Abstract
The nucleus pulposus (NP) cells adapt to their physiologically hyperosmotic microenvironment through Tonicity-responsive enhancer binding protein (TonEBP/nuclear factor of activated T-cell5 [NFAT5])-mediated osmoregulation. Primary cilia in different organs serve diverse roles including osmosensing, but its contribution to NP cell osmoadaptive response is unknown. A high percentage of cultured primary NP cells possessed primary cilia that changed length in response to osmotic stimuli. Stable silencing of Intraflagellar Transport 88 (Ift88) or Kinesin Family Member 3 A (Kif3a) to inhibit the formation of primary cilia did not affect hyperosmotic upregulation of TonEBP. While ShKif3a blocked hyperosmotic increase of TonEBP-Transactivation Domain (TAD) activity, overall the knockdown of either gene did not alter the hyperosmotic status of proximal promoter activities and transcription of key TonEBP targets. On the other hand, a small decrease in TonEBP level under hypoosmotic condition was attenuated by Ift88 or Kif3a knockdown. Noteworthy, none of the TonEBP target genes were responsive to hypoosmotic stimulus in control and Ift88 or Kif3a knockdown cells, suggesting the primary role of TonEBP in the hyperosmotic adaptation of NP cells. Similarly, in Kif3a null mouse embryonic fibroblasts (MEFs), the overall TonEBP-dependent hyperosmotic responses were preserved. Unlike NP cells, TonEBP targets were responsive to hypoosmolarity in wild-type MEFs, and these responses remained intact in Kif3a null MEFs. Together, these results suggest that primary cilia are dispensable for TonEBP-dependent osmoadaptive response.
Subject terms: Cell signalling, Organelles
Introduction
The nucleus pulposus (NP) is a gelatinous center-most structure of the intervertebral disc. It is surrounded by concentric layers of annulus fibrosus and sandwiched between cartilaginous endplates. The extracellular matrix of NP consists of collagens and abundant proteoglycans that are bound by sulfated glycosaminoglycan chains1. The high negative charge density of glycosaminoglycan molecules draws water and cations, in particular, Na+, into the NP tissue. This contributes to the elevated osmotic swelling pressure within the tissue compartment and provides the tissue with its ability to resist daily spinal loadings1,2. The NP matrix is, therefore, hyperosmotic with estimated tissue osmolarity raging from 430 to 496 mOsm/kg H2O3,4. With aging and degeneration of the tissue, the aberrant changes in cell phenotype lead to altered extracellular matrix composition, decreased osmotic pressurization of the NP, and compromised the mechanical function of the tissue1,5,6. Therefore, adaptation to their osmodynamic niche for survival and function is crucial for the resident NP cells.
Tonicity-responsive enhancer binding protein (TonEBP/nuclear factor of activated T-cell5 [NFAT5]) is a Rel homology transcription factor that has been well characterized for its osmolarity-dependent function in mammalian cells including NP cells7–14. In response to a hyperosmotic stimulus, TonEBP transcriptionally controls the expression of several osmoprotective genes including taurine transporter (TauT), betaine-GABA transporter (BGT-1), sodium/myo-inositol co-transporter (SMIT), and aldose reductase (AR), all of which are essential regulators of intracellular levels of non-ionic osmolytes15–18. Additionally, TonEBP transcriptionally increases the expression of heat shock protein-70 (HSP-70) to maintain proper protein folding, cellular trafficking, and degradation of misfolded proteins under hyperosmotic conditions19,20. It was recently shown that TonEBP is highly expressed in NP as well as notochord cells of developing mouse embryo, supporting its importance in NP development and maintenance21.
Primary cilia found in most types of mammalian cells have diverse roles, including modulation of key signaling pathways22–28. They can also function as a sensory organelle that relays extracellular stimuli, such as changes in osmolarity, to intracellular signaling pathways29–38. In Caenorhabditis elegans, OSM-9, a homolog of mammalian transient receptor potential channel vanilloid subfamily (TRPV), and OCR-2, OSM-9/capsaicin receptor related TRPV channel, that are localized in primary cilia are important for sensing extracellular hyperosmotic stimulus35. Similarly, primary cilia of mammalian cholangiocytes and articular chondrocytes are shown to link extracellular hypoosmotic stimulus to intracellular calcium signaling pathway36,37. In a recent report, Siroky et al. showed that renal epithelial cells without primary cilia have weakened induction of TonEBP target genes AR/Akr1b3 as well as Bgt1 under hyperosmotic conditions38. Although the role of TonEBP in modulating osmoresponse in NP cells has been well studied, it is unknown whether primary cilia contribute to this process. The objective of this study was to investigate if primary cilia function as osmosensory organelles in NP cells. Specifically, we examined if primary cilia control TonEBP-mediated osmoadaptive response through loss-of-function studies measuring the expression of TonEBP and its target genes after inhibition of primary cilia formation. Furthermore, we confirmed our findings in NP cells using Kif3a null mouse embryonic fibroblasts (MEFs) that are completely devoid of primary cilia.
Results
The length of primary cilia in NP cells is responsive to changes in extracellular osmolarity
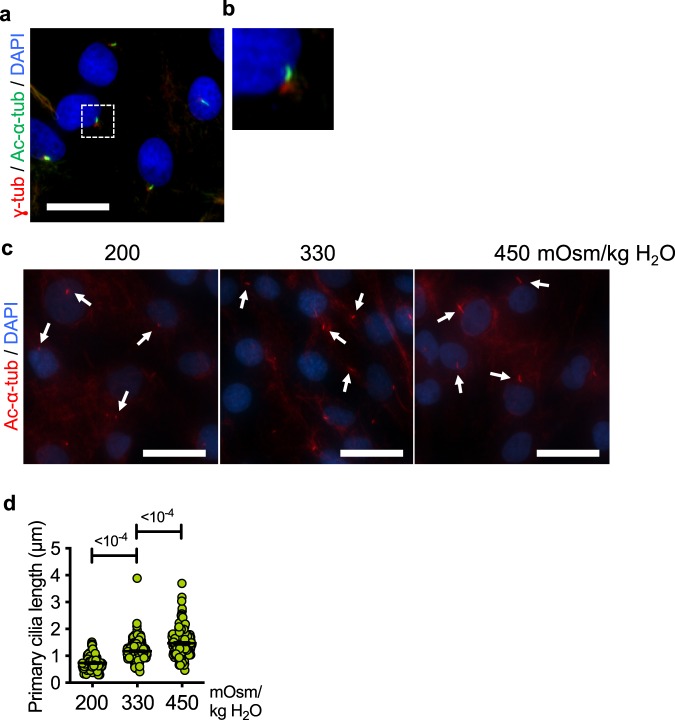
Primary cilia were visualized in cultured primary rat NP cells by co-immunostaining acetylated α-tubulin and γ-tubulin, labeling ciliary axoneme and basal bodies, respectively (Fig. 1a,b). Previous studies showed that the length of primary cilia in different types of cells changed in response to extracellular stimuli39–41. To examine if primary cilia in NP cells respond to extracellular osmotic stimulus, we cultured NP cells under different osmotic conditions and measured the length of the cilia. The average length of primary cilia was significantly shorter under hypoosmotic condition (200 mOsm/kg H2O) compared to isoosmotic (330 mOsm/kg H2O) condition (Fig. 1c,d; p < 0.0001 for both 200 mOsm/kg H2O and 450 mOsm/kg H2O groups). On the other hand, the length of primary cilia increased under hyperosmotic condition (450 mOsm/kg H2O), suggesting that the primary cilia in NP cells are sensitive to changes in extracellular osmolarity.
Figure 1.
NP cell primary cilia modulate their lengths in response to changes in extracellular osmolarity. (a) Immunofluorescence staining of acetylated α-tubulin (green) and γ-tubulin (red) to mark primary cilia axoneme and basal bodies, respectively, in primary rat NP cells. Scale bar = 30 μm. (b) Zoomed-in image of a primary cilium from the area demarcated by the white square in panel (a). (c,d) Primary cilia of rat NP cells cultured under different osmotic conditions for 24 h were visualized by immunofluorescence staining of acetylated α-tubulin. (c) The lengths of primary cilia increase in response to increased osmolarity (450 mOsm/kg H2O) compared to isoosmotic control (330 mOsm/kg H2O) conditions, whereas they appear shorter under hypoosmotic conditions (200 mOsm/kg H2O). White arrows mark primary cilia. Scale bar = 50 μm. (d) Quantification of primary cilium length was done using ImageJ software. (n = 3 experiments; at least 150 cells/group) Data are represented as scatter plots (mean ± SEM). One-way ANOVA with Dunnett’s multiple comparison test was used to determine statistical significance.
