Abstract
The retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) are a family of cytosolic pattern recognition receptors that play a critical role in binding viral RNA and triggering antiviral immune responses. The RLR LGP2 (or DHX58) is a known regulator of the RIG-I signaling pathway; however, the underlying mechanism by which LGP2 regulates RIG-I signaling is poorly understood. To better understand the effects of LGP2 on RIG-I-specific signaling and myeloid cell responses, we probed RIG-I signaling using a highly specific RIG-I agonist to compare transcriptional profiles between WT and Dhx58−/− C57BL\6 bone marrow-derived dendritic cells. Dhx58−/− cells exhibited a marked increase in the magnitude and kinetics of type I interferon (IFN) induction and a broader antiviral response as early as 1 h post-treatment. We determined that LGP2 inhibited RIG-I-mediated IFN-β, IRF-3, and NF-κB promoter activities, indicating a function upstream of the RLR adaptor protein mitochondrial antiviral signaling. Mutational analysis of LGP2 revealed that RNA binding, ATP hydrolysis, and the C-terminal domain fragment were dispensable for inhibiting RIG-I signaling. Using mass spectrometry, we discovered that LGP2 interacted with the E3 ubiquitin ligase TRIM25. Finally, we determined that LGP2 inhibited the TRIM25-mediated K63-specific ubiquitination of the RIG-I N-terminus required for signaling activation.
Keywords: RIG-I-like receptors, ubiquitination, LGP2, TRIM25
Introduction
Pattern recognition receptor signaling is essential for regulating immune responses to virus infection. The retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) are a family of cytosolic RNA helicases that, upon recognition of pathogen associated molecular patterns (PAMPs) such as non-self RNAs, trigger a robust antiviral defense response characterized by the production of type I interferon (IFN), pro-inflammatory cytokines, and expression of hundreds of antiviral effector genes.
The RLRs are composed of 3 structurally related proteins: Laboratory of Genetics and Physiology 2 (LGP2), RIG-I, and melanoma differentiation-associated gene 5 (MDA5). All 3 RLRs contain a DExD/H box helicase domain with ATP hydrolysis and RNA binding activities (Bamming and Horvath 2009; Bruns and others 2013) and a C-terminal domain (CTD) that aids in RNA substrate recognition and prevents signaling activation (Saito and others 2007; Cui and others 2008; Takahasi and others 2009). RIG-I and MDA5 bind distinct RNA ligands, allowing for recognition of different viruses (Loo and Gale 2011).
Upon binding RNA, RIG-I, and MDA5 undergo several post-translational modifications to reach an activated state, including dephosphorylation by PP1 (Wies and others 2013) and K63-ubiquitination by TRIM25 in the case of RIG-I (Gack and others 2007). Once activated, RIG-I and MDA5 translocate to mitochondrial membranes where they interact with mitochondrial antiviral signaling (MAVS), the central RLR signaling adaptor protein, through 2 N-terminal caspase activation and recruitment domains (CARDs) to initiate downstream signaling.
LGP2 (also known as DHX58) lacks N-terminal CARDs, which are important for mediating protein–protein interaction and signaling activation, but does possess a DExD/H box helicase domain and CTD (Rothenfusser and others 2005; Komuro and Horvath 2006; Satoh and others 2010). The LGP2 helicase domain shares 31–34% identity with the RIG-I helicase domain and 41–43% identity with the MDA5 helicase domain (Yoneyama and others 2005; Bruns and Horvath 2012). The CTD of LGP2 shares 29% homology with the RIG-I CTD and 33% with the MDA5 CTD (Takahasi and others 2009; Bruns and Horvath 2012).
Like RIG-I and MDA5, the helicase domain of LGP2 is capable of binding RNA and hydrolyzing ATP, while the CTD contains an RNA binding motif that serves to recognize specific RNA ligands (Cui and others 2001, 2008; Bamming and Horvath 2009; Bruns and others 2013). However, it is believed that LGP2 is capable of binding a more diverse set of RNA substrates than either RIG-I or MDA5, including 5′-triphosphate single-stranded RNA (ssRNA), short (<2 kb) double-stranded RNA (dsRNA), and long (>2 kb) dsRNA (Li and others 2009; Takahasi and others 2009; Bruns and others 2013).
While LGP2 is known to enhance MDA5 signaling through previously described mechanisms (Bruns and others 2013, 2014; Childs and others 2013), the role of LGP2 in regulating RIG-I-specific signaling is still not well understood. LGP2 has been identified as both a positive and a negative regulator of RIG-I signaling during virus infection (Rothenfusser and others 2005; Venkataraman and others 2007; Satoh and others 2010). One study found that IFN-β and pro-inflammatory cytokine protein levels were depressed in Dhx58−/− bone marrow-derived dendritic cells (BM-DCs) upon infection with a diverse panel of viruses and that this positive regulation is dependent on the ATP hydrolysis function of LGP2 (Satoh and others 2010). Others, however, have demonstrated a negative regulatory function of LGP2 in cells infected with Hepatitis C virus (HCV) or Sendai virus (SeV) or treated with poly(I:C) (Yoneyama and others 2005; Saito and others 2007; Venkataraman and others 2007).
These results could be indicative of a more complex set of roles for LGP2. Indeed, functions for LGP2 have been described in several noncanonical cellular processes. LGP2 was found to play a role in the inhibition of Dicer-mediated RNA interference (RNAi), a process in which small interfering RNAs (siRNAs) bind and target viral RNA for degradation (Komuro and others 2016; van der Veen and others 2018). Dicer is an endoribonuclease known to generate siRNAs by cleaving viral dsRNA. The CTD of LGP2 interacts with Dicer and prevents Dicer cleavage of dsRNA. In addition, LGP2 has been found to promote CD8+ T cell survival and fitness during virus infection and inhibit apoptosis of cancer cells subjected to ionizing radiation (Suthar and others 2012; Widau and others 2014).
In this study, we examine the effects of LGP2 on RIG-I-specific signaling and myeloid cell responses and reveal how LGP2 inhibits RIG-I signaling. We first probed RIG-I signaling in wild type (WT) and Dhx58−/− murine BM-DCs (pure C57BL\6 background; (Suthar and others 2012)) using a highly specific and well-characterized RIG-I agonist (PAMP RNA; Saito and others 2008; Schnell and others 2012) and found that as early as 1 h post-treatment, Dhx58−/− BM-DCs displayed a marked increase in the expression kinetics and magnitude of type I interferon (IFN) genes, as well as a broader antiviral response, characterized by genes encoding pro-inflammatory cytokines, chemokines, and interferon-stimulated genes (ISGs).
Using reporter-based assays, we found that LGP2 inhibited RIG-I-mediated IFN-β, interferon regulatory factor 3 (IRF-3), and nuclear factor-κB (NF-κB) promoter activities, indicating that LGP2 functions upstream of the RLR adaptor protein MAVS. Furthermore, we found that the RNA binding and ATP hydrolysis enzymatic functions of LGP2, as well as the CTD fragment of LGP2 alone, were dispensable for negatively regulating RIG-I signaling. Using mass spectrometry, we discovered that LGP2 interacted with TRIM25, an E3 ubiquitin ligase that post-translationally modifies RIG-I into a signaling “active” form by K63-ubiquitinating the RIG-I CARDs. Finally, we found that LGP2 inhibits TRIM25-mediated K63-specific ubiquitination of the RIG-I N-terminal CARDs. These results demonstrate that LGP2 negatively regulates RIG-I activation by blocking RIG-I ubiquitination and this is critical for dampening RIG-I signaling and antiviral gene expression.
Materials and Methods
Cells
Dendritic cells were derived from C57BL\6 WT and Dhx58−/− mouse bone marrow cells cultured in 1 × RPMI (Corning Cellgro) containing 10% FBS, 2 mM L-glutamine (Corning Cellgro), 1 mM sodium pyruvate (Corning Cellgro), 1 × Nonessential Amino Acids (Corning Cellgro), 1 × antibiotics (penicillin, streptomycin, amphotericin B; Corning Cellgro), and 20 ng/mL GM-CSF cytokine. Media was changed on days 3 and 6, and cells were harvested on day 8 post-bone marrow extraction. The Dhx58−/− mice were generated on a pure C57BL\6 background as previously described (Suthar and others 2012). HEK293 cells were maintained in 1 × Dulbecco's modification of Eagle's medium (DMEM; Corning Cellgro) containing 10% FBS, 25 mM HEPES Buffer (Corning Cellgro), 2 mM L-glutamine, 1 mM sodium pyruvate, 1 × Nonessential Amino Acids, and 1 × antibiotics. LGP2-deficient HEK293 cells were generated by a CRISPR/Cas9 system and characterized as previously described by Curt Horvath's laboratory (Parisien and others 2018).
