Abstract
Single-domain antibodies (VHHs or nanobodies), developed from heavy chain-only antibodies of camelids, are gaining attention as next-generation therapeutic agents. Despite their small size, the high affinity and specificity displayed by VHHs for antigen molecules rival those of IgGs. How such small antibodies achieve that level of performance? Structural studies have revealed that VHHs tend to recognize concave surfaces of their antigens with high shape-complementarity. However, the energetic contribution of individual residues located at the binding interface has not been addressed in detail, obscuring the actual mechanism by which VHHs target the concave surfaces of proteins. Herein, we show that a VHH specific for hen egg lysozyme, D3-L11, not only displayed the characteristic binding of VHHs to a concave region of the surface of the antigen, but also exhibited a distribution of energetic hot-spots like those of IgGs and conventional protein-protein complexes. The highly preorganized and energetically compact interface of D3-L11 recognizes the concave epitope with high shape complementarity by the classical lock-and-key mechanism. Our results shed light on the fundamental basis by which a particular VHH accommodate to the concave surface of an antigens with high affinity in a specific manner, enriching the mechanistic landscape of VHHs.
Subject terms: Proteins, X-ray crystallography, Thermodynamics, Biophysical chemistry
Introduction
Immunoglobulin molecules of the IgG family are widely employed as therapeutic agents and increasingly produced in a variety of formats as drug conjugates or bispecific antibodies1. In addition, fragments of antibodies and antibody-like scaffolds of small size devoid of the Fc fragment are increasingly considered as promising alternatives because of their unique pharmacokinetic profile2. Because of their smaller size, they are amenable to straightforward modifications such as in fusion-proteins or in drug-conjugates by protein engineering technologies. Among them, miniature single-domain antibodies derived from the variable region of the heavy chain-only antibodies of camelids have gained relevance due to their high therapeutic potential and the robust techniques employed to generate them3,4. Indeed, these miniature antibodies, termed VHHs, have already entered the pharmaceutical market. For example, the bivalent VHH dimer known as caplacizumab was approved in 2018 for the treatment of thrombosis in Europe5. VHHs are also valuable sensors for a range of applications6,7, in affinity chromatography8–10, as tools in cell-biology11,12, or as co-crystallization chaperons13,14.
The usefulness of VHHs relies on their ability to specifically recognize antigens with high affinity despite their small size. An important mechanistic feature of VHHs consists in their ability to target clefts on the surface of the antigen molecules3,15,16. VHHs comprise only one domain, and for that reason it is less likely that they will adopt the type of large and flat surface displayed by the variable fragments (Fvs) of conventional antibodies. Instead, because of their small size, VHHs tend to interact with the concave surface of antigen molecules17. The long complementarity determining region 3 (CDR3) of VHHs often protrudes from the body of the antibody and docks into clefts located on the surface of antigens with high shape complementarity, in a manner that mimics some human and bovine antibodies15. It is believed that in this way, VHHs compensate for the limitations of their small size, while maintaining the high affinity and specificity that constitute the hallmarks of antibodies17. This mechanism has been observed in a structural study that compared the binding mode of VHHs with that of Fvs using hen egg lysozyme (HEL) as a model antigen18. Several more studies have revealed additional details of how VHHs target the concave surface of antigen molecules19–27. However, the energetic contribution of each individual residue to the strength of the antibody-antigen complex has not been examined in detail, and therefore the mechanism by which VHHs favor concave surfaces of antigens is still incomplete.
Herein, we sought to reveal the molecular and energetic basis for antigen recognition by a VHH to an unprecedented level of detail. For that purpose, a VHH termed D3-L11 recognizing the active site of HEL18 was selected. The characteristic roles of the residues located at the antigen-antibody interface were examined using site-directed mutagenesis with high-resolution calorimetric techniques, NMR, and X-ray crystallography. We explain the fundamental basis by which D3-L11 accommodate to an antigen cleft with high affinity in a specific manner.
Results
Energetic basis for the antibody-antigen interaction
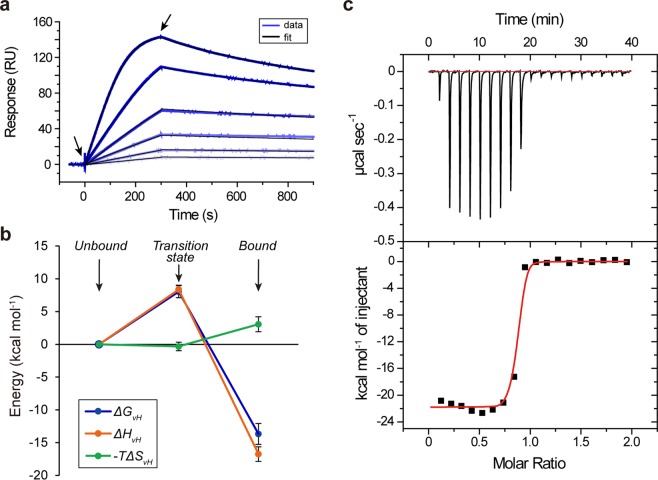
The kinetic parameters of the interaction between the VHH antibody termed D3-L11 and its antigen HEL were determined by surface plasmon resonance (SPR) analysis at 25 °C (Fig. 1a). The values of the association rate constant (kon = 6.1 ± 1.6 × 106 M−1 s−1) and dissociation rate constant (koff = 7.9 ± 1.7 × 10−4 s−1) were determined as described in the methods section, resulting in a dissociation constant in the sub-nanomolar range (KD = 0.13 ± 0.04 nM). These values were consistent with the data reported in the literature for this antibody18.
Figure 1.
D3-L11 recognizes HEL with high affinity. (a) Binding of VHH to immobilized HEL at 25 °C monitored by SPR. The sensorgrams corresponded to the signal obtained from two-fold dilution series between 2 nM (darkest blue) and 0.125 nM (lightest blue). The black traces corresponded to the best fit to a 1:1 binding model. (b) Energy diagram of the interaction of D3-L11 with HEL with respect to the unbound state. Left, center, and right, corresponded to the unbound, transition, and equilibrium states, respectively. Errors bars corresponded to the standard error. (c) Titration of D3-L11 with HEL by ITC. The top panel shows the injection data (red, baseline). The bottom panel corresponded to the binding isotherm, (red, non-linear fitting). The slope of the binding isotherm was too steep for a precise estimation of the dissociation constant. In contrast, this condition favors a precise determination of the change of enthalpy.
