored circles correspond to the 4 major subnetworks defined in Fig. 2.
3.4. Gene expression with age exhibits tissue specificity
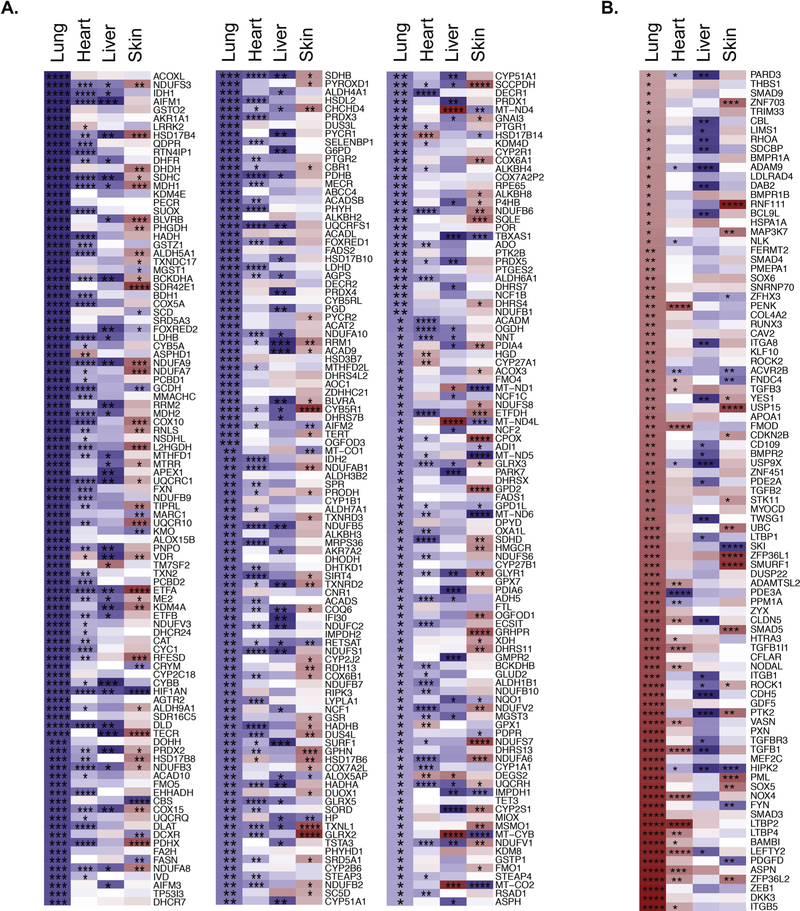
To address whether the changes in gene expression we observe relate to mechanisms with potential relevance to age-associated fibrotic pulmonary disease susceptibility, or are simply changes universal to aging cells, we repeated the Spearman correlation analysis with age on RNAseq data from other tissues known to experience age-associated fibrotic disease [40–42], notably heart, liver and skin (Tables S5–S7). We then compared age-dependent expression patterns across all four tissues using gene members from GO terms described in Fig. 3, namely “oxidoreductase activity” (GO:0016491) and “response to transforming growth factor beta” (GO:0071559) (Fig. 4). With respect to expression of oxidation/reduction effectors, the aging lung demonstrates repression of substantial numbers of genes when qualitatively compared to heart, liver or skin (Fig. 4A). Similarly, expression of TGF-beta 1 pathway associated genes are upregulated with age in lung when compared to heart, liver or skin (Fig. 4B). Thus, the coordinated changes in gene expression with age in lung are tissue-specific, and do not arise solely as a function of aging cells.
Fig. 4. Age-dependent gene expression displays tissue specificity.
A. 297 gene subset from the GO term 0016491 (Reduction Oxidation Activity) exhibiting significant negative correlation with age in lung compared to heart, liver, and skin. B. 91 gene subset from the GO term 0071559 (Response to TGFB Activity) exhibiting significant positive correlation with age in lung compared to heart, liver, and skin. Box color corresponds to Spearman Rho value; blue indicates negative correlation (decrease with age); red indicates positive correlation (increase with age). Associated p-values are denoted by stars (* = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001).
4. Discussion
Age currently represents the best predictor of fibrotic pulmonary disease risk [43–46]. The goal of the present study was to examine gene expression changes occurring in disease-free lung as a function of age in order to identify coordinately regulated gene clusters that might inform our understanding of the link between age and fibrotic pulmonary disease susceptibility. Following unbiased analyses, we identified changes in the transcriptome of aging healthy lung highlighting clusters of genes encompassing reduction/oxidation effectors and the TGF-beta 1 signaling axis that progressively shift with age.
