Abstract
Aim:
The aim of this study was to analyze the Clostridium difficile and their toxins in cancerous tissues in comparison to their adjacent healthy tissues in patients with colorectal cancer (CRC) in Iran.
Background:
Intestinal infection or colonization by microbial pathogens and their released metabolites may have a role in the exacerbation of CRC.
Methods:
A total of 60 biopsy samples from 30 cancerous and 30 adjacent healthy tissues were collected from patients with CRC. Biopsies were homogenized and cultured in cycloserine cefoxitin fructose agar-agar medium to investigate the presence of C. difficile. DNA was extracted, PCR was performed on pure colonies for bacteria detection, and toxin genes were evaluated in each bacterium positive cases. Real-time PCR was performed on extracted DNA for quantitative comparison of Clostridium difficile in healthy and tumor tissues in CRC patients.
Results:
Clostridium difficile was isolated from 18 of the cancerous tissue (60%) and 6 of their healthy adjacent tissue (20%) in the culture medium, but toxin genes were positive just in one sample in both groups. Real-time PCR showed the colonization in all samples.
Conclusion:
This study showed a higher prevalence of Clostridium difficile in cancerous lesions in comparison to healthy tissues. We suggest that the investigation of the rate of CD of colorectal cancer patients before surgery is critical for patients. Further studies with more samples size to study the importance of this bacterium and its toxins in the investigation of colorectal cancer patients survey is recommended.
Key Words: CRC, Intestinal microbiota, Toxin, Clostridium difficile
Introduction
Clostridium difficile, a gram-positive spore-forming anaerobe, is one of the major concerns in healthcare-associated environments and is the leading cause of antibiotic-associated diarrhea (AAD), colitis, toxic megacolon and pseudomembranous colitis (1, 2). The incidence and severity of Clostridium difficile infection (CDI) has been increased during the past two decades, and about 20-30% of patients with AAD experienced laboratory-confirmed CDI (3).
Colorectal Cancer disease (CRC) is the third-highest cancer morbidity in the world. The main symptoms might include abdominal pain, weight loss, change in bowel habits, bleeding, and anemia. Majority of colorectal cancer cases occur in persons without a family history of colorectal cancer. Although old age is one of the risk factors for colorectal cancer, it seems to be increasing among younger persons (4, 5). Although the pathogenesis of CRC is accurately understood, previous studies confirmed the crucial role of intestinal microbiota on the onset of this disease (6- 8). Antibiotic therapies can alter the typical composition of gut microbiota, which in turn may favor colonization of various pathobionts in the mucosal sites of the intestinal lumen (9).
Several studies have highlighted the individual role of specific bacterial pathogens in exacerbation of CRC (10-12). Some studies have been focused on CDI, shown the association of this infection with excess morbidity and mortality along with with the elevated risk of hospitalization, stop of complementation therapy after surgery and increased systemic costs in CRC patients (13). It was reported that up to 17% of the CRC patients are infected by C. difficile (14). Moreover, colonic involvement, chemotherapies, and use of antibiotics reported being as the main risk factors associated with the development of CDI among CRC patients (13).
The majority of commensal microorganisms, collectively known as microbiota that resides in the human body are colonized in niches adjacent to epithelial surfaces of the gastrointestinal tract (15, 16). The diverse and abundant intestinal bacteria play a crucial role in the development and maturation of the immune system early in life, as well as in protection against pathogen colonization (17, 18). However, intestinal infection or colonization by pathogens or a pathobiont, and their released metabolites may alter the composition of the gut microbiota (19, 20). There is limited data regarding the fecal carriage and intestinal colonization of C. difficile among CRC patients. Thus, the main focus of this study was to estimate the prevalence of C. difficile in the gut of Iranian patients with CRC referred to the surgery clinic.
Methods
Patients and sample collection
Colonic biopsies were collected from 30 patients with CRC, who under surgery for CRC in Bahman Hospital in Tehran from September 2016 to June 2017. All CRC patients had a definite diagnosis based on colonoscopy and pathologic reports. The patients with others organ malignancy or exposed to antibiotic therapy within three months before sample collection, as well as those who had undertaken radiotherapy and chemotherapy before the surgical resection were excluded.
Bacterial culture conditions
The colon biopsies were transported to the laboratory in thioglycolate broth and homogenized with a suitable tissue grinder. Cent microliter of the homogenized biopsy was cultured in the CCFA (under anaerobic conditions at 37oC for 48h) for detection of C. difficile. The gram-positive isolates with characteristic colony morphology were considered as C. difficile isolates and selected for further identification by specific primers (21).