Plasmids
Plasmids used in these experiments include Flag-RIG-I (pEF-Bos), HA-RIG-I (pEF-Tak), Flag-N-RIG (pEF-Bos), Flag-LGP2 (p3xFLAG-CMV10), Flag-LGP2 MI-MVI (p3xFLAG-CMV10), Flag-LGP2 1–546, 1–350, 1–176 (p3xFLAG-CMV10; supplied by Curt Horvath), Flag-LGP2 1–159, 1–121, 122–678, 160–678, 177–678, CTD (547–678) (p3xFLAG-CMV10; generated as described below), Myc-LGP2, HA-TRIM25 (pCAGGS), HA-ubiquitin (Ub; pRK5), HA-K63-Ub (pRK5), p125-luc, pRL-CMV, pEF-Bos empty vector, and pEF-Tak empty vector.
LGP2 deletion mutants were generated from the p3xFLAG-CMV10 construct containing full-length (FL) LGP2. FL LGP2 construct was amplified using PCR with primers specific to the region of interest: LGP2 1–121, For 5′-GATGACAAGCTTGCGGCCGCGATGGAG-3′, Rev 5′-GAACCGGCTCATCTAGATTACTCCACGTGCTCCTCCTCCTC-3′; LGP2 1–159, For 5′-GATGACAAGCTTGCGGCCGCGATGGAG-3′, Rev 5′-GAACCGGCTCATCTAGATTACGGCTGTGCCCTCTGGAGTTT-3′; LGP2 122–678, 5′-GAACCGGCTCAGCGGCCGCGATGCTCACTGTCTTCTCCCTGATC-3′, Rev 5′-CCGGGATCCTCTAGATTAGTCCAGGGA-3′; LGP2 160–678, For 5′-GAACCGGCTCAGCGGCCGCGATGCTACCCCAGGTGCTGGGTCTC-3′, Rev 5′-CCGGGATCCTCTAGATTAGTCCAGGGA-3′; LGP2 177–678, For 5′-GAACCGGCTCAGCGGCCGCGATGAAACTCGATGGGGCCATCAAC-3′, Rev 5′-ACAAGGCTGGTGGGCACTGGAGTG-3′; LGP2 CTD (547–678), For 5′-GAACCGGCTCAGCGGCCGCGATGTTCCCAGTGGAGCACGTGCAGCTAC-3′, Rev 5′-CCGGGATCCTCTAGATTAGTCCAGGGA-3′. Vector and PCR fragments were digested and gel purified. Desired fragments were extracted from gel using the 5 Prime Agarose GelExtract Mini Kit and PCR fragments ligated into linearized p3xFLAG-CMV10 vector. Vectors containing LGP2 deletion mutants were transformed into NEB 5-α competent Escherichia coli cells. Successfully transformed colonies were grown up in LB Broth and plasmids harvested using E.Z.N.A Endotoxin-free Plasmid DNA Miniprep Kit (Omega) as per the manufacturers' instructions.
Reagents and antibodies
Mirus TransIT-293, Mirus TransIT-mRNA, MG132 (Fisher), DMSO (Fisher), N-ethylmaleimide (Sigma), Dual-Luciferase Reporter Assay System (Promega), SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific), Anti-FLAG M2 Magnetic Beads (Sigma), Dynabeads Protein G magnetic beads (Novex), Mouse Cytokine Magnetic 20-Plex Panel (Invitrogen), ProcartaPlex Mouse IFN alpha/IFN beta Panel (Affymetrix eBioscience), Ms anti-Flag 1:1000 (Sigma), Rb anti-GAPDH 1:2500 (Cell Signaling), Rb anti-RIG-I 1:1000 (Gale Lab), Rb anti-LGP2 1:100 (IBL), Rb anti-MDA5 1:1000 (IBL), Ms anti-TRIM25 1:2000 (BD Bioscience), Rb anti-HA Tag 1:1000 (Cell Signaling), Rb anti-Myc 1:5000 (Novus), Ms anti-β-actin 1:1000 (Cell Signaling). The RIG-I agonist (PAMP RNA) was generated as previously described (Saito and others 2008; Schnell and others 2012). Briefly, 5′-ppp RNA products were generated using a synthetic DNA oligonucleotide template (Integrated DNA Technologies; HCV 3′ untranslated region poly-U/UC sequence) using the T7 MEGAshortscript Kit (Ambion) as per the manufacturer's instructions. Following in vitro transcription, DNA templates were removed with DNAse treatment, RNA was precipitated using ethanol and ammonium acetate as described by the manufacturer, and then resuspended in nuclease-free water. RNA concentration was determined by absorbance using a NanoDrop spectrophotometer.
RNA-Seq analysis
WT and Dhx58−/− DCs were CD11c+-purified using EasySep Mouse CD11c Positive Selection Kit II (Stemcell) as per the manufacturer's protocol and transfected with 20 ng PAMP RNA, or left untreated. Cells were plated in biological triplicates for each condition/time point. PAMP RNA-treated cells and time-matched mock-treated cells were collected at 1, 3, and 6 h postagonist transfection in Buffer RLT (Qiagen). Total RNA was isolated (QIAGEN RNeasy Plus Mini Kit) according to the manufacturer's protocol. RNA quality was assessed using an Agilent 2000 Bioanalyzer capillary electrophoresis, and all RNA integrity (RIN) scores were greater than 8. mRNA sequencing libraries were prepared, and the quality of the libraries was verified using DNA-1000 Kits (Agilent Bioanalyzer) and quantified using the Qubit 2.0 Fluorometer (Life Technologies). Libraries were clustered and sequenced on an Illumina HiSeq 2500 System (100 bp single end reads). Sequencing reads were mapped to the mouse reference genome mm10. Reads were normalized, and differential expression analysis performed using DESeq2 (Love and others 2014). The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through the GEO accession number GSE132696.
Cytokine secretion
Cell supernatants from WT and Dhx58−/− DCs used in RNA-Seq were analyzed for cytokines using multiplex bead assays—Mouse Cytokine Magnetic 20-Plex Panel (Invitrogen) and ProcartaPlex Mouse IFN alpha/IFN beta Panel (Affymetrix eBioscience) —according to the manufacturers' protocols.
Flow cytometry
WT and Dhx58−/− DCs were transfected with 20 ng PAMP RNA or left untreated. Cells were plated in quadruplicate for each condition/time point. PAMP RNA-treated cells and time-matched mock-treated cells were collected at 24 h postagonist transfection. Cells were blocked with Anti-mouse CD16/CD32 Fc block (TONBO Biosciences) and stained for surface markers CD11c-PE/Cy7 (TONBO Biosciences), CD80-FITC (TONBO Biosciences), CD86-PE (TONBO Biosciences), and MHC I-Alexa647 (TONBO Biosciences). Viability was determined by staining with Ghost Dye 780 (TONBO Biosciences). Cells were run on BD LSR II. Cells were gated for single cells, CD11c+ expression, and viability before looking at surface marker expression.
Signaling assays
RIG-I-dependent signaling was determined using luciferase assay, using reagents from Promega's Dual-Luciferase Reporter Assay System. 1 × 105 HEK293 cells were transfected with 50 ng p125-luc containing the luciferase gene under the control of the IFN-β promoter region, 20 ng pRL-CMV containing the renilla gene under the control of the CMV promoter, and 100 ng other indicated plasmid DNA, using 1 × Opti-MEM reduced serum media (Life Technologies) and Mirus TransIT-293 transfection reagent, for 24 h. Cells were subsequently transfected with 100 ng PAMP RNA for 6 h using 1 × Opti-MEM and Mirus TransIT-mRNA transfection reagent. Lysates were collected in 1 × Passive Lysis Buffer, and results were read on a SynergyH1 Hybrid Reader (BioTek). Luciferase measurements were normalized to Renilla expression, which serves as a transfection control. Firefly luciferase values were divided by Renilla values to produce a normalized value (relative luciferase units; RLU). Technical triplicates were tested for each sample under each condition.
Immunoprecipitation and immunoblotting
1–1.5 × 106 HEK293 cells were transfected with 500 ng indicated plasmid DNA using 1 × Opti-MEM reduced serum media (Life Technologies) and Mirus TransIT-293 transfection reagent. Twenty-four hours post-transfection, cells were transfected with 100 ng PAMP RNA using 1 × Opti-MEM and Mirus TransIT-mRNA transfection reagent. Lysates were collected in modified RIPA buffer (10 mM Tris, 150 mM NaCl, 1% NA-deoxycholate, and 1% Triton X-100) containing protease and phosphatase inhibitors and deubiquitinase inhibitor, N-Ethylmaleimide (Sigma), for ubiquitination assays. In coexpression assays, immunoprecipitation (IP) was performed with either anti-Flag (Sigma) or anti-HA (Pierce) magnetic beads. In endogenous assays, IP was performed with magnetic Protein G Dynabeads (Life Technologies) conjugated to the appropriate antibody. Proteins were eluted in 2 × loading buffer (0.25 M Tris, 40% glycerol, 20% β-ME, 9.2% SDS, and 0.04% Bromophenol Blue). IP supernatants and whole cell lysate controls were run on polyacrylamide gel using SDS-PAGE and transferred to nitrocellulose membrane for immunoblotting. Blocking was performed in 5% milk in 0.1% PBST. Primary antibodies were prepared in 0.1% PBST containing 10% FBS. Secondary antibodies were prepared in 0.1% PBST containing 1% FBS. Blots were developed using Thermo Scientific SuperSignal West Femto Maximum Sensitivity Substrate and a Bio-Rad ChemiDoc XRS+.