Inhibition of primary cilia formation in NP cells does not affect TonEBP expression in hyperosmotic condition
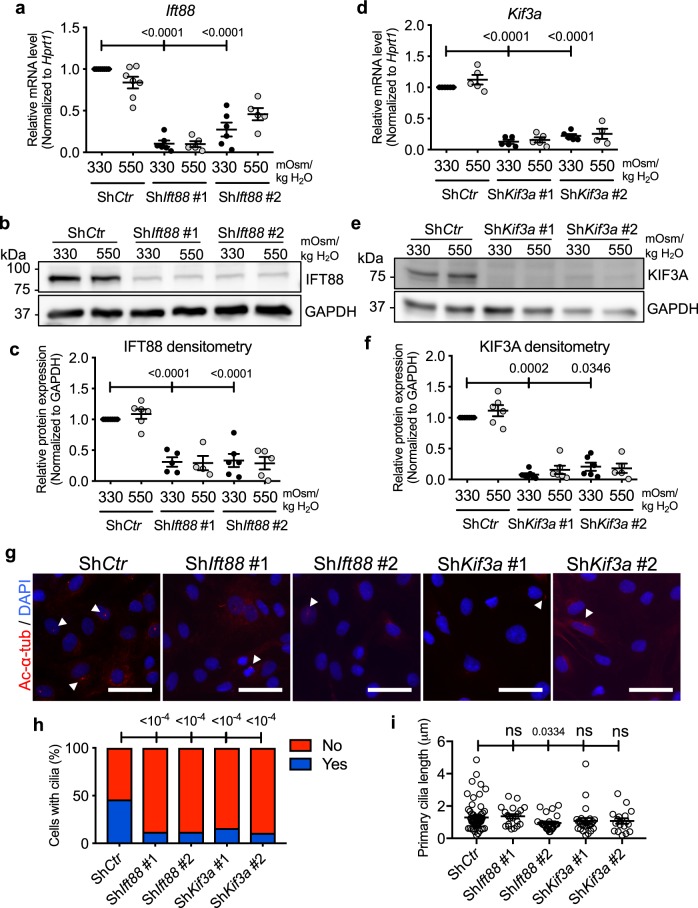
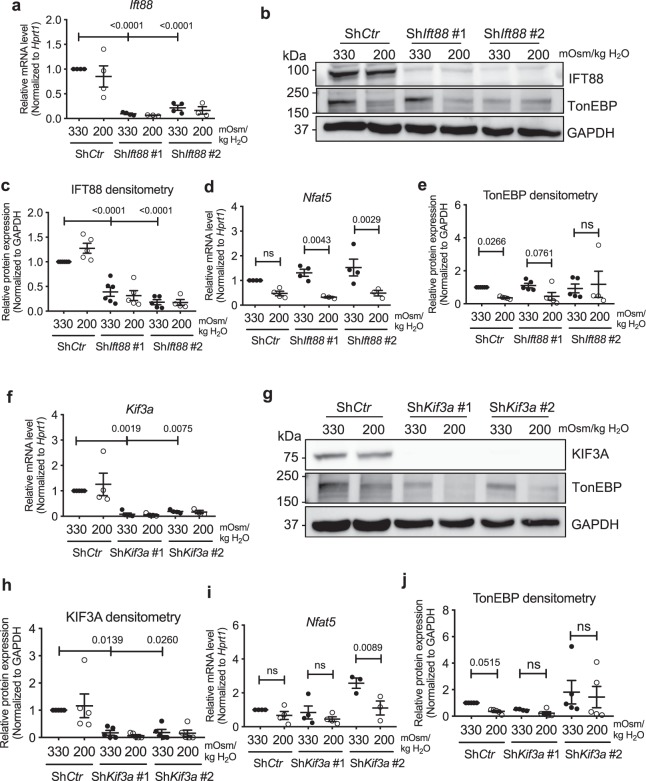
The function of primary cilia as an osmosensor has been observed in C. elegans as well as in some types of mammalian cells, including renal tubular epithelial cells, articular chondrocytes, and cholangiocytes35–38. NP cells reside in an osmotically active microenvironment due to high proteoglycan content of the NP matrix and dynamic loading of the spine. We examined if primary cilia of the NP cells play a role in sensing extracellular osmolarity and mediating cellular osmotic response. We inhibited formation of primary cilia in NP cells by performing stable knockdown of Ift88, a component of complex B of the intraflagellar transport (IFT) particles42–45 (Fig. 2a–c; Supp. Fig. S1–1), or Kif3a, a motor subunit of kinesin 246 (Fig. 2d–f), both of which are necessary for ciliogenesis. Lentiviral transduction of NP cells with shRNA against Ift88 or Kif3a resulted in a significant decrease in the transcript and protein levels of IFT88 (Fig. 2a–c; p < 0.0001 for all statistical comparisons) or KIF3A (Fig. 2d–f; p < 0.0001 for ShKif3a #1 and #2 isoosmotic groups in Fig. 2d; p = 0.0002 for ShKif3a #1 isoosmotic group, p = 0.346 for ShKif3a #2 isoosmotic group in Fig. 2f; Supp. Fig. S1–1), respectively. Stable silencing of either gene resulted in a decreased number of cells with primary cilia (Fig. 2g). Quantification of the number of cells with primary cilia confirmed this result (Fig. 2h; p < 0.0001 for all groups). Overall, the lengths of primary cilia that remained after the stable silencing of Ift88 or Kif3a were not significantly different from that of the control cells (Fig. 2i; p = 0.0334 for ShIft88 #2, all other groups were statistically not significant).
Figure 2.
Stable knockdown of Ift88 or Kif3a inhibits formation of NP cell primary cilia. (a) Ift88 mRNA levels in NP cells transduced with control (ShCtr) or two different ShIft88 clones were measured by qRT-PCR to confirm the knockdown (n ≥ 5). (b) Western blot image showing significant reduction of IFT88 protein levels after the knockdown of Ift88. (c) Densitometry analyses of Western blots confirm significant knockdown of IFT88 (n ≥ 5). (d–f) qRT-PCR, Western blot, and corresponding densitometry analyses, show significant downregulation of KIF3A after stable knockdown using two different ShKif3a clones (n ≥ 4). (g) Acetylated α-tubulin immunofluorescence staining after lentiviral transduction of ShIft88 or ShKif3a shows inhibition of primary cilia formation in majority of rat NP cells. Scale bar = 75 μm. White arrowheads point to primary cilia. (h,i) Quantitation of percentage of NP cells with primary cilia and primary cilium length after stable silencing of Ift88 or Kif3a (n = 3; at least 150 cells/group). Data are represented as scatter plots (mean ± SEM). ns = not significant. One-way ANOVA or Kruskal-Wallis test with Sidak’s, Holm-Sidak’s, or Dunn’s multiple comparison test was used based on the distribution of the data to determine statistical significance. For statistical comparison of the percentages of NP cells with primary cilia, Fisher’s exact test was used. Western blot images were cropped and acquired under same experimental conditions. See Supplementary Fig. S1–1 for un-cropped Western blot images.
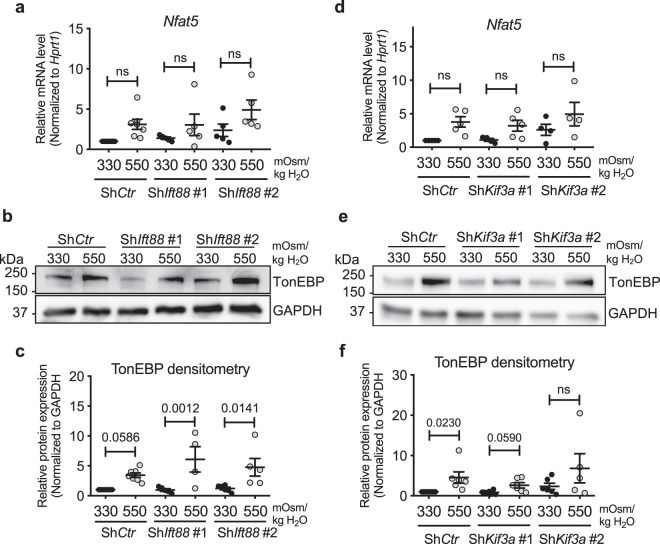
To determine if inhibition of primary cilia formation resulted in dysregulation of cellular osmoresponse, we stably silenced Ift88 or Kif3a and measured the expression of TonEBP/NFAT5, a transcription factor crucial for osmoregulation in NP cells8–11. While there was a trend of increase in TonEBP/Nfat5 transcript levels under hyperosmotic conditions (550 mOsm/kg H2O), this increase did not reach a statistical significance in both control and Ift88 knockdown cells (Fig. 3a; all analyses were statistically not significant). Hyperosmotic increase in TonEBP protein levels was unaffected by Ift88 knockdown (Fig. 3b,c; p = 0.0586 for ShCtr 550 mOsm/kg H2O, p = 0.0012 for ShIft88 #1 550 mOsm/kg H2O, p = 0.0141 for ShIft88 #2 550 mOsm/kg H2O; Supp. Fig. S1–1). Similarly, when Kif3a was stably silenced, the trend of increase in TonEBP/Nfat5 transcript levels and the upregulation of TonEBP protein under hyperosmotic condition were maintained (Fig. 3d–f; Statistical non-significance for all analyses in Fig. 3d; p = 0.023 for ShCtr 550 mOsm/kg H2O, p = 0.059 for ShKif3a #1 550 mOsm/kg H2O; Supp. Fig. S1–1). Taken together, our data suggest that inhibition of primary cilia formation in NP cells does not affect their TonEBP expression.
Figure 3.
Knockdown of Ift88 or Kif3a in NP cells does not affect hyperosmotic upregulation of TonEBP. (a) TonEBP/Nfat5 mRNA levels in NP cells with Ift88 knockdown (n ≥ 5). (b) Western blot image showing increased TonEBP expression in response to hyperosmolarity (550 mOsm/kg H2O) independently of Ift88 knockdown. (c) Densitometry analyses of TonEBP with Ift88 knockdown (n ≥ 4). (d) TonEBP/Nfat5 mRNA levels in NP cells with Kif3a knockdown (n ≥ 3). (e) Western blot image showing that hyperosmotic induction of TonEBP is maintained after Kif3a knockdown. (f) Densitometry analyses of TonEBP after Kif3a knockdown (n ≥ 4). Data are represented as scatter plots (mean ± SEM). ns = not significant. One-way ANOVA or Kruskal-Wallis test with Sidak’s or Dunn’s multiple comparison test was used based on the distribution of the data to determine statistical significance. Western blot images were cropped and acquired under same experimental conditions. See Supplementary Fig. S1-1 for un-cropped Western blot images.