Thermodynamic parameters were calculated by the van’t Hoff approximation (constant values within a narrow range of temperatures) as described in materials and methods. Binding is driven by a favorable change of enthalpy (ΔHvH = −17 ± 1 kcal mol−1) and opposed by the change of entropy (−TΔSvH = 3.1 ± 1.1 kcal mol−1) resulting in a large and favorable change of free energy (ΔGvH = −14 ± 2 kcal mol−1) (Supplementary Information Figs S1 and S2). The thermodynamic parameters in the transition state were determined by the Eyring approximation using the association rate constants (Supplementary Information Fig. S3). The values of change of enthalpy, change of entropy and change of free energy at the transition state were 8.4 ± 0.6 kcal mol−1, 0.3 ± 0.7 cal K−1 mol−1, and 8.0 ± 0.9 kcal mol−1, respectively. The values of all three thermodynamic parameters throughout the reaction coordinate are represented schematically in Fig. 1b.
Collectively, the interaction between D3-L11 and HEL is strongly dependent on the enthalpic component suggesting that non-covalent interactions drive the recognition of the antigen by D3-L11. The overwhelming influence of the change of enthalpy in the formation of the antibody-antigen complex was corroborated by isothermal titration calorimetry (ITC), yielding a large value for the change of enthalpy (ΔH° = −21.4 ± 0.6 kcal mol−1) (Fig. 1c). The high affinity of the complex between D3-L11 and HEL precluded a more detailed analysis of the entropy and free energy by this technique. The difference of ~4 kcal mol−1 between the calorimetric enthalpy and the van’t Hoff enthalpy calculated from SPR data (corresponding to a 20% of the total value) is likely related to the methodology to calculate it (a direct determination in ITC vs. the van’t Hoff method in SPR) and/or the presence of detergent in the buffer employed for SPR. Despite this modest difference, the principle that the binding is driven by the change of enthalpy and opposed by the entropy component is consistent in both techniques.
Individual contribution of VHH’s interfacial residues to the binding affinity
To analyze the contribution of the side chains of each residue at the binding interface, first the crystal structure of the D3-L11·HEL complex was determined at 1.65 Å resolution (Supplementary Information Table S1). Overall, the crystal structure was essentially identical to that previously reported (PDB ID: 1ZVY). The root mean square deviation (RMSD) value between Cα atoms of VHH and between Cα atoms of HEL in each crystal structure as calculated with COOT28 was 0.49 Å and 0.54 Å, respectively (Supplementary Information Fig. S4). It is observed that the CDR3 region of D3-L11 protruded into a cleft of HEL, comprising 68% of the buried surface area of the binding interface (491 Å2 of a total of 719 Å2) with high complementarity (Sc = 0.78). The list of residues appearing at the antibody-antigen interface with a buried surface area greater than 10 Å2 as identified with the PISA server29 are listed on the Supplementary Information Table S2.
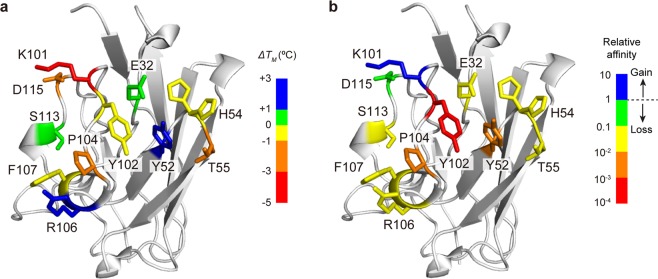
We prepared single alanine-mutants corresponding to each of these residues (Supplementary Information Fig. S5). The structural integrity of each of the mutants was verified by circular dichroism (Supplementary Information Fig. S6). The thermal stability of D3-L11 was monitored by differential scanning calorimetry (DSC), confirming that all the mutants were stable at physiological temperatures (Supplementary Information Fig. S7). The mid-point temperature (TM) at which thermal unfolding of wild-type (WT) or mutant antibodies ocurred was determined to be in the range 62–69 °C (Table 1). The heat map in Fig. 2a identifies the locations of the residues whose Ala mutations were prepared, as a function of their effect in the thermal stability (ΔTM = − ). Positive and negative values indicate a gain or loss of stability, respectively. The CD and calorimetric data thus verified the integrity of the mutants for the follow-up experiments.
Table 1.
Thermal stability of D3-L11 determined by DSC.
| TM (°C)a | ΔTM (°C) (TMmutant − TMWT) |
Order of stability (WT, reference) |
|
|---|---|---|---|
| WT | 66.79 ± 0.01 | 0 | 0 |
| E32A | 67.26 ± 0.02 | 0.47 | +1 |
| Y52A | 68.79 ± 0.01 | 2.00 | +4 |
| H54A | 66.78 ± 0.01 | −0.01 | −1 |
| T55A | 63.74 ± 0.01 | −3.05 | −4 |
| K101A | 62.43 ± 0.02 | −4.36 | −7 |
| Y102A | 66.38 ± 0.01 | −0.41 | −2 |
| P104A | 63.54 ± 0.02 | −3.25 | −5 |
| R106A | 68.04 ± 0.01 | 1.25 | +3 |
| F107A | 66.24 ± 0.01 | −0.55 | −3 |
| S113A | 67.59 ± 0.01 | 0.80 | +2 |
| D115A | 62.92 ± 0.02 | −3.87 | −6 |
aValues ± S.E. of the curve fitting.
Figure 2.
Site-directed mutagenesis. The figure shows the distribution of residues mutated to alanine. Mutated residues are depicted with sticks. (a) Residues are colored according to the effect of the mutation on ΔTM. Blue and red indicate greatest stabilizing and destabilizing effect, respectively. (b) Residues are colored depending on the effect of the mutation on the affinity, quantified with respect to WT antibody. Blue indicates a gain of affinity. Green to red signals an increasing loss of affinity.