RNA immunoprecipitation
HEK293 cells were transfected with 1 μg/μL unlabeled or biotin-labeled PAMP RNA using 1 × Opti-MEM and Mirus TransIT-mRNA transfection reagent. Lysates were collected 3 h later in RIPA buffer containing RNase, protease, and phosphatase inhibitors (Cell Signaling). IP was performed with streptavidin-conjugated magnetic beads (Pierce). Proteins were eluted in 2 × loading buffer. IP supernatants and whole cell lysate controls were run on polyacrylamide gel through SDS-PAGE and transferred to nitrocellulose membrane for immunoblotting.
Mass spectrometry
1 × 106 HEK293 cells were transfected with 500 ng Flag-LGP2 plasmid DNA using 1 × Opti-MEM reduced serum media (Life Technologies) and Mirus TransIT-293 transfection reagent. Twenty-four hours post-transfection, cells were transfected with 100 ng PAMP RNA using 1 × Opti-MEM and Mirus TransIT-mRNA transfection reagent. Lysates were collected in modified RIPA buffer containing protease and phosphatase inhibitors. IP was performed for Flag-LGP2 with anti-Flag (Sigma) magnetic beads. Samples were eluted in 8 M urea and run on a LTQ Orbitrap XL Mass Spectrometer (Thermo Scientific) to identify associated proteins.
Statistical analysis
mRNA-sequencing samples were submitted in triplicate for each condition tested, with triplicate time-matched, mock-treated controls. Differentially expressed genes (DEGs) were identified through DESeq2 analysis, and thresholds were set at fold change >2 and P < 0.01. Supernatants used for protein secretion analysis were collected from the same cells used for mRNA-seq. Samples were run in biological triplicates and analyzed by 2-way ANOVA followed by Sidak's test for multiple comparisons, P < 0.05. Flow cytometry samples were run in biological quadruplicates and analyzed by 2-way ANOVA followed by Sidak's test for multiple comparisons, P < 0.05. Flow cytometry data are representative of at least 3 separate experiments. Luciferase assays were run in biological triplicates for each condition tested. RLU were calculated by normalizing luciferase expression readings to Renilla transfection control expression within the same sample. Data were analyzed by 1-way ANOVA followed by either Tukey's or Dunnett's test for multiple comparisons, P < 0.05. All luciferase assay and immunoblot data are representative of at least 3 separate experiments. Statistical analysis was performed using GraphPad Prism, version 6 software.
Results
LGP2 is a negative regulator of RIG-I-mediated innate immune responses in BM-DCs
The mechanism underlying LGP2-mediated regulation of RIG-I signaling is not well defined. To understand the impact of LGP2 on RIG-I antiviral signaling, we probed RIG-I signaling using a previously characterized, highly specific RIG-I agonist derived from the polyU/UC region of the HCV 3′ UTR (PAMP RNA; (Saito and others 2008)) and performed transcriptomic analysis. Specifically, we transfected WT and Dhx58−/− BM-DCs from mice of a pure C57BL\6 genetic background (Suthar and others 2012) with PAMP RNA and harvested biological triplicate mock and treated cells at 1, 3, and 6 h post-treatment followed by mRNA sequencing (mRNA-Seq; Fig. 1A). We compared gene expression changes between treated and time-matched mock controls and determined differential gene expression by 2-fold change and P < 0.01.
FIG. 1.
LGP2 negatively regulates RIG-I-mediated antiviral transcriptional responses in BM-DCs. (A) WT and Dhx58−/− BM-DCs were treated with PAMP RNA and collected in biological triplicate with time-matched, mock-treated cells and submitted for mRNA-sequencing. Transcripts with a fold change >2, P < 0.01 were identified for further analysis. (B) Heat map of genes differentially regulated in WT and Dhx58−/− (KO) BM-DCs post-treatment. Venn diagrams illustrate numbers of differentially expressed and shared genes. (C) Differentially expressed and shared genes in WT and Dhx58−/− BM-DCs plotted to illustrate ratio of expression (Dhx58−/−/WT). Genes differentially expressed in both WT and Dhx58−/− cells (orange); genes differentially expressed only in Dhx58−/− cells (red); genes differentially expressed only in WT cells (green). (D) Heat map of genes induced in WT and Dhx58−/− (KO) BM-DCs at 1 h post-treatment, with gene clusters indicated by vertical colored bars. Purple, genes induced only in Dhx58−/− BM-DCs; gray, shared genes with similar expression in WT and Dhx58−/− cells; blue, shared genes with higher induction in Dhx58−/− cells; pink, shared genes highly expressed in WT BM-DCs but even more highly expressed in Dhx58−/− cells. (E) Individual analysis of select transcripts at 1 h post-treatment with PAMP RNA or mock treated (M). Data shown are average normalized transcript counts of biological triplicates ± SD of WT (blue) and Dhx58−/− (red) analyzed by Sidak's test, P < 0.01. (F) Ddx58 (RIG-I) transcripts at 1 h post-treatment with PAMP RNA or mock treated (M). Data shown are average normalized transcript counts of biological triplicates ± SD of WT (blue) and Dhx58−/− (red) analyzed by Sidak's test, P < 0.01. (G) Analysis of Ddx58 (RIG-I) gene expression in WT (blue) and Dhx58−/− (red) BM-DCs treated with PAMP RNA (P, solid line) or mock treated (M, dashed line). Data shown are average normalized transcript counts of biological triplicates ± SD. (H) Analysis of Dhx58 (LGP2) gene expression in WT BM-DCs treated with PAMP RNA (black) or mock treated (white). Data shown are average normalized transcript counts of biological triplicates ± SD analyzed by Tukey's test, P < 0.01. **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant. BM-DC, bone marrow-derived dendritic cell; DEG, differentially expressed gene; PAMP, pathogen associated molecular pattern; RIG-I, retinoic acid-inducible gene I. Color images are available online.
Through this analysis, we observed that the genetic ablation of LGP2 led to enhanced and broader transcriptional responses following PAMP RNA treatment. We found that at 1 h post-treatment Dhx58−/− cells exhibited 44 DEGs (WT = 0, Shared = 30), at 3 h post-treatment Dhx58−/− cells exhibited 1374 DEGs (WT = 102, Shared = 950), and at 6 h post-treatment Dhx58−/− cells exhibited 2763 DEGs (WT = 136, Shared = 2024; Fig. 1B, C). Furthermore, many of the shared genes at 1, 3, and 6 h post-treatment were induced to a greater extent in Dhx58−/− cells compared with WT cells (Fig. 1C, orange circles).
We next focused on the transcriptional differences at 1 h post-treatment (Fig. 1D). Using hierarchical clustering, we defined 4 gene clusters that were differentially enhanced in WT and Dhx58−/− BM-DCs: (1) Genes only expressed in Dhx58−/− cells (purple), which include antiviral effectors (Gbp5), chemokines (Ccl2, Ccl3, Cxcl1), and a transcription factor (Jun); (2) Genes expressed to similar levels in both WT and Dhx58−/− cells (gray), which include antiviral effectors (Mx2, Oasl1), a cytokine (Tnfsf15), a transcription factor (Fos, otherwise known as AP-1), and a cell cycle-related gene (Ccno); (3) Shared genes induced to a higher level in Dhx58−/− cells (blue), which include type I IFNs (Ifna1, Ifna2, Ifna5), antiviral effectors (Isg15, Ifit2, Rsas2), and pro-inflammatory cytokines/chemokines (Tnf, Cxcl2); and (4) Shared genes highly expressed in WT cells but exceeded in Dhx58−/− cells (pink), which include type I IFNs (Ifnb1, Ifna4), antiviral effectors (Ifit1, Ifit3), and pro-inflammatory cytokines/chemokines (Il6, Cxcl10, Ccl4). A select set of genes known to promote an antiviral state, including type I IFNs (Ifnb1, Ifna2, Ifna4), pro-inflammatory cytokines (Il6), and ISGs (Ifit1, Ifit2, Ifit3, Isg15), were significantly increased in Dhx58−/− BM-DCs compared to WT cells at the 1 h time point (Fig. 1E).
