Abstract
Background
The oral cavity of humans is inhabited by several hundreds of bacterial species and other microorganisms such as fungi and archaeal methanogens. Regarding methanogens, data have been obtained from oral cavity samples collected in Europe, America and Asia. There is no study published on the presence of methanogens in the oral cavity in persons living in Africa. The objective of our study was to bring new knowledge on the distribution of oral methanogens in persons living in Mali, Africa.
Methods
A total of 31 patients were included in the study during a 15-day collection period in September. Bacterial investigations consisted in culturing the bacteria in 5% sheep blood–enriched Columbia agar and PolyViteX agar plates. For archaeal research, we used various methods including culture, molecular biology and fluorescent in situ hybridization (FISH).
Results
Eight of 31 (26%) oral samples collected in eight patients consulting for stomatology diseases tested positive in polymerase chain-reaction (PCR)-based assays for methanogens including five cases of Methanobrevibacter oralis and one case each of Methanobrevibacter smithii, Methanobrevibacter massiliense and co-infection Methanobrevibacter oralis and Methanobrevibacter massiliense.
Conclusions
In this pilot study, we are reporting here the first characterization of methanogens in the oral cavity in eight patients in Mali. These methanogen species have already been documented in oral specimens collected from individuals in Europe, Asia, North America and Brazil.
Keywords: Methanobrevibacter oralis, Methanobrevibacter smithii, Methanobrevibacter massiliense, Methanogen, Oral cavity, Mali, Africa
Introduction
The oral cavity microbiota is considered to be of heterogeneous origin, including various endo- and exogenous species [1–3]. The oral cavity of humans is inhabited by several hundreds of bacterial species and other microorganisms including unicellular eukaryotes and prokaryotes including fungi and Archaea. Some of these microorganisms have a key role in the development of oral diseases, mainly dental caries and periodontitis [2]. Periodontitis is an inflammatory disease resulting from the polymicrobial infection of the subgingival dental plaque by oral bacteria [4]. Archaea are microorganisms classified among one domain of life, different from the ones including bacteria and eukaryotes [5]. Archaea and more specifically methanogen-producing archaea (herein referred as methanogens) are part of the oral microbiota [6] . Methanogens are archaea with a hydrogenotrophic metabolism requiring the presence of hydrogen (H2) to reduce CO2 to methane, a process called methanogenesis [7]. Methanogenesis is a unique metabolic process by which carbon dioxide (CO2) is reduced to methane (CH4) using hydrogen (H2) produced by anaerobic bacterial fermentation as an electron donor [7]. Indeed, six species of methanogens belonging to the genera Methanobrevibacter have been documented in the oral cavity by polymerase chain-reaction (PCR)-based methods and culture, including Methanobrevibacter oralis and Methanobrevibacter smithii, Methanosphaera stadtmanea, Methanosarcina mazeii, Methanobacterium curvum/congolense and Thermoplasmata [8, 9]. Previous studies have shown that M. oralis is highly dominant with a prevalence greater than 40%, while other methanogens have been detected with a lower prevalence of 10 to 20% [8, 10, 11]. In particular, M. oralis has been isolated from the dental plaques of healthy subjects in the oral cavity [12]. One step forward, a recent study disclosed a significant correlation between PCR-detection of methanogens in the oral cavity and tobacco smoking in oral disease-free individuals, illustrating the potential influence of environmental factors of the repertoire of oral cavity methanogens [13]. A molecular study has also shown that methanogens constitute an essential part of the microbiota of the dental root and could participate in the endodontic community in the necrotic root canal [14].
Also, methanogens have been implicated in oral cavity pathologies including Methanobrevibacter massiliense and M. oralis in cases of periimplantitis [4, 15].
All the studies related to the repertoire of methanogens in the oral cavity have been conducted in specimens collected in individuals in Europe, Asia, North America and Brazil. No study issued from individuals in Africa, living unknown whether the methanogens previously documented in the oral cavity are universally distributed over continents or restricted to certain geographic populations. In order to further study this question, we embarked in studying the presence of methanogens and associated bacteria in oral cavity specimens collected from volunteer patients in Mali, Africa.
