Abstract
Pandoraea sputorum (P. sputorum), an emerging pathogen, is able to trigger a pronounced pro-inflammatory response that results in lung dysfunction in cystic fibrosis (CF) patients. All previous P. sputorum isolates have been obtained from the respiratory samples of CF patients, with no reported cases of P. sputorum bacteremia. For the first time, we report P. sputorum isolates recovered twice from the blood cultures of a patient with liver cancer who had undergone allogeneic liver transplantation. These isolates were successfully identified by combining mass spectrometry and molecular techniques based on 16S rRNA sequencing methods. At the onset of the P. sputorum bacteremia, the patient’s peripheral T, B and NK cell counts were 181.68/μL, 59.57/μL and 70.66/μL, respectively. The serum procalcitonin level, C-reactive protein level and peripheral neutrophil granulocyte percentage were 0.56 ng/mL, 61.00 mg/L and 96.8%, respectively. We found these isolates to be susceptible to ciprofloxacin and piperacillin/tazobactam and to be intermediate to amikacin. Previous studies have found P. sputorum isolates to be resistant. All of the data combined showed that compromised immune function from allogeneic liver transplantation plus immunosuppressive therapy contributes to the occurrence of P. sputorum bacteremia. Furthermore, the P. sputorum isolates demonstrated characteristic resistance profiles.
Keywords: Pandoraea sputorum, bacteremia, liver cancer, allogeneic liver transplantation
Introduction
The genus Pandoraea was first described in 2000 to accommodate microorganisms from pseudomonas rRNA homology group II.1 To date, the genus Pandoraea comprises 11 named species, namely Pandoraea sputorum, Pandoraea pulmonicula, Pandoraea pnomenusa, Pandoraea apista, Pandoraea norimbergensis,2 Pandoraea faecigallinarum, Pandoraea oxalativorans, Pandoraea terrae, Pandoraea thiooxydans, Pandoraea vervacti and Pandoraea fibrosis sp. nov.3,4 Pandoraea species are emerging pathogens that cause chronic lung infection in cystic fibrosis (CF) patients due to their ability to trigger a pronounced pro-inflammatory response, which results in lung dysfunction.5–7 P. pulmonicula has also been demonstrated to invade A549 human lung epithelial cells.8
Pandoraea species can be recovered from a variety of specimens, such as sputum, blood, urine, lung tissue and wounds. To date, P. sputorum has only been isolated from the respiratory tract samples of five CF patients from Australia, Spain, France and Argentina.9–13 In Australia, a 32-year-old man with end-stage CF was diagnosed with a pulmonary P. sputorum infection via a combination of 16S rRNA sequencing and biochemical profiling.9 The first case of pulmonary P. sputorum infection diagnosed in Spain was also reported to be a CF patient. In this case, P. sputorum was identified by a combination of 16S rRNA sequencing and mass spectrometry and was reported to be susceptible to piperacillin-tazobactam, cotrimoxazole and imipenem.10 The patient’s lung function improved after treatment with piperacillin/tazobactam and imipenem.10 The second case was also a CF patient with a persistent 5-year P. sputorum lung infection. During the 5-year period, eight P. sputorum strains initially identified as various non-fermentative gram-negative bacilli (NFGNB) were re-identified as P. sputorum via a combination of 16S rRNA sequencing and mass spectrometry.11 The patient’s lung function improved significantly after treatment with azithromycin and 7% hypertonic saline for 1 year.11 In France, P. sputorum recovered twice from the respiratory samples of a 13-year-old boy with CF was susceptible to imipenem and trimethoprim/sulfamethoxazole (TST). However, his lung function failed to improve after treatment using these antibiotics.12 In Argentina, five P. sputorum isolates from a 9-year-old CF patient first identified as NFGNB by biochemical profiling were also re-identified as P. sputorum by a combination of 16S rRNA sequencing and mass spectrometry.13 The five isolates were susceptible to imipenem and TST, but the patient’s lung function declined after a combination treatment using TST, colistin and ciprofloxacin.13 Although these CF patients had a common lung dysfunction due to a pronounced pro-inflammatory response, not enough evidence that P. sputorum was the real contributor to their clinical condition exists. Specifically, other microorganisms, such as Pseudomonas aeruginosa and Staphylococcus aureus, were also co-isolated from the patients’ respiratory tracts.