We next evaluated whether the differences in gene induction at 1 h post-treatment were attributable to differences in basal expression of RIG-I. We found that Ddx58 (RIG-I) transcripts were expressed at similar levels in WT and Dhx58−/− BM-DCs (Fig. 1F), confirming previous findings at the protein level in these cells by our group (Suthar and others 2012). While other antiviral effector genes were robustly elevated above mock at 1 h post-treatment (Fig. 1E), RIG-I transcripts were not significantly induced in either WT or Dhx58−/− cells at this time (Fig. 1F). This suggests that the increased RIG-I signaling observed in the absence of LGP2 is not due to basal expression differences of RIG-I. RIG-I mRNA expression was induced at 3 and 6 h post-treatment in both WT and Dhx58−/− BM-DCs (Fig. 1G).
We also evaluated the kinetics of Dhx58 (LGP2) expression in WT BM-DCs and found that LGP2 mRNA was present under mock conditions (normalized read counts = 103.6) and that LGP2 expression was induced between 1 and 3 h post-treatment (Fig. 1H). Combined, these findings indicate that LGP2 is important for negatively regulating early events within the RIG-I signaling cascade and that basal levels of LGP2 are sufficient for enacting this regulation.
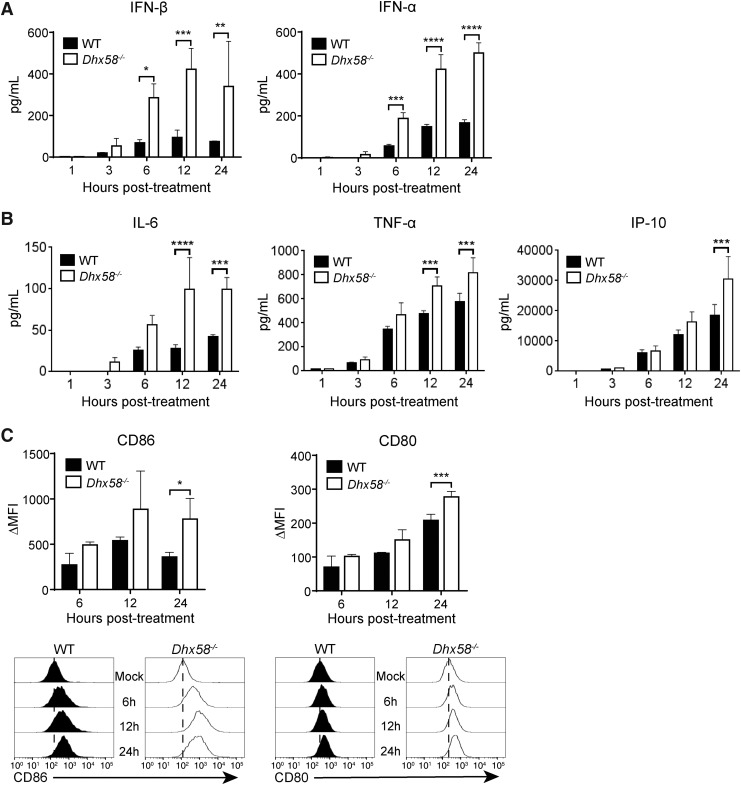
Next, we evaluated the impact of LGP2 on cytokine production and costimulatory molecule expression following RIG-I signaling activation. We performed multiplex bead assays to determine the secretion of type I IFN and pro-inflammatory cytokines by WT and Dhx58−/− BM-DCs in matching samples used in the mRNA-sequencing analysis. Protein secretion of IFN-β, IFN-α (Fig. 2A), and pro-inflammatory cytokines, including IL-6, TNF-α, and IP-10 (Fig. 2B), was elevated in Dhx58−/− BM-DCs compared to WT cells. In addition, we observed that protein expression of the costimulatory markers CD86 and CD80 was higher on the surface of Dhx58−/− BM-DCs compared to WT cells (Fig. 2C), suggesting more pronounced activation of BM-DCs in the absence of LGP2. Taken together, these results illustrate that LGP2 functions as a negative regulator in the early stages of RIG-I-mediated antiviral immune signaling in BM-DCs.
FIG. 2.
LGP2 negatively regulates downstream antiviral immune responses. Protein secretion (pg/mL) of (A) type I IFNs and (B) pro-inflammatory cytokines/chemokines in the supernatant of PAMP RNA-treated WT (black) and Dhx58−/− (white) BM-DCs. Data shown are biological triplicates ± SD analyzed by Sidak's test, P < 0.05. (C) Surface expression of costimulatory markers CD86 and CD80 in PAMP RNA-treated WT (black) and Dhx58−/− (white) BM-DCs. Data are shown as average ΔMFI (mean fluorescence intensity) of biological quadruplicates ± SD. Data were analyzed by Sidak's test, P < 0.05. Representative histograms are shown below. Data shown are representative of at least 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
LGP2 is a negative regulator of RIG-I signaling in human cells
To dissect the mechanism by which LGP2 negatively regulates RIG-I signaling, we performed reporter-based assays in cultured human cells (HEK293 cells). To ensure LGP2 functions in a similar manner in this model, we assessed IFN-β promoter-driven luciferase production with increasing amounts of overexpressed LGP2. We found that PAMP RNA-induced IFN-β promoter activity was inhibited in a dose-dependent manner by LGP2 (Fig. 3A), with inhibition observed with as little as 800 pg of overexpressed LGP2 plasmid and maximum inhibitory effect with 100 ng of LGP2 plasmid.
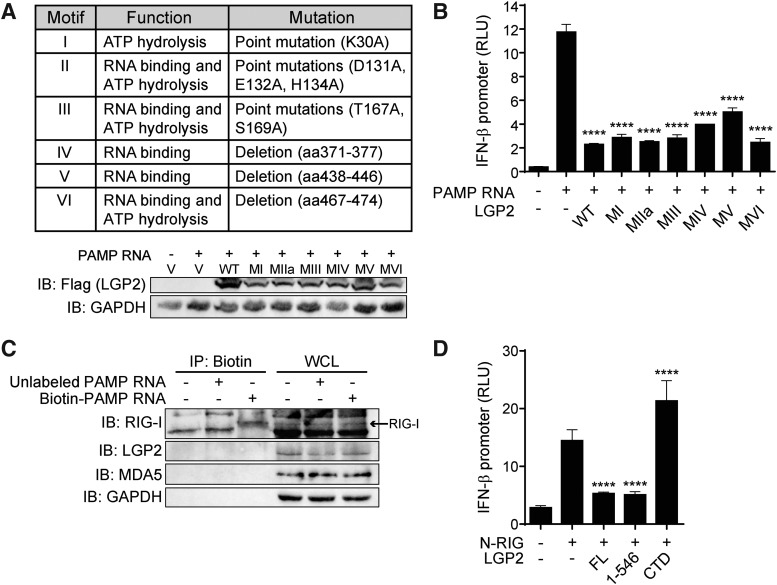
FIG. 3.
LGP2 is a negative regulator of RIG-I signaling in human cells, and the CTD is not required for this function. (A) Left: HEK293 cells were transfected with LGP2 plasmid at increasing doses (1.28 pg, 6.4 pg, 32 pg, 160 pg, 800 pg, 4 ng, 20 ng, and 100 ng) and treated with 100 ng PAMP RNA. Right: HEK293 cells were transfected with LGP2 plasmid at increasing doses (100 pg, 1 ng, 10 ng, and 100 ng) and infected with 50HA SeV. Lysates were collected at 6 h post-treatment/infection. Data shown are average RLU of biological triplicates ± SD analyzed by Dunnett's test with comparisons made to PAMP RNA-treated, or SeV-infected, vector control (LGP2 −), P < 0.05. (B) WT (black) and LGP2 knockout (white) HEK293 cells were transfected with 100 ng PAMP RNA and lysates collected at 6 h post-treatment. Data shown are average RLU of biological triplicates ± SD analyzed by Tukey's test, P < 0.05. (C) Flag-tagged LGP2 deletion mutants with lengths indicated by number of amino acids. Defined motifs of the DExD/H box helicase domain are labeled I-VI. IB illustrates deletion mutant expression in HEK293 cells (indicated by asterisks). (D) HEK293 cells were transfected with FL LGP2 or LGP2 deletion mutants and treated with 100 ng PAMP RNA. Lysates were collected at 6 h post-treatment. Data shown are average RLU of biological triplicates ± SD analyzed by Dunnett's test with comparisons made to PAMP RNA-treated vector control (LGP2 −), P < 0.05. Data shown are representative of at least 3 independent experiments. *P < 0.05; ***P < 0.001; ****P < 0.0001. CTD, C-terminal domain; IB, immunoblot; FL, full length.
