Abstract
Objective:
To investigate the association of specific amino acid positions, residues and haplotypes of HLA-DRB1 in black South Africans with autoantibody-positive RA.
Methods:
High resolution HLA-DRB1 genotyping was performed in 266 autoantibody-positive RA black South Africans and 362 ethnically and geographically matched controls. The alleles were converted to specific amino acid residues at polymorphic sites for downstream analyses. Logistic regression models were used to test whether variability at site, specific amino acid residues and haplotypes (constructed from positions 11, 71, and 74) were associated with RA.
Results:
Of the 29 amino acid positions examined, positions 11, 13 and 33 (permutation P = 3.4e−26, 1.2e−27 and 2.1e−28, respectively) showed the strongest association with RA. Univariate analyses of individual amino acid residues showed valine at position 11 (OR= 5.1 (±95% CI: 3.7–7.0)) and histidine at position 13 (OR=6.1 (±95% CI: 4.2–8.6)) conferred the highest risk. The valine containing haplotypes of position 11, 71, 74, V_K_A conferred the most risk (OR=4.52 (±95% CI: 2.68–7.61)) and conversely the haplotype with serine at this position, S_K_R conferred the most protection, (OR=0.83 (±95%CI: 0.61–1.15)).
Conclusion:
Autoantibody-positive RA in black South Africans is associated with histidine at position 13 and valine at position 11 of HLA-DRB1 and haplotypes with valine at position 11 conferred the highest risk; conversely, serine at position 11 conveyed protection.
Keywords: valine, serine, haplotypes, anti citrullinated peptide antibody
The genetic association of HLA-DRB1 alleles that include the ‘shared epitope’ (SE), a conserved sequence of amino acids (QKRAA, QRRAA, or RRRAA) at positions 70–74 of DRB1 with rheumatoid arthritis (RA) is well established in most populations (1, 2) including black South Africans (4, 5).
Approximately 90% of black South Africans with RA carry at least one allele bearing the SE motif (6). Moreover, in a genome wide association study using the Immunochip SNP array, the HLA region showed the strongest association with RA in this population (7). These findings contrast with studies in other populations of African ancestry, where the frequency of the SE in RA is much lower, 42% in African Americans (8) and only 30% in west Africans from the Cameroon (9).
Using a more refined and novel approach to study the role of the major histocompatibility complex region in RA, Raychaudhuri et al found that just 5 amino acids in 3 HLA proteins, HLA-DRB1, HLA-DP1 and HLA-B, explain most of the genetic association in anti-citrullinated peptide antibody (ACPA) positive Caucasian RA patients (10). The strongest association was with position 11 of HLA-DRB1 and less so with the previously associated SE positions, 70 and 74. Certain haplotypes of amino acid residues at positions 11, 71 and 74 were shown to be associated with risk or protection. Specifically, haplotypes with a valine residue at position 11 were associated with a 4-fold increased risk of RA and conversely, haplotypes with a serine residue at this position showed a reduced risk (10).
In African Americans, like in Caucasians, valine at amino acid position 11 of DRB1 conferred the strongest risk for RA, in addition, an association with position 57 was also observed, but no association was identified with positions 71 and 74 (11).
In the absence of studies on amino acid substitutions in sub-Saharan African populations, we sought to determine the association of specific amino acid positions, residues and haplotypes in the HLA-DRB1 region in black South Africans with antibody-positive RA.
Materials and Methods
RA patients fulfilling the 1987 American College of Rheumatology classification criteria for RA, ≥18 years at disease onset and who were antibody-positive rheumatoid factor ((RF) and/or ACPA+ (n=266) were recruited from the Rheumatology Clinic, Chris Hani Baragwanath Academic Hospital, Soweto, South Africa. The control participants (n=362) were ethnically and geographically matched and consisted of either hospital staff members or patients presenting to the Accident and Emergency Department for minor trauma, but with no history of joint symptoms or autoimmune diseases. Black ethnicity was defined on the basis of participants self-reporting all 4 grandparents as being black South Africans. Written consent was obtained from all participants. The study was approved by the Human Research Ethics Committee (Medical) of the University of the Witwatersrand (M10707).