Primary cilia do not modulate TonEBP activity and TonEBP target gene expression in NP cells under hyperosmotic conditions
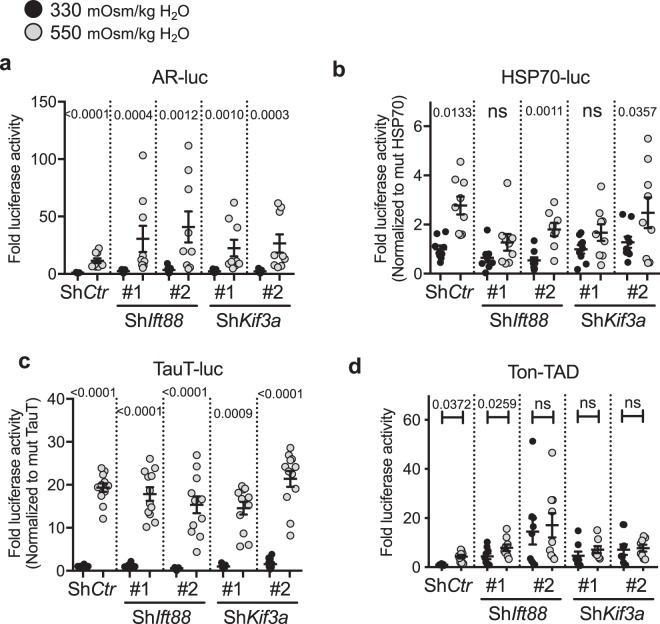
Hyperosmotic stimulus leads to the increased transcriptional activity of TonEBP in the nucleus9,11,47. We investigated if primary cilia controlled TonEBP activity without affecting its expression. Changes in proximal promoter activities of well-characterized TonEBP target genes including, AR (aldose reductase), HSP-70 and TauT (taurine transporter) were measured in NP cells transfected with shRNA against Ift88 or Kif3a (Fig. 4a–c). These promoters contain highly conserved TonEBP-binding sites that are active in NP cells10,15,19,20,47,48. Hyperosmotic (550 mOsm/kg H2O) increase in the AR (Fig. 4a; p < 0.0001) and TauT (Fig. 4c; p < 0.0001) promoter activities was not affected by either ShIft88 (p = 0.0004 for clone #1, p = 0.0012 for clone #2 for AR-luc; p < 0.0001 for both clones for TauT-luc) or ShKif3a (p = 0.001 for clone #1, p = 0.0003 for clone #2 for AR-luc; p = 0.0009 for clone #1 and p < 0.0001 for clone #2 for TauT-luc). Similarly, the HSP-70 promoter activity increased under hyperosmotic conditions with or without ShIft88 or ShKif3a. While one of the ShIft88 and ShKif3a clone groups did not reach a statistical significance, the trend of hyperosmotic induction of HSP-70 promoter activity was maintained (Fig. 4b; p = 0.0011 for ShIft88 #2, p = 0.0357 for ShKif3a #2). In addition, we measured the activity of the TonEBP transactivation domain (TAD) in response to the hyperosmotic stimulus (550 mOsm/kg H2O) with or without ShIft88 and ShKif3a. While ShIft88 #2 inhibited the hyperosmotic increase of TonEBP-TAD activity, the other clone had no effect (Fig. 4d; p = 0.0259 for ShIft88 #1). Both clones of ShKif3a prevented a further increase of TonEBP-TAD activity in response to the hyperosmotic stimulus. However, the average level of TonEBP-TAD activity under hyperosmotic conditions was similar or higher in knockdown cells when compared to ShCtr group, suggesting that the overall TonEBP-TAD activity was unaffected by ShIft88 or ShKif3a (Fig. 4d).
Figure 4.
ShIft88 or ShKif3a does not affect the promoter activities of TonEBP target genes under hyperosmotic conditions. (a–c) Proximal promoter activities of AR (a), HSP-70 (b), and TauT (c) in response to hyperosmotic stimuli (550 mOsm/kg H2O) with or without ShIft88 or ShKif3a. The promoter activities of TonEBP target genes are mostly unaffected by ShIft88 or ShKif3a. (d) ShKif3a as well as one of the ShIft88 clones prevent further upregulation of TonEBP-TAD (Ton-TAD) activity in response to hyperosmolarity. Interestingly, the levels of TonEBP-TAD activity under isoosmotic conditions are increased with ShIft88 or ShKif3a. Data are represented as scatter plots (mean ± SEM) (n ≥ 3 with 3 technical replicates per biological replicate). ns = not significant. One-way ANOVA or Kruskal-Wallis test with Sidak’s or Dunn’s multiple comparison test was used based on the distribution of the data to determine statistical significance.
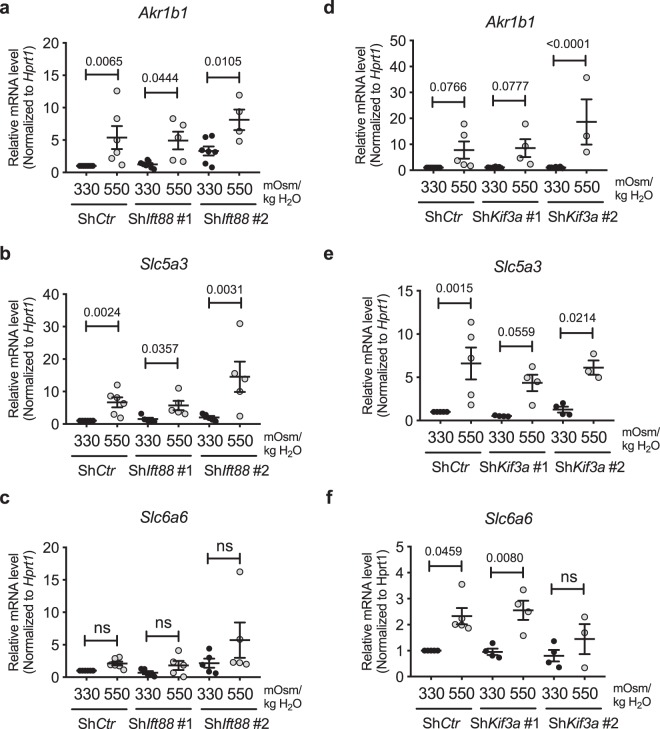
To confirm that TonEBP-mediated hyperosmotic response was independent of primary cilia, we measured mRNA levels of TonEBP targets in NP cells with Ift88 or Kif3a knockdown. AR/Akr1b1, TauT/Slc6a6 as well as sodium/myo-inositol co-transporter (SMIT/Slc5a3) are osmolarity-dependent target genes of TonEBP15–18. Ift88 knockdown did not affect the hyperosmotic (550 mOsm/kg H2O) increase in the expression of AR/Akr1b1 and SMIT/Slc5a3 genes (Fig. 5a,b; p = 0.0065 for ShCtr, p = 0.0444 for ShIft88 #1, p = 0.0105 for ShIft88 #2 in Fig. 5a; p = 0.0024 for ShCtr, p = 0.0357 for ShIft88 #1, p = 0.0031 for ShIft88 #2 in Fig. 5b). While the increase in TauT/Slc6a6 levels in response to hyperosmotic stimulus did not reach a statistical significance, the trend of increase was unaffected by Ift88 knockdown (Fig. 5c). Likewise, stable silencing of Kif3a did not affect the induction of SMIT/Slc5a3 and AR/Akr1b1 in response to hyperosmolarity (550 mOsm/kg H2O) (Fig. 5d, e; p = 0.0766 for ShCtr, p = 0.0777 for ShKif3a #1, p < 0.0001 for ShKif3a #2 in Fig. 5d; p = 0.0015 for ShCtr, p = 0.0559 for ShKif3a #1, p = 0.0214 for ShKif3a #2 in Fig. 5e). In addition, the hyperosmotic increase of TauT/Slc6a6 expression was preserved with ShKif3a, except one of the clones (Fig. 5f; p = 0.0459 for ShCtr, p = 0.008 for ShKif3a #1). Taken together, the gene expression data showed that inhibition of primary cilia formation in NP cells did not affect the overall TonEBP transcriptional activity or the target gene expression under hyperosmotic conditions.
Figure 5.
Hyperosmotic upregulation of TonEBP target gene expression is not affected by inhibition of primary cilia formation in NP cells. (a–c) mRNA levels of TonEBP targets, AR/Akr1b1, SMIT/Slc5a3 and TauT/Slc6a6, showing that stable silencing of Ift88 does not affect their response to hyperosmotic stimuli (550 mOsm/kg H2O) (n ≥ 5). (d–f) mRNA levels of AR/Akr1b1, SMIT/Slc5a3 and TauT/Slc6a6 demonstrate that Kif3a knockdown also does not influence hyperosmotic upregulation of these genes (n ≥ 3). Data are represented as scatter plots (mean ± SEM). ns = not significant. One-way ANOVA or Kruskal-Wallis test with Sidak’s or Dunn’s multiple comparison test was used based on the distribution of the data to determine statistical significance.