Similar LGP2 dose-dependent inhibition of IFN-β promoter activity was observed in cells infected with Sendai virus (SeV; Fig. 3A), a virus shown to specifically activate RIG-I signaling (Loo and others 2008; Loo and Gale 2011), and in cells where RIG-I was overexpressed (data not shown). In addition, we observed that IFN-β promoter activity was significantly increased in LGP2-deficient HEK293 cells compared with WT cells (Fig. 3B). Combined, these results confirm our findings in BM-DCs and demonstrate that LGP2 is a negative regulator of RIG-I signaling following PAMP RNA treatment.
Following activation, RIG-I interacts with the central adaptor protein MAVS, which leads to the activation of latent transcription factors, namely IRF-3 and NF-κB. The minimal IFN-β promoter contains binding sites for both IRF-3 (PRD I and PRD III) and NF-κB (PRD II) (Lenardo and others 1989; Visvanathan and Goodbourn 1989; Schafer and others 1998). Thus, we evaluated the specific activities of these promoter sites to determine whether LGP2 attenuates RIG-I signaling downstream of MAVS by inhibiting activation of IRF-3 or NF-κB transcription factors. To this end, we utilized luciferase constructs driven by either IRF-3 or NF-κB alone. We observed a dose-dependent inhibition of both IRF-3- and NF-κB-specific promoter activities with increasing amounts of overexpressed LGP2 in PAMP RNA-treated cells (Supplementary Fig. S1A, B). This was also true for cells infected with SeV (Supplementary Fig. S1C, D). These findings strongly suggest that LGP2 inhibits RIG-I signaling upstream of IRF-3 and NF-κB activation.
The CTD of LGP2 is dispensable for inhibition of RIG-I signaling
We next attempted to define the motifs within LGP2 that are responsible for inhibiting RIG-I signaling. To this end, we generated a panel of LGP2 deletion mutants (Fig. 3C). The helicase domain of LGP2 contains 6 defined motifs responsible for RNA binding and ATP hydrolysis (Figs. 3C, (I–VI) and 4A) (Bamming and Horvath 2009; Bruns and others 2013). In addition, previous studies have implicated the CTD of LGP2 as a regulatory domain that inhibits RIG-I through interaction with the RIG-I helicase domain (Saito and others 2007). When transfected into HEK293 cells, LGP2 1–121 and the LGP2 CTD (amino acids 547–678) failed to inhibit IFN-β promoter activity. LGP2 1–159, 1–176, 1–546, 122–678, 160–678, and 177–678 reduced IFN-β promoter activity compared to empty vector control (LGP2 −, PAMP RNA +), but none inhibited signaling as effectively as FL LGP2 (Fig. 3D). These results indicate that the first 121 amino acids within the N-terminus and the CTD of LGP2 are not sufficient for the negative regulation of RIG-I. However, it appears that multiple regions within the helicase domain are involved in the inhibition of RIG-I signaling.
RNA binding and ATP hydrolysis are dispensable for negative regulation
Previous studies have proposed that LGP2 binds and sequesters RNA ligands to prevent RIG-I activation (Rothenfusser and others 2005; Yoneyama and others 2005). In addition, LGP2 hydrolyzes ATP to increase its affinity for particular RNA substrates, allowing it to enhance MDA5 signaling (Bruns and others 2013).
To determine whether these functions play a role in the negative regulation of RIG-I, we evaluated a panel of 6 LGP2 constructs, each with mutations within one of the motif domains that result in the loss of motif function (MI-MVI, Fig. 4A) (Bamming and Horvath 2009). We found that all 6 mutants retained the ability to inhibit IFN-β promoter activity when introduced into HEK293 cells (Fig. 4B). Of particular significance, the mutations introduced in motif III ablate both the RNA binding and ATP hydrolysis activities of LGP2 (Bamming and Horvath 2009; Bruns and others 2013), yet this MIII mutant was able to inhibit IFN-β promoter activity almost as effectively as WT LGP2, indicating that neither binding PAMP RNA nor ATP hydrolysis is required for inhibition of RIG-I signaling. These results align with the findings of a previous study using alternate agonists (eg, poly(I:C), SeV) which shows that inhibition by LGP2 is independent of its RNA-binding ability (Bamming and Horvath 2009).
FIG. 4.
RNA binding and ATP hydrolysis are dispensable for negative regulation. (A) LGP2 helicase domain motifs, their associated functions, and the mutations made to generate functionally deficient motifs. IB illustrates LGP2 motif mutant expression in HEK293 cells. V, empty vector plasmid control. (B) HEK293 cells were transfected with LGP2 motif mutants (MI-MVI) and treated with 100 ng PAMP RNA. Lysates were collected at 6 h post-treatment. Data shown are average RLU of biological triplicates ± SD analyzed by Dunnett's test with comparisons made to PAMP RNA-treated vector control (LGP2 −), P < 0.05. (C) HEK293 cells were transfected with 1 μg either unlabeled or biotin-labeled PAMP RNA. Lysates were collected at 6 h post-treatment, and IP was performed with streptavidin beads that bind the biotin-labeled RNA. IB was performed for indicated proteins. WCL, whole cell lysate (input control). (D) HEK293 cells were transfected with N-RIG (10 ng) and FL LGP2, LGP2 1–546, or LGP2 CTD for 24 h. Data shown are average RLU of biological triplicates ± SD analyzed by Dunnett's test with comparisons made to N-RIG + vector control (LGP2 −), P < 0.05. Data shown are representative of at least 3 independent experiments. ****P < 0.0001. RLU, relative luciferase units; IP, immunoprecipitation.
To further confirm our findings, we immunoprecipitated biotin-labeled PAMP RNA from transfected cells and found that while RIG-I was pulled down as expected, LGP2 did not bind the PAMP RNA (Fig. 4C), indicating that LGP2 is not negatively regulating RIG-I signaling by sequestering the PAMP RNA ligand. We next evaluated the effects of LGP2 on a constitutively active RIG-I mutant (N-RIG), which consists of only the 2 N-terminal CARDs and lacks the RNA binding helicase domain. We found that FL LGP2 and LGP2 lacking the CTD (aa 1–546), but not the CTD fragment alone (aa 547–678), attenuated N-RIG-induced IFN-β promoter activity (Fig. 4D), indicating that RIG-I binding to RNA is also dispensable for LGP2 inhibition of RIG-I activity. Combined, these findings indicate that LGP2 negatively regulates RIG-I signaling independent of the ability of LGP2 to bind RIG-I ligands but is instead influencing the N-terminal signaling domains.
LGP2 associates with TRIM25
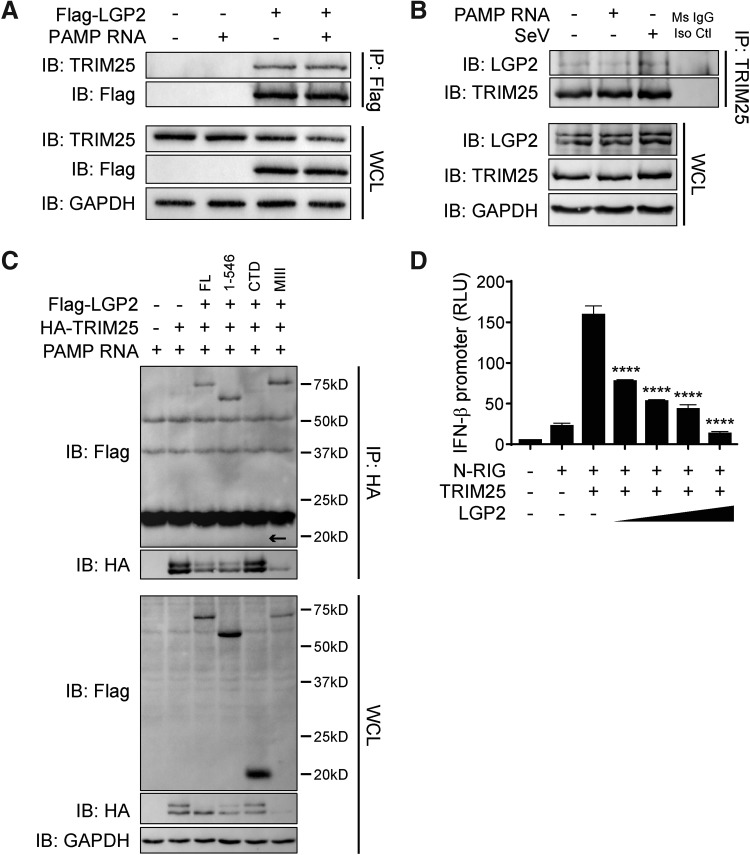
Our data strongly suggest that LGP2 functions after RIG-I binds RNA but before MAVS activation of downstream transcription factors. To more precisely determine the step within the RIG-I signaling pathway that LGP2 is inhibiting, we performed mass spectrometry analysis on immunoprecipitated Flag-LGP2. Through this analysis, we identified tripartite motif-containing protein 25 (TRIM25), an E3 ubiquitin ligase, responsible for the K63-ubiquitination of the RIG-I N-terminus (Gack and others 2007). We confirmed this interaction by overexpressing LGP2 in HEK293 cells and found that LGP2 associates with endogenous TRIM25 (Fig. 5A). Treatment with PAMP RNA did not appear to enhance the interaction between TRIM25 and LGP2, intimating that LGP2 and TRIM25 interact in unstimulated cells. Furthermore, we immunoprecipitated endogenous TRIM25 and confirmed an association with endogenous LGP2 in cells untreated, treated with PAMP RNA, or infected with SeV (Fig. 5B).