Rheumatoid factor (composite IgM, IgG, IgA) was assayed by nephelometry (Siemens Healthcare Diagnostics, BN Prospec Nephelometer, Newark, USA). The ACPA status was determined using the anti-CCP2 antibody immunofluorimetric assay (Phadia AB, Uppsala, Sweden). The tests were considered positive when the values were >15 IU/ml for the RF test and 10 U/m for the anti-CCP2 test.
HLA-DRB1 genotyping and conversion of classical alleles to amino acid residues
Four-digit high resolution HLA typing was performed by DNA sequencing of exon 2, using the AlleleSEQR HLA-DRB1 reagent kit and protocol (Atria Genetics, South San Francisco, CA) as previously described (7). The sequences were analyzed using Assign software (Conexio Genomics, Fremantle, Western Australia, Australia), which enables assignment of genotypes based on a library file of HLA-DRB1 alleles (12). This method detects all of the SE positive alleles.
The SNP to HLA tool was used to impute the amino acid polymorphisms using a reference set of HLA alleles in the context of adjacent SNPs genotyped directly on the Immunochip array. Phased and imputed haplotypes generated using BEAGLE, implemented within the SNP2HLA program, were used to assign the amino acid residues at positions 11, 71, and 74 (13).
Statistical analysis
Amino acid position analysis:
To determine whether a specific amino acid position, containing “n” alternate residues, was significantly associated with RA, logistic regression models were fit with the sum of “n-1” indicator variables of the presence/absence of particular amino acid residues. This was repeated for all positions with polymorphic residues, e.g., valine, serine and leucine. To determine if additional variation was explained by other positions after accounting for the most statistically significant positions (e.g. 11, 13, 33), conditional logistic regression was used. In these models, the P-value of the test where additional explanatory covariance is explained by a given position was obtained by comparing two models: 1) a base model with only the most significant position residues, and 2) the base model plus additional residues from the position in question. The P-value was determined by considering the difference in residual deviance between two models as χ2 with degrees of freedom equal to the difference in number residues between the two models. We also performed a sensitivity analysis using a dummy variable for sex coded as {0,1: male, female) for the positions (11, 13) corresponding to HLA-DRB1 allele *04:01, which was the only allele showing indications of population structure (see results).
Haplotype analysis:
Logistic regression was used to fit the RA case control data to the haplotype information. The log likelihood ratio test was applied to assess the signal from the haplotypes. As 12 haplotypes were observed for positions 11, 71 and 74, a conservative test type 1 error correction was made so that only P -values < 0.004 were considered statistically significant. Univariate logistic models were also fit to the data to test for frequency differences between cases and controls.
Results
Most patients were females (89.7%) with a mean (±SD) age of 55 (10.8) years and established disease with a mean (±SD) symptom duration of 7.2 (8.9) years. All patients were antibody-positive, either for RF (n=234/248) or ACPA (n=195/217). Not all patients were tested for both autoantibodies.
Twenty-eight different 4-digit HLA-DRB1 alleles were identified in the controls and 26 in the cases were identified, of which 17 alleles occurred at low frequencies (<0.05). Compared to the control group, the frequencies of 4 alleles were significantly higher in the patient groups, namely, *0401, *0404, *0405 and *1001, whilst the frequencies of 3 alleles were significantly lower, namely, *1101, *1301 and *1302 (Table 1). The only allele with frequency differences observed between males and females was *0401 (c2 = 8.71, DF = 1, P=0.003).
Table 1.
