Abstract
Background
A pandemic influenza viral strain, influenza A/California/07/2009 (pdmH1N1), has been considered to be a potential issue that needs to be controlled to avoid the seasonal emergence of mutated strains.
Materials and methods
In this study, aptamer-antibody complementation was implemented on a multiwalled carbon nanotube-gold conjugated sensing surface with a dielectrode to detect pandemic pdmH1N1. Preliminary biomolecular and dielectrode surface analyses were performed by molecular and microscopic methods. A stable anti-pdmH1N1 aptamer sequence interacted with hemagglutinin (HA) and was compared with the antibody interaction. Both aptamer and antibody attachments on the surface as the basic molecule attained the saturation at nanomolar levels.
Results
Aptamers were found to have higher affinity and electric response than antibodies against HA of pdmH1N1. Linear regression with aptamer-HA interaction displays sensitivity in the range of 10 fM, whereas antibody-HA interaction shows a 100-fold lower level (1 pM). When sandwich-based detection of aptamer-HA-antibody and antibody-HA-aptamer was performed, a higher response of current was observed in both cases. Moreover, the detection strategy with aptamer clearly discriminated the closely related HA of influenza B/Tokyo/53/99 and influenza A/Panama/2007/1999 (H3N2).
Conclusion
The high performance of the abovementioned detection methods was supported by the apparent specificity and reproducibility by the demonstrated sensing system.
Keywords: influenza pandemic, membrane protein, aptasensor, immunosensor, dielectrode sensor
Introduction
Influenza, a severe illness caused by the enveloped spherical or filamentous influenza viruses, has a diameter of 80 to 120 nm and leads to respiratory diseases.1–4 Among the primary types of influenza (A, B, and C), influenza A followed by influenza B causes a higher death rate in humans. Two major glycoproteins on the surface, neuraminidase (NA) and hemagglutinin (HA), play vital roles in emerging influenza viruses due to their importance in host cell interactions. HA is the predominant surface protein for influenza viral infection needed for membrane fusion with host cells to mediate early-stage infection.1,2,5 When the influenza virus initially infects the host cell, HA binds to the glycan residues, namely, α-2,3- and α-2,6-sialic acids of bird and human cells, respectively.3,4,6 The surface protein, HA, of influenza virus binds to a terminal of the sialic acid residues and forms the glycoconjugate on a host cell surface, inducing the uptake of the viral infection. Since HA and NA are the predominant surface proteins, the type of influenza is used to determine HA and NA. Due to the emergence of new viruses, it is difficult to identify and discriminate the influenza strains at earlier stages. This disease has had a large impact and a significant death rate at the level of several million people worldwide.
Anti-HA is one of the most commonly used probes in most sensors because HA is the predominant surface antigen and occupies approximately 80% of the membrane of influenza viruses. Although the anti-HA antibody is efficient for detecting the influenza virus, it can only differentiate influenza A and B viruses, and early detection is difficult. It is mandatory to generate a detection method to effectively identify HA of the influenza virus at earlier stages. This research detected influenza virus by the aptamer selected against HA of influenza virus pdmH1N1 (A/California/07/2009) and compared it with antibody-mediated detection. A/California/07/2009 is an important strain that emerges due to the reassortment of different viruses infected with swine, humans and avians.7 For an additional impact, the sandwich patterns of aptamer-HA-antibody and antibody-HA-aptamer were employed to detect HA at the lower level.
The above probe, aptamer, is an artificial antibody consisting of either DNA or RNA, generated by Systematic Evaluation of Ligands and Exponential enrichment (SELEX) with three simple mandatory steps, such as binding, separation and amplification.8 Aptamers have advantages over antibodies, including high binding affinity, ease of preparation, high stability, cost-effectiveness and non-immunogenicity.9 Even though aptamer applications have been demonstrated widely in all biological fields, considerable research has focused on sensor development due to its selective binding to the target. Moreover, the target binds with a few bases of aptamer sequence and is able to differentiate the closed related biomolecules. In the case of influenza, aptamers can differentiate the subtypes of influenza viruses, which helps to identify the emergence of new viral types. In general, aptamers binding with targets occur by adapting the folding of aptamers under particular ionic conditions to form specific 3D structures, such as pseudoknots, hair folds and convex rings. With this specific secondary structure, aptamers can bind with the targets specifically and yield higher sensitivity.10 Since antibodies and aptamers have the potential characteristics of being able to contribute significantly in the field of biosensors, the complementation of these biomolecules shows the good improvement of the detection method. In most cases, aptamers and antibodies have different binding sites on the analyte, making it possible to explore the antibody and aptamer sandwich pattern to detect the analyte and to boost the detection limit.11 This research focused on the sandwich-mediated interaction of aptamer-HA-antibody and antibody-HA-aptamer on the multiwalled carbon nanotube (MWCNT)-modified interdigitated dielectrode sensor (IDE) to diagnose and discriminate the influenza viruses at a lower level.
Carbon nanotubes have been widely used in several sensors due to their higher surface-to-volume ratio, ability to be supercapacitors, participation in high electron mobility and good electrical conductivity. Due to the large surface area of carbon nanotubes, they have elevated the loading capacity of biomolecules by passive adsorption/covalent crosslinking, while their small band gap and excellent conductivity are useful for conducting electrons between the electrode surface and the biomolecule. The coverage on the surface and orientation provided by graphene on the electrode significantly enhance its performance with electrochemical behavior.12,