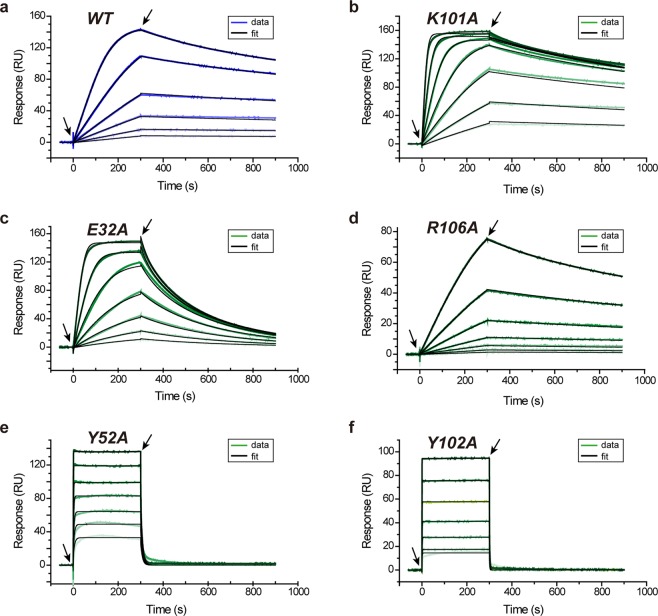
The binding interaction between each mutant and its antigen HEL was evaluated by SPR (Figs 2b and 3, Table 2, and Supplementary Information Fig. S8). Among the mutants, only K101A showed higher affinity for HEL than the WT antibody (1.4-fold tighter binding corresponding to a favorable ΔΔG° = −0.2 kcal mol−1) despite being the mutant with the lowest stability overall (ΔTM = −4.4 ± 0.1 °C). Except K101A, all other mutants displayed lower (or much lower) affinity for the antigen than WT antibody. From a kinetic point of view, the meager performance of these mutants was generally linked to a faster dissociation of the antibody-antigen complex (greater values of koff).
Figure 3.
Binding of mutants. SPR sensorgrams corresponding to the binding of few D3-L11 mutants to HEL at 25 °C. Data for additional mutants are found in Supplementary Information Fig. S8. (a) WT (same as Fig. 1a). (b) K101A, corresponding to the only mutant in which the affinity increased with respect to WT. (c–f) Mutants E32A, R106A, Y52A, and Y102A showing less affinity for HEL. Experimental data is shown in blue (WT) or green (mutants). The color darkens as the concentration of VHH gradually increased in the experiment. Other details as in Fig. 1a. The concentrations employed were: (a) 2 nM to 0.125 nM (b–d) 16 nM to 0.25 nM, (e) 1024 to 16 nM, and (f) 2048 to 32 nM, in all cases in two-fold dilutions.
Table 2.
Kinetic and thermodynamic parameters obtained from SPR.
|
k
on
(×106 M−1 s−1) |
k
off
(×10−3 s−1) |
K
D
(nM) |
Relative affinity (KDmut/KDWT) |
|
|---|---|---|---|---|
| WTa | 6.10 ± 1.62 | 0.785 ± 0.170 | 0.129 ± 0.044 | 1 |
| E32Ab | 6.43 ± 0.02 | 12.0 ± 0.5 | 1.87 ± 0.01 | 15 |
| Y52Ab | 2.77 ± 0.02 | 128 ± 9 | 46.1 ± 0.5 | 358 |
| H54Ab | 3.48 ± 0.01 | 9.21 ± 0.39 | 2.65 ± 0.02 | 21 |
| T55Ab | 3.77 ± 0.04 | 11.3 ± 1.1 | 2.99 ± 0.04 | 23 |
| K101Ab | 7.54 ± 0.01 | 0.687 ± 0.008 | 0.0911 ± 0.0002 | 0.7 |
| Y102Ab | 2.32 ± 0.02 | 686 ± 75 | 295 ± 4 | 2,299 |
| P104Ab | 17.4 ± 0.6 | 226 ± 77 | 13.0 ± 0.6 | 101 |
| R106Ab | 1.11 ± 0.00 | 1.83 ± 0.05 | 1.65 ± 0.01 | 13 |
| F107Ab | 8.99 ± 0.08 | 14.7 ± 1.4 | 1.63 ± 0.02 | 13 |
| S113Ab | 5.17 ± 0.11 | 8.31 ± 1.6 | 1.61 ± 0.04 | 13 |
| D115Ab | 6.11 ± 0.01 | 0.956 ± 0.013 | 0.156 ± 0.000 | 1.2 |
aValues ± S.D. from seven independent experiments.
bValues ± S.E of the fitting to the sensorgrams.
Based on the mutagenesis analysis, residues Y52, Y102 and P104 were identified as hot-spot residues bearing a large influence in binding (>100-fold loss of affinity) and therefore resulting in a large energetic contribution (ΔΔG > 2.7 kcal mol−1). The effect of mutation in the hot-spot residues was so significant that the sensorgrams displayed a box-shape indicating a very fast dissociation rate. The values of KD were also determined from their responses in equilibrium (Supplementary Information Fig. S9). The values obtained were 144 ± 22 nM, 635 ± 66 nM and 44.8 ± 19.0 nM for Y52A, Y102A and P104A, respectively. These values were uniformly greater than those obtained by global analysis by 2-3.5-fold (i.e. weaker affinity than that obtained by global analysis), but the order of affinities calculated by either method (Y102A < Y52A < P104A << others) was uniform in both calculations.
Thermodynamic dissection of hot-spot tyrosine residues
The thermodynamic basis of the contribution of the two hot-spot tyrosine residues to the binding and to the transition state were next determined by the van’t Hoff and Eyring approximations, respectively (Supplementary Information Figs S10–S12). The thermodynamic parameters are given in Table 3. The removal of the aromatic side-chain of Tyr52 or Tyr102 resulted in large losses of change of free energy with respect to the WT antibody. In Y52A, the change of free energy was significantly diminished (ΔGvH = −10 ± 1 kcal mol−1, corresponding to KD = 46 ± 19 nM at 25 °C), whereas in Y102A it was even less favorable (ΔGvH = −8.7 ± 4.0 kcal mol−1 corresponding to KD = 413 ± 667 nM at 25 °C). The values obtained when calculations were carried out with dissociation constants obtained from the Scatchard plots were consistent with the data presented just above within experimental error (Supplementary Information Figs S13–15 and Supplementary Information Table S3).
Table 3.