The absence of primary cilia has a minor effect on hypoosmotic downregulation of TonEBP and its targets in NP cells
The tissue osmotic conditions of the NP compartment undergo diurnal changes due to dynamic loading and unloading of the spine. During disc degeneration, the breakdown of the proteoglycan matrix by proteases decreases the tissue osmolarity49–51. Consequently, NP cells need to maintain their function and integrity under both hyper- and hypo-osmotic conditions52,53. We, therefore, assessed the effects of inhibiting primary cilia formation in NP cells on TonEBP expression under hypoosmotic conditions (200 mOsm/kg H2O). NP cells with Ift88 (Fig. 6a–c; p < 0.0001 for all analyses in Fig. 6a,c; Supp. Figs S1 and 2) or Kif3a (Fig. 6f–h; p = 0.0019 for ShKif3a #1, p = 0.0075 for ShKif3a #2 in Fig. 6f; p = 0.0139 for ShKif3a #1, p = 0.026 for ShKif3a #2 in Fig. 6h) knockdown showed significant hypoosmotic decrease in TonEBP/Nfat5 transcript, compared to control cells that showed similar decreasing trend (Fig. 6d,i; p = 0.0043 for ShIft88 #1, p = 0.0029 for ShIft88 #2 in Fig. 6d; p = 0.0089 for ShKif3a #2 in Fig. 6i). On the other hand, a small but significant decrease in TonEBP protein levels seen in control cells under the hypoosmotic condition was attenuated following knockdown of Ift88 (Fig. 6b,e; p = 0.0266 for ShCtr, p = 0.0761 for ShIft88 #1) or Kif3a (Fig. 6g,j; p = 0.0515 for ShCtr; Supp. Figs S1 and 2). Although the changes in mRNA and protein levels in response to hypoosmotic stimuli were somewhat opposing in cells lacking primary cilia, these results suggested that primary cilia may play a minor role in maintaining TonEBP levels in NP cells under hypoosmotic conditions.
Figure 6.
Inhibition of primary cilia formation in NP cells have minimal effect on hypoosmotic downregulation of TonEBP levels. (a–c) qRT-PCR, Western blot and densitometry analyses demonstrating IFT88 levels under hypoosmotic conditions (200 mOsm/kg H2O) after stable knockdown of Ift88. (d) TonEBP/Nfat5 expression in NP cells with Ift88 knockdown under hypoosmotic conditions (n ≥ 3). (d) Hypoosmotic decrease in TonEBP protein levels is attenuated by Ift88 knockdown (n ≥ 4). (f,g) KIF3A mRNA and protein levels in NP cells with Kif3a knockdown. (h) TonEBP/Nfat5 mRNA expression in NP cells with Kif3a knockdown under hypoosmotic conditions (n ≥ 3). (i) Hypoosmotic downregulation of TonEBP is blunted after Kif3a knockdown (n ≥ 4). Data are represented as scatter plots (mean ± SEM). ns = not significant. One-way ANOVA or Kruskal-Wallis test with Sidak’s or Dunn’s multiple comparison test was used based on the distribution of the data to determine statistical significance. See Supplementary Figs S1 and 2 for un-cropped Western blot images.
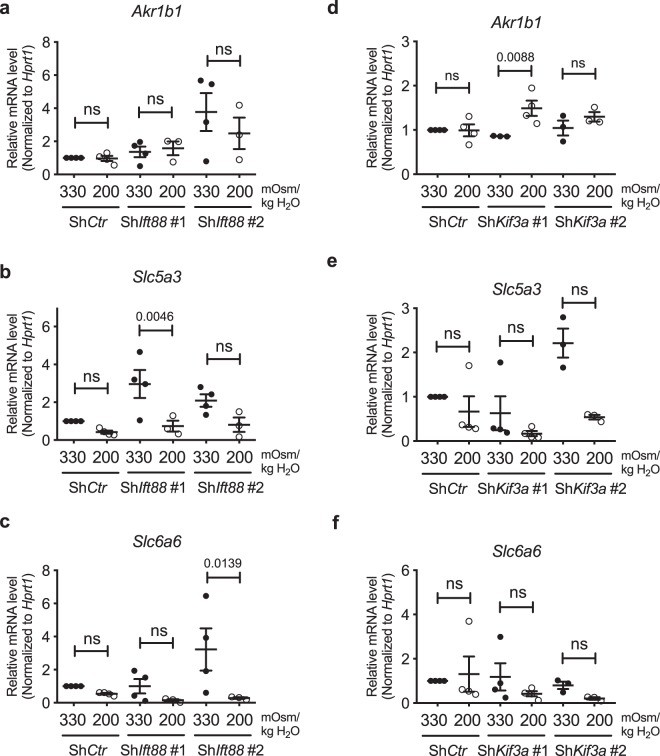
We then measured mRNA levels of TonEBP targets to investigate if the small changes in TonEBP levels in cells without primary cilia had functional implications in overall hypoosmotic cellular response. AR/Ark1b1, SMIT/Slc5a3, and TauT/Slc6a6 were not affected by stable knockdown of Ift88 or Kif3a (Fig. 7a–f; all statistical analyses were not significant in Fig. 7a,e,f; p = 0.0046 for ShIft88 #1 in Fig. 7b; non-significant for ShCtr and ShIft88 #1, p = 0.0139 for ShIft88 #2 in Fig. 7c; non-significant for ShCtr and ShKif3a #2, p = 0.0088 for ShKif3a #1 in Fig. 7d). Interestingly, none of these osmotic targets showed a significant decrease in their expression under hypoosmotic conditions, and this trend was not affected by either Ift88 or Kif3a knockdown (Fig. 7a–f). Taken together, our results showed that primary cilia may have a small role in modulating hypoosmotic levels of TonEBP, but this did not lead to any appreciable effect on target gene expression.
Figure 7.
TonEBP target gene expression under hypoosmotic conditions is unaffected by inhibition of primary cilia formation in NP cells. (a–f) mRNA levels of AR/Akr1b1, SMIT/Slc5a3 and TauT/Slc6a6 under hypoosmotic conditions (200 mOsm/kg H2O) following knockdown of Ift88 (a–c) or Kif3a (d–f). Stable silencing of either Ift88 or Kif3a does not affect the trend of downregulation in SMIT/Slc5a3 and TauT/Slc6a6 under hypoosmotic conditions (n ≥ 3). Data are represented as scatter plots (mean ± SEM). ns = not significant. One-way ANOVA or Kruskal-Wallis test with Sidak’s or Dunn’s multiple comparison test was used based on the distribution of the data to determine statistical significance.
TonEBP expression and activity are not mediated by primary cilia in mouse embryonic fibroblasts
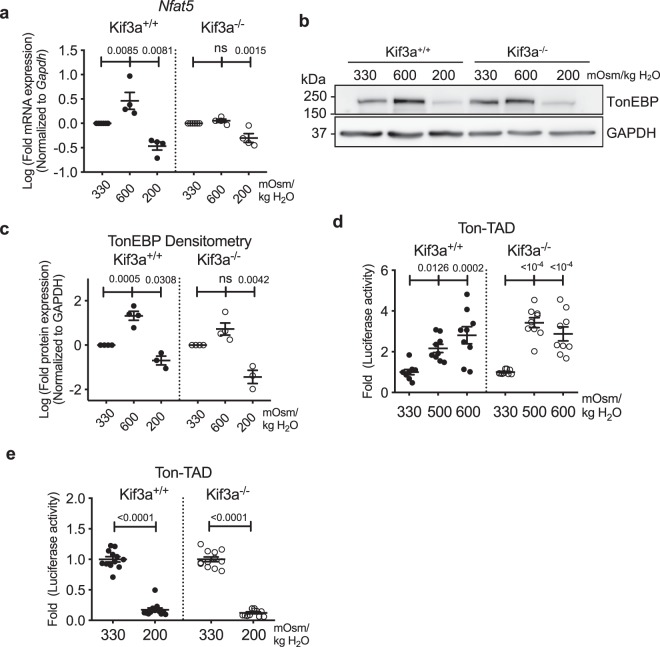
Similar to NP cells, mouse embryonic fibroblasts (MEFs) induce TonEBP-dependent osmotic response under hyperosmotic conditions8. Parallel experiments were performed using Kif3a null MEFs that completely lack primary cilia to further confirm that primary cilia do not control TonEBP-dependent osmotic response54. As reported in other studies, TonEBP mRNA (Fig. 8a; p = 0.0085) and protein (Fig. 8b,c; p = 0.0005) levels were significantly upregulated in wild-type MEFs under hyperosmotic conditions (600 mOsm/kg H2O). On the other hand, the increase of TonEBP expression in Kif3a null MEFs under hyperosmotic conditions did not reach a statistical significance (Fig. 8a–c). In response to hypoosmotic conditions (200 mOsm/kg H2O), both wild-type and Kif3a null MEFs downregulated TonEBP transcript and protein levels (Fig. 8a–c; p = 0.0081 for wild-type MEFs, p = 0.0015 for Kif3a null MEFs in Fig. 8a; p = 0.0308 for wild-type MEFs, p = 0.0042 for Kif3a null MEFs in Fig. 8c; Supp. Figs S1 and 3).
Figure 8.
The absence of primary cilia in Kif3a null MEFs has a small effect on hyperosmotic induction of TonEBP without influencing its TAD activity (a) TonEBP/Nfat5 gene expression in wild-type and Kif3a null (Kif3a−/−) MEFs under hyper- (600 mOsm/kg H2O) and hypoosmotic (200 mOsm/kg H2O) conditions. (b,c) Western blot image and corresponding densitometry analyses showing TonEBP levels under different osmotic conditions in wild-type and Kif3a null MEFs. Kif3a null MEFs show slightly attenuated hyperosmotic increase but unaffected hypoosmotic decrease in TonEBP expression. (d,e) TonEBP-TAD (Ton-TAD) activity in wild-type and Kif3a null MEFs under hyperosmotic (d) and hypoosmotic (e) conditions (n = 4). Similar to wild-type cells, Kif3a null MEFs show increase and decrease in TonEBP-TAD activity under hyperosmotic and hypoosmotic conditions, respectively (n ≥ 3 with 3 technical replicates per biological replicate). Data are represented as scatter plots (mean ± SEM). ns = not significant. One-way ANOVA or Kruskal-Wallis test with Sidak’s or Dunn’s multiple comparison test was used based on the distribution of the data to determine statistical significance. Western blot images were cropped and acquired under same experimental conditions. See Supplementary Figs S1 and 3 for un-cropped Western blot images.