Aging has also been strongly associated with changes in redox homeostasis. For example, mitochondrial dysfunction is an established feature of aging that remains incompletely understood, but is widely considered a major source of age-related oxidative stress [47,48]. We found that genes encoding components of mitochondrial complexes are downregulated with age in healthy lung (Fig. 3A, Tables S2–S3). Dysfunction of mitochondrial complexes I, III and IV are established mechanisms governing elevated ROS production in cells [49–51]. An increase in oxidant production, combined with a reduced ability to quench oxidants, results in increased oxidative stress linked to aging [10,11,14,17,18,32]. Here we document effectors of reduction/oxidation activity are significantly enriched among genes repressed with age in disease-free lung. These genes exhibit a significant negative correlation with age and are included in the GO term “Oxidoreductase activity” (GO:0016491) (Table 2 and Fig. 3A). Of particular interest, numerous genes within this term are hydrogen peroxide metabolizing enzymes. For example, our analysis identified catalase (CAT), peroxiredoxins (PRDXs) and glutathione peroxidases as exhibiting significant negative correlation with age (Fig. 3A), all of which are key regulators of cellular hydrogen peroxide homeostasis [52]. PRDXs utilize the thioredoxin/thioredoxin reductase (TXN/TXNRD) pathway to maintain their active reduced state. Our data show that mRNA expression of PRDX1–5, as well as TXN2 and TXNRD2–3, all negatively correlate with age in lung (Fig. 3A). The catalytic cycles of CAT, PRDXs, and TXNs all require NADPH in order to remain active. NADPH is generated in the first step of the pentose phosphate pathway by glucose-6-phosphate dehydrogenase (G6PD), which also exhibits significant negative correlation with age.
In contrast to the decreases in expression of these aforementioned oxidant metabolizing enzymes, our analysis shows that expression of NOX4, an NADPH oxidase that directly produces hydrogen peroxide, positively correlates with age (Fig. 3C, Table S1). These findings are of particular interest given the strong established link between NOX4 activation and lung fibrosis. Active myofibroblasts express NOX4, and resultant hydrogen peroxide has been implicated in death of epithelial cells [26,27]. Rodents treated with NOX4 inhibitors, Cpd 88 [4-[(benzyloxy) methyl]-2-(2-chlorophenyl)-5-(pyrazin-2-ylmethyl)-1H-pyrazolo [4,3-c]pyridine-3,6(2H,5H)-dione] or GKT137831, were strongly protected from the development of bleomycin induced fibrosis [27,53]. Furthermore, GKT137831 reversed increases in age-associated persistent lung fibrosis [27]. It is important to acknowledge that the prior observations regarding NOX4 have been made predominantly in fibroblasts [26–28]. The GTEx RNAseq data for human lung were generated using homogenized lung tissue and therefore we cannot conclude that the increased NOX4 we observe with age originates from fibroblasts specifically. Additional analysis, notably single cell transcriptomes in cells derived from the aging human lung will be required to formally link NOX4 levels to specific cell subsets. That said, increased NOX4 expression would be expected to result in increased hydrogen peroxide production, regardless of the cell-type expressing the protein. Therefore, the fact that we see increased NOX4 expression with age in lung suggests a potential source of oxidative stress with age, independent of the cell-type responsible.
Collectively, these observations support the hypothesis that age-dependent dysregulation of genes associated with redox homeostasis may contribute to increases in oxidative stress and suggest altered H2O2 metabolism as culminating in enhanced oxidative stress with age.