Thermodynamic binding parameters (SPR)a.
| ΔGvH | ΔΔGvHb | ΔHvH | ΔΔHvHb | −TΔSvH | −TΔΔSvHb | ΔCp,vH | ΔG‡ | ΔH‡ | −TΔS‡ | |
|---|---|---|---|---|---|---|---|---|---|---|
| kcal mol−1 | kcal mol−1 | kcal mol−1 | kcal mol−1 | kcal mol−1 | kcal mol−1 | kcal K−1 mol−1 | kcal mol−1 | kcal mol−1 | kcal mol−1 | |
| WT | −14 ± 2 | — | −17 ± 1 | 3.1 ± 1.1 | — | −0.024 ± 0.186 | 8.0 ± 0.9 | 8.4 ± 0.6 | −0.32 ± 0.66 | |
| Y52A | −10 ± 1 | 4.0 ± 2.2 | −11 ± 1 | 6.0 ± 1.4 | 0.68 ± 0.54 | −2.4 ± 1.2 | −0.16 ± 0.07 | 8.8 ± 0.2 | 4.3 ± 0.1 | −4.5 ± 0.1 |
| Y102A | −8.7 ± 4.0 | 5.3 ± 4.5 | −12 ± 3 | 5.0 ± 3.2 | 3.2 ± 2.9 | 0.1 ± 3.1 | 0.15 ± 0.48 | 8.8 ± 1.3 | 6.7 ± 0.9 | −2.1 ± 0.9 |
aValues ± S.E of the curve fitting based on van’t Hoff or Eyring plots.
bΔΔGvH = ΔGvH,mutant − ΔGvH,WT, ΔΔHvH = ΔHvH, mutant − ΔHvH, WT; −TΔΔSvH = (−TΔSvH, mutant) − (−TΔSvH,WT).
Importantly, the meager affinity between HEL and the mutated antibodies was caused by a significant loss in the change of enthalpy with respect to wild type antibody. In Y52A and Y102A the loss of enthalpy with respect to WT antibody was 6.0 ± 1.4 kcal mol−1, and 5.0 ± 3.2 kcal mol−1, respectively (Table 3). The loss of enthalpy was not compensated by a favorable change of entropy of similar magnitude, leading to weaker binding. Analogous conclusions were obtained when the analysis was carried out with the parameters from the binding at equilibrium (Supplementary Information Table S3). Furthermore, similar observations were made when the analysis was carried with the orthogonal technique of ITC (Supplementary Information Figs S16–S17 and Supplementary Information Table S4). The large loss of enthalpy with respect to WT antibody in Y52A (ΔΔH° = 9.1 ± 1.4 kcal mol−1) and Y102A (ΔΔH° = 4.8 ± 3.3 kcal mol−1) is consistent with the conclusion made above by SPR, although similarly to WT, the value of enthalpy obtained by ITC in mutant Y52A was slightly above the error limits of the value of enthalpy calculated by SPR.
In the transition state, the change of free energy and the change of enthalpy were unfavorable and remarkably similar to each other (ΔG‡ = 8.0 ± 0.9 kcal mol−1; ΔH‡ = 8.4 ± 0.6 kcal mol−1) (Table 3). Predictably, the change of free energy of the mutants with respect to WT is more unfavorable in the modified antibodies, suggesting that loss of affinity in equilibrium is already influenced by the events at the transition state. When the energetic terms were dissected it was observed that, contrary to the situation in equilibrium, the change of enthalpy (ΔH‡) observed in both Y52A and Y102A was stabilizing (ΔΔH‡ = −4.1 kcal mol−1 and −1.7 kcal mol−1, respectively). In other words, the deleterious effects at this stage of the reaction coordinate in the mutants were of entropic nature, suggesting more unfavorable rearrangement of VHH and/or water molecules in the mutants than in the WT antibody.
Structural analysis
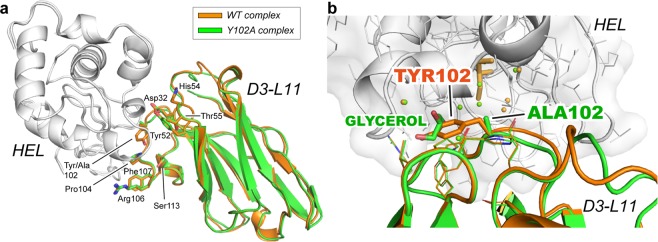
The large loss of affinity of Y102A (~2,300-fold) with respect to WT prompted a more detailed structural analysis by X-ray crystallography. The complex Y102A·HEL was determined at 1.50 Å and 1.55 Å resolution, respectively. The complex of the mutant with the antigen superimposed onto the complex with WT antibody is shown in Fig. 4a. A close-up view of the mutated residue and its environment is depicted in Fig. 4b. Except for the presence of a molecule of cryoprotectant (glycerol) filling up the vacancy of the side-chain of Tyr102, no other significant differences with respect to WT antibody were observed, such as in the conformation of the neighboring residues of Ala102, or in residues contributing to the affinity (Glu32, Tyr52, His54, Thr55, Pro104, Arg106 and Phe107). Similarly, the shape complementarity remained high as shown by the little change in the Sc parameter (Sc = 0.76). Although the mutation Y102A considerably impaired the binding affinity for the antigen, the conformation of the antibody-antigen interface remained largely unaffected.
Figure 4.
Crystal structure of the antibody-antigen complex. (a) Superimposed structures of the complex WT·HEL with that of the mutant Y102A·HEL. Alignment was carried out on the antigen. WT, Y102A, and HEL are shown in orange, green and gray, respectively. The side chains of residues influencing binding (loss of affinity >10-fold) are depicted with sticks. (b) Close-up view of the environment around Tyr102. The position of the side chains of the residues contributing to the affinity of the complex were not altered by the mutation. The vacancy generated by the removal of the side chain at position 102 was filled by a molecule of cryoprotectant (glycerol).
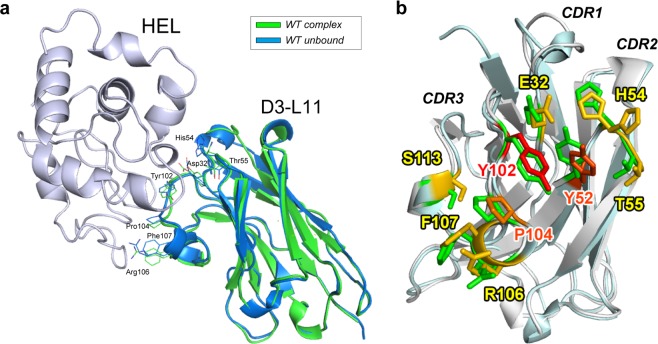
The X-ray structure of WT D3-L11 in the absence of the antigen was also determined, achieving a resolution of 1.15 Å. The coordinates of unbound D3-L11 were aligned to those of the antibody in complex with HEL using the Cα atoms of the residues of the framework region (FR) (Fig. 5). The conformation of the residues contributing to the affinity did not change significantly in the unbound form compared to the antibody-antigen complex (RMSD = 0.76 Å among main chain α-carbons). Collectively, the X-ray data suggests that the interface residues of VHH contributing to binding the antigen adopted a rigid conformation.
Figure 5.