Although nine cases of bacteremia caused by P. pnomenusa, P. apista, P. norimbergensis and unclassfied Pandoraea at species level have been described,14–16 no case has reported P. sputorum bacteremia. We report, for the first time, a bloodstream infection with P. sputorum in a patient with liver cancer who had undergone allogeneic liver transplantation.
Case Presentation
A 43-year-old male patient who was diagnosed with grade II hepatocellular carcinoma was admitted to Zhongnan Hospital’s organ transplant center for allogeneic liver transplantation. After transplantation on July 16, 2018, he was sent to the center’s intensive care unit for further treatment. To prevent graft rejection, the patient was treated with Tacrolimus FK506, Mycophenolate mofetil and prednisone to suppress lymphocyte proliferation. On July 18, the patient’s peripheral T, B and NK cell counts were 181.68/μL (reference interval, 805~4459/μL), 59.57/μL (reference interval, 240~1317/μL) and 70.66/μL (reference interval, 210~1514/μL), respectively. P. sputorum bacteremia was detected based on the culture of blood samples collected on July 17 and 20, respectively. On July 20, the patient’s serum procalcitonin (PCT) and C-reactive protein (CRP) levels were 0.56 ng/mL (reference interval, <0.05 ng/mL) and 61.00 mg/L (reference interval, <10 mg/L), respectively, and his peripheral neutrophil granulocyte percentage (Neu%) was 96.8% (reference interval, 40-75%). Imipenem (500 mg every 8 h) and ceftriaxone/tazobactam (2000 mg every 12 h) were then administered used. The infection was controlled on July 31, with very low serum PCT levels (<0.05 ng/mL) and a normal peripheral Neu% (73.67%).
Laboratory Identification Of P. sputorum And Antibiotic Susceptibility Tests
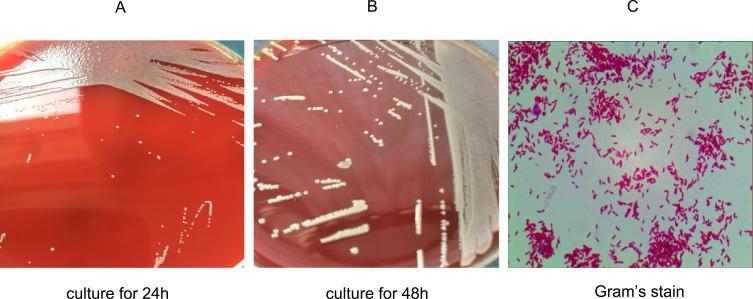
Two isolates were obtained from the liver cancer patient’s cultured blood samples and processed according to the laboratory’s protocol. Briefly, the cultured blood samples were transferred and incubated on Columbia blood agar plates in a 5% CO2 incubator at 37°C. Several tiny, moist and pale colonies appeared on the surface of the plates after 24 h of incubation and transformed into big, moist and ginger colonies over 48 h of incubation (Figure 1). Gram staining revealed that the isolates were gram-negative. The initial biochemical tests for the two isolates revealed positive results for oxidase reaction and nitrate reduction and negative results for urease activity, providing little information for identification. In addition, the strains could not be accurately identified using the Vitek 2 gram-negative identification card, which contains 47 biochemical tests in the Vitek 2 Compact system (bio-Merieux, France), a widely used automated bacteria identification platform based on biochemical tests.17 The reports obtained from the Vitek 2 Compact system indicated a 34% possibility of Pseudomonas fluorescens, a 33% possibility of Sphingomonas paucimobilis and a 33% possibility of Achromobacter xylosoxidans.
Figure 1.
Morphology and Gram’s staining of P. sputorum. Culture blood samples were transferred to Columbia blood plates and incubated in the 5% CO2 incubator at 37°C for (A) 24 h and (B) 48 h. Gram’s staining showed that P. sputorum is (C) a gram-negative bacilli.