Methods
Patients and clinical sample collection
This case-series study was reviewed and approved by the Faculty of Medicine, Pharmacy and Odonto-Stomatology Ethics Committee, Bamako, Mali under N°2015/132/CE/FMPOS. This study was conducted in the Stomatology Department of the University Hospital of Bamako, where patients are consulting for oral cavity pathologies with the exclusion of orthodontia. The objective of the study and the sampling protocol were explained to the patients before asking for their consent to participate. A consent document given to each patient and all the patients included in the study, was signed by the patient or one of his parents for children under 18 years. All parental consents were written. Patients who did not sign the contentment form, were excluded from this study. Patients with, gingivitis, periodontitis (either of pulp or peridontal origin) or with dental abscesses who signed the consent form, were included in the study. This is a preliminary and explorative study for a series of pathological clinical cases. The patients were recruited regardless of their recent antibiotics usage status due to its prevalent pre-operative usage in the studied population. This study concerns the presence of methanogens archaea for these cases. Each case was diagnosed by a dentist, before taking samples. Dental X-Ray were made to make the different diagnoses. The classification of periodontal diseases adopted by the dentist was the classification of Armitage et al. [16]. A total of 31 samples of abscesses in 2 cases and dental plaque in 29 cases, were prospectively collected at the University Hospital of Odontology and Stomatology of Bamako in agreed patients who came for odontology consultations. Subgingival plaque samples were collected from each patient by using sterile curettes. The two abscesses were swabbed. These samples were put in a transport medium preserving methanogen viability. This transport medium consisted of (per liter): KCl 0.2 g, CaCl2 0.1 g, MgCl2 0.1 g, KH2PO4 0.2 g, Na2HPO4 1.15 g, NaCl 3 g, ascorbic acid 1 g, uric acid 0.1 g, and glutathione 0.1 g [17]. The pH was adjusted to 7.5 using KOH 10 M and kept at 4 °C. The samples were sent to the IHU Méditerranée Infection for investigating methanogens and bacteria.
Culture of bacteria
Culture of bacteria was performed at 37 °C in 5% sheep blood-enriched Columbia agar and PolyViteX agar (bioMérieux, Marcy l’Etoile, France) under aerobic and anaerobic atmosphere, for 48 h. All microbial colonies that grew on agar plates were identified by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF-MS) using a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) [18, 19]. The bacteria were tested for their capability in hydrogen production using the following protocol. Briefly, bacteria were cultured in a BD Difco™ Brain heart infusion (Fisher Scientific, USA) [20] at 37 °C in Hungate tubes (Dutscher, Issy-les-Moulineaux, France). All tubes were gassed with nitrogen and incubated at 37 °C for 72 h. Hydrogen was detected by gas chromatography (Perkin Elmer, Toulouse, France) as previously described [21].
PCR-sequencing-based detection of methanogens
Methanogens were searched by PCR-sequencing. A 0.3-g quantity of acid-washed beads (B106 mm, Sigma, Saint-Quentin Fallavier, France) was added in each tube containing 250 μL of oral cavity sample (abscess or dental plaque), the suspension was shaken to achieve a mechanical lysis in a FastPrep BIO 101 apparatus (Qbiogene, Strasbourg, France) at level 6.5 for 2 min. Then 200 μL of buffer TL and OB Protease Solution (1.5 mL) from the E.Z.N.A. Tissue DNA Kit (OMEGA, bio-tek, Norcross, USA) were added. The mixture was incubated overnight at 56 °C. After a second cycle of mechanical lysis, the mixture was incubated for 60 min at 70 °C. Extracted DNA was eluted with 100 μL of elution buffer and the DNA was stored at − 20 °C. A sterile PBS was used as a negative control for each batch of DNA extraction (a negative control for eight samples). Amplification of the archaeal 16S rRNA gene (primers used: SDArch0333aS15, 5′-TCCAGGCCCTACGGG-3′ and SDArch0958aA19, 5′-YCCGGCGTTGAMTCCAATT-3′) and the methyl-coenzyme M reducer (mcrA) gene (primers used: mcrAFor, 5’GCTCTACGACCAGATMTGGCTTGG-3′ and mcrARev, 5′ CCGTAGTACGTGAAGTCATCCAGCA − 3′) genes was performed as previously described [22]. Sequencing reactions (Sangers’ method) were carried-out using the Big- Dye Terminator, version 1.1, cycle sequencing kit DNA according to the manufacturer’s instructions (Applied Biosystems, Foster City, USA). Nucleotide sequences were assembled using Chromas Pro software, version 1.7 (Technelysium Pty Ltd., Tewantin, Australia) and compared to the GenBank database by similarity search using the BLASTN program (http://www.ncbi.nlm.nih.gov/blast/).