HLA-DRB1 four digit alleles with RA (Odds Ratio (OR)) and 95% Confidence Interval (CI))
| HLA DRB1 allele | Cases (n=261)* Allele count (allele frequency) | Controls (n=362) Allele count (allele frequency) | RA Association Multivariable OR (95% Cl) | P value |
|---|---|---|---|---|
| 0101 | 1 (0.0) | 2 (0.0) | ||
| 0102 | 26 (0.04) | 30 (0.04) | ||
| 0201 | 1 (0.0) | 0 (0.0) | ||
| 0301 | 28 (0.05) | 63 (0.08) | ||
| 0302 | 50 (0.09) | 73 (0.10) | ||
| 0401 | 63 (0.12) | 24 (0.03) | 4.0 (2.5–6.5) | <0.000 1 |
| 0402 | 1 (0.0) | 0 (0.0) | ||
| 0404 | 67 (0.13) | 15 (0.02) | 6.9 (3.9–12.2) | <0.000 1 |
| 0405 | 14 (0.02) | 4 (0.01) | 4.96 (1.6–15.2) | 0.0018 |
| 0408 | 1 (0.0) | 0 (0.0) | ||
| 0410 | 3 (0.0) | 2 (0.0) | ||
| 0701 | 30 (0.06) | 41 (0.06) | ||
| 0804 | 13 (0.02) | 30 (0.04) | ||
| 0901 | 8 (0.02) | 10 (0.01) | ||
| 1001 | 27 (0.05) | 21 (0.02) | 1.8 (1.0–3.3) | 0.039 |
| 1101 | 41 (0.08) | 101 (0.14) | 0.5 (0.4–0.8) | 0.0008 |
| 1102 | 12 (0.02) | 36 (0.05) | ||
| 1114 | 2 (0.0) | 1 (0.0) | ||
| 1201 | 19 (0.04) | 28 (0.04) | ||
| 1202 | 1 (0.0) | 4 (0.0) | ||
| 1301 | 40 (0.08) | 92 (0.13) | 0.6 (0.4–0.8) | 0.004 |
| 1302 | 21 (0.04) | 47 (0.06) | 0.6 (0.4–1.0) | 0.06 |
| 1303 | 6 (0.01) | 14 (0.02) | ||
| 1336 | 0 (0.0) | 1 (0.0) | ||
| 1340 | 0 (0.0) | 1 (0.0) | ||
| 1401 | 2 (0) | 8 (0.01) | ||
| 1404 | 1 (0.0) | 0 (0.0) | ||
| 1417 | 2 (0.0) | 0 (0.0) | ||
| 1501 | 10 (0.02) | 14 (0.02) | ||
| 1503 | 32 (0.06) | 61 (0.08) | ||
| 1506 | 0 (0.0) | 1 (0.0) | ||
| Total | 522 | 724 |
5 cases were not typed successfully
Of the 29 amino acid positions examined, amino acid position 11, 13 and 33 were the most highly associated with RA (p=3.4×10−26, 1.2×10−27 and 2.1×10−28, respectively). Position 71 was less significantly associated (p = 2.6×10−05) and there was no significant association with position 74 (p = 0.15). The relationship of the amino acid positions 11 and 13 were corrected for sex, which exhibit some apparent population structure at the *0401 allele, and the effects remained highly significant (1.3×10−23,p = 1.2×10−23, respectively). After conditioning on position 11, the effects of the other positions were diminished (position 13: p = 0.015; position 33: p = 0.0043) (Table 2). Conditioning on position 13 rendered all amino acid positions non-significant, which indicates the strongest signal of association with RA in Black South Africans arises from positions 11 and 13.
Table 2.
RA associations with variation at amino acids before and after conditioning on the most highly associated amino acid positions 11, 13 and 33.