To investigate if cilia controlled osmolarity-dependent TonEBP activity in MEFs, we measured the activation status of TonEBP-TAD in wild-type and Kif3a null MEFs under different osmotic conditions. Both wild-type and Kif3a null MEFs increased and decreased TonEBP-TAD activities under hyperosmotic conditions (600 mOsm/kg H2O) (Fig. 8d; p = 0.0126 for 500 mOsm/kg H2O, p = 0.0002 for 600 mOsm/kg H2O in wild-type MEFs; p < 0.0001 for both hyperosmotic conditions in Kif3a null MEFs), and hypoosmotic conditions (200 mOsm/kg H2O) (Fig. 8e; p < 0.0001 for both wild-type and Kif3a null MEFs), respectively. These results suggested that the effect of osmolarity on TonEBP-TAD activity remained unaffected in the absence of primary cilia.
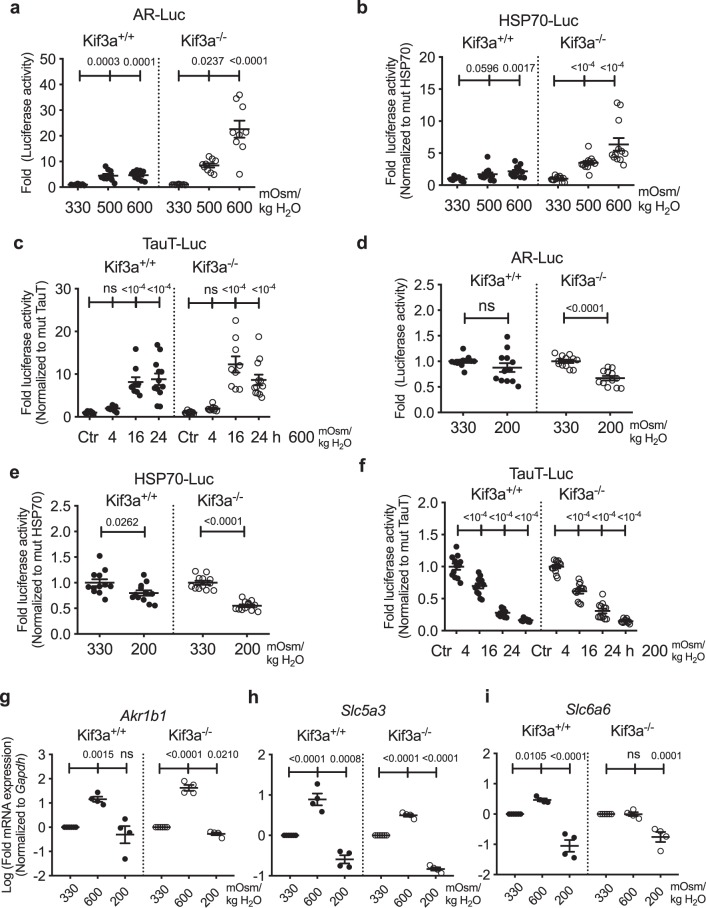
We then measured the activities of TonEBP responsive reporters and the level of TonEBP target genes in wild-type and Kif3a null MEFs to determine if the presence of primary cilia affected TonEBP transcriptional activity. Hyperosmotic stimulus (600 mOsm/kg H2O) significantly increased AR and HSP-70 promoter activities in both wild-type and Kif3a null MEFs (Fig. 9a,b; p = 0.0003 for 500 mOsm/kg H2O, p = 0.0001 for 600 mOsm/kg H2O for wild-type MEFs, p = 0.0237 for 500 mOsm/kg H2O, p < 0.0001 for 600 mOsm/kg H2O for Kif3a null MEFs in Fig. 9a; p = 0.0596 for 500 mOsm/kg H2O, p = 0.0017 for 600 mOsm/kg H2O for wild-type MEFs, p < 0.0001 for hyperosmolarity in Kif3a null MEFs in Fig. 9b). Likewise, the temporal response of TauT promoter to the hyperosmotic stimulus was similar in both wild-type and Kif3a null MEFs, where a robust activation at 16 h post-treatment was observed (Fig. 9c; p < 0.0001 for 16 h and 24 h in both wild-type and Kif3a null MEFs). Interestingly, the AR promoter activity was insensitive to the hypoosmotic stimulus (200 mOsm/kg H2O) in wild-type cells, while it was significantly downregulated in Kif3a null MEFs (Fig. 9d; p < 0.0001). Both wild-type and Kif3a null MEFs showed a hypoosmotic reduction in HSP70 promoter activity (Fig. 9e; p = 0.0262 for wild-type, p < 0.0001 for Kif3a null MEFs). A gradual and time-dependent decrease in TauT promoter activity was also observed under hypoosmotic conditions (Fig. 9f; p < 0.0001).
Figure 9.
The absence of primary cilia does not affect TonEBP activity and target gene expression in MEFs (a–c) Proximal promoter activities of AR, HSP-70 and TauT in wild-type and Kif3a null MEFs show that hyperosmotic induction (600 mOsm/kg H2O) of these promoter activities are intact in null MEFs. (d–f) AR promoter activity is unaffected in wild-type MEFs but decreased in Kif3a null MEFs under hypoosmotic conditions (200 mOsm/kg H2O). Hypoosmotic decrease in the promoter activities of HSP-70 and TauT are maintained in Kif3a null MEFs. (n ≥ 3 with 3 technical replicates per biological replicate). (g,h) The changes in expression of AR/Akr1b1 (g) and SMIT/Slc5a3 (h) under different osmotic conditions are similar between wild-type and Kif3a null MEFs (n = 4). (i) Hyperosmotic induction of TauT/Slc6a6 gene expression observed in wild-type MEFs is absent in Kif3a null MEFs. TauT/Slc6a6 gene expression was downregulated in both wild-type and Kif3a null MEFs under hypoosmotic conditions (n = 4). Data are represented as scatter plots (mean ± SEM). ns = not significant. One-way ANOVA or Kruskal-Wallis test with Sidak’s or Dunn’s multiple comparison test was used based on the distribution of the data to determine statistical significance.
A qRT-PCR analysis was performed to measure the expression of select TonEBP target genes in MEFs under different osmotic conditions. AR/Akr1b1 was significantly upregulated in both wild type and Kif3a null MEFs under hyperosmotic conditions (600 mOsm/kg H2O) (Fig. 9g; p = 0.0015 for wild-type, p < 0.0001 for null MEFs). In agreement with its promoter activity, AR/Akr1b1 expression was not changed in wild-type MEFs but was significantly decreased in Kif3a null MEFs in response to the hypoosmotic stimulus (200 mOsm/kg H2O) (Fig. 9g; p = 0.021). SMIT/Slc5a3 expression in both wild-type and Kif3a null MEFs was significantly increased and decreased under hyperosmotic and hypoosmotic conditions, respectively (Fig. 9h; p < 0.0001 for hyperosmotic condition in wild-type and Kif3a null MEFs, p = 0.0008 for hypoosmotic condition in wild-type MEFs, p < 0.0001 for hypoosmotic condition in Kif3a null MEFs). Interestingly, unlike in wild-type MEFs, TauT/Slc6a6 mRNA levels were not induced by hyperosmolarity in Kif3a null MEFs (Fig. 9i; p = 0.0105 in wild-type MEFs). The hypoosmotic decrease in TauT/Slc6a6 levels was maintained in both wild-type and Kif3a null MEFs (Fig. 9i; p < 0.0001 in wild-type, p = 0.0001 in null MEFs). Taken together, the osmotic regulation of TonEBP activity in MEFs remained largely unchanged in the absence of primary cilia.
Discussion
Primary cilia serve diverse physiological functions that are well-conserved from C. elegans to vertebrates. Their sensory function in various cell types and organ systems is essential in multiple biological processes such as development and maintenance of tissue homeostasis. There are only a few studies examining the presence of primary cilia in the disc tissues55. However, few studies have investigated the role of primary cilia in the intervertebral disc56. A few studies have suggested that primary cilia can function as osmosensors in mammalian cells36–38. In rat cholangiocytes or porcine articular chondrocytes, removal of primary cilia inhibits intracellular Ca2+ increase in response to hypoosmotic stimuli36,37. In addition, renal epithelial cells depleted of primary cilia cannot induce TonEBP target genes, AR/Akr1b3, and Bgt1, under hyperosmotic conditions38. These cell types are frequently exposed to dynamic changes in extracellular osmolarity, similar to those experienced by the NP cells. Importantly, since NP tissue is physiologically hyperosmotic, osmoadaptation is a key survival mechanism for the resident cells. We, therefore, investigated whether primary cilia in NP cells have an osmosensory function and if the activity of TonEBP, a key osmoregulatory transcription factor, is dependent on primary cilia. We show, for the first time, that NP cell primary cilia do not have the osmoregulatory function, although they alter their lengths in response to changes in extracellular osmolarity.
In many cellular pathways that are mediated by primary cilia, the localization of signaling molecules to primary cilia is important for activating downstream signaling pathways22,23,57. Therefore, the changes in the length of primary cilia are thought to be reflective of their functional modification. Indeed, the length of osteocyte primary cilia has been shown to correlate with their sensitivity to the mechanical stimuli58. In addition, several known human ciliopathies present with primary cilia of abnormal lengths59. In mouse femoral chondrocytes, primary cilia shorten in response to changes in extracellular osmolarity40. Likewise, in kidney epithelial cells as well as bone mesenchymal cells, fluid sheer-mediated bending of primary cilia results in decreased intracellular cAMP, which in turn causes shortening of primary cilia41. It is, therefore, possible that the changes in the length of primary cilia in NP cells under different osmotic conditions indicate altered intracellular signaling pathways. However, the pathway mediating the length of cilia in response to osmotic stimuli may be completely independent of the TonEBP-mediated osmotic response.