Fluorescent in situ hybridization detection of methanogens
We used a fluorescent in situ hybridization (FISH) protocol derived from a FISH protocol for bacteria. We used PCR-negative samples for control. A 10 μL-volume of the sample was deposited on a glass slide, allowed to dry in ambient air then fixed with 20 μL of 4% paraformaldehyde for 30 min. Subsequently, a 20 μL-volume of a 10 g/L solution of lysozyme (Sigma) was deposited on the sample and incubated for 30 min at 37 °C further incubated with 5 μL of proteinase K (Sigma) for 5 min at 37 °C. After washing the slide with distilled water, a 10 μL volume of a solution containing 1 μL of the probe Arch915 labeled with Alexa fluor-546 and specific for Archaeal 16S rRNA gene (10 μmoL/L), 1 μL of the mcrA probe (10 μmoL/L), 5 μL of hybridization buffer [4x sodium salt citrate (SSC), 10% of sulfate de dextrane, 1 mM ethylene-diamine-tetraacetic-acid (EDTA), 25% of formamide, 300 ng/mL salmon sperm DNA and 1 x solution de Denhardt] (Sigma-Aldrich), 1 μL of a solution containing 0.1% Tween 20 and 0.1% Triton X-100 (Euromedex, Souffelweyersheim, France) and 2 μL of distilled water. The glass slide was covered with a coverslip glued with Fixogum adhesive (Marabu, Bietigheim-Bissingen, Germany) and incubated at 65 °C for 10 min, then at 37 °C for 20 h. Once the hybridization process was completed, the slide was immersed in a series of SSC baths (4X, 2X, 1X and 0.5X) for 5 min in each bath at room temperature. Finally, the slide was covered with a coverslip and observed at 100X magnification with a fluorescence microscope (Leica DMI 6000, Nanterre, France).
Results
In this study, oral cavity samples were collected from volunteer patients consulting in a tertiary Odontology Department in Bamako, Mali for chronic periodontitis, gingivitis or cellulitis. A total of 31 patients were included in the study during a 15-day collection period in September 2017. Patients included five children, 10 women and 16 men aged from 4 to 63 years. Pathologies were abscesses of pulp origin in 2 cases and dental plaque in 29 cases (12 gingivitis and 17 periodontitis). We did not include any control patients in this case-series study. The samples consisting in 29 dental plaque and two abscesses, were stored in a transport medium [17].
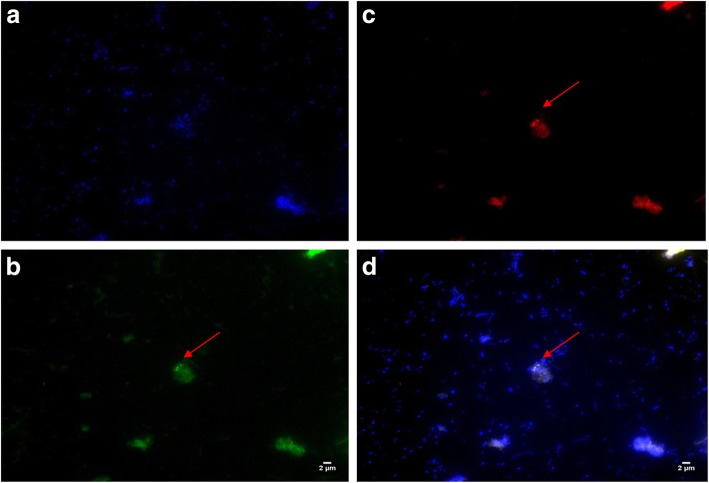
Eight samples (26%) were positive for methanogens by PCR and FISH including five M. oralis, one M. smithii, one M. massiliense and one co-infection with M. oralis and M. massiliense [12, 22], two methanogens which have been previously documented in cases of periodontitis [23–25] and peri-implantitis [10]. These observations were authenticated by the negativity of the negative controls introduced in the different experiments as well as the fact that concordant observations were made using different observation tools. In particular, bacterial culture allowed the isolation of nine different bacterial species Delftia acidovorans, Microbacterium