Total DNA extraction and Polymerase chain reaction (PCR)
InstaGene matrix extraction kit (Bio-Rad, USA) was used for DNA extraction of C. difficile genome (22). Extracted DNA was used as a template for PCR amplification. For molecular identification and confirmation of C. difficile isolates, PCR was accomplished by a conserved gene of PaLoc (pathogenicity locus) which is called cdd3. For confirming of toxigenic C. difficile isolates, PCR was also performed using specific primers for tcdA, tcdB, cdtA, and cdtB as described previously (23, 24). The nucleotide sequences of the used primers and PCR process for each PCR assay are shown in table 1.
Table 1.
Primer sequences and PCR conditions of studied genes
| Toxin gene | Primers | Oligonucleotide sequences (5' – 3') | PCR conditions | references | |
|---|---|---|---|---|---|
| cdd3 | Time6 Struppi6 |
TCCAATATAATAAATTAGCATTCC GGCTATTACACGTAATCCAGATA |
94°C 5 min, 40 cycles (94°C 1 min; 53°C 1 min;72°C 45 sec), 72°C 5 min | (23, 24) | |
| tcdA | TA1 TA2 |
ATGATAAGGCAACTTCAGTGG TAAGTTCCTCCTGCTCCATCAA |
94°C 5 min, 35 cycles (94°C 1 min; 50°C 1 min;72°C 30 sec), 72°C 5 min | ||
| tcdB | TB1 TB2 |
GACCTGCTTCAATTGGAGAGA GTAACCTACTT CATAACACCAG |
94°C 5 min, 35 cycles (94°C 1 min; 50°C 1 min;72°C 30 sec), 72°C 5 min | ||
| 16S rRNA gene | 27F 1525R |
AGAGTTTGATCCTGGCTCAG AAGGAGGTGWTCCARCC | 94°C 5 min, 40 cycles (94°C 1 min; 60°C 1 min;72°C 45 sec), 72°C 5 min | (25) | |
| C.difficile 16srRNA | C.diff-F C.diff-R | TTGAGCGATTTACTTCGGTAAAGA CCATCCTGTACTGGCTCACCT | 94°C 5 min, 40 cycles (94°C 20 sec; 60°C 1 min, 72°C 5 min), 72°C 5 min | (26) | |
Real-Time Quantitative PCR
To estimate the relative amount of C. difficile over the total amount of bacteria, the DNA from each sample was assayed by real-time quantitative PCR (qPCR); the estimation of the total number of 16S rRNA gene copies in all samples was performed with bacterial primers 27F and 1525R targeting the 16S rRNA gene, using a previously reported protocol (25). The value of C. difficile was assessed with specific primers which form an amplicon of 151 bp (26), targeting a fragment of the 16S rRNA gene. qPCR was performed in a Rotor-Gene Q apparatus (Applied QIAGEN), amplification program was as follows: 35 cycles of 95 °C for 5 s and 60 °C for 34 s with an initial cycle of 95 °C for 10 min, and a primer pair-specific annealing temperature for 60 s. A melting curve was used to evaluate the presence of primers-dimers. C. difficile (ATCC 10898) DNA was used as a standard for qPCR quantification. Reactions were performed in duplicates in 20 μl final volume. PCR results were analyzed by comparing the CT values of the samples, representing the threshold cycles; CT is a relative measure of the concentration of the target gene in the PCR reaction; lower CT values indicate high amounts of targeted nucleic acid, while higher CT values indicate smaller amounts of the target nucleic acid. The presence of C.difficile has been calculated as the ratio between the CT value of C.difficile 16S rRNA gene and the CT value of the total bacterial community 16S rRNA gene amplicons.
Statistical analysis
Data analysis was performed using SPSS software version 21 (SPSS Inc., USA). Statistical differences between the groups were analyzed by T-test, and the results were considered to be significant at a P-value of ≤0.05. The Real-time PCR data were analyzed with one-way analysis of variance (one-way ANOVA) by Prism graft pad soft way.
Results
Patients
A total of sixty colon biopsies samples from 30 CRC patients were evaluated in this study. Study participants consisted of 14 females (46.6%) and 16 males (53.4%) patients within the age range of 19 to 79 years old and a mean age of 58±12.2 years.