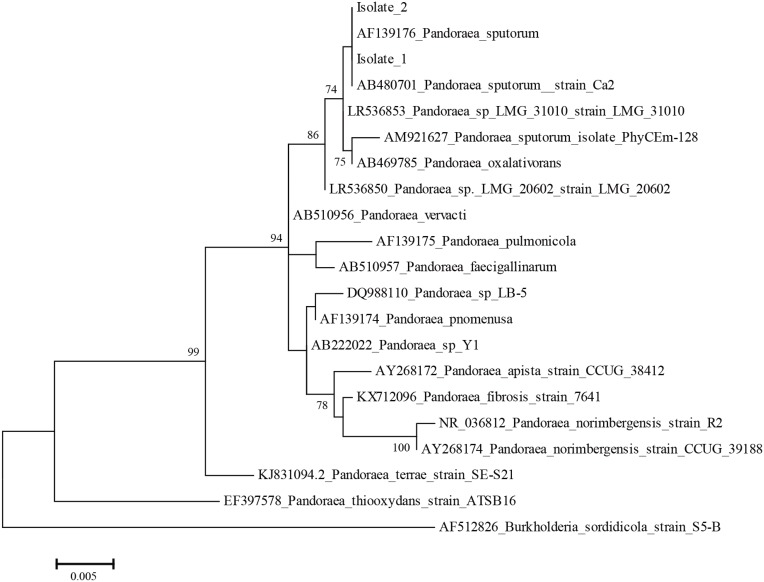
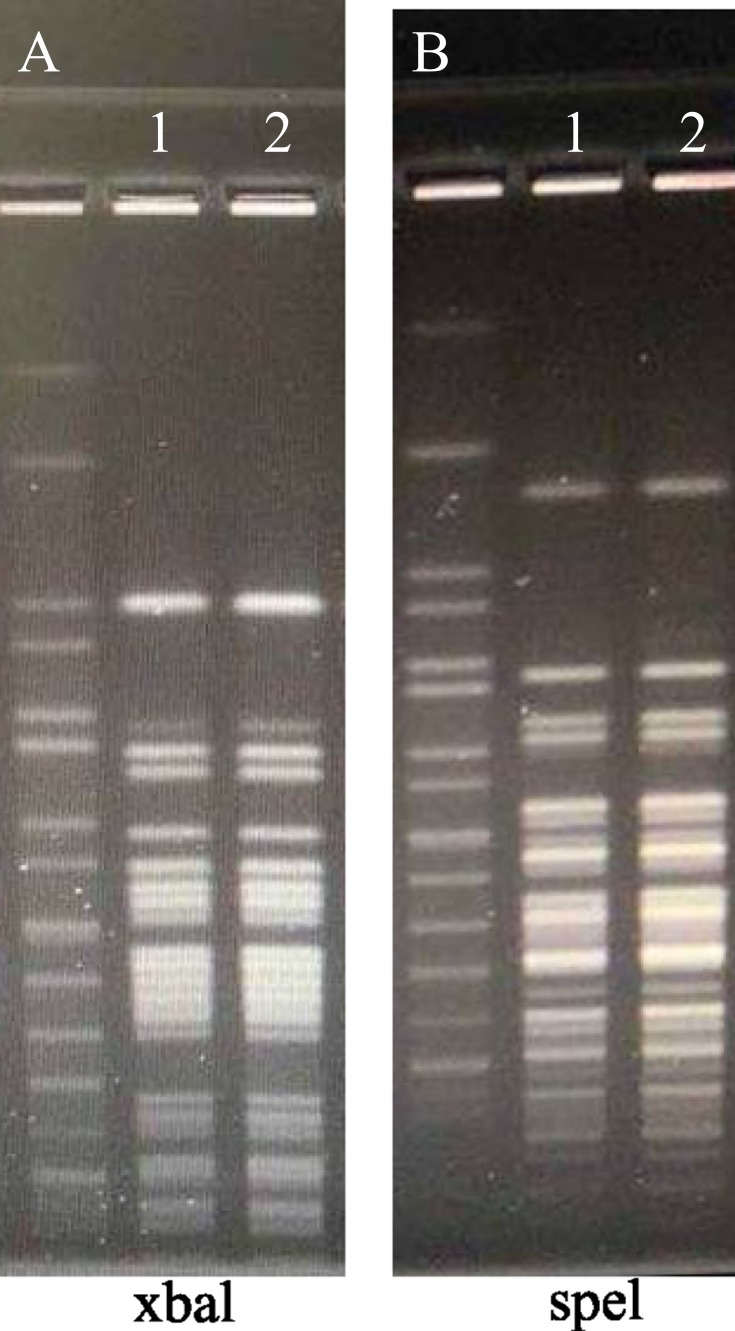
The two isolates were then identified by mass spectrometry using an Ultraflex MALDI-TOF instrument and MALDIBiotyper 3.1 software (Bruker Daltonics, Bremen, Germany). At the species level, these isolates were identified as P. sputorum with a score of >2. The results were further confirmed by 16S rRNA sequencing. For this purpose, the 16S rRNA gene of the two isolates was amplified, followed by sequencing and blasting via the GenBank database.16 Phylogenetic analysis based on the 16S rRNA sequences showed that the two isolates were 100% similar to strains of P. sputorum, namely AF139176 and AB480701 (Figure 2). The genetic relatedness of the sequenced isolates was further determined by a pulsed-field gel electrophoresis (PFGE) analysis.18 The chromosomal DNA of the bacteria was digested with restriction endonucleases xbal and spel, followed by PFGE. The DNA restriction patterns of the isolates were then compared to determine their relatedness. The two isolates had identical digested DNA patterns (Figure 3), suggesting that they were derived from a common source.
Figure 2.