FIG. 5.
LGP2 associates with TRIM25. (A) HEK293 cells were transfected with Flag-LGP2 or empty vector control (−) and treated with 100 ng PAMP RNA. Lysates were collected at 6 h post-treatment, and IP was performed for Flag-LGP2. IB was performed for indicated proteins. WCL, whole cell lysate (input control). (B) HEK293 cells were transfected with 100 ng PAMP RNA or infected with 50HA SeV. Lysates were collected at 6 h post-treatment/infection. IP was performed for endogenous TRIM25 using Protein G beads conjugated to anti-TRIM25 antibody or a mouse IgG isotype control antibody (Ms IgG Iso Ctl). IB was performed for indicated proteins. (C) HEK293 cells were transfected with HA-TRIM25 and FL Flag-LGP2, Flag-LGP2 1–546, Flag-LGP2 CTD fragment, or Flag-LGP2 MIII motif mutant and treated with 100 ng PAMP RNA. Lysates were collected at 6 h post-treatment, and IP was performed for HA-TRIM25. IB was performed for indicated proteins. (D) HEK293 cells were transfected with N-RIG (10 ng), TRIM25, and increasing doses of LGP2 (10, 50, 100, and 200 ng) for 24 h. Data shown are average RLU of biological triplicates ± SD analyzed by Dunnett's test with comparisons made to N-RIG+TRIM25+vector control (LGP2 −), P < 0.05. Data shown are representative of at least 3 independent experiments. ****P < 0.0001.
We also observed that the helicase domain of LGP2 (aa 1–546) and the RNA binding/ATP hydrolysis-inactive MIII mutant associated with TRIM25, but that the CTD of LGP2, which did not inhibit RIG-I signaling, did not interact (Fig. 5C). Notably, in instances where LGP2 successfully associated with TRIM25, expression of TRIM25 was noticeably decreased, whereas when the noninteracting LGP2 CTD fragment was coexpressed, TRIM25 levels are similar to that seen when exogenous LGP2 is absent. Finally, we found that LGP2 efficiently inhibits TRIM25-enhanced N-RIG-mediated IFN-β promoter activity in a dose-dependent manner (Fig. 5D).
Previous studies have established that following RIG-I binding to nonself RNA, TRIM25 interacts with RIG-I (Gack and others 2007). Given that LGP2 also interacts with TRIM25, we next investigated whether LGP2 interacts with RIG-I. Previous studies have demonstrated that LGP2 and RIG-I associate when both proteins are ectopically expressed (Komuro and Horvath 2006; Saito and others 2007), a finding which we were able to independently confirm (Supplementary Fig. S2A). We also found that overexpressed LGP2 interacted with endogenous RIG-I, in the presence or absence of stimulatory PAMP RNA (Supplementary Fig. S2B). However, we were unable to demonstrate an interaction between endogenous LGP2 and endogenous RIG-I, in contrast to our finding with endogenous LGP2 and TRIM25 (Supplementary Fig. S2C and Fig. 5B). This may indicate that LGP2 interacts with RIG-I under conditions when both proteins are expressed at high levels, such as late times during signaling activation, or that the interaction between LGP2 and RIG-I is an artifact of overexpression. Nonetheless, our findings demonstrate that FL LGP2 associates with TRIM25 and that the CTD of LGP2 is dispensable for mediating this interaction.
LGP2 suppresses K63-ubiquitination of RIG-I
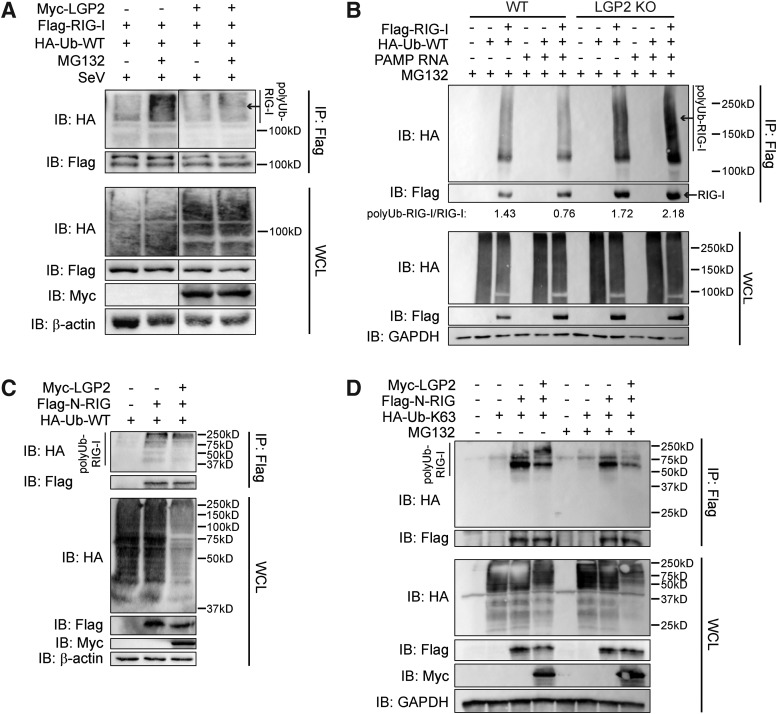
The observed association of LGP2 and TRIM25 led us to evaluate the effects of LGP2 on RIG-I ubiquitination. Upon binding nonself RNA, RIG-I undergoes K63-ubiquitination on its N-terminus to reach a fully active state (Gack and others 2007). To determine the impact of LGP2 on the ubiquitination state of RIG-I, we coexpressed RIG-I, LGP2, and ubiquitin within HEK293 cells. We observed decreased total ubiquitination of RIG-I in these cells (Fig. 6A, IP lane 3) compared with cells transfected with RIG-I and a vector control (Fig. 6A, IP lane 1). To determine whether this decrease was due to increased degradation of RIG-I, we performed this experiment in the presence of MG132, a proteasomal inhibitor. We found that coexpression of LGP2 with RIG-I resulted in a noticeable reduction of ubiquitinated RIG-I even in cells treated with MG132 (Fig. 6A, IP lane 2 vs. 4).
FIG. 6.
LGP2 inhibits K63-ubiquitination of RIG-I. (A) HEK293 cells were transfected with Myc-LGP2, Flag-RIG-I, and HA-ubiquitin (Ub-WT). Cells were treated with MG132 or DMSO control for 2 h and, subsequently, infected with Sendai virus for 8 h. IP was performed for Flag-RIG-I. IB was performed for indicated proteins. Noncontiguous panels are from the same blot. WCL, whole cell lysate (input control). (B) WT and LGP2 knockout HEK293 cells were transfected with Flag-RIG-I and HA-ubiquitin (Ub-WT). Cells were treated with MG132 for 2 h and, subsequently, transfected with 100 ng PAMP RNA. Lysates were collected at 6 h post-treatment, and IP was performed for Flag-RIG-I. IB was performed for indicated proteins. Ratio of poly-ubiquitinated RIG-I (polyUb-RIG-I) to Flag-RIG-I was determined by densitometry and is indicated below IP blots. (C) HEK293 cells were transfected with Myc-LGP2, Flag-N-RIG, and HA-ubiquitin (Ub-WT) for 24 h. IP was performed for Flag-N-RIG. IB was performed for indicated proteins. (D) HEK293 cells were transfected with Myc-LGP2, Flag-N-RIG, and HA-Ub-K63 for 24 h. Cells were treated with MG132 or DMSO control, and IP was performed for Flag-N-RIG. IB was performed for indicated proteins. Data shown are representative of at least 3 independent experiments.
In addition, we observed that the ratio of polyubiquitinated RIG-I to total RIG-I (polyUb-RIG-I/RIG-I) was increased in cells lacking LGP2 compared with WT cells (Fig. 6B, IP lane 6 vs. 12), even under basal conditions (Fig. 6B, IP lane 3 vs. 9). In cells deficient in LGP2, levels of exogenous RIG-I were higher than in WT cells under basal conditions (Fig. 6B, WCL lane 3 vs. 9). Overexpression of RIG-I can induce signaling slightly in the absence of an agonist, and as RIG-I is itself an ISG, this further illustrates the dysregulation of RIG-I signaling in the absence of LGP2. Combined, these findings strongly suggest that LGP2 is negatively regulating the activation of RIG-I rather than destabilizing RIG-I protein expression in HEK293 cells.