Crystal structure of unbound VHH. (a) Comparison of unbound D3-L11 (blue) with that in complex with HEL (green). HEL is shown in light purple. Overlaid structures using the three CDR loops. Backbone with the side chains of residues showing influence in binding (affinity loss >10-fold). (b) Enlarged view of the VHH binding interface from panel (a). The unbound and bound antibodies are depicted in cyan and gray, respectively; side chains of residues are shown with the same color gradient as that in Fig. 3.
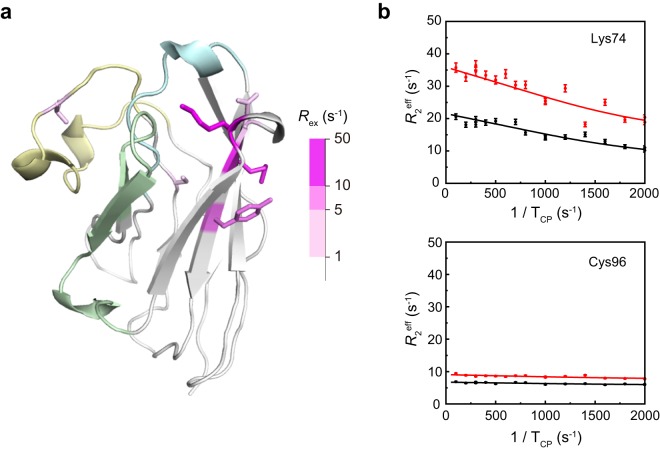
To clarify the conformational fluctuation of these residues, the R2 dispersion parameters were determined by NMR. This method is employed to characterize fluctuations in the microsecond to millisecond scale, which is within the relevant time scale for protein function. Only six residues displayed values of Rex greater than 1 s−1 (Lys74, Asn77, Met78, Tyr80, Cys96 and Val103) (Fig. 6). Those residues with significant fluctuations (Rex > 5 s−1) were exclusively located in the FR3 (Lys74, Met78, Tyr80). Except for Val103, residues in the CDRs showed little fluctuation, suggesting that the CDRs were little dynamic in solution, in agreement with the conclusions extracted from the crystallographic data.
Figure 6.
Fluctuation of residues of D3-L11 (main chain). (a) Structure of D3-L11 displaying the residues with the greatest fluctuations (sticks in magenta gradient). The key recognition regions CDR1, CDR2, and CDR3 are colored in cyan, green, and yellow, respectively. Other regions are shown in gray. (b) R2 dispersion profiles of Lys74 and Cys96 collected at 14.1 T (black) and 17.6 T (red) are shown. The residues were selected as representative examples of a residue displaying a large fluctuation, and that of a residue with no fluctuation.
Discussion
The combination of thermodynamic, structural and dynamic information is a powerful tool to accurately describe interaction phenomena like that in the recognition of antigens by antibodies. Herein we report that the recognition mode of D3-L11 for its antigen HEL shared some features in common with Fv from conventional antibodies30 and some features in common with other VHHs31,32. The present study further dissected the roles of each residue at the binding interface by alanine-scan mutagenesis.
Previous studies have suggested a dominant role for the CDR3 in the recognition of antigens by antibodies of the VHH class17. The CDR3 region stems from recombination of all of V, D and J gene segments, and somatic hypermutation, becoming the most diverse CDR in length and sequence in antibodies33. The analysis of D3-L11 carried herein supports this general idea, and embraces the concept of three-dimensional proximity around a core of key residues acting as energetic hot-spots. Of the three hot-spots residues identified in D3-L11, two of them are located in the CDR3 and one in the CDR2. These three residues are clustered together in the three-dimensional structure, and uniformly surrounded by a constellation of residues of lesser but still appreciable importance for binding the antigen. This uniform layer comprises residues belonging to each of the three CDR regions of the VHH.
This kind of arrangement is commonly observed in the paratope of Fvs and in the interaction surface of protein-protein complexes30,34–36. However, an important difference between Fvs and D3-L11 is observed. Whereas Fvs generally recognize planar epitopes37–39, D3-L11 recognize a cleft on the surface of the antigen with high shape complementarity (Sc > 0.75), a signature characteristic of numerous other VHHs18. The results presented herein demonstrated that hot-spot residues coalescing on a small geometric region are characteristic not only when interacting with the planar surface of antigens such as in Fvs, but can also occur in a VHH recognizing a concave surface of the antigen.
In contrast, this arrangement of residues is not common among other VHHs. Generally, VHHs of known structure display long, extended CDR3 (or less commonly, CDR1)40 that interact with the antigen epitope independently from other regions of the VHH moiety21,23–25. For example, a previous study of a VHH exhibiting a protruding CDR3 indicated that the most determining residues were present in the extended CDR3, but not outside of it21. The constellation of residues of VHH in that study is clearly different from that of D3-L11, even if there are also some similarities in other parts of the CDR3. In the majority of VHHs with long CDR3, and D3-L11 is not an exception, this loop covers a relatively hydrophobic surface of the framework region 2 (FR2) of the VHH molecule. These miniature antibodies utilize the long CDR3 loop to naturally act as a part of the single domain antibody, rather than as an independent element. The equivalent surface in Fv is located between the VL and VH regions41.
The structural data showed only minor differences in the conformation of the main chain between unbound and bound D3-L11, suggesting that preorganization of the interaction surface is a key aspect of the mechanism of recognition. The rigidity of the residues comprising the interaction surface would ensure a highly cooperative mode of interaction of the three CDR loops, enhancing the affinity for the antigen. Similar structural characteristics were observed for a VHH interacting with a shallow concave surface of its antigen26. In contrast, in VHHs where CDR3 protrudes to access deep concave sites the structural data revealed large differences between the complexed and unbound structures21,27. Mixed of VHHs binding concave sites of antigens without protruding CDRs show slight reorganization of the residues of these CDRs loops upon binding20,22,27. In our case, because both HEL42 and D3-L11 (this study) are rigid, the interaction could be explained with classical mechanism of lock-and-key43. The results of thermodynamic analysis including dominant enthalpic component throughout the association process and reduced enthalpy upon hot-spot mutagenesis are consistent with this model. Despite the little structural change of the interface residues, binding is strongly opposed by the change of entropy suggesting an intricate role for the solvent.