Phylogenetic analysis. Phylogenetic trees based on 16S rRNA gene sequences indicated the phylogenetic positions of isolates 1 and 2 and of other Pandoraea species. Burkholderia sordidicola S5-BT12826 was used as the outgroup. Bootstrap values (>70%) are shown for appropriate nodes. The scale bars represent the number of nucleotide substitutions per site.
Figure 3.
Relatedness of the two isolates as determined by pulsed-field gel electrophoresis. Chromosomal DNA of the two isolates was digested with restriction endonucleases xbal and spel, followed by pulsed-field gel electrophoresis. (A) Lane 1, Marker; Lane 2, Isolate 1; Lane 3, Isolate 2. (B) Lane 1, Marker; Lane 2, Isolate 1; Lane 3, Isolate 2.
The minimum inhibitory concentrations (MICs) were obtained using the antibiotic sensitivity analysis system ARIX 2x (Thermo Fisher, USA). The MIC values of 18 antibiotics are presented in Table 1. Subsequently, the antibiotic susceptibility pattern was characterized according to the standards established for other non-Enterobacteriaceae by the Clinical and Laboratory Standards Institute (28th edition). These two isolates were resistant to aztreonam, cefepime, ceftazidime, gentamicin, meropenem and tobramycin, but susceptible to ceftriaxone, ciprofloxacin, imipenem, levofloxacin, minocycline, piperacillin, piperacillin-tazobactam, tetracycline, ticarcillin/clavulanic acid, tigecycline and trimethoprim. In addition, they were intermediate to amikacin.
Table 1.