Bacterial isolates and confirmation with PCR
Clostridium difficile was isolated from 24/60 (40%) cases and control samples of 30 patients by culture, 18/30 (60%) positive cultures were belonging to CRC specimens and 6/30(20%) isolates from healthy tissues. This difference was statistically significant (P=0.044). All isolates were confirmed by PCR and were positive for cdd3 genes. Just two of the isolates were positive for toxins and were encoded by tcdA, tcdB, and both positive toxins belong to one patient in its healthy and tumor tissues. But no one was positive for binary toxin genes.
Real-time PCRs
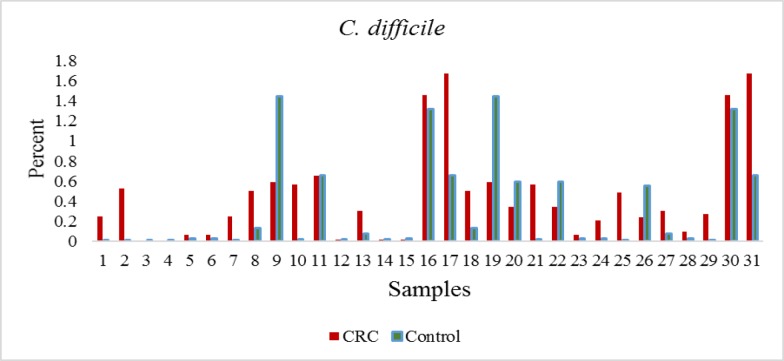
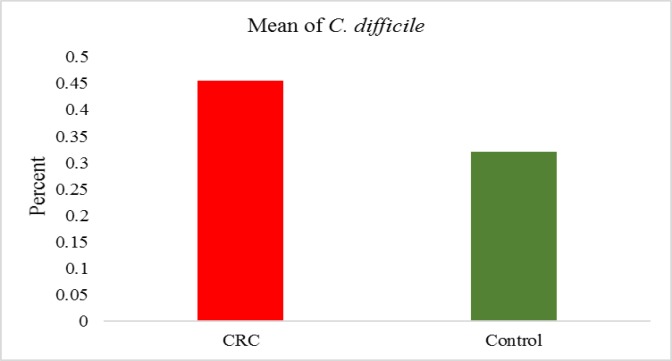
The quantification of C. difficile expressed as the ratio between the CT value of C. difficile 16S rRNA gene, and the CT value of the total bacterial community 16S rRNA gene amplicons is reported. The Real-time PCR showed all of the samples were positive for C. difficile. Consequently, the ratio values indicate higher C. difficile abundance in the tumor tissue samples (figure 1 and 2). But no significant differences between tumor samples and healthy tissue were observed CRC patients (p<0.076).
Figure 1.
The percentage of C. difficile in CRC patients by real time PCR
Figure 2.
The percentage and average presence of C. difficile in cancerous and normal tissues versus total bacteria
Discussion
CRC is becoming an emergent disease in the developed and recently developing countries, in a relatively short period. CRC is disabling for young patients, generating a substantial burden on health-care systems in the world (12). Lifestyle is an important issue in impairing of the microbiota of the human gastrointestinal (27). The interplay of microbiota with immune systems has a significant effect on instruction and regulation of the mucosal immunity. Excessive and dysregulation of the mucosal immune response in CRC patients can be linked to abnormal and abrogated microbial communities (Dysbiosis) in the gut of CRC patients (28). A lack of diversity of the gut microbiome and colonization of pathogenic bacteria can be reasons of dysbiosis (29).
In recent studies, showed patients, who cured with broad-spectrum antibiotics, hospitalized and immunocompromised, are at increased risk for the CDI. Because of the presence of the same these risk factors in CRC Patients, CDI can quickly be developed in CRC patients (30). The accurate colonization role of C. difficile in CRC patients has not been determined until now. C difficile can produce some toxins (enterotoxin A, cytotoxin B, and binary toxin), which can initiate an inflammatory response in the colon (31). Chronic inflammatory could be one of the initiation pathways toward the CRC by DNA damage (32).