In conclusion, the paratope of the VHH antibody D3-L11 recognized its epitope by a lock-and-key mechanism, and is organized similarly to that of a conventional antibody (Fvs). Although this mechanism is not common among VHHs, the high affinity of D3-L11 indicates that mimicking Fvs may also work for a VHH to achieve its high affinity and specificity, the two hallmarks of antibodies. Because the long CDR3 covers the surface that emerges in the absence of the VL domain of IgGs, diversity in this loop is fully employed to generate a preorganized interface on the convex surface of the VHH. This organization is suitable to recognize concave surface of antigens, complementing the more common mechanism of protrusion of CDR observed in other VHHs. Diversity in binding modes enable VHHs to accommodate to various antigen molecules and thus brings VHHs advantages over other cleft binders, achieving high affinity and specificity.
Materials and Methods
Cloning, expression and purification of D3-L11
A synthetic gene of D3-L11 optimized for Escherichia coli (Genscript) was cloned in pRA244 between NcoI and SacII restriction sites. The construct also contained a pelB signal peptide at the N-terminus, and a His6-tag at the C-terminus. For expression, E. coli strain BL21(DE3) carrying the expression vector of D3-L11 were grown in 1 L of LB medium containing 50 μg/mL ampicillin at 28 °C and 120 rpm. Expression was induced by addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside when the optical density at 600 nm reached 0.5 after which the temperature was reduced to 20 °C overnight. The cells were harvested by centrifugation (7,000 × g for 15 min) at 4 °C. The cell pellet was resuspended in buffer A (20 mM TRIS-HCl, 500 mM NaCl, pH 8.0) supplemented with 5 mM imidazole, after which it was lysed with an ultrasonic disruptor (UD-201, TOMY) for 15 min. The cell lysate was centrifuged (40,000 × g for 30 min) at 4 °C. The supernatant was filtered through a membrane of a nominal pore size of 0.45 μm and subsequently loaded onto a 1 mL of Ni-NTA agarose column (Qiagen) equilibrated with buffer A. After a washing step with Buffer A containing 100 mM imidazole, VHHs were eluted from the column with buffer A supplemented with 500 mM imidazole. The eluate was dialyzed against buffer A, and subjected to size-exclusion chromatography (SEC) using a HiLoad 26/600 superdex 75 pg column (GE Healthcare) equilibrated with a buffer containing 20 mM TRIS-HCl, 150 mM NaCl, and 1 mM EDTA at pH 7.4.
For crystallization of the unbound form of D3-L11, the gene encoding the antibody was cloned into a Champion pET-SUMO vector bearing a His6-SUMO-tag. The protein was expressed as above. After the affinity chromatography step, the His6-SUMO-tag was cleaved-off with Ulp1 protease overnight at 4 °C in 20 mM TRIS-HCl, 150 mM NaCl at pH 8.0. The protein was separated from the protease, from the cleaved tag, and from the uncleaved protein by immobilized metal-affinity chromatography. The flow-thorough was concentrated and subjected to SEC using a HiLoad 16/600 superdex 75 pg column as described above.
Preparation of the antigen
HEL was purchased from Wako Pure Chemical (Cat. No. 126-02671, Japan) and solubilized in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) at the desired concentration and used without further purification.
Circular dichroism
The secondary structure of D3-L11 was examined in a CD J-820 spectrometer (Jasco, Japan) with a 1-mm quartz cuvette. Measurements were performed in a buffer containing 20 mM TRIS-HCl, 150 mM NaCl, 1 mM EDTA, pH 7.4 at a protein concentration of 10 μM. The spectrum of each sample was recorded five times at a speed of 50 nm/min and at 25 °C.
Differential scanning calorimetry
Thermal stability of D3-L11 and mutants (20 μM) was monitored with a VP-Capillary DSC instrument (MicroCal) in PBS. Samples were scanned at a speed of 1 °C/min from 10 to 100 °C. Data analysis was performed with Origin 7.0 using a non-two-state denaturation model.
Surface plasmon resonance
The binding of D3-L11 to HEL was determined by SPR using a Biacore T200 instrument (GE Healthcare). HEL (6.5 μg/mL) in 10 mM sodium acetate at pH 5.5 was immobilized onto a Series S CM5 Sensor Chip at 200 RU as previously described45,46. Measurements were performed at 25 °C in PBS buffer supplemented with the detergent Tween 20 (0.005%). Contact and dissociation times were 300 and 600 sec, respectively. The range of concentrations of D3-L11 was adjusted depending on the mutation studied and indicated where appropriate. Regeneration was conducted with 1.0 M Arg-HCl at pH 4.4 for 60 sec. To obtain the thermodynamic parameters the experiments were conducted at several temperatures. Data analysis was conducted with the instrument’s BiaEvaluation Software. Global fitting analysis and Scatchard plots were performed with a 1:1 binding model. The thermodynamic parameters were determined with the BiaEvaluation Software employing non-linear regressions (van’t Hoff plot) or linear regressions (Eyring plots). The van’t Hoff analysis of WT was not conducted with the data obtained from the Scatchard plot because the association step did not reach a plateau under the experimental conditions tested. In the experiments with the mutants at 30 °C, non-specific binding significantly affected the sensorgrams, and thus these data were excluded from the analysis.
Isothermal titration calorimetry
ITC measurements were carried out in an iTC200 instrument (Microcal) at 25 °C and a stirring rate of 1000 rpm in PBS buffer. The WT antibody or its mutants (10 μM) were titrated with HEL (100 μM) by injecting one-time 0.5 μL followed by 18 injections of 2.0 μL each with a fixed time between injections of 120 sec. The binding isotherm was fitted by non-linear regression with the program Origin 7.0 to a 1:1 binding model to obtain the thermodynamic parameters of the interaction.
Crystallization
For the crystallization of the antibody·antigen complexes, WT or Y102A antibody was mixed with ca. 5-fold excess of HEL in PBS, followed by dialysis against a buffer containing 20 mM TRIS-HCl, 100 mM NaCl, pH 8.0 overnight. The antibody-antigen complex was separated from excess HEL by SEC using a HiLoad 16/600 superdex 75 pg column equilibrated in the same buffer. The protein sample was concentrated to 13 mg/mL (WT-HEL complex) or 8.3 mg/mL (Y102A-HEL complex) in an Amicon Ultra-4 unit (molecular weight cutoff 10 kDa). Preliminary crystallization experiments were explored by mixing an equal volume of precipitant solution and protein solution in an Oryx8 instrument (Douglas Instruments) using commercial sparse matrix screens (Hampton Research) at 20 °C. Each complex (WT·HEL or Y102A·HEL) was crystallized in a solution containing 100 mM sodium nitrate and 16% PEG-3350 (pH 8.0). The complex between HEL and the mutant Y102A was independently crystalized in a buffer containing 100 mM lithium chloride and 18% PEG-3350. Crystals of WT D3-L11 (10 mg/mL) in the unbound form were obtained in a solution containing 100 mM Tris-HCl and 2.15 M ammonium sulfate at pH 7.0.