Antibiotic Susceptibility Patterns Of P. Sputorum Isolates Described In This And Previous Studies
| Antibiotic | This Study | Martinez-Lamas L. et al (2011)10 | Fernandez-Olmos A. et al (2012)11 | Puges M. et al (2015)12 | Martina P.F. et al (2017)13 |
|---|---|---|---|---|---|
| Amikacin | I (32) | R (≥256) | R (NR) | NR | R (≥64) |
| Aztreonam | R (>64) | R (≥16) | NR | R (NR) | R (NR) |
| Cefepime | R (>32) | R (≥16) | NR | R (NR) | R (≥64) |
| Ceftazidime | R (>64) | R (≥256) | R (NR) | NR | R (≥64) |
| Ciprofloxacin | S (≤0.5)* | NR | R (NR) | I (NR) | R (≥4) |
| Ceftriaxone | S (≤0.5) | NR | NR | NR | NR |
| Gentamicine | R (>16) | NR | R (NR) | NR | R (≥64) |
| Imipenem | S (≤0.5)# | S (1.5) | S (4) | NR | S (≤0.25) |
| Levofloxacin | S (≤1) | NR | NR | S (NR) | NR |
| Meropenem | R (16) | R (≥8) | R (NR) | R (>32) | R (≥16) |
| Minocyline | S (≤1) | NR | NR | S (1) | NR |
| Piperacillin. | S (≤16) | NR | NR | S (NR) | NR |
| Piperacillin/tazobactam | S (≤8/4) | S (16) | S (≤16/4) | S (NR) | R (≥128) |
| Tetracycline | S (≤4) | NR | NR | NR | NR |
| Ticarcillin/clavulanic acid | S (≤8/2) | NR | NR | S (NR) | NR |
| Tigecycline | S (≤1) | NR | NR | S (NR) | NR |
| Tobramycin | R (>16) | R (≥256) | R (NR) | NR | NR |
| Trimethoprim | S (≤2/38) | NR | NR | NR | NR |
Note: Minimum inhibitory concentrations for certain antibiotics are listed in brackets.
Abbreviations: R, resistant; I, intermediate; S, sensitive; NR, not reported; Ref, reference.
Discussion
Previous studies have reported that P. sputorum only colonizes on the surface of the respiratory tract in CF patients.7,13,14 However, no studies to date have reported a patient with P. sputorum bacteremia. We present a case study of a patient with P. sputorum bacteremia. The results suggest that P. sputorum, similar to P. pnomenusa, P. apista and P. norimbergensis, is not only capable of colonizing on the surface of the respiratory tract and hampering lung function, but also potentially invasive, causing bacteremia and even sepsis. Serum PCT, CRP or peripheral Neu% levels were found to be elevated in the patient, suggesting that P. sputorum in the blood can also trigger a pronounced pro-inflammatory and inflammatory response. Therefore, lung dysfunction due to chronic colonization of P. sputorum on the surface of the airways and lungs may be due to the overwhelming inflammatory response.7,14
A subsequent review of the patient’s medical record revealed that he had suffered from a severe underlying disease, namely liver cancer due to chronic hepatitis B virus infection, and that he had undergone allogeneic liver transplantation and immunosuppressive treatment. It is well known that cancer is generally associated with compromised immune function and that surgery and immunosuppressive therapy can further decrease immunity temporarily.19–21 Analysis of the patient’s lymphocyte subsets showed that he had very low levels of peripheral T, B and NK lymphocytes. Therefore, it is reasonable to suggest that compromised immune function contributes to the occurrence of P. sputorum bacteremia.
As P. sputorum is capable of causing bacteremia, the accurate identification of P. sputorum is important. We show that both the initial biochemical tests and the Vitek 2 Compact system failed to correctly identify P. sputorum, even providing incorrect results. However, the mass spectrometry assays accurately identified P. sputorum and the results were further confirmed by 16S rRNA sequencing. These results echoed the finding of a previous study that conventional phenotypic methods are unreliable in identifying Pandoraea species, which are often misidentified as Burkholderia, Stenotrophomonas or Ralstonia species.11,12,22 In another study, various NFGNB isolates that were initially incorrectly identified were re-identified as P. sputorum using a combination of 16S rRNA sequencing and mass spectrometry.13 These results indicate that P. sputorum can be accurately identified using a combination of conventional biochemical tests, mass spectrometry and sequencing.