| Amino acid position | RA associated p value | p value after conditioning on position 11 | p value after conditioning on position 13 | p value after conditioning on position 33 |
|---|---|---|---|---|
| 33 | 2.14E-28 | 0.0043 | N/A | -- |
| 13 | 1.16E-27 | 0.015 | -- | 0.0072 |
| 11 | 3.39E-26 | -- | 0.49 | 0.0093 |
| 12 | 7.74E-17 | N/A | N/A | 0.0027 |
| 10 | 7.88E-16 | 0.004 | 0.27 | 0.0013 |
| 47 | 8.30E-14 | 0.35 | 0.39 | 0.0094 |
| 67 | 3.11E-13 | 0.27 | 0.27 | 0.048 |
| 70 | 5.78E-10 | 0.007 | 0.28 | 0.0053 |
| 37 | 2.30E-05 | 0.43 | 0.47 | 0.69 |
| 71 | 2.58E-05 | 0.7 | 0.84 | 0.19 |
| 58 | 3.84E-05 | 0.32 | 0.38 | 0.062 |
| 32 | 0.0003 | 0.96 | 0.072 | 0.54 |
| 40 | 0.04 | 0.004 | 0.27 | 0.004 |
| 26 | 0.08 | 0.48 | 0.72 | 0.012 |
| 31 | 0.08 | 0.004 | 0.27 | 0.001 |
| 73 | 0.1 | 0.16 | 0.13 | 0.43 |
| 38 | 0.12 | 0.009 | 0.28 | 0.013 |
| 74 | 0.15 | 0.53 | 0.27 | 0.68 |
| 16 | 0.18 | 0.68 | N/A | 0.84 |
| 86 | 0.19 | 0.32 | 0.88 | 0.67 |
| 57 | 0.33 | 0.73 | 0.67 | 0.38 |
| 60 | 0.56 | 0.58 | 0.65 | 0.30 |
| 30 | 0.57 | 0.69 | 0.68 | 0.32 |
| 28 | 0.72 | 0.46 | 0.056 | 0.00044 |
| 9 | 0.73 | N/A | 0.49 | 0.17 |
| 85 | 0.86 | 0.32 | 0.91 | 0.15 |
| 14 | 0.95 | N/A | N/A | 0.49 |
| 25 | 0.95 | N/A | N/A | 0.49 |
| 78 | 0.96 | N/A | 0.95 | 0.22 |
N/A means that the effect at the position in question was completely collinear with the conditioned variable, i.e., there were no polymorphic residues to test with case/ control status after conditioning on a position.
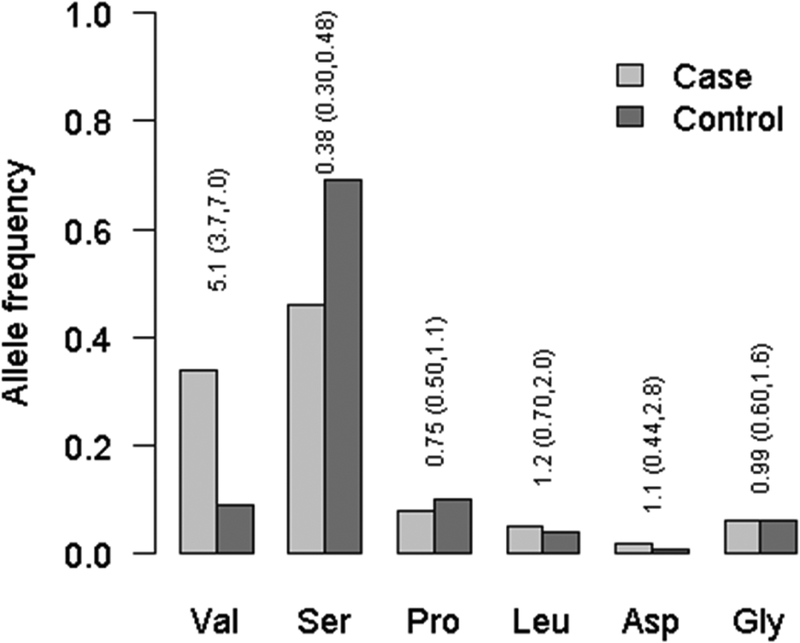
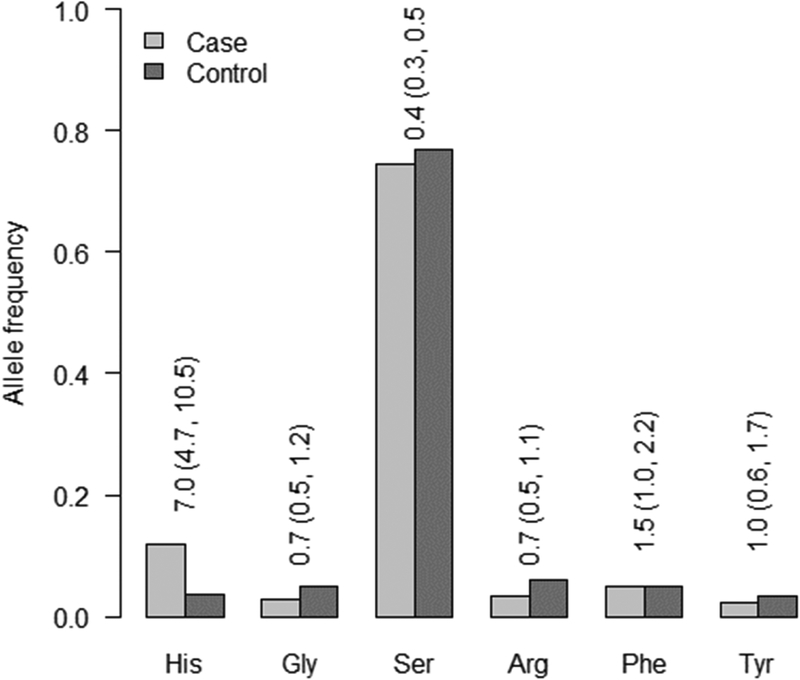
At position 11, with proline as the referent amino acid residue, valine, conferred the strongest risk (OR=5.1 (±95%CI:3.74–6.97) and conversely, serine had a lower frequency in the RA group compared to the control group (Figure 1). At position 13 the strongest risk residue was histidine (OR=6.1 (±95%CI: 4.2–8.6)) and the serine residue appeared protective.