Data collection and refinement
Data were collected in beamlines BL5A and AR-NW12 at the Photon Factory (Tsukuba, Japan) under cryogenic conditions (100 K). The diffraction images were processed with the program MOSFLM and subsequently merged and scaled with the program SCALA of the CCP4 suite47. The structures were determined using the method of molecular replacement with the program PHASER48. The coordinates were refined with the program REFMAC549, and manually improved with COOT28. Validation was carried out with PROCHECK50. No residues were found in the disallowed regions of the Ramachandran plot. Residues were numbered sequentially. Kabat sequence numbering is shown in Supplementary Information Table S5. Data collection and refinement statistics are given in Supplementary Information Table S1.
R2 dispersion (NMR)
Three-dimensional spectra of HNCO, HN(CA)CO, HNCA, HN(CO)CA, HNCACB, and HN(CO)CACB were measured on an AVANCE DRX600 spectrometer (Bruker BioSpin) for sequential assignments of the backbone 1H, 13C, and 15N chemical shifts51 of free D3-L11 using the protein dissolved at 0.5 mM in NMR buffer (95% H2O/5% D2O, 20 mM sodium phosphate [pH 7.4], 143 mM NaCl). NMR data were processed and analyzed as previously described52. 15N effective R2 relaxation rates were measured at 37 °C on AVANCE DRX600 and AVANCE DMX750 spectrometers (Bruker BioSpin) using the 1H continuous-wave Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence53. Effective R2 rates were calculated as described previously54. Relaxation dispersion data whose R2 values changed by <1 s−1 over the entire range of τcp were excluded. By using the program GLOVE55, the relaxation dispersion curves were fitted to a two-state exchange model.
Accession numbers
The coordinates and structure factors of unbound D3-L11 (entry code 6JB9), WT D3-L11 in complex with HEL (entry code 6JB8), and Y102A D3-L11 in complex with HEL (entry codes 6JB2 and 6JB5) have been deposited in the Protein Data Bank.
Supplementary information
Acknowledgements
This work was supported by JSPS KAKENHI Grant Numbers 15K06962, JP25249115, JP13J09564, JP16H02420, JP17H05893, JP18K14892.
Author contributions
All authors conceived the experiments; H.A., H.T., M.K., S.Y. and J.M.M.C. conducted the experiments, all authors analyzed the results; H.A. and J.M.M.C. wrote the manuscript; all authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hiroki Akiba and Hiroko Tamura.
Contributor Information
Jose M. M. Caaveiro, Email: jose@phar.kyushu-u.ac.jp
Kouhei Tsumoto, Email: tsumoto@bioeng.t.u-tokyo.ac.jp.
Supplementary information
is available for this paper at 10.1038/s41598-019-50722-y.
References
- 1.Kaplon Hélène, Reichert Janice M. Antibodies to watch in 2019. mAbs. 2018;11(2):219–238. doi: 10.1080/19420862.2018.1556465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 3.Konning D, et al. Camelid and shark single domain antibodies: structural features and therapeutic potential. Curr Opin Struct Biol. 2017;45:10–16. doi: 10.1016/j.sbi.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 4.Hassanzadeh-Ghassabeh G, Devoogdt N, De Pauw P, Vincke C, Muyldermans S. Nanobodies and their potential applications. Nanomedicine-Uk. 2013;8:1013–1026. doi: 10.2217/Nnm.13.86. [DOI] [PubMed] [Google Scholar]
- 5.Arbabi-Ghahroudi M. Camelid single-domain antibodies: Historical perspective and future outlook. Front Immunol. 2017;8:1589. doi: 10.3389/fimmu.2017.01589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goode J, Dillon G, Millner PA. The development and optimisation of nanobody based electrochemical immunosensors for IgG. Sensor Actuat B-Chem. 2016;234:478–484. doi: 10.1016/j.snb.2016.04.132. [DOI] [Google Scholar]
- 7.Bever CS, et al. VHH antibodies: emerging reagents for the analysis of environmental chemicals. Anal Bioanal Chem. 2016;408:5985–6002. doi: 10.1007/s00216-016-9585-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klooster R, et al. Improved anti-IgG and HSA affinity ligands: clinical application of VHH antibody technology. J Immunol Methods. 2007;324:1–12. doi: 10.1016/j.jim.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Adams H, et al. Specific immuno capturing of the staphylococcal superantigen toxic-shock syndrome toxin-1 in plasma. Biotechnol Bioeng. 2009;104:143–151. doi: 10.1002/bit.22365. [DOI] [PubMed] [Google Scholar]
- 10.Godar M, et al. Dual anti-idiotypic purification of a novel, native-format biparatopic anti-MET antibody with improved in vitro and in vivo efficacy. Sci Rep. 2016;6:31621. doi: 10.1038/srep31621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helma J, Cardoso MC, Muyldermans S, Leonhardt H. Nanobodies and recombinant binders in cell biology. J Cell Biol. 2015;209:633–644. doi: 10.1083/jcb.201409074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beghein E, Gettemans J. Nanobody technology: A versatile toolkit for microscopic imaging, protein-protein interaction analysis, and protein function exploration. Front Immunol. 2017;8:771. doi: 10.3389/fimmu.2017.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koide S. Engineering of recombinant crystallization chaperones. Curr Opin Struct Biol. 2009;19:449–457. doi: 10.1016/j.sbi.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manglik A, Kobilka BK, Steyaert J. Nanobodies to study G protein-coupled receptor structure and function. Annu Rev Pharmacol Toxicol. 2017;57:19–37. doi: 10.1146/annurev-pharmtox-010716-104710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muyldermans S, Smider VV. Distinct antibody species: structural differences creating therapeutic opportunities. Curr Opin Immunol. 2016;40:7–13. doi: 10.1016/j.coi.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henry KA, MacKenzie CR. Antigen recognition by single-domain antibodies: structural latitudes and constraints. MAbs. 2018;10:815–826. doi: 10.1080/19420862.2018.1489633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zavrtanik U, Lukan J, Loris R, Lah J, Hadzi S. Structural Basis of Epitope Recognition by Heavy-Chain Camelid Antibodies. J Mol Biol. 2018;430:4369–4386. doi: 10.1016/j.jmb.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 18.De Genst E, et al. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci USA. 2006;103:4586–4591. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veesler D, et al. Crystal structure and function of a DARPin neutralizing inhibitor of lactococcal phage TP901-1: comparison of DARPin and camelid VHH binding mode. J Biol Chem. 2009;284:30718–30726. doi: 10.1074/jbc.M109.037812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitz KR, Bagchi A, Roovers RC, van Bergen en Henegouwen PM, Ferguson KM. Structural evaluation of EGFR inhibition mechanisms for nanobodies/VHH domains. Structure. 