To further define this phenotype, we coexpressed LGP2 and ubiquitin with constitutively active N-RIG, which contains the TRIM25 ubiquitination site (Gack and others 2007). Ubiquitination of N-RIG was reduced in the presence of LGP2 (Fig. 6C, IP lane 2 vs. 3), supporting a role for LGP2 in negatively regulating RIG-I activation. Finally, we utilized a ubiquitin mutant that specifically forms K63-linked chains on proteins (Ub-K63). We observed diminished K63-specific ubiquitination of N-RIG in the presence of LGP2 both without and with MG132 treatment (Fig. 6D, lane 3 vs. 4 and lane 7 vs. 8, respectively), which strongly suggests that LGP2 inhibits RIG-I-specific signaling by preventing the K63-ubiquitinaiton of the RIG-I N-terminal CARD through an association with the E3 ubiquitin ligase TRIM25. Inhibition of this post-translational modification prevents RIG-I activation and dampens subsequent innate immune responses during virus infection (Fig. 7).
FIG. 7.
Molecular mechanism of LGP2 negative regulation of RIG-I signaling. Upon binding nonself RNA, RIG-I undergoes conformational changes and post-translational modifications to reach a signaling active state that can interact with MAVS to initiate downstream antiviral signaling. LGP2 negatively regulates RIG-I signaling by inhibiting the TRIM25-mediated K63-ubiquitination step of RIG-I activation. MAVS, mitochondrial antiviral signaling. Color images are available online.
Discussion
In this study, we have elucidated an as yet undescribed role for LGP2 in negatively regulating RIG-I antiviral signaling and BM-DC activation. First, we used primary cells from WT and Dhx58−/− mice and compared the transcriptional responses following RIG-I activation. We found that as early as 1 h post-treatment, Dhx58−/− BM-DCs displayed a more robust antiviral response than WT BM-DCs that was characterized by enhanced expression of type I IFNs, pro-inflammatory cytokines, and antiviral effector genes. This was not due to differences in basal expression of RIG-I between WT and Dhx58−/− BM-DCs. Furthermore, LGP2 is basally expressed in WT BM-DCs and is transcriptionally induced between 1 and 3 h post-treatment, which is subsequent to the induction of type I IFN gene expression (Fig. 1E), indicating that LGP2 functions at early times following RIG-I signaling activation rather than at later time points as suggested by previous studies (Rothenfusser and others 2005). We confirmed that increased type I IFN mRNA expression corresponded to increased type I IFN protein secretion and, subsequently, led to enhanced costimulatory molecule expression on cells lacking LGP2.
Second, we modeled LGP2 function in cultured human cells and confirmed that LGP2 is a negative regulator of RIG-I signaling at a point between RIG-I and MAVS in the signaling cascade. We performed structure function studies and found that the LGP2 CTD was insufficient and dispensable for inhibiting RIG-I signaling. Furthermore, we determined that the PAMP RNA binding and ATP hydrolysis functions of the LGP2 helicase domain were similarly dispensable for reducing RIG-I activity.
Finally, we discovered that LGP2 interacted with the E3 ligase TRIM25 and that LGP2 inhibited TRIM25-mediated K63-ubiquitination of RIG-I. These findings reveal that LGP2 functions at early times to inhibit RIG-I signaling and elucidate the mechanistic underpinnings for how LGP2 negatively impacts RIG-I activation.
Previous studies suggest that LGP2 may compete for and sequester RIG-I ligands to inhibit RIG-I signaling (Rothenfusser and others 2005; Yoneyama and others 2005). However, our in vitro studies demonstrated that neither LGP2 binding to PAMP RNA nor RIG-I binding to PAMP RNA was required for LGP2 inhibition of RIG-I signaling. Furthermore, other studies propose that the CTD of LGP2 directly binds RIG-I and inhibits signaling activation (Saito and others 2007; Pippig and others 2009). It has been shown that the CTD of RIG-I performs an autoregulatory role and that the CTD of LGP2 shares homology with the RIG-I CTD (Saito and others 2007). However, in our studies, we found that the LGP2 CTD fragment (aa 547–678) was insufficient and dispensable for inhibiting RIG-I. It should be noted that our LGP2 CTD deletion mutant is shorter than the CTD fragments used in previous studies - aa 476–678 (Saito and others 2007) and aa 537–678 (Pippig and others 2009). These previous CTD fragments include portions of the LGP2 helicase domain (Cui and others 2008; Li and others 2009; Pippig and others 2009; Takahasi and others 2009). Thus, it is still plausible that these previous versions of the LGP2 CTD may interact with TRIM25 and inhibit RIG-I signaling. However, our LGP2 CTD mutant does not contain potentially interfering fragments from the helicase domain and thus likely represents a truer illustration of LGP2 CTD function.
Our LGP2 deletion mutants containing parts of the helicase domain, with the exception of LGP2 1–121, retained the ability to attenuate RIG-I signaling. However, it is worth noting that no single deletion mutant was as effective as FL LGP2. This likely indicates the importance of a complete, intact tertiary structure and requires further investigation. One report has evaluated the structure of FL LGP2 in the context of RNA binding (Murali and others 2008), but little is known about how this structure influences protein–protein interactions or regulation of RIG-I ubiquitination. It is plausible that the LGP2 helicase domain forms long-distance interactions within LGP2 and that this allows for more efficient binding to TRIM25 and inhibition of RIG-I signaling activation. It is also plausible that LGP2 requires post-translational modifications to interact with TRIM25. Given that post-translational modifications are responsible for both induction and repression of RIG-I and MDA5 activity, it would be informative to determine whether there are similar post-translational modifications, including phosphorylation, ubiquitination, or sumoylation, that regulate the activity of LGP2.
Indeed, post-translational modifications to LGP2 could be a contributing factor to its inhibitory mechanism if LGP2 is itself a target of TRIM25-mediated ubiquitination. It is possible that LGP2 competes with RIG-I for binding and/or ubiquitination by TRIM25. However, considering that the TRIM25 binding site on RIG-I is located within the CARDs (Gack and others 2007), it seems unlikely that LGP2, which lacks CARDs, would be bound by the same region of TRIM25, making competition for binding unlikely. If LGP2 is binding to a different region of TRIM25, it may be that LGP2 limits TRIM25 activation, which requires deubiquitination by USP15 (Pauli and others 2014), or that interaction with LGP2 promotes TRIM25 degradation. Such mechanisms could account for the decrease in HA-TRIM25 expression observed in Fig. 5C.
Alternatively, LGP2 may directly disrupt the E3 ligase function of TRIM25. For instance, if LGP2 binds the N-terminal RING domain of TRIM25, which is responsible for binding E2 ubiquitin-conjugating enzymes (Napolitano and others 2011), it could prevent this necessary step for TRIM25-mediated ubiquitination of target proteins. MAVS is another ubiquitination target of TRIM25. In this case, TRIM25 K48-ubiquitinates the MAVS CARD, targeting it for proteasomal degradation, which is reported to potentiate IRF-3 signaling (Castanier and others 2012). Disruption of MAVS ubiquitination was shown to hinder IRF-3, but not NF-κB signaling. In our studies, we observe inhibition of both IRF-3- and NF-κB-specific signaling in the presence of LGP2; however, the effect on IRF-3 signaling is more dramatic. Thus, it seems plausible that LGP2 is also affecting other substrates of TRIM25-mediated ubiquitination, including MAVS.
This role of LGP2 in interfering with E3 ligase activity may not be limited to inhibition of TRIM25. Another study described an interaction between LGP2 and the E3 ubiquitin ligase TRAF family proteins (Parisien and others 2018). In this case, LGP2 bound TRAFs within the domain responsible for interacting with ubiquitination target proteins. It was determined that LGP2 interfered with TRAF E3 ligase function, preventing TRAF auto-K63-ubiquitination and thus inhibiting downstream antiviral signaling.
LGP2 has been implicated in regulating a diverse array of cellular processes, including regulating RLR signaling, depressing RNAi processes (Komuro and others 2016; van der Veen and others 2018), promoting CD8+ T cell responses during virus infection (Suthar and others 2012), and inhibiting apoptosis of cancer cells subjected to ionizing radiation (Suthar and others 2012; Widau and others 2014). In addition, RLR agonists are being developed and considered as potential broad-spectrum antiviral therapeutics (Martinez-Gil and others 2013; Olagnier and others 2014) and as adjuvants to enhance immunogenicity during vaccination (Saito and others 2008; Chakravarthy and others 2010). Therefore, it is crucial to understand how the RLR signaling pathway is regulated to successfully augment immunogenicity and also prevent cytokine-mediated tissue damage (Clyde and others 2006; Sun and others 2012; Oldstone and Rosen 2014). As a key regulator of RLR signaling, LGP2 is central to understanding the regulation of innate and adaptive immune responses. A more complete knowledge of LGP2 function would aid in developing new antiviral therapies and vaccines or additional treatments to improve the efficacy of existing anticancer therapies.