oxydans, Pseudomonas putida, Citrobacter freundii, Brevundimonas aurantiaca, Rhizobium radiobacter, Klebsiella pneumoniae, Microbacterium kitamiense and Peptoniphilus harei (Table 1). By using the gas chromatography method, we detected the presence of hydrogen in the culture of two bacteria previously known as hydrogenogen bacteria including Citrobacter freundii [26] and Klebsiella pneumoniae [27]. However, Peptoniphilus harei and Pseudomonas putida are not known to produce hydrogen. PCR-sequencing confirmed the presence of M. oralis in patients’ n° 2, 3, 4, 6 and 7; of M. smithii in patient n° 5 whose clinical data is unavailable [28] and one co-infection M. oralis and M. massiliense in patient n°1. All these methanogens were identified on the basis of complete sequence identity with references, were detected in association with previously isolated bacteria. The clinical characteristics of the patients and the results are reported in Table 1. These eight PCR-positive samples were all positive by FISH (Fig. 1).
Table 1.
Detailed clinical information of patients with archaea detected in Odonto stomatology, department, Bamako, Mali. NA: Not Available
| Patient | Age range | Locality | Clinical data | Culture | PCR | FISH | Tobacco |
|---|---|---|---|---|---|---|---|
|
1 A 1 B |
[60–70] | Badalabougou | Generalized chronic periodontitis |
Delftia acidovorans, Microbacterium oxydans Pseudomonas putida, Citrobacter freundii, Brevundimonas aurantiaca, Rhizobium radiobacter |
Methanobrevibacter oralis Methanobrevibacter massilense |
Positive | + |
| 2 | [30–40] | Djelibougou | Gingivitis | Pseudomonas putida, Citrobacter freundii, Microbacterium oxydans, | Methanobrevibacter oralis | Positive | – |
| 3 | [60–70] | Darsalam | Moderate generalized chronic periodontitis | Pseudomonas putida, Citrobacter freundii, Brevundimonas aurantiaca, | Methanobrevibacter oralis | Positive | + |
| 4 | [1–10] | Bacodjicoroni | Circumscribed cellulitis of the primary molar 75 (pulpal necrosis) | Citrobacter freundii, Brevundimonas aurantiaca, Delftia acidovorans, Pseudomonas putida, | Methanobrevibacter oralis | Positive | – |
| 5 | [10–20] | Yirimadio | N.A | Klebsiella pneumoniae, Brevundimonas aurantiaca, Microbacterium oxydans, Pseudomonas putida, Delftia acidovorans, | Methanobrevibacter smithii | Positive | – |
| 6 | [60–70] | Daoudabougou | Generalized chronic periodontitis | Brevundimonas aurantiaca, Delftia acidovorans, Citrobacter freundii, Pseudomonas putida, Rhizobium radiobacter, Microbacterium oxydans, Microbacterium kitamiense | Methanobrevibacter oralis | Positive | – |
| 7 | [20–30] | Niarela | Gingivitis | Rhizobium radiobacter, Pseudomonas putida, Peptoniphilus harei, Delftia acidovorans, Klebsiella pneumoniae, | Methanobrevibacter oralis | Positive | – |
| 8 | [40–50] | Korofina nord | Gingivitis |
Pseudomonas putida, Brevundimonas aurantiaca, Delftia acidovorans, |
Methanobrevibacter massilense | Positive | + |
Fig. 1.