Figure 1.
Amino acid allele frequencies of HLA-DRB1 at position 11 between black South African cases and controls. The odds ratios (95% CI) reported are for univariate tests of allelic associations with RA.
The two haplotypes constructed based on positions 11, 71, 74, with a valine residue at position 11 (V_K_A (OR (CI) 4.52 (2.68–7.61)) and V_R_A (OR (CI) 4.44 (3.01–6.56)) were the most strongly associated with risk and three haplotypes with serine at position 11 conferred protection (Table 3).
Table 3.
The univariate association of HLA-DR1 haplotypes constructed from amino acid at positions 11, 71 and 74.
| Haplotype | Haplotype frequencies | Haplotype associations OR (95%CI) | |
|---|---|---|---|
| Case | Control | ||
| V_K_A | 0.12 | 0.02 | 4.52 (2.68–7.61)** |
| V_R_A | 0.22 | 0.06 | 4.44 (3.01–6.56)** |
| D_R_E | 0.02 | 0.01 | 1.44 (0.53–3.94) |
| L_R_A | 0.05 | 0.05 | 1.08 (0.62–1.87) |
| G_R_Q | 0.05 | 0.06 | 0.99 (0.59–1.66) |
| S_K_R | 0.15 | 0.18 | 0.83 (0.61–1.15) |
| P_A_A | 0.08 | 0.1 | 0.77 (0.51–1.16) |
| S_R_A | 0.13 | 0.20 | 0.59 (0.43–0.82)** |
| S_K_A | 0.01 | 0.02 | 0.55 (0.19–1.57) |
| S_E_A | 0.14 | 0.24 | 0.52 (0.38–0.71)** |
| S_R_L | 0.02 | 0.04 | 0.47 (0.22–0.98)* |
| S_R_E | 0.004 | 0.01 | 0.36 (0.07–1.73) |
V=Valine, S=Serine, A=Alanine, R=Arginine, E=Glutamic acid, K=Lysine, D=Aspartic acid
p<0.004 (Bonferroni adjusted for 12 tested)
Haplotypes containing valine at position 11 and histidine at position 13 showed that histidine is in perfect linkage with valine, but the valine residue at position 11 may occur with either histidine residues (haplotype frequency (cases and controls) = 0.15) or phenylalanine residues (0.038) at position 13.
Discussion
In the present study, we validated several of the previously observed associations of HLA-DRB1 alleles, amino acid positions, residues and haplotypes with antibody-positive RA in black South Africans. Amino acid positions, 11, 13, and 33 were associated with the highest risk for RA. Due to the high proximity and linkage among the amino acid residue polymorphisms it was difficult to locate the exact source of the signal. Using conditional tests and permutation p-values we evaluated the evidence that the source of the signal was coming from a particular site. Conditioning on position 13 completely nullified the effects of position 33. However, conditioning on position 33 left some residual signal at positions 11 and 13. Finally, conditioning on position 11, left some residual signal at positions 13 and 33. These results suggest that the signal in polymorphism from position 33, a biallelic site, is probably stemming from linkage with the signal coming from positions 13 and 11.