Our Ift88 and Kif3a knockdown studies showed that the expression of TonEBP and its target genes was unaffected by the absence of primary cilia in NP cells. Under hyperosmotic conditions, TonEBP inhibits excessive sodium influx by increasing the levels of intracellular non-ionic osmolytes including taurine, sodium/myo-inositol, and betaine by inducing transcription of TauT/Slc6a6, Bgt1, SMIT/Slc5a3, and AR/Akr1b115–18. Therefore, disruption of TonEBP activity under hyperosmotic condition can compromise cell survival. The fact that both Ift88 and Kif3a knockdown did not affect the hyperosmotic induction of TonEBP and its target genes suggests that this was the result of the absence of primary cilia rather than a specific effect of the knockdown of either gene. Interestingly, despite the decrease in TonEBP protein levels, the expression of its transcriptional targets, TauT/Slc6a6, SMIT/Slc5a3, and AR/Akr1b1 was not significantly affected under hypoosmotic conditions. This may be due to the fact that the osmoregulatory role of TonEBP is more important under hyperosmotic rather than hypoosmotic condition. Nonetheless, neither Ift88 nor Kif3a silencing significantly affected how these genes were regulated in response to hypoosmotic stimuli, strongly suggesting the lack of primary cilia’s involvement in this process. In addition, the proximal promoter activities of select TonEBP targets, as well as TonEBP-TAD activity, were mostly unaffected by ShIft88 or ShKif3a regardless of the osmotic status, suggesting that primary cilia play a minimal role, if any, in osmotic response of NP cells.
Our parallel studies using MEFs further confirmed that primary cilia do not control TonEBP-dependent osmotic response. The use of Kif3a null MEFs was advantageous mainly because they completely lack primary cilia, and because MEFs have TonEBP-dependent osmoregulatory pathways similar to that of NP cells8,54. Interestingly, unlike NP cells, Kif3a null MEFs were unable to upregulate TauT mRNA in response to the hyperosmotic stimulus. This was intriguing since TauT promoter activity was not affected by the absence of primary cilia in Kif3a null MEFs. This is perhaps due to the high sensitivity of luciferase reporter assay and the fact that it measures the activities of shorter promoter fragments. We cannot exclude the possibility of involvement of either a regulatory sequence outside the analyzed promoter fragment or a primary cilia-associated post-transcriptional regulation. Nonetheless, other osmoresponsive TonEBP targets, SMIT/Slc5a3 and AR/Akr1b1, were upregulated under the hyperosmotic condition in Kif3a null MEFs, indicating that the overall cellular osmotic response to hyperosmotic stimuli is intact. Furthermore, we did not observe any obvious increase in Kif3a null MEF cell death in response to hyper- or hypo-osmotic stimuli, indicating that these cells were able to maintain their cellular homeostasis without primary cilia. The results of our Kif3a null MEFs are particularly interesting because the importance of primary cilia in modulating cell signaling during embryonic development is well-known.
Our data clearly demonstrate that TonEBP-mediated osmoregulation is not controlled by primary cilia in both NP cells and MEFs. The previously reported osmoregulatory and osmosensing function of primary cilia in cholangiocytes36 and kidney epithelial cells38 are therefore likely cell-type specific. Although the in vitro culture system is a simplification of the complex in vivo extracellular environment, the osmoregulatory role of primary cilia in other cell types was shown to be preserved in in vitro settings, suggesting that primary cilia are indispensable in these cell types. On the other hand, the results of our studies indicate that NP cells are capable of initiating TonEBP-mediated cellular osmoresponse without primary cilia. A recent study showed that mouse NP tissue cultured in an ex vivo setting decreased their TRPV4 expression in response to cyclic hyperosmolarity60. Future studies examining the localization and function of ion channels such as TRPV4, TRM3, or PC-1 and PC-2 that have been associated with cilia osmosensing would be helpful in better understanding how NP cells sense osmotic stimuli.
Materials and Methods
Reagents and plasmids
Lentiviral ShIft88 #1 (TRCN0000178064), ShIft88 #2 (TRCN0000182620), ShKif3a #1 (TRCN0000339512), ShKif3a #2 (TRCN0000339514), and control shRNA plasmids were purchased from Sigma. psPAX2 (12260) and pMD2.G (12259) developed by Dr. Didier Trono were obtained from Addgene. Luciferase reporter plasmids were kindly provided by Dr. Takashi Ito, Osaka University (wild type and TonE-mutant TauT-Luc)15, Dr. H Moo Kwon, University of Maryland (wild type and TonE-mutant HSP70-Luc)20, and Dr. Joan D. Ferraris, NIH (GAL4dbd-548-1531, GAL4dbd, and AR-Luc)47,48. Backbone GAL4dbd contains no TAD but only expresses the GAL4dbd. pFR-Luc reporter (Stratagene) contains the yeast GAL4-binding site, upstream of a minimal promoter driving the firefly luciferase gene.
Cell culture and treatments
All procedures regarding collection of animal tissues was performed as per approved protocols by Institutional Animal Care and Use Committee (IACUC) of the Thomas Jefferson University, in accordance with the IACUC’s relevant guidelines and regulations. Rat NP cells were isolated using a method described by Risbud et al.61. After isolation, cells were maintained in Dulbecco’s Modified Eagles Medium (DMEM) (Corning, 10-013-CV) with 10% fetal bovine serum (FBS) (Sigma-Aldrich, F6178) supplemented with antibiotics in flask in normoxia (20.9% pO2). NP cells isolated from two rats were pulled into one T25 flask (P0) and cultured till confluency before passaging (P1). From this point, cells were either plated directly into experiment-specific plates (P2) or passaged. Approximately total of 16 rats were used for the study. Cells from different isolations were used for replication experiments. Cells up to P4 were used for the experiments. Wild type and Kif3a−/− MEFs54 developed by Dr. Pao-Tien Chuang were kindly provided by Dr. Natalia Riobo, Thomas Jefferson University. For hyper- or hypoosmotic treatment, cells were cultured in DMEM with 10% FBS, containing either no additional osmolytes (330 mOsm/kg H2O), additional NaCl (60-135 mM to final osmolarity ranging 450–600 mOsm/kg H2O) or additional dH2O (to final osmolarity of 200 mOsm/kg H2O) for 4–24 h. For the hyperosmolarity experiments using NP cells, osmolarity ranging 450 to 550 mOsm/kg H2O have been used. While the estimated osmolarity of NP tissue in vivo ranges from 430 to 496 mOsm/kg H2O3,4, the above osmolarity range was chosen based on the previous reports on hyperosmotic TonEBP activation in NP cells8,11,62. Similarly, 600 mOsm/kg H2O was used for MEFs experiments based on the previous report on TonEBP/NFAT5 activity in MEFs, where the peak TonEBP-responsive promoter activity was observed at ~610 mOsm/kg H2O63.
Real-time RT-PCR analysis
Total RNA was extracted from NP cells using RNAeasy mini columns (Qiagen). The purified, DNA-free RNA was converted to cDNA using RNA to cDNA EcoDryTM Premix (Clontech). Template cDNA and gene specific primers were added to SYBR green master mix (Applied Biosystems) and mRNA expression was quantified using the Step One Plus Real-Time PCR System (Applied Biosystems). Hprt1 was used to normalize gene expression. All the primers used were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA).
Transfections and dual luciferase assay
Cells were plated on 48-well plates (2 × 104 cells/well) one day before transfection. Cells were transfected with 250 ng of wild type TauT, mutant TauT, wild type HSP70, mutant HSP70, or AR reporter plasmid and 250 ng of pRL-TK plasmid. For measuring TonEBP-TAD activity, cells were transfected with 200 ng of pFR-Luc, 200 ng of GAL4dbd-548-1531 (Ton-TAD) and 100 ng of pRL-TK plasmids. For each transfection, plasmids were premixed with the transfection reagent Lipofectamine 2000 (Invitrogen). The treatments were done so that the cells were lysed 48 h after the transfection. Dual-LuciferaseTM reporter assay system (Promega) was used for sequential measurements of firefly and Renilla luciferase activities using TECAN Infinite200 Pro microplate reader (TECAN). At least three independent transfections were performed and all analyses were carried out in triplicate.
Immunofluorescence microscopy
NP cells were plated on poly-L-lysine-coated glass coverslips. After treatments, cells were fixed and permeabilized with 4% paraformaldehyde at room temperature for 15 minutes, washed with PBS and then blocked with 5% normal goat serum in PBS with 0.3% Triton X-100 (Sigma Aldrich, T8787) for 1 h at room temperature. Cells on coverslip were then incubated with anti-acetyl-α-tubulin (Lys40) (D20G3) XP® antibody (Cell Signaling Technology, #5335, 1:700), or anti-acetylated-α-tubulin (Sigma, T6793, 1:300) and anti-γ-tubulin antibody (Abcam, ab11317, 1:1000) for co-staining in blocking buffer at 4 °C overnight, washed with PBS, and then incubated with Alexa Fluor®-594 conjugated anti-rabbit, or Alexa Fluor®-594 conjugated anti-mouse, or Alexa Fluor®-488 conjugated anti-rabbit secondary antibody (Jackson ImmunoResearch Lab, Inc.), at a dilution of 1:700 for 1 h at room temperature in dark. Then the coverslips were washed with PBS and mounted with ProLong® Gold Antifade Mountant with DAPI (Thermo Fisher Scientific, P36934). Mounted slides were visualized using a Zeiss AxioImager A2 (Carl Zeiss, Germany), or Zeiss LSM510 confocal microscope (Carl Zeiss, Germany). Three independent experiments were performed for quantitative analysis of cilia length and incidence using ImageJ software (http://rsb.info.nih.gov/ij/). Briefly, each cilium was traced using Segmented Lines of the Line Selection Tools, and then the length of the tracing was measured using Measure function. The lengths of at least 150 primary cilia per treatment group were measured.