Detection of Methanobrevibacter oralis from a chronic periodontitis specimen using FISH. a Blue color represents DAPI fluorescence staining any DNA b Green color represents mcrA probe staining the methanogen mcrA gene c Red color represents ARC915 fluorescence staining the archaeal DNA d Fluorescent in situ hybridization combining mcrA fluorescence, ARC915 fluorescence and DAPI: the arrow points to archaea. Scale bar, 2 μm
Discussion
We are reporting the first characterization of methanogens in the oral cavity in patients living in Africa, more precisely in Mali and we are reporting the first case of co-infection by M. oralis and M. massiliense in the same patient. The observed 20% prevalence of M. oralis is lower than the > 40% prevalence previously reported in various studies [8, 9]. This observation most probably relies on the fact that in Mali, there is a systematic prescription of metronidazole before patient care and oral cavity sampling. Indeed, previous studies have shown that methanogens are sensitive to metronidazole and other imidazoles [29]. Previous studies also showed that M. oralis was significantly associated with periodontal disease in terms of abundance when comparing patients and controls, as well as diseased and healthy sites in the same patient [23].
The repertoire of three methanogen species here reported in oral cavity specimens collected in patients in Mali, Africa is in agreement with the repertoire recently reported from oral cavity specimens collected in individuals in France, Europa [30]; as well as in North America and Brazil, Asia [4, 8, 14]. Accordingly, two of these methanogens species have also been detected in ancient oral cavity specimens recovered from 100 specimens samples dating from the fourteenth and nineteenth century collected in various archaeological sites in France [31].
Altogether, these data suggest that the oral cavity of populations is hosting an universal core methanogen repertoire comprising of at least M. oralis, M. smithii and M. massiliense which is expected to be recovered from oral cavity specimens collected throughout continents and historical periods; albeit with variations in the relative abundance of the various species in relation with the various environments to which populations are exposed.
Conclusions
We are reporting here the first characterization of methanogens in the oral cavity in Africa in in particularly in Mali in eight patients. In addition, we are reporting on one case co-infection with M. oralis and M. massiliense in one patient diagnosed with generalized chronic periodontitis. Further, we observe that M. oralis, M. smithii and M. massiliense are presents in Africa as in other continents, this observation suggests that these three archaea methanogens are frequently represented in situations of periodontitis, whatever the continent.
Limitations
This a preliminary, pilot study.
The fact that only 31 patients were investigated does not allow to draw an actual picture of prevalence and repertoire of oral cavity methanogens in Mali.
The fact that we were not able to isolate methanogens by culture is one limitation of the present study, as the viability of the three methanogens here detected was not confirmed.
Based on the proof-of-concept results here reported, further studies designed to assess the viability of methanogens may prospectively include more individuals in additional African countries to overpass these limitations.
Acknowledgements
Not applicable.
Abbreviations
- EDTA
Ethylene-diamine-tetraacetic-acid
- FISH
Fluorescent in situ hybridization
- MALDI-TOF-MS
Matrix-assisted laser desorption-ionization time-of-flight mass spectrometry
- PCR
Polymerase chain-reaction
- SSC
Sodium salt citrate
Authors’ contributions
ES performed detection of methanogens, analyzed data and drafted the manuscript. OD designed study, analyzed data and drafted the manuscript. GA designed study, analyzed data and drafted the manuscript. ST, OD, BK and HK collected the samples. MD designed study, analyzed data, drafted and approved the final manuscript. All authors read and approved the final manuscript.
Funding
ES benefits a PhD grant from the Fondation Méditerranée Infection, Marseille, France. This work was supported by the French Government under the “Investissements d’avenir” (Investments for the Future) program managed by the Agence Nationale de la Recherche (ANR, fr: National Agency for Research), (reference: Méditerranée Infection 10-IAHU-03). This work was supported by Région Sud (Provence Alpes Côte d’Azur) and European funding FEDER PA 0000319 PRIMMI. These funding bodies had no role in the design of the study, the collection, analysis and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author.
Ethics approval and consent to participate
This study was reviewed and approved by the Faculty of Medicine, Pharmacy and Odonto-Stomatology Ethics Committee, Bamako, Mali under N°2015/132/CE/FMPOS. A consent document given to each patient and all the patients included in the study, was signed by the patient or one of his parents for children under 18 years. All parental consents were written.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Ogobara Doumbo is deceased. This paper is dedicated to his memory.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Elisabeth Sogodogo, Email: elisabethsogodogo@yahoo.fr.
Ogobara Doumbo, Email: okd@icermali.org.