TGF-beta 1 (TGFB1) is a potent pro-fibrotic growth factor whose increased expression is a hallmark of fibrotic diseases. Recent work has linked enhanced ROS levels to activation of the TGF-beta 1 pathway [54]. Our analyses identify enrichment of TGFB1 signaling pathway members among those exhibiting positive correlation with age in disease-free lung, suggesting a reciprocal connection between TGFB1 and reduction/oxidation effectors. This observation supports prior evidence linking enhanced TGFB1 signaling to the activation of NOX424,55 and increased oxidative stress [54]. The release and activation of latent TGFB1 is mediated by several different molecules including proteases, thrombospondin-1, reactive oxygen species, and the integrins (ITG), ITGB6 and ITGB8 [56]. It is plausible that a progressive imbalance in age-dependent ROS mitigation, marked by dysregulation of redox effectors, could drive TGF-beta 1 pathway activation, while TGFB1 in turn promotes expression of NOX424,54,55, pointing to a feedforward loop whereby NOX4-derived hydrogen peroxide and TGFB1 amplify one another. Supporting this idea, elevated ROS have been proposed to modulate TGF-beta 1 expression in epithelial cells, and concomitantly participate in the activation of latent TGF-beta 1 in the ECM, thus incrementing TGF-beta 1 bioavailability [57]. Additionally, activation of latent TGF-beta 1 is enhanced by latent transforming growth factor beta binding proteins (LTBPs) [58], three of four of which (LTBP1, 2 & 4) exhibit positive correlation with age in lung (Fig. 3C). Therefore, a chronic ROS imbalance in aging lung is consistent with TGFB1 activation. Although the lungs evaluated herein were disease-free, the age-dependent gene expression patterns suggest a shift toward a pro-fibrotic pulmonary microenvironment, potentially explaining age-related fibrosis susceptibility [59].
While the GTEx RNAseq dataset represents a unique and robust resource to investigate gene expression in disease-free human tissue, there are notable limitations that deserve acknowledgment. First, samples assessed were collected post-mortem. While reasonable steps were taken to limit time between death and sampling, variation does exist sample to sample, and it is possible that such variation could impact sample quality. Second, older individuals are overrepresented in the dataset, which may impact our analysis by artificially weighting the correlations. Third, although we do filter out those samples described as suffering from chronic interstitial lung disease at time of death, the completeness of the medical information across samples varies, and therefore we cannot definitively rule out some included samples suffered a related undiagnosed condition. Fourth, it is possible that manner of death could impact sample quality and/or expression profiles, and we did not control for this variable given incomplete data in the available records. Despite these limitations, given the impracticality of performing lung biopsies on healthy individuals, the GTEx data represent a highly valuable resource for describing alterations in human tissue transcriptional networks with age.
Overall, our transcriptome analysis of disease-free lung suggests that age-associated fibrotic pulmonary disease susceptibility may be linked to chronic, progressive disruption of redox homeostasis and enhanced TGFB1 signaling. We propose that over a lifetime, modest yet progressive shifts in the expression of genes impacting functionally linked networks increase susceptibility to fibrotic pulmonary disease in response to repeated injuries or insults. While not everyone develops fibrotic pulmonary disease, the strong association between lung fibrosis and age illustrates that susceptibility dramatically increases as a function of age. The analyses of healthy lung tissues in the present study offer a theoretical mechanism explaining how this association may arise. Mitochondrial dysfunction and/or increases in NOX4 result in chronic elevated ROS [24,47,48,55], which in turn drives TGFB1 expression and downstream signaling. Enhanced TGFB1 activity induces changes to the ECM, and also promotes additional mitochondrial dysfunction and ROS production [54], thus creating a feedforward amplification loop that over time enhances susceptibility to fibrosis. As described in Fig. 4, the transcriptional changes we report exhibit tissue specificity, indicating the observed expression changes are not simply a function of aging cells. Comparing transcriptional profiles between tissues with differing age-associated disease susceptibilities may allow for the identification of features critical to disease initiation. We propose that the constellation of specific transcriptional changes illuminated herein has the potential to serve as a biomarker for fibrotic pulmonary disease susceptibility, and may ultimately help explain the mechanisms linking age with the increased incidence of fibrotic lung disease. That said, additional studies incorporating patients with fibrotic lung disease, analyses of sex-dependent differences, along with experimental studies to determine the nature of associations between oxidation/reduction and the TGFB1 signaling axis will be required to solidify these claims.
Supplementary Material
Acknowledgments
This work was supported by an internal grant (University of Vermont Department of Pathology and Laboratory Medicine (DJS), NIH R35 HL135828, Parker B. Francis fellowships (VA and JvdV), NIH R01HL05646 (AvdV).
Abbreviations
- ILD
interstitial lung disease
- ECM
extracellular matrix
- IPF
idiopathic pulmonary fibrosis
- Redox
reduction/oxidation
- ROS
reactive oxygen species
- TGF-beta 1
transforming growth factor beta 1
- ALFN
aging lung functional network
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.freeradbiomed.2019.07.011.