The antibiotic susceptibility tests conducted in this study demonstrated that the P. sputorum isolates were resistant to six antibiotics, namely aztreonam, cefepime, ceftazidime, gentamicin, meropenem and tobramycin. Their resistance to over three classes of antibiotics allows them to be classified as multi-drug resistant. Interestingly, these isolates were susceptible to imipenem and 10 other antibiotics, which included ciprofloxacin and piperacillin/tazobactam, reflecting the resistant effect observed in Martina’s report.13 In addition, the P. sputorum isolate was intermediate to amikacin in this study. Previous studies have shown P. sputorum isolates to be resistant.10,11,13 Therefore, the characteristic patterns of antibiotic susceptibility of the P. sputorum isolates collected in this study were revealed. A combined treatment using imipenem and ceftriaxone/tazobactam successfully eliminated P. sputorum in the blood and improved the patient’s clinical condition. Thus, imipenem can be considered to be a powerful antibiotic for eliminating P. sputorum.12,13
This is the first study on P. sputorum bloodstream infection and P. sputorum infection in a non-CF patient. Compromised immune function from allogeneic liver transplantation plus immunosuppressive therapy contributes to the presence of P. sputorum bacteremia. Although the P. sputorum isolates in this study exhibited multi-drug resistance, they also demonstrated characteristic resistance profiles.
Acknowledgments
The authors are grateful to Dr. Tiangang Liu and Dr. Aisi Fu of the Wuhan University School of Pharmaceutical Sciences for their help in sequencing the isolates of P. sputorum.
Ethics Statement
This study was approved by the Ethics Committee of the Zhongnan Hospital of Wuhan University. In the case of the liver transplantation, written informed consent was obtained. The liver donor was a patient resident at Zhongnan Hospital, Wuhan University who died of brain death. Fully understanding the medical value of organ donation, all of the deceased’s immediate relatives agreed to donate his liver and voluntarily signed an informed consent form for organ donation. After fully evaluating the will of the deceased’s family, the donated organs and the legal and ethical norms, the Organ Donation and Transplantation Ethics Committee of Zhongnan Hospital agreed to take the deceased’s organs and carry out a follow-up liver transplantation. Written informed consent has been provided by the patient and the institutional approved us to publish the case report.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Coenye T, Falsen E, Hoste B, et al. Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pulmonicola sp. nov., Pandoraea pnomenusa sp. nov., Pandoraea sputorum sp. nov. and Pandoraea norimbergensis comb. nov. Int J Syst Evol Microbiol. 2000;50(Pt 2):887–899. doi: 10.1099/00207713-50-2-887 [DOI] [PubMed] [Google Scholar]
- 2.Daneshvar MI, Hollis DG, Steigerwalt AG, et al. Assignment of CDC weak oxidizer group 2 (WO-2) to the genus Pandoraea and characterization of three new Pandoraea genomospecies. J Clin Microbiol. 2001;39:1819–1826. doi: 10.1128/JCM.39.5.1819-1826.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.See-Too WS, Ambrose M, Malley R, et al. Pandoraea fibrosis sp. nov., a novel Pandoraea species isolated from clinical respiratory samples. Int J Syst Evol Microbiol. 2019;69:645–651. doi: 10.1099/ijsem.0.003147 [DOI] [PubMed] [Google Scholar]
- 4.Green H, Jones AM. Emerging Gram-negative bacteria: pathogenic or innocent bystanders. Curr Opin Pulm Med. 2018;24:592–598. doi: 10.1097/MCP.0000000000000517 [DOI] [PubMed] [Google Scholar]
- 5.Jorgensen IM, Johansen HK, Frederiksen B, et al. Epidemic spread of Pandoraea apista, a new pathogen causing severe lung disease in cystic fibrosis patients. Pediatr Pulmonol. 2003;36:439–446. doi: 10.1002/ppul.10383 [DOI] [PubMed] [Google Scholar]