Gérard Aboudharam, Email: gerard.aboudharam@univ-amu.fr.
Bourema Kouriba, Email: kouriba@icermali.org.
Ousseynou Diawara, Email: usseynu@gmail.com.
Hapssa Koita, Email: hapssakoita68@gmail.com.
Souleymane Togora, Email: souleymanetogora@yahoo.fr.
Michel Drancourt, Email: michel.drancourt@univ-amu.fr.
References
- 1.Zawadzki PJ, Perkowski K, Padzik M, Mierzwińska-Nastalska E, Szaflik JP, Conn DB, et al. Examination of Oral microbiota diversity in adults and older adults as an approach to prevent spread of risk factors for human infections. Biomed Res Int. 2017;2017:8106491. doi: 10.1155/2017/8106491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, Romero H, Simón-Soro A, Pignatelli M, et al. The oral metagenome in health and disease. ISME J. 2012;6:46–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atanasova KR, Yilmaz Ö. Prelude to Oral microbes and chronic diseases: past, present and future. Microbes Infect Inst Pasteur. 2015;17:473–483. doi: 10.1016/j.micinf.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamabe K, Maeda H, Kokeguchi S, Tanimoto I, Sonoi N, Asakawa S, et al. Distribution of Archaea in Japanese patients with periodontitis and humoral immune response to the components. FEMS Microbiol Lett. 2008;287:69–75. doi: 10.1111/j.1574-6968.2008.01304.x. [DOI] [PubMed] [Google Scholar]
- 5.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huynh HTT, Nkamga VD, Drancourt M, Aboudharam G. Genetic variants of dental plaque Methanobrevibacter oralis. Eur J Clin Microbiol Infect Dis. 2015;34:1097–1101. doi: 10.1007/s10096-015-2325-x. [DOI] [PubMed] [Google Scholar]
- 7.Triantafyllou K, Chang C, Pimentel M. Methanogens, methane and gastrointestinal motility. J Neurogastroenterol Motil. 2014;20:31–40. doi: 10.5056/jnm.2014.20.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen-Hieu T, Khelaifia S, Aboudharam G, Drancourt M. Methanogenic archaea in subgingival sites: a review. APMIS. 2013;121:467–477. doi: 10.1111/apm.12015. [DOI] [PubMed] [Google Scholar]
- 9.Wilson Michael. The Human Microbiota in Health and Disease. Boca Raton, FL : CRC Press, Taylor & Francis Group, 2019.: Garland Science; 2018. [Google Scholar]
- 10.Faveri M, Gonçalves LFH, Feres M, Figueiredo LC, Gouveia LA, Shibli JA, et al. Prevalence and microbiological diversity of Archaea in peri-implantitis subjects by 16S ribosomal RNA clonal analysis. J Periodontal Res. 2011;46:338–344. doi: 10.1111/j.1600-0765.2011.01347.x. [DOI] [PubMed] [Google Scholar]
- 11.Li CL, Jiang YT, Liu DL, Qian J, Liang JP, Shu R. Prevalence and quantification of the uncommon Archaea phylotype Thermoplasmata in chronic periodontitis. Arch Oral Biol. 2014;59:822–828. doi: 10.1016/j.archoralbio.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 12.Ferrari A, Brusa T, Rutili A, Canzi E, Biavati B. Isolation and characterization of Methanobrevibacter oralis sp. nov. Curr Microbiol. 1994;29:7–12. doi: 10.1007/BF01570184. [DOI] [Google Scholar]
- 13.Grine G, Terrer E, Boualam MA, Aboudharam G, Chaudet H, Ruimy R, et al. Tobacco-smoking-related prevalence of methanogens in the oral fluid microbiota. Sci Rep. 2018;8:9197. doi: 10.1038/s41598-018-27372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brzezińska-Błaszczyk E, Pawłowska E, Płoszaj T, Witas HW, Godzik U, Agier J. Presence of archaea and selected bacteria in infected root canal system. Can J Microbiol. 2018;64:317–326. doi: 10.1139/cjm-2017-0531. [DOI] [PubMed] [Google Scholar]