In this study, 60 CRC patients were introduced for assessment of C. difficile colonization. Forty percent of samples in this study were positive for C. difficile with culture methods, whereas 100% of samples were positive with real-time PCR but with different percentage. C. difficile is identified as the most common bacteria in the colon. Pathogenesis of C. difficile is dependent on toxin production, and the toxins are a crucial role in pathogenicity. In our study, Rate of toxins positive C. difficile isolates was low (3.3%) but, other studies have been stated higher percent of toxigenic C. difficile in CRC patients (14). One reason for this observation may be related to our method, because isolates from culture were an examination for toxin, while C. difficile were positive in all the samples by Real-time PCR. Zheng et al. showed 16.1 percentage of preoperative CRC patients were C. difficile positive with 19% toxigenic C. difficile (14). However, in contrast to our results, a study reported rate of C. difficile colonization in admitted children in the hematologic ward was reported to be 25.6%, with a 92.6% of toxigenic strains (33). The previous study revealed 20.5% of toxigenic C. difficile colonization in cancer patients, and they concluded that CDI risk could increase 4.8-fold in cancer patients (34). Several studies suggested that more generally colon involvement, are risk factors for CDI and the risk of developing CDI is more in post-surgery cancer patients (14, 33) So, screening of C. difficile for every patient with colon complication and risk factors was recommended. This fact shows a requirement of a rapid test for CDI detection and starts an appropriate treatment promptly.
The impact of C. difficile on CRC is not well clear, but Patients admitted to hospital with CRC have many of these risk factors and may be predisposed to C. difficile. Our results elucidate that 100% of CRC patients were C. difficile positive. In CRC patients, it has been described that CDI can increase morbidity and mortality rate in post-surgery infected. So, early detection and treatment CDI is important and problematics issue in CRC patients, but more studies are needed to determine the risk factors causing the transformation from C. difficile colonization to CDI in CRC patients.
C. difficile is a common bacterium in the Colon of CRC patients but, after the hospitalized and the treatments which induce immunodeficiency the occurrence of CDI in CRC patients have been scarcely explored. Also, antibiotic resistance can challenge the treatment of CDI in CRC patients in the future. So, c. difficile monitoring is a crucial issue before starting chemotherapy and radiography in CRC patients.
Conflict of interests
The authors declare that they have no conflict of interest.
References
- 1.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA) Clin Infect Dis. 2018;66:e1–48. doi: 10.1093/cid/cix1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farooq PD, Urrunaga NH, Tang DM, von Rosenvinge EC. Pseudomembranous colitis. Dis Mon. 2015;61 doi: 10.1016/j.disamonth.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asha NJ, Tompkins D, Wilcox MH. Comparative analysis of prevalence, risk factors, and molecular epidemiology of antibiotic-associated diarrhea due to Clostridium difficile, Clostridium perfringens, and Staphylococcus aureus. J Clin Microbiol. 2006;44:2785–91. doi: 10.1128/JCM.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 5.Bishehsari F, Mahdavinia M, Vacca M, Malekzadeh R, Mariani-Costantini R. Epidemiological transition of colorectal cancer in developing countries: environmental factors, molecular pathways, and opportunities for prevention. World J Gastroenterol. 2014;20 doi: 10.3748/wjg.v20.i20.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hope ME, Hold GL, Kain R, El-Omar EM. Sporadic colorectal cancer-role of the commensal microbiota. FEMS Microbiol lett. 2005;244:1–7. doi: 10.1016/j.femsle.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 7.Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PloS One. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16s rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas LV, Ockhuizen T. New insights into the impact of the intestinal microbiota on health and disease: a symposium report. Br J Nutr. 2012;107:S1–13. doi: 10.1017/S0007114511006970. [DOI] [PubMed] [Google Scholar]
- 11.Rowland IR. The role of the gastrointestinal microbiota in colorectal cancer. Curr Pharm Des. 2009;15:1524–7. doi: 10.2174/138161209788168191. [DOI] [PubMed] [Google Scholar]
- 12.Jahani-Sherafat S, Alebouyeh M, Moghim S, Ahmadi Amoli H, Ghasemian-Safaei H. Role of gut microbiota in the pathogenesis of colorectal cancer; a review article. Gastroenterol Hepatol Bed Bench. 2018;11:101–10913. [PMC free article] [PubMed] [Google Scholar]
- 13.Lugito NP, Shin A, Kelly CP. A 21 Year-Old Male colorectal cancer Clostridium difficile and intestinal amebiasis infection. Indonesian J Cancer. 2014;8 [Google Scholar]