Haplotypes containing valine at position 11 and histidine at position 13 showed that histidine is in perfect linkage with valine, but the valine residue at position 11 may occur with either histidine residues (haplotype frequency (cases and controls) = 0.15) or phenylalanine residues (0.038) at position 13. Because there remains variability in high risk allele containing haplotypes at these amino acid positions, covariance between the positions and case-control status remains.
Unlike the finding in RA patients of Caucasian ancestry, we found no association with positions 71 and 74, after conditioning for position 11 (or 13 and 33). We also did not observe an association with position 57, previously reported in African-Americans (11). Possible reasons for a lack of association include inter-ethnic differences in specific HLA-DRB1 allele frequencies, for example, the lack of association of a Caucasian risk allele, DRB1 *0408 in our cohort. Association differences between black South Africans and African Americans are not totally unexpected as we previously showed that black South Africans are not only genetically distinct from Caucasians but also from West Africans, who are ancestrally related to African-Americans (14). Finally, the present study is relatively small compared to other studies, and thus underpowered to detect potentially small effect sizes of positions 71, 74 and 57.
As in previous studies, among the residues at position 11, valine conferred the highest risk for RA, when analyzed either in terms of individual amino acid residues at position 11 or as part of the position 11, 71, 74 haplotypes. Conversely, serine at position 11 or as part of the haplotype was protective. It should be noted that the absolute frequencies of valine at position 11 and valine containing haplotypes were lower in both our patients and controls compared to those reported in Caucasians, whereas the frequencies of serine at position 11 and serine containing haplotypes were more common in our cohort. These differences are due to the lower frequency of HLA DRB1*0401, *0404, *0405, *0408 and *1001, which have valine at position 11 and the two-fold higher frequency of HLA-DRB1 *1301 and *1302 alleles, which have serine at position 11 in black South Africans.
Among the residues at position 13, histidine conferred nearly double the risk of RA compared to valine at position 11. The serine residue at 13 also had a similarly protective effect as observed at 11 (Figure 2). Mechanistically, the role of positions 11 and 13 and the specific amino acid residues at these positions is not clear. However, position 11 locates to the peptide binding region of HLA class II molecule, suggesting a likely role in antigen presentation. It is the only position with variable residues in the pocket 6 of the beta chain. The increased risk associated with valine at position 11 is thought to be related to its hydrophobic polar state, in contrast to serine which is hydrophilic and highly polar (15). Moreover, a recent study suggests that this interaction is mediated by ACPA positive status (16).
Figure 2.
Amino acid allele frequencies of HLA-DRB1 at position 13 between black South African RA cases and controls. The odds ratios (95% CI) reported are for univariate tests of allelic associations with RA.
The importance of valine at position 11 has been further elucidated in recent studies showing worse radiographic (17) and non-radiographic outcomes in RA patients carrying this amino acid residue at position 11. Worse radiographic damage, higher all-course mortality and poorer response to TNF inhibitor therapy is associated with valine at position 11 of HLA-DRB1 (18). In addition, there is an association with higher swollen joint counts and C reactive protein levels (16).
One of the limitations of the present study was the relatively small sample size. Although adequate to detect the large effects of amino acid positions 11 and 13, the sample size may not have been powered to detect the smaller effects of other amino acid positions.
In conclusion, this work provides further evidence of the role of HLA class II region in genetic susceptibility to RA in black South Africans and more specifically amino acid positions 11 and 13 of HLA DRB1. Further studies are needed to validate these findings, characterize antigen(s) that bind to these sites, and the role of the amino acid residues with respect to response to traditional and commercial disease modifying anti-rheumatic drugs.
Acknowledgements
This work was made possible by grants from Carnegie Corporation of New York (B8749), the Connective Tissue Diseases Fund, University of the Witwatersrand and the Medical Research Council of South Africa to Mohammed Tikly. Michele Ramsay received financial support from the National Research Foundation of South Africa. Richard J Reynolds was supported by NIH grant K01 AR060848.