Protein extraction and western blotting
Following treatment, cells were immediately placed on ice and washed with ice-cold PBS. All the wash buffers and the final cell lysis/re-suspension buffers included 1X completeTM Mini Protease Inhibitor Cocktail (Roche, 11836153001), NaF (5 mM) (Sigma Aldrich, 201154) and Na3VO4 (200 μM) (Sigma Aldrich, S6508). Total cell proteins were resolved by electrophoresis on 8–12% SDS-polyacrylamide gels and transferred by electroblotting to PVDF membranes (EMD Millipore, IPVH00010). Dual-color Lane marker (Bio-Rad, 161-0394) was used as a protein ladder to guide identification of the band sizes. The membranes were blocked with 5% non-fat dry milk in TBST (1% Tween 20 in TBS) and incubated overnight at 4 °C in 5% non-fat dry milk in TBST with the antibodies against IFT88 (Proteintech, #13967-1-AP, 1:500), KIF3A (D7G3) (Cell Signaling Technology, #8507, 1:1000), TonEBP/NFAT5 (Novus Biologicals, NB120-3446, 1:1000), or GAPDH (Novus Biologicals, NB300-221, 1:3000). Immunolabeling was detected using the AmershamTM ECLTM Prime Western Blotting Detection Reagent (Thermo Fisher Scientific, 45-002-401). All Western blot experiments were performed at least three independent times.
Lentiviral particle production and viral transduction
HEK 293 T cells (ATCC, CRL-3216) were plated in 10 cm plates (5 × 106 cells/plate) in DMEM with 10% heat-inactivated FBS one day before transfection. Cells were transfected with 9 μg of ShCtr, ShIft88, or ShKif3a plasmids along with 6 μg psPAX2 and 3 μg pMD2.G using Lipofectamine 2000 (Invitrogen). After 6 h, transfection medium was replaced with DMEM with 10% heat-inactivated FBS and penicillin-streptomycin. Lentiviral medium was harvested at 48 to 60 h post-transfection, and mixed with 7% PEG 6000 (Sigma Aldrich, 81253) solution and incubated overnight at 4 °C to precipitate virus particles. PEG solution was removed from virus medium before transduction by centrifugation at 1,500 × g for 30 min to pellet virus particles. NP cells were plated in DMEM with 10% heat-inactivated FBS one day before transduction. Cells were transduced with fresh DMEM with 10% heat-inactivated FBS containing viral particles along with 8 μg/ml polybrene (Sigma Aldrich, H9268). After 16 h, the medium was removed and replaced with DMEM with 10% FBS. Cells were harvested for protein extraction 4–5 days after transduction to ensure maximum knockdown efficiency without affecting cell viability. At least three independent experiments were performed.
Statistical analysis
All experiments were performed at least three independent times. Data are presented as the mean ± SE. Differences between multiple groups were assessed by one-way ANOVA or Kruskal-Wallis test depending on the distribution of the data with Sidak’s, Holm-Sidak’s, or Dunn’s multiple comparison test for post-hoc analyses using Prism7 (GraphPad Software). Some data were log-transformed before statistical analyses to account for non-Gaussian distribution. P < 0.05 was considered statistically significant.
Supplementary information
Acknowledgements
This study is supported by grants from the National Institutes of Health R01 AR055655, R01 AR064733 and R01AR074813.
Author contributions
H.C. conducted the experiments, analyzed data, and wrote the manuscript. V.M. conducted the experiments and analyzed data. M.V.R. and I.M.S. conceived the study, secured funding, and contributed to interpretation of the results. All authors reviewed the results, contributed to editing the manuscript, and approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-51939-7.
References
- 1.Silagi ES, Shapiro IM, Risbud MV. Glycosaminoglycan synthesis in the nucleus pulposus: Dysregulation and the pathogenesis of disc degeneration. Matrix Biol. 2018;71–72:368–379. doi: 10.1016/j.matbio.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson ZI, Shapiro IM, Risbud MV. Extracellular osmolarity regulates matrix homeostasis in the intervertebral disc and articular cartilage: Evolving role of TonEBP. Matrix Biol. 2014;40:10–16. doi: 10.1016/j.matbio.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishihara H, Warensjo K, Roberts S, Urban JP. Proteoglycan synthesis in the intervertebral disk nucleus: the role of extracellular osmolality. Am. J. Physiol. 1997;272:C1499–506. doi: 10.1152/ajpcell.1997.272.5.C1499. [DOI] [PubMed] [Google Scholar]
- 4.van Dijk B, Potier E, Ito K. Culturing bovine nucleus pulposus explants by balancing medium osmolarity. Tissue Eng. Part C. Methods. 2011;17:1089–96. doi: 10.1089/ten.tec.2011.0215. [DOI] [PubMed] [Google Scholar]
- 5.Choi H, et al. A novel mouse model of intervertebral disc degeneration shows altered cell fate and matrix homeostasis. Matrix Biol. 2018;70:102–122. doi: 10.1016/j.matbio.2018.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, et al. Early onset of disc degeneration in SM/J mice is associated with changes in ion transport systems and fibrotic events. Matrix Biol. 2018;70:123–139. doi: 10.1016/j.matbio.2018.03.024. [DOI] [PubMed] [Google Scholar]
- 7.Hiyama A, et al. Activation of TonEBP by calcium controls {beta}1,3-glucuronosyltransferase-I expression, a key regulator of glycosaminoglycan synthesis in cells of the intervertebral disc. J. Biol. Chem. 2009;284:9824–34. doi: 10.1074/jbc.M807081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gajghate S, et al. Osmolarity and intracellular calcium regulate aquaporin2 expression through TonEBP in nucleus pulposus cells of the intervertebral disc. J. Bone Miner. Res. 2009;24:992–1001. doi: 10.1359/jbmr.090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai T-T, et al. TonEBP/OREBP is a regulator of nucleus pulposus cell function and survival in the intervertebral disc. J. Biol. Chem. 2006;281:25416–24. doi: 10.1074/jbc.M601969200. [DOI] [PubMed] [Google Scholar]
- 10.Tsai T-T, et al. MEK/ERK signaling controls osmoregulation of nucleus pulposus cells of the intervertebral disc by transactivation of TonEBP/OREBP. J. Bone Miner. Res. 2007;22:965–74. doi: 10.1359/jbmr.070322. [DOI] [PubMed] [Google Scholar]
- 11.Johnson ZI, Shapiro IM, Risbud MV. RNA Sequencing Reveals a Role of TonEBP Transcription Factor in Regulation of Pro-inflammatory Genes in Response to Hyperosmolarity in Healthy Nucleus Pulposus Cells. J. Biol. Chem. 2016;291:26686–26697. doi: 10.1074/jbc.M116.757732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi H, et al. COX-2 expression mediated by calcium-TonEBP signaling axis under hyperosmotic conditions serves osmoprotective function in nucleus pulposus cells. J. Biol. Chem. 2018;293:8969–8981. doi: 10.1074/jbc.RA117.001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S, Kwon M, Woo S. TonEBP transcriptional activator in the cellular response to increased osmolality. Pflugers Arch. Eur. J. Physiol. 2002;444:579–585. doi: 10.1007/s00424-002-0849-2. [DOI] [PubMed] [Google Scholar]
- 14.Libert S, et al. Involvement of TonEBP/NFAT5 in osmoadaptative response of human retinal pigmented epithelial cells to hyperosmolar stress. Mol. Vis. 2016;22:100–15. [PMC free article] [PubMed] [Google Scholar]
- 15.Ito T, et al. Expression of taurine transporter is regulated through the TonE (tonicity-responsive element)/TonEBP (TonE-binding protein) pathway and contributes to cytoprotection in HepG2 cells. Biochem. J. 2004;382:177–82. doi: 10.1042/BJ20031838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyakawa H, et al. Cis- and trans-acting factors regulating transcription of the BGT1 gene in response to hypertonicity. Am. J. Physiol. 1998;274:F753–61. doi: 10.1152/ajprenal.1998.274.4.F753. [DOI] [PubMed] [Google Scholar]
- 17.Rim JS, et al. Transcription of the sodium/myo-inositol cotransporter gene is regulated by multiple tonicity-responsive enhancers spread over 50 kilobase pairs in the 5′-flanking region. J. Biol. Chem. 1998;273:20615–21. doi: 10.1074/jbc.273.32.20615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko BC, Ruepp B, Bohren KM, Gabbay KH, Chung SS. Identification and characterization of multiple osmotic response sequences in the human aldose reductase gene. J. Biol. Chem. 1997;272:16431–16437. doi: 10.1074/jbc.272.26.16431. [DOI] [PubMed] [Google Scholar]
- 19.Gogate SS, Fujita N, Skubutyte R, Shapiro IM, Risbud MV. Tonicity enhancer binding protein (TonEBP) and hypoxia-inducible factor (HIF) coordinate heat shock protein 70 (Hsp70) expression in hypoxic nucleus pulposus cells: Role of Hsp70 in HIF-1α degradation. J. Bone Miner. Res. 2012;27:1106–1117. doi: 10.1002/jbmr.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woo SK, Lee SD, Na KY, Park WK, Kwon HM. TonEBP/NFAT5 stimulates transcription of HSP70 in response to hypertonicity. Mol. Cell. Biol. 2002;22:5753–60. doi: 10.1128/MCB.22.16.5753-5760.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tessier, S., Madhu, V., Johnson, Z. I., Shapiro, I. M. & Risbud, M. V. NFAT5/TonEBP controls early acquisition of notochord phenotypic markers, collagen composition, and sonic hedgehog signaling during mouse intervertebral disc embryogenesis. Dev. Biol., 10.1016/j.ydbio.2019.07.004 (2019). [DOI] [PMC free article] [PubMed]