- 6.Degand N, Lotte R, Deconde Le Butor C, et al. Epidemic spread of Pandoraea pulmonicola in a cystic fibrosis center. BMC Infect Dis. 2015;15:583. doi: 10.1186/s12879-015-1327-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kokcha S, Bittar F, Reynaud-Gaubert M, et al. Pandoraea pulmonicola chronic colonization in a cystic fibrosis patient, France. New Microbes New Infect. 2013;1:27–29. doi: 10.1002/2052-2975.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caraher E, Collins J, Herbert G, et al. Evaluation of in vitro virulence characteristics of the genus Pandoraea in lung epithelial cells. J Med Microbiol. 2008;57:15–20. doi: 10.1099/jmm.0.47544-0 [DOI] [PubMed] [Google Scholar]
- 9.Pimentel JD, MacLeod C. Misidentification of Pandoraea sputorum isolated from sputum of a patient with cystic fibrosis and review of Pandoraea species infections in transplant patients. J Clin Microbiol. 2008;46:3165–3168. doi: 10.1128/JCM.00855-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Lamas L, Rabade Castedo C, Martin Romero Dominguez M, et al. Pandoraea sputorum colonization in a patient with cystic fibrosis. Arch Bronconeumol. 2011;47:571–574. doi: 10.1016/j.arbres.2011.06.015 [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Olmos A, Morosini MI, Lamas A, et al. Clinical and microbiological features of a cystic fibrosis patient chronically colonized with Pandoraea sputorum identified by combining 16S rRNA sequencing and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012;50:1096–1098. doi: 10.1128/JCM.05730-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puges M, Debelleix S, Fayon M, Megraud F, Lehours P. Persistent infection because of Pandoraea sputorum in a young cystic fibrosis patient resistant to antimicrobial treatment. Pediatr Infect Dis J. 2015;34:1135–1137. doi: 10.1097/INF.0000000000000843 [DOI] [PubMed] [Google Scholar]
- 13.Martina PF, Martinez M, Frada G, et al. First time identification of Pandoraea sputorum from a patient with cystic fibrosis in Argentina: a case report. BMC Pulm Med. 2017;17:33. doi: 10.1186/s12890-017-0500-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stryjewski ME, LiPuma JJ, Messier RH Jr., Reller LB, Alexander BD. Sepsis, multiple organ failure, and death due to Pandoraea pnomenusa infection after lung transplantation. J Clin Microbiol. 2003;41:2255–2257. doi: 10.1128/jcm.41.5.2255-2257.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falces-Romero I, Gutierrez-Arroyo A, Romero-Gomez MP. Catheter-associated bacteremia by Pandoraea pnomenusa in an infant with acute lymphoblastic leukemia. Med Clin (Barc). 2016;147:132. doi: 10.1016/j.medcli.2016.04.009 [DOI] [PubMed] [Google Scholar]
- 16.Coenye T, Liu L, Vandamme P, LiPuma JJ. Identification of Pandoraea species by 16S ribosomal DNA-based PCR assays. J Clin Microbiol. 2001;39:4452–4455. doi: 10.1128/JCM.39.12.4452-4455.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakasone I, Kinjo T, Yamane N, Kisanuki K, Shiohira CM. Laboratory-based evaluation of the colorimetric VITEK-2 Compact system for species identification and of the Advanced Expert System for detection of antimicrobial resistances: VITEK-2 Compact system identification and antimicrobial susceptibility testing. Diagn Microbiol Infect Dis. 2007;58:191–198. doi: 10.1016/j.diagmicrobio.2006.12.008 [DOI] [PubMed] [Google Scholar]
- 18.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang JW, Ren ZG, Lu HF, et al. Optimal immunosuppressor induces stable gut microbiota after liver transplantation. World J Gastroenterol. 2018;24:3871–3883. doi: 10.3748/wjg.v24.i34.3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasic T, Viola A. Prostate cancer-induced immunodysfunction: a lesson from organ cultures. Immunol Lett. 2005;100:98–102. doi: 10.1016/j.imlet.2005.06.021 [DOI] [PubMed] [Google Scholar]
- 21.Schneider C, von Aulock S, Zedler S, Schinkel C, Hartung T, Faist E. Perioperative recombinant human granulocyte colony-stimulating factor (Filgrastim) treatment prevents immunoinflammatory dysfunction associated with major surgery. Ann Surg. 2004;239:75–81. doi: 10.1097/01.sla.0000103062.21049.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev. 2010;23:299–323. doi: 10.1128/CMR.00068-09 [DOI] [PMC free article] [PubMed] [Google Scholar]