References
- [1].Rezende F, et al. , Knock out of the NADPH oxidase Nox4 has no impact on life span in mice, Redox Biol. 11 (2017) 312–314, 10.1016/j.redox.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Harman D, Aging: a theory based on free radical and radiation chemistry, J. Gerontol 11 (1956) 298–300. [DOI] [PubMed] [Google Scholar]
- [3].Beckman KB, Ames BN, The free radical theory of aging matures, Physiol. Rev 78 (1998) 547–581, 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- [4].Harman D, Free radical theory of aging, Mutat. Res 275 (1992) 257–266. [DOI] [PubMed] [Google Scholar]
- [5].Harman D, Free-radical theory of aging. Increasing the functional life span, Ann. N. Y. Acad. Sci 717 (1994) 1–15. [DOI] [PubMed] [Google Scholar]
- [6].Harman D, Free radical theory of aging: an update: increasing the functional life span, Ann. N. Y. Acad. Sci 1067 (2006) 10–21, 10.1196/annals.1354.003. [DOI] [PubMed] [Google Scholar]
- [7].Harman D, About “Origin and evolution of the free radical theory of aging: a brief personal history, 1954–2009”, Biogerontology 10 (2009) 783, 10.1007/s10522-009-9253-z. [DOI] [PubMed] [Google Scholar]
- [8].Ivanova DG, Yankova TM, The free radical theory of aging in search of a strategy for increasing life span, Folia Med. (Plovdiv) 55 (2013) 33–41. [DOI] [PubMed] [Google Scholar]
- [9].Koltover VK, Free radical timer of aging: from chemistry of free radicals to systems theory of reliability, Curr. Aging Sci 10 (2017) 12–17. [DOI] [PubMed] [Google Scholar]
- [10].Liochev SI, Reflections on the theories of aging, of oxidative stress, and of science in general. Is it time to abandon the free radical (oxidative stress) theory of aging? Antioxid. Redox Signal 23 (2015) 187–207, 10.1089/ars.2014.5928. [DOI] [PubMed] [Google Scholar]
- [11].Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G, The hallmarks of aging, Cell 153 (2013) 1194–1217, 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Meiners S, Eickelberg O, Konigshoff M, Hallmarks of the ageing lung, Eur. Respir. J 45 (2015) 807–827, 10.1183/09031936.00186914. [DOI] [PubMed] [Google Scholar]
- [13].Finkel T, Holbrook NJ, Oxidants, Oxidative stress and the biology of ageing, Nature 408 (2000) 239–247, 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- [14].Zhang Y, et al. , A new role for oxidative stress in aging: the accelerated aging phenotype in Sod1(-/)(−) mice is correlated to increased cellular senescence, Redox Biol. 11 (2017) 30–37, 10.1016/j.redox.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Aunan JR, Cho WC, Soreide K, The biology of aging and cancer: a brief overview of shared and divergent molecular hallmarks, Aging Dis. 8 (2017) 628–642, 10.14336/AD.2017.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Beeh KM, et al. , Glutathione deficiency of the lower respiratory tract in patients with idiopathic pulmonary fibrosis, Eur. Respir. J 19 (2002) 1119–1123. [DOI] [PubMed] [Google Scholar]
- [17].Kurundkar A, Thannickal VJ, Redox mechanisms in age-related lung fibrosis, Redox Biol. 9 (2016) 67–76, 10.1016/j.redox.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Thannickal VJ, Mechanistic links between aging and lung fibrosis, Biogerontology 14 (2013) 609–615, 10.1007/s10522-013-9451-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Raghu G, Weycker D, Edelsberg J, Bradford WZ, Oster G, Incidence and prevalence of idiopathic pulmonary fibrosis, Am. J. Respir. Crit. Care Med 174 (2006) 810–816, 10.1164/rccm.200602-163OC. [DOI] [PubMed] [Google Scholar]
- [20].Wynn TA, Ramalingam TR, Mechanisms of fibrosis: therapeutic translation for fibrotic disease, Nat. Med 18 (2012) 1028–1040, 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bray N, Lung disease: resetting the redox balance in lung fibrosis, Nat. Rev. Drug Discov 13 (2014) 415, 10.1038/nrd4344. [DOI] [PubMed] [Google Scholar]