- 14.Zheng y, Luo Y, Lv Y, Huang C, Sheng Q, et al. Clostridium difficile colonization in preoperative colorectal cancer patients. oncotarget. 2017;8:11877–86. doi: 10.18632/oncotarget.14424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balram B, Battat R, Al-Khoury A, D’Aoust J, Afif W, Bitton A, et al. Risk factors associated with Clostridium difficile infection in inflammatory bowel disease: a systematic review and meta-analysis. J Crohn's Colitis. 2018;13:27–38. doi: 10.1093/ecco-jcc/jjy143. [DOI] [PubMed] [Google Scholar]
- 16.Marchesi JR, Adams DH, Fava F, Hermes GDA, Hirschfield GM, Hold G, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330–9. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazar V, Ditu LM, Pircalabioru GG, Gheorghe I, Curutiu C, Holban AM, et al. Aspects of gut microbiota and immune system interactions in infectious diseases, immunopathology, and cancer. Front Immunol. 2018;9:1830. doi: 10.3389/fimmu.2018.01830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S, Covington A, Pamer EG. The intestinal microbiota: Antibiotics, colonization resistance, and enteric pathogens. Immunol Rev. 2017;279:90–105. doi: 10.1111/imr.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rolhion N, Chassaing B. When pathogenic bacteria meet the intestinal microbiota. Biol Sci. 2016:371. doi: 10.1098/rstb.2015.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong D, Ni Q, Wang C, Zhang L, Li Z, Jiang C, et al. Effects of intestinal colonization by Clostridium difficile and Staphylococcus aureus on microbiota diversity in healthy individuals in China. BMC Infect Dis. 2018;18 doi: 10.1186/s12879-018-3111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shayganmehr F-S, Alebouyeh M, Azimirad M, Aslani MM, Zali MR. Association of tcdA+/tcdB+ Clostridium difficile genotype with emergence of multidrug-resistant strains conferring metronidazole resistant phenotype. Iranian Biomed J. 2015;19:143. doi: 10.7508/ibj.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azimirad M, Dezfulian A, Alebouyeh M, Esfehani RB, Shahrokh S, Zali MR. Infection with enterotoxigenic Staphylococcus aureus as a concern in patients with gastroenteritis. J Glob Antimicrob Resist. 2017;9:111–4. doi: 10.1016/j.jgar.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Spigaglia P, Mastrantonio P. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J Clin Microbiol. 2002;40:3470–5. doi: 10.1128/JCM.40.9.3470-3475.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Persson S, Torpdahl M, Olsen K. New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin Microbiol Infect. 2008;14:1057–64. doi: 10.1111/j.1469-0691.2008.02092.x. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Mang-Kun Chen, Bing-Ya Yang, Xian-Jie Huang, Xue-Rui Zhang, Liang-Qiang He, et al. Use of 16S rRNA gene-targeted group-specific primers for realtime PCR analysis of predominant bacteria in mouse feces. Appl Environ Microbiol J. 2015:81. doi: 10.1128/AEM.01906-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penders J. Vink C. Driessen C. London N. Thijs C. Stobberingh E.E. Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol Lett. 2005;243:141–7. doi: 10.1016/j.femsle.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 27.Azimirad M, Rostami-Nejad M, Rostami K, Naji T, Zali MR. The susceptibility of celiac disease intestinal microbiota to Clostridium difficile infection. Am J Gastroenterol. 2015;110:1740–1. doi: 10.1038/ajg.2015.360. [DOI] [PubMed] [Google Scholar]
- 28.Uronis JM, Mühlbauer M, Herfarth HH, Rubinas TC, Jones GS, Jobin C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One. 2009;4:e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchesi JR, Dutilh BE, Hall N, Peters W, Roelofs R, Boleij A, et al. Towards the human colorectal cancer microbiome. PloS One. 2011;6:e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hung YP, Lee JC, Lin HJ, Liu HC, Wu YH, Tsai PJ, et al. Clinical impact of Clostridium difficile colonization. J Microbiol Immunol Infect. 2015;48:241–8. doi: 10.1016/j.jmii.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 31.Singh H, Nugent Z, Yu BN, Lix LM, Targownik LE, Bernstein CN. Higher incidence of Clostridium difficile infection among individuals with inflammatory bowel disease. Gastroenterology. 2017;153:430–8. doi: 10.1053/j.gastro.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 32.Terzic´ J, Grivennikov S, Karin E, Karin M. Inflammation and cancer. Gastroenterology. 2010;138:2101–14. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 33.Armin S, Shamsian S, Drakhshanfar H. Colonization with Clostridium difficile in Children with Cancer. Iran J Pediatr. 2013;23:473–6. [PMC free article] [PubMed] [Google Scholar]
- 34.Fang WJ, Jing DZ, Luo Y, Fu CY, Zhao P, Qian J, et al. Clostridium difficile carriage in hospitalized cancer patients: a prospective investigation in eastern China. BMC Infect Dis. 2014;14:523. doi: 10.1186/1471-2334-14-523. [DOI] [PMC free article] [PubMed] [Google Scholar]