- 22.Rohatgi R, Milenkovic L, Scott MP. Patched1 Regulates Hedgehog Signaling at the Primary Cilium. Science (80-.). 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 23.Dorn KV, Hughes CE, Rohatgi R. A Smoothened-Evc2 complex transduces the Hedgehog signal at primary cilia. Dev. Cell. 2012;23:823–35. doi: 10.1016/j.devcel.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huangfu D, et al. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 25.Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl. Acad. Sci. USA. 2005;102:11325–30. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haycraft CJ, et al. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park TJ, Haigo SL, Wallingford JB. Ciliogenesis defects in embryos lacking inturned or fuzzy function are associated with failure of planar cell polarity and Hedgehog signaling. Nat. Genet. 2006;38:303–11. doi: 10.1038/ng1753. [DOI] [PubMed] [Google Scholar]
- 28.Simons M, et al. Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nat. Genet. 2005;37:537–43. doi: 10.1038/ng1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McEwen DP, et al. Hypomorphic CEP290/NPHP6 mutations result in anosmia caused by the selective loss of G proteins in cilia of olfactory sensory neurons. Proc. Natl. Acad. Sci. USA. 2007;104:15917–22. doi: 10.1073/pnas.0704140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bronshteín AA, Minor AV. Regeneration of olfactory flagella and restoration of the electroolfactogram following application of triton X-100 to the olfactory mucosa of frogs. Tsitologiia. 1977;19:33–9. [PubMed] [Google Scholar]
- 31.Menco BPM. The fine-structural distribution of G-protein receptor kinase 3, beta-arrestin-2, Ca2+/calmodulin-dependent protein kinase II and phosphodiesterase PDE1C2, and a Cl(−)-cotransporter in rodent olfactory epithelia. J. Neurocytol. 2005;34:11–36. doi: 10.1007/s11068-005-5045-9. [DOI] [PubMed] [Google Scholar]
- 32.Narita K, Kawate T, Kakinuma N, Takeda S. Multiple primary cilia modulate the fluid transcytosis in choroid plexus epithelium. Traffic. 2010;11:287–301. doi: 10.1111/j.1600-0854.2009.01016.x. [DOI] [PubMed] [Google Scholar]
- 33.Elias RV, Sezate SS, Cao W, McGinnis JF. Temporal kinetics of the light/dark translocation and compartmentation of arrestin and alpha-transducin in mouse photoreceptor cells. Mol. Vis. 2004;10:672–81. [PubMed] [Google Scholar]
- 34.Strissel KJ, et al. Recoverin undergoes light-dependent intracellular translocation in rod photoreceptors. J. Biol. Chem. 2005;280:29250–5. doi: 10.1074/jbc.M501789200. [DOI] [PubMed] [Google Scholar]
- 35.Tobin DM, et al. Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons. Neuron. 2002;35:307–318. doi: 10.1016/S0896-6273(02)00757-2. [DOI] [PubMed] [Google Scholar]
- 36.Gradilone SA, et al. Cholangiocyte cilia express TRPV4 and detect changes in luminal tonicity inducing bicarbonate secretion. Proc. Natl. Acad. Sci. USA. 2007;104:19138–43. doi: 10.1073/pnas.0705964104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phan MN, et al. Functional characterization of TRPV4 as an osmotically sensitive ion channel in porcine articular chondrocytes. Arthritis Rheum. 2009;60:3028–3037. doi: 10.1002/art.24799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siroky BJ, et al. Primary cilia regulate the osmotic stress response of renal epithelial cells through TRPM3. Am. J. Physiol. Physiol. 2017;312:F791–F805. doi: 10.1152/ajprenal.00465.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGlashan SR, et al. Mechanical loading modulates chondrocyte primary cilia incidence and length. Cell Biol. Int. 2010;34:441–446. doi: 10.1042/CBI20090094. [DOI] [PubMed] [Google Scholar]
- 40.Rich DR, Clark AL. Chondrocyte primary cilia shorten in response to osmotic challenge and are sites for endocytosis. Osteoarthritis Cartilage. 2012;20:923–30. doi: 10.1016/j.joca.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 41.Besschetnova TY, et al. Identification of Signaling Pathways Regulating Primary Cilium Length and Flow-Mediated Adaptation. Curr. Biol. 2010;20:182–187. doi: 10.1016/j.cub.2009.11.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pazour GJ, et al. Chlamydomonas IFT 88 and Its Mouse Homologue, Polycystic Kidney Disease Gene Tg 737, Are Required for Assembly of Cilia and Flagella. J. Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murcia NS, et al. The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development. 2000;127:2347–2355. doi: 10.1242/dev.127.11.2347. [DOI] [PubMed] [Google Scholar]
- 44.Yoder BK, et al. Polaris, a protein disrupted in orpk mutant mice, is required for assembly of renal cilium. Am. J. Physiol. Renal Physiol. 2002;282:F541–52. doi: 10.1152/ajprenal.00273.2001. [DOI] [PubMed] [Google Scholar]
- 45.Haycraft CJ, et al. Intraflagellar transport is essential for endochondral bone formation. Development. 2007;134:307–16. doi: 10.1242/dev.02732. [DOI] [PubMed] [Google Scholar]
- 46.Lin F, et al. Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc. Natl. Acad. Sci. USA. 2003;100:5286–91. doi: 10.1073/pnas.0836980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferraris JD, et al. Activity of the TonEBP/OREBP transactivation domain varies directly with extracellular NaCl concentration. Proc. Natl. Acad. Sci. USA. 2002;99:739–44. doi: 10.1073/pnas.241637298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferraris JD, Williams CK, Martin BM, Burg MB, García-Pérez A. Cloning, genomic organization, and osmotic response of the aldose reductase gene. Proc. Natl. Acad. Sci. USA. 1994;91:10742–6. doi: 10.1073/pnas.91.22.10742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borthakur A, et al. T1ρ magnetic resonance imaging and discography pressure as novel biomarkers for disc degeneration and low back pain. Spine (Phila. Pa. 1976). 2011;36:2190–6. doi: 10.1097/BRS.0b013e31820287bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian Y, et al. Inflammatory Cytokines Associated with Degenerative Disc Disease Control Aggrecanase-1 (ADAMTS-4) Expression in Nucleus Pulposus Cells through MAPK and NF-κB. Am. J. Pathol. 2013;182:2310–2321. doi: 10.1016/j.ajpath.2013.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, et al. TNF-α and IL-1β promote a disintegrin-like and metalloprotease with thrombospondin type I motif-5-mediated aggrecan degradation through syndecan-4 in intervertebral disc. J. Biol. Chem. 2011;286:39738–49. doi: 10.1074/jbc.M111.264549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilke HJ, Neef P, Caimi M, Hoogland T, Claes LE. New in vivo measurements of pressures in the intervertebral disc in daily life. Spine (Phila. Pa. 1976). 1999;24:755–62. doi: 10.1097/00007632-199904150-00005. [DOI] [PubMed] [Google Scholar]
- 53.Boos N, Wallin A, Gbedegbegnon T, Aebi M, Boesch C. Quantitative MR imaging of lumbar intervertebral disks and vertebral bodies: influence of diurnal water content variations. Radiology. 1993;188:351–4. doi: 10.1148/radiology.188.2.8327677. [DOI] [PubMed] [Google Scholar]
- 54.Chen M-H, et al. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23:1910–28. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Donnelly E, Williams R, Farnum C. The primary cilium of connective tissue cells: imaging by multiphoton microscopy. Anat. Rec. (Hoboken). 2008;291:1062–73. doi: 10.1002/ar.20665. [DOI] [PubMed] [Google Scholar]
- 56.Zheng L, et al. Ciliary parathyroid hormone signaling activates transforming growth factor-β to maintain intervertebral disc homeostasis during aging. Bone Res. 2018;6:21. doi: 10.1038/s41413-018-0022-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piperi C, Basdra EK. Polycystins and mechanotransduction: From physiology to disease. World J. Exp. Med. 2015;5:200. doi: 10.5493/wjem.v5.i4.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spasic M, Jacobs CR. Lengthening primary cilia enhances cellular mechanosensitivity. Eur. Cell. Mater. 2017;33:158–168. doi: 10.22203/eCM.v033a12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McIntyre JC, Williams CL, Martens JR. Smelling the roses and seeing the light: Gene therapy for ciliopathies. Trends Biotechnol. 2013;31:355–363. doi: 10.1016/j.tibtech.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palacio-Mancheno PE, Evashwick-Rogler TW, Laudier DM, Purmessur D, Iatridis JC. Hyperosmolarity induces notochordal cell differentiation with aquaporin3 upregulation and reduced N-cadherin expression. J. Orthop. Res. 2017;36:788–798. doi: 10.1002/jor.23715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Risbud MV, et al. Nucleus pulposus cells express HIF-1 alpha under normoxic culture conditions: a metabolic adaptation to the intervertebral disc microenvironment. J. Cell. Biochem. 2006;98:152–9. doi: 10.1002/jcb.20765. [DOI] [PubMed] [Google Scholar]
- 62.Liu C, et al. Lack of evidence for involvement of TonEBP and hyperosmotic stimulus in induction of autophagy in the nucleus pulposus. Sci. Rep. 2017;7:4543. doi: 10.1038/s41598-017-04876-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Go WY, Liu X, Roti MA, Liu F, Ho SN. NFAT5/TonEBP mutant mice define osmotic stress as a critical feature of the lymphoid microenvironment. Proc. Natl. Acad. Sci. USA. 2004;101:10673–8. doi: 10.1073/pnas.0403139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.