Footnotes
Disclosures
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
REFERENCES
- 1.Wordsworth BP. HLA class II antigens in susceptibility to rheumatoid arthritis. Br J Rheumatol. 1991;30:151–2. [DOI] [PubMed] [Google Scholar]
- 2.Deighton CM, Walker DJ, Griffiths ID, Roberts DF. The contribution of HLA to rheumatoid arthritis. Clin Genet. 1989;36:178–82. [DOI] [PubMed] [Google Scholar]
- 3.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–13. [DOI] [PubMed] [Google Scholar]
- 4.Mody GM, Hammond MG, Naidoo PD. HLA associations with rheumatoid arthritis in African blacks. J Rheumatol. 1989;16:1326–8. [PubMed] [Google Scholar]
- 5.Pile KD, Tikly M, Bell JI, Wordsworth BP. HLA-DR antigens and rheumatoid arthritis in black South Africans: a study of ethnic groups. Tissue Antigens. 1992;39:138–40. [DOI] [PubMed] [Google Scholar]
- 6.Meyer PW, Hodkinson B, Ally M, Musenge E, Wadee AA, Fickl H, et al. HLA-DRB1 shared epitope genotyping using the revised classification and its association with circulating autoantibodies, acute phase reactants, cytokines and clinical indices of disease activity in a cohort of South African rheumatoid arthritis patients. Arthritis Res Ther. 2011. ;13:R160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Govind N, Choudhury A, Hodkinson B, Ickinger C, Frost J, Lee A, et al. Immunochip identifies novel, and replicates known, genetic risk loci for rheumatoid arthritis in black South Africans. Mol Med. 2014;20:341–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes LB, Morrison D, Kelley JM, Padilla MA, Vaughan LK, Westfall AO, et al. The HLA-DRB1 shared epitope is associated with susceptibility to rheumatoid arthritis in African Americans through European genetic admixture. Arthritis Rheum. 2008;58:349–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singwe-Ngandeu M, Finckh A, Bas S, Tiercy JM, Gabay C. Diagnostic value of anti-cyclic citrullinated peptides and association with HLA-DRB1 shared epitope alleles in African rheumatoid arthritis patients. Arthritis Res Ther. 2010;12:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, Jia X, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynolds RJ, Ahmed AF, Danila MI, Hughes LB, Consortium for the Longitudinal Evaluation of African Americans with Early Rheumatoid Arthritis I, Gregersen PK, et al. HLA-DRB1-associated rheumatoid arthritis risk at multiple levels in African Americans: hierarchical classification systems, amino acid positions, and residues. Arthritis Rheumatol. 2014;66:3274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cano P, Klitz W, Mack SJ, Maiers M, Marsh SG, Noreen H, et al. Common and well-documented HLA alleles: report of the Ad-Hoc committee of the american society for histocompatiblity and immunogenetics. Hum Immunol. 2007;68:392–417. [DOI] [PubMed] [Google Scholar]
- 13.Jia X, Han B, Onengut-Gumuscu S, Chen WM, Concannon PJ, Rich SS, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One. 2013;8:e64683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.May A, Hazelhurst S, Li Y, Norris SA, Govind N, Tikly M, et al. Genetic diversity in black South Africans from Soweto. BMC Genomics. 2013;14:644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Achkar JP, Klei L, de Bakker PI, Bellone G, Rebert N, Scott R, et al. Amino acid position 11 of HLA-DR beta1 is a major determinant of chromosome 6p association with ulcerative colitis. Genes Immun. 2012;13:245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling SF, Viatte S, Lunt M, Van Sijl AM, Silva-Fernandez L, Symmons DP, et al. HLA-DRB1 Amino Acid Positions 11/13, 71, and 74 are associated with inflammation level, disease activity, and the Health Assessment Questionnaire score in patients with Inflammatory polyarthritis. Arthritis Rheumatol. 2016;68:2618–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Steenbergen HW, Raychaudhuri S, Rodriguez-Rodriguez L, Rantapaa-Dahlqvist S, Berglin E, Toes RE, et al. Association of valine and leucine at HLA-DRB1 position 11 with radiographic progression in rheumatoid arthritis, independent of the shared epitope alleles but not independent of anti-citrullinated protein antibodies. Arthritis Rheumatol. 2015;67:877–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viatte S, Plant D, Han B, Fu B, Yarwood A, Thomson W, et al. Association of HLA-DRB1 haplotypes with rheumatoid arthritis severity, mortality, and treatment response. JAMA. 2015;313:1645–56. [DOI] [PMC free article] [PubMed] [Google Scholar]