- [22].Iyer SS, et al. , Oxidation of extracellular cysteine/cystine redox state in bleomycin-induced lung fibrosis, Am. J. Physiol. Lung Cell Mol. Physiol 296 (2009) L37–L45, 10.1152/ajplung.90401.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kinnula VL, Redox imbalance and lung fibrosis, Antioxid. Redox Signal 10 (2008) 249–252, 10.1089/ars.2007.1912. [DOI] [PubMed] [Google Scholar]
- [24].Richter K, Konzack A, Pihlajaniemi T, Heljasvaara R, Kietzmann T, Redox-fi-brosis: impact of TGFbeta1 on ROS generators, mediators and functional consequences, Redox Biol. 6 (2015) 344–352, 10.1016/j.redox.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cantin AM, Hubbard RC, Crystal RG, Glutathione deficiency in the epithelial lining fluid of the lower respiratory tract in idiopathic pulmonary fibrosis, Am. Rev. Respir. Dis 139 (1989) 370–372, 10.1164/ajrccm/139.Z370. [DOI] [PubMed] [Google Scholar]
- [26].Carnesecchi S, et al. , A key role for NOX4 in epithelial cell death during development of lung fibrosis, Antioxid. Redox Signal 15 (2011) 607–619, 10.1089/ars.2010.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hecker L, et al. , Reversal of persistent fibrosis in aging by targeting Nox4-Nrf2 redox imbalance, Sci. Transl. Med 6 (2014) 231–247, 10.1126/scitranslmed.3008182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hecker L, et al. , NADPH oxidase-4 mediates myofibroblast activation and fibro-genic responses to lung injury, Nat. Med 15 (2009) 1077–1081, 10.1038/nm.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liu R, Choi J, Age-associated decline in gamma-glutamylcysteine synthetase gene expression in rats, Free Radic. Biol. Med 28 (2000) 566–574. [DOI] [PubMed] [Google Scholar]
- [30].Santos-Silva MA, et al. , Redox imbalance and pulmonary function in bleomycin-induced fibrosis in C57BL/6, DBA/2, and BALB/c mice, Toxicol. Pathol 40 (2012) 731–741, 10.1177/0192623312441404. [DOI] [PubMed] [Google Scholar]
- [31].Zhang H, et al. , Nrf2-regulated phase II enzymes are induced by chronic ambient nanoparticle exposure in young mice with age-related impairments, Free Radic. Biol. Med 52 (2012) 2038–2046, 10.1016/j.freeradbiomed.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhou L, Zhang H, Davies KJA, Forman HJ, Aging-related decline in the induction of Nrf2-regulated antioxidant genes in human bronchial epithelial cells, Redox Biol. 14 (2018) 35–40, https://doi.org/10.1016Zj.redox.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wong AK, Krishnan A, Troyanskaya OG, GIANT 2.0: genome-scale integrated analysis of gene networks in tissues, Nucleic Acids Res. 46 (2018) W65–W70, 10.1093/nar/gky408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Goya J, et al. , FNTM: a server for predicting functional networks of tissues in mouse, Nucleic Acids Res. 43 (2015) W182–W187, 10.1093/nar/gkv443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Huang JK, et al. , Systematic evaluation of molecular networks for discovery of disease genes, Cell Syst. 6 (2018) 484–495, 10.1016/j.cels.2018.03.001e485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Newman ME, Modularity and community structure in networks, Proc. Natl. Acad. Sci. U. S. A 103 (2006) 8577–8582, 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bastian M, Heymann S, Jacomy M, Gephi: an Open Source Software for Exploring and Manipulating Networks. aaai.Org, (2009). [Google Scholar]
- [38].Reimand J, Arak T, Vilo J.g, Profiler-a web server for functional interpretation of gene lists (2011 update), Nucleic Acids Res. 39 (2011) W307–W315, 10.1093/nar/gkr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chen J, Bardes EE, Aronow BJ, Jegga AG, ToppGene Suite for gene list enrichment analysis and candidate gene prioritization, Nucleic Acids Res. 37 (2009) W305–W311, 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Delire B, et al. , Aging enhances liver fibrotic response in mice through hampering extracellular matrix remodeling, Aging (Albany NY) 9 (2016) 98–113, 10.18632/aging.101124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Biernacka A, Frangogiannis NG, Aging and cardiac fibrosis, Aging Dis. 2 (2011) 158–173. [PMC free article] [PubMed] [Google Scholar]