2013;21:1214–1224. doi: 10.1016/j.str.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kromann-Hansen T, et al. A camelid-derived antibody fragment targeting the active site of a serine protease balances between inhibitor and substrate behavior. J Biol Chem. 2016;291:15156–15168. doi: 10.1074/jbc.M116.732503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossey I, et al. Potent single-domain antibodies that arrest respiratory syncytial virus fusion protein in its prefusion state. Nat Commun. 2017;8:14158. doi: 10.1038/ncomms14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruse AC, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gulati S, et al. Targeting G protein-coupled receptor signaling at the G protein level with a selective nanobody inhibitor. Nat Commun. 2018;9:1996. doi: 10.1038/s41467-018-04432-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desmyter A, Decanniere K, Muyldermans S, Wyns L. Antigen specificity and high affinity binding provided by one single loop of a camel single-domain antibody. J Biol Chem. 2001;276:26285–26290. doi: 10.1074/jbc.M102107200. [DOI] [PubMed] [Google Scholar]
- 27.Garza JA, Taylor AB, Sherwood LJ, Hart PJ, Hayhurst A. Unveiling a Drift Resistant Cryptotope within Marburgvirus Nucleoprotein Recognized by Llama Single-Domain Antibodies. Front Immunol. 2017;8:1234. doi: 10.3389/fimmu.2017.01234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 30.Akiba H, Tsumoto K. Thermodynamics of antibody-antigen interaction revealed by mutation analysis of antibody variable regions. J Biochem. 2015;158:1–13. doi: 10.1093/jb/mvv049. [DOI] [PubMed] [Google Scholar]
- 31.Fanning SW, Walter R, Horn JR. Structural basis of an engineered dual-specific antibody: conformational diversity leads to a hypervariable loop metal-binding site. Protein Eng Des Sel. 2014;27:391–397. doi: 10.1093/protein/gzu033. [DOI] [PubMed] [Google Scholar]
- 32.Kubala MH, Kovtun O, Alexandrov K, Collins BM. Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. 2010;19:2389–2401. doi: 10.1002/pro.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell LS, Colwell LJ. Comparative analysis of nanobody sequence and structure data. Proteins. 2018;86:697–706. doi: 10.1002/prot.25497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol. 2002;12:14–20. doi: 10.1016/S0959-440X(02)00283-X. [DOI] [PubMed] [Google Scholar]
- 35.Moreira IS, Fernandes PA, Ramos MJ. Hot spots–a review of the protein-protein interface determinant amino-acid residues. Proteins. 2007;68:803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 36.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 37.Pons J, Rajpal A, Kirsch JF. Energetic analysis of an antigen/antibody interface: alanine scanning mutagenesis and double mutant cycles on the HyHEL-10/lysozyme interaction. Protein Sci. 1999;8:958–968. doi: 10.1110/ps.8.5.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Urrutia M, Smith-Gill SJ, Mariuzza RA. Dissection of binding interactions in the complex between the anti-lysozyme antibody HyHEL-63 and its antigen. Biochemistry. 2003;42:11–22. doi: 10.1021/bi020589+. [DOI] [PubMed] [Google Scholar]
- 39.Dall’Acqua W, et al. A mutational analysis of binding interactions in an antigen-antibody protein-protein complex. Biochemistry. 1998;37:7981–7991. doi: 10.1021/bi980148j. [DOI] [PubMed] [Google Scholar]
- 40.Dong J, et al. A single-domain llama antibody potently inhibits the enzymatic activity of botulinum neurotoxin by binding to the non-catalytic alpha-exosite binding region. J Mol Biol. 2010;397:1106–1118. doi: 10.1016/j.jmb.2010.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sircar A, Sanni KA, Shi J, Gray JJ. Analysis and modeling of the variable region of camelid single-domain antibodies. J Immunol. 2011;186:6357–6367. doi: 10.4049/jimmunol.1100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buck M, et al. Structural determinants of protein dynamics: analysis of 15N NMR relaxation measurements for main-chain and side-chain nuclei of hen egg white lysozyme. Biochemistry. 1995;34:4041–4055. doi: 10.1021/bi00012a023. [DOI] [PubMed] [Google Scholar]
- 43.Csermely P, Palotai R, Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem Sci. 2010;35:539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makabe K, et al. Tumor-directed lymphocyte-activating cytokines: refolding-based preparation of recombinant human interleukin-12 and an antibody variable domain-fused protein by additive-introduced stepwise dialysis. Biochem Biophys Res Commun. 2005;328:98–105. doi: 10.1016/j.bbrc.2004.12.141. [DOI] [PubMed] [Google Scholar]
- 45.Kiyoshi M, et al. Affinity improvement of a therapeutic antibody by structure-based computational design: generation of electrostatic interactions in the transition state stabilizes the antibody-antigen complex. PLoS One. 2014;9:e87099. doi: 10.1371/journal.pone.0087099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakamoto S, et al. Contributions of interfacial residues of human Interleukin15 to the specificity and affinity for its private alpha-receptor. J Mol Biol. 2009;389:880–894. doi: 10.1016/j.jmb.2009.04.050. [DOI] [PubMed] [Google Scholar]
- 47.Evans P. Scaling and assessment of data quality. Acta Crystallogr D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 48.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 50.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK - a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 51.Ikura M, Kay LE, Bax A. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry. 1990;29:4659–4667. doi: 10.1021/bi00471a022. [DOI] [PubMed] [Google Scholar]
- 52.Yanaka S, Moriwaki Y, Tsumoto K, Sugase K. Elucidation of potential sites for antibody engineering by fluctuation editing. Sci Rep. 2017;7:9597. doi: 10.1038/s41598-017-10246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loria JP, Rance M, Palmer AG. A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J Am Chem Soc. 1999;121:2331–2332. doi: 10.1021/ja983961a. [DOI] [Google Scholar]
- 54.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 55.Sugase K, Konuma T, Lansing JC, Wright PE. Fast and accurate fitting of relaxation dispersion data using the flexible software package GLOVE. J Biomol NMR. 2013;56:275–283. doi: 10.1007/s10858-013-9747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.