- 15.Belkacemi S, Mazel A, Tardivo D, Tavitian P, Stephan G, Bianca G, et al. Peri-implantitis-associated methanogens: a preliminary report. Sci Rep. 2018;8:9447. doi: 10.1038/s41598-018-27862-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 17.Khelaifia S, Raoult D, Drancourt M. A versatile medium for cultivating Methanogenic Archaea. PLoS One. 2013;8:e61563. doi: 10.1371/journal.pone.0061563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier P-E, Rolain JM, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 19.Seng P, Rolain J-M, Fournier PE, La Scola B, Drancourt M, Raoult D. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol. 2010;5:1733–1754. doi: 10.2217/fmb.10.127. [DOI] [PubMed] [Google Scholar]
- 20.McKay LF, Holbrook WP, Eastwood MA. Methane and hydrogen production by human intestinal anaerobic bacteria. Acta Pathol Microbiol Immunol Scand [B] 1982;90:257–260. doi: 10.1111/j.1699-0463.1982.tb00114.x. [DOI] [PubMed] [Google Scholar]
- 21.Khelaifia S, Lagier J-C, Nkamga VD, Guilhot E, Drancourt M, Raoult D. Aerobic culture of methanogenic archaea without an external source of hydrogen. Eur J Clin Microbiol Infect Dis. 2016;35:985–991. doi: 10.1007/s10096-016-2627-7. [DOI] [PubMed] [Google Scholar]
- 22.Grine G, Boualam MA, Drancourt M. Methanobrevibacter smithii, a methanogen consistently colonising the newborn stomach. Eur J Clin Microbiol Infect Dis. 2017;36:2449–2455. doi: 10.1007/s10096-017-3084-7. [DOI] [PubMed] [Google Scholar]
- 23.Huynh HTT, Pignoly M, Nkamga VD, Drancourt M, Aboudharam G. The repertoire of Archaea cultivated from severe periodontitis. PLoS One. 2015;10:e0121565. doi: 10.1371/journal.pone.0121565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huynh HTT, Pignoly M, Drancourt M, Aboudharam G. A new methanogen “Methanobrevibacter massiliense” isolated in a case of severe periodontitis. BMC Res Notes. 2017;10:657. doi: 10.1186/s13104-017-2980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA. Methanogenic Archaea and human periodontal disease. Proc Natl Acad Sci U S A. 2004;101:6176–6181. doi: 10.1073/pnas.0308766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamilton C, Hiligsmann S, Beckers L, Masset J, Wilmotte A, Thonart P. Optimization of culture conditions for biological hydrogen production by Citrobacter freundii CWBI952 in batch, sequenced batch and semicontinuous operating mode. Int J Hydrog Energy. 2010;35:1089–1098. doi: 10.1016/j.ijhydene.2009.10.073. [DOI] [Google Scholar]
- 27.Kanazuru T, Sato EF, Nagata K, Matsui H, Watanabe K, Kasahara E, et al. Role of hydrogen generation by Klebsiella pneumoniae in the oral cavity. J Microbiol Seoul Korea. 2010;48:778–783. doi: 10.1007/s12275-010-0149-z. [DOI] [PubMed] [Google Scholar]
- 28.Belay N, Johnson R, Rajagopal BS, Conway de Macario E, Daniels L. Methanogenic bacteria from human dental plaque. Appl Environ Microbiol. 1988;54:600–603. doi: 10.1128/aem.54.2.600-603.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khelaifia S, Drancourt M. Susceptibility of archaea to antimicrobial agents: applications to clinical microbiology. Clin Microbiol Infect. 2012;18:841–848. doi: 10.1111/j.1469-0691.2012.03913.x. [DOI] [PubMed] [Google Scholar]
- 30.Bringuier A, Khelaifia S, Richet H, Aboudharam G, Drancourt M. Real-time PCR quantification of Methanobrevibacter oralis in periodontitis. J Clin Microbiol. 2013;51:993–994. doi: 10.1128/JCM.02863-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huynh HTT, Nkamga VD, Signoli M, Tzortzis S, Pinguet R, Audoly G, et al. Restricted diversity of dental calculus methanogens over five centuries. France Sci Rep. 2016;6:25775. doi: 10.1038/srep25775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author.