- [42].Luckhardt TR, Thannickal VJ, Systemic sclerosis-associated fibrosis: an accelerated aging phenotype? Curr. Opin. Rheumatol 27 (2015) 571–576, 10.1097/BOR.0000000000000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Araki T, Katsura H, Sawabe M, Kida K, A clinical study of idiopathic pulmonary fibrosis based on autopsy studies in elderly patients, Intern. Med 42 (2003) 483–489. [DOI] [PubMed] [Google Scholar]
- [44].Blasco MA, Telomere length, stem cells and aging, Nat. Chem. Biol 3 (2007) 640–649, 10.1038/nchembio.2007.38. [DOI] [PubMed] [Google Scholar]
- [45].King TE Jr., Tooze JA, Schwarz MI, Brown KR, Cherniack RM, Predicting survival in idiopathic pulmonary fibrosis: scoring system and survival model, Am. J. Respir. Crit. Care Med 164 (2001) 1171–1181, 10.1164/ajrccm.164.7.2003140. [DOI] [PubMed] [Google Scholar]
- [46].Reichel W, Garcia-Bunuel R, Pathologic findings in progeria: myocardial fibrosis and lipofuscin pigment, Am. J. Clin. Pathol 53 (1970) 243–253. [DOI] [PubMed] [Google Scholar]
- [47].Cui H, Kong Y, Zhang H, Oxidative stress, mitochondrial dysfunction, and aging, J. Signal Transduct (2012) 646354, 10.1155/2012/646354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kudryavtseva AV, et al. , Mitochondrial dysfunction and oxidative stress in aging and cancer, Oncotarget 7 (2016) 44879–44905, 10.18632/oncotarget.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jain M, et al. , Mitochondrial reactive oxygen species regulate transforming growth factor-beta signaling, J. Biol. Chem 288 (2013) 770–777, 10.1074/jbc.M112.431973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yoon YS, Lee JH, Hwang SC, Choi KS, Yoon G, TGF beta1 induces prolonged mitochondrial ROS generation through decreased complex IV activity with senescent arrest in Mv1Lu cells, Oncogene 24 (2005) 1895–1903, 10.1038/sj.onc.1208262. [DOI] [PubMed] [Google Scholar]
- [51].Byun HO, et al. , GSK3 inactivation is involved in mitochondrial complex IV defect in transforming growth factor (TGF) beta1-induced senescence, Exp. Cell Res 318 (2012) 1808–1819, 10.1016/j.yexcr.2012.04.012. [DOI] [PubMed] [Google Scholar]
- [52].Winterbourn CC, The biological chemistry of hydrogen peroxide, Methods Enzymol. 528 (2013) 3–25, 10.1016/B978-0-12-405881-1.00001-X. [DOI] [PubMed] [Google Scholar]
- [53].Jarman ER, et al. , An inhibitor of NADPH oxidase-4 attenuates established pulmonary fibrosis in a rodent disease model, Am. J. Respir. Cell Mol. Biol 50 (2014) 158–169, 10.1165/rcmb.2013-0174OC. [DOI] [PubMed] [Google Scholar]
- [54].Liu RM, Desai LP, Reciprocal regulation of TGF-beta and reactive oxygen species: a perverse cycle for fibrosis, Redox Biol. 6 (2015) 565–577, 10.1016/j.redox.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ghatak S, et al. , Transforming growth factor beta1 (TGFbeta1)-induced CD44V6-NOX4 signaling in pathogenesis of idiopathic pulmonary fibrosis, J. Biol. Chem 292 (2017) 10490–10519, 10.1074/jbc.M116.752469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Annes JP, Munger JS, Rifkin DB, Making sense of latent TGFbeta activation, J. Cell Sci 116 (2003) 217–224. [DOI] [PubMed] [Google Scholar]
- [57].Gorowiec MR, et al. , Free radical generation induces epithelial-to-mesenchymal transition in lung epithelium via a TGF-beta1-dependent mechanism, Free Radic. Biol. Med 52 (2012) 1024–1032, https://doi.Org/10.1016/j.freeradbiomed.2011.12.020. [DOI] [PubMed] [Google Scholar]
- [58].Yoshinaga K, et al. , Perturbation of transforming growth factor (TGF)-beta1 association with latent TGF-beta binding protein yields inflammation and tumors, Proc. Natl. Acad. Sci. U. S. A 105 (2008) 18758–18763, 10.1073/pnas.0805411105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sicard D, et al. , Aging and anatomical variations in lung tissue stiffness, Am. J. Physiol. Lung Cell Mol. Physiol 314 (2018) L946–L955, 10.1152/ajplung.00415.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.