Supplementary Material
Acknowledgments
The authors acknowledge the expertise of the Yerkes Genomics Core in performing mRNA-sequencing, the Emory Integrated Proteomics Core in performing mass spectrometry, and the Pediatrics Department Flow Core for the upkeep and use of their flow cytometry and multiplex bead assay machines. The authors thank the EVC Flow Cytometry Core, which is supported, in part, by the National Institutes of Health grant P30A050509.
This work was funded, in part, by National Institutes of Health grants U19AI083019 (M.S.S), R56AI110516 (M.S.S), R21AI113485 (M.S.S.), U01AI131566 (M.S.S.), and R01GM111652 (C.M.H.) and Children's Health care of Atlanta, Emory Vaccine Center, Junior Faculty Focused award and The Georgia Research Alliance (M.S.S), and P510D11132 to Yerkes National Primate Research Center. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Author Disclosure Statement
There are no conflicts of interest with regard to the material contained within this article.
Supplementary Material
References
- Bamming D, Horvath CM. 2009. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J Biol Chem 284(15):9700–9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns AM, Horvath CM. 2012. Activation of RIG-I-like receptor signal transduction. Crit Rev Biochem Mol Biol 47(2):194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns AM, Leser GP, Lamb RA, Horvath CM. 2014. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol Cell 55(5):771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns AM, Pollpeter D, Hadizadeh N, Myong S, Marko JF, Horvath CM. 2013. ATP hydrolysis enhances RNA recognition and antiviral signal transduction by the innate immune sensor, laboratory of genetics and physiology 2 (LGP2). J Biol Chem 288(2):938–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanier C, Zemirli N, Portier A, Garcin D, Bidere N, Vazquez A, Arnoult D. 2012. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol 10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy KV, Bonoiu AC, Davis WG, Ranjan P, Ding H, Hu R, Bowzard JB, Bergey EJ, Katz JM, Knight PR, Sambhara S, Prasad PN. 2010. Gold nanorod delivery of an ssRNA immune activator inhibits pandemic H1N1 influenza viral replication. Proc Natl Acad Sci U S A 107(22):10172–10177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs KS, Randall RE, Goodbourn S. 2013. LGP2 plays a critical role in sensitizing mda-5 to activation by double-stranded RNA. PLoS One 8(5):e64202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyde K, Kyle JL, Harris E. 2006. Recent advances in deciphering viral and host determinants of dengue virus replication and pathogenesis. J Virol 80(23):11418–11431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. 2008. The C-terminal regulatory domain is the RNA 5'-triphosphate sensor of RIG-I. Mol Cell 29(2):169–179 [DOI] [PubMed] [Google Scholar]
- Cui Y, Li M, Walton KD, Sun K, Hanover JA, Furth PA, Hennighausen L. 2001. The Stat3/5 locus encodes novel endoplasmic reticulum and helicase-like proteins that are preferentially expressed in normal and neoplastic mammary tissue. Genomics 78(3):129–134 [DOI] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446(7138):916–920 [DOI] [PubMed] [Google Scholar]
- Komuro A, Homma Y, Negoro T, Barber GN, Horvath CM. 2016. The TAR-RNA binding protein is required for immunoresponses triggered by Cardiovirus infection. Biochem Biophys Res Commun 480(2):187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A, Horvath CM. 2006. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J Virol 80(24):12332–12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenardo MJ, Fan CM, Maniatis T, Baltimore D. 1989. The involvement of NF-kappa B in beta-interferon gene regulation reveals its role as widely inducible mediator of signal transduction. Cell 57(2):287–294 [DOI] [PubMed] [Google Scholar]
- Li X, Ranjith-Kumar CT, Brooks MT, Dharmaiah S, Herr AB, Kao C, Li P. 2009. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J Biol Chem 284(20):13881–13891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M., Jr. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol 82(1):335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Gale M., Jr. 2011. Immune signaling by RIG-I-like receptors. Immunity 34(5):680–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Gil L, Goff PH, Hai R, Garcia-Sastre A, Shaw ML, Palese P. 2013. A Sendai virus-derived RNA agonist of RIG-I as a virus vaccine adjuvant. J Virol 87(3):1290–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali A, Li X, Ranjith-Kumar CT, Bhardwaj K, Holzenburg A, Li P, Kao CC. 2008. Structure and function of LGP2, a DEX(D/H) helicase that regulates the innate immunity response. J Biol Chem 283(23):15825–15833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano LM, Jaffray EG, Hay RT, Meroni G. 2011. Functional interactions between ubiquitin E2 enzymes and TRIM proteins. Biochem J 434(2):309–319 [DOI] [PubMed] [Google Scholar]
- Olagnier D, Scholte FE, Chiang C, Albulescu IC, Nichols C, He Z, Lin R, Snijder EJ, van Hemert MJ, Hiscott J. 2014. Inhibition of dengue and chikungunya virus infections by RIG-I-mediated type I interferon-independent stimulation of the innate antiviral response. J Virol 88(8):4180–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldstone MB, Rosen H. 2014. Cytokine storm plays a direct role in the morbidity and mortality from influenza virus infection and is chemically treatable with a single sphingosine-1-phosphate agonist molecule. Curr Top Microbiol Immunol 378:129–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien JP, Lenoir JJ, Mandhana R, Rodriguez KR, Qian K, Bruns AM, Horvath CM. 2018. RNA sensor LGP2 inhibits TRAF ubiquitin ligase to negatively regulate innate immune signaling. EMBO Rep 19(6): pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli EK, Chan YK, Davis ME, Gableske S, Wang MK, Feister KF, Gack MU. 2014. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci Signal 7(307):ra3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pippig DA, Hellmuth JC, Cui S, Kirchhofer A, Lammens K, Lammens A, Schmidt A, Rothenfusser S, Hopfner KP. 2009. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res 37(6):2014–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol 175(8):5260–5268 [DOI] [PubMed] [Google Scholar]
- Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A 104(2):582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr. 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454(7203):523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. 2010. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A 107(4):1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer SL, Lin R, Moore PA, Hiscott J, Pitha PM. 1998. Regulation of type I interferon gene expression by interferon regulatory factor-3. J Biol Chem 273(5):2714–2720 [DOI] [PubMed] [Google Scholar]
- Schnell G, Loo YM, Marcotrigiano J, Gale M., Jr. 2012. Uridine composition of the poly-U/UC tract of HCV RNA defines non-self recognition by RIG-I. PLoS Pathog 8(8):e1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jin C, Zhan F, Wang X, Liang M, Zhang Q, Ding S, Guan X, Huo X, Li C, Qu J, Wang Q, Zhang S, Zhang Y, Wang S, Xu A, Bi Z, Li D. 2012. Host cytokine storm is associated with disease severity of severe fever with thrombocytopenia syndrome. J Infect Dis 206(7):1085–1094 [DOI] [PubMed] [Google Scholar]
- Suthar MS, Ramos HJ, Brassil MM, Netland J, Chappell CP, Blahnik G, McMillan A, Diamond MS, Clark EA, Bevan MJ, Gale M., Jr. 2012. The RIG-I-like receptor LGP2 controls CD8(+) T cell survival and fitness. Immunity 37(2):235–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K, Kumeta H, Tsuduki N, Narita R, Shigemoto T, Hirai R, Yoneyama M, Horiuchi M, Ogura K, Fujita T, Inagaki F. 2009. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem 284(26):17465–17474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen AG, Maillard PV, Schmidt JM, Lee SA, Deddouche-Grass S, Borg A, Kjaer S, Snijders AP, Reis ESC. 2018. The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells. EMBO J 37(4): pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol 178(10):6444–6455 [DOI] [PubMed] [Google Scholar]
- Visvanathan KV, Goodbourn S. 1989. Double-stranded RNA activates binding of NF-kappa B to an inducible element in the human beta-interferon promoter. EMBO J 8(4):1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widau RC, Parekh AD, Ranck MC, Golden DW, Kumar KA, Sood RF, Pitroda SP, Liao Z, Huang X, Darga TE, Xu D, Huang L, Andrade J, Roizman B, Weichselbaum RR, Khodarev NN. 2014. RIG-I-like receptor LGP2 protects tumor cells from ionizing radiation. Proc Natl Acad Sci U S A 111(4):E484–E491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wies E, Wang MK, Maharaj NP, Chen K, Zhou S, Finberg RW, Gack MU. 2013. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity 38(3):437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr., Akira S, Yonehara S, Kato A, Fujita T. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol 175(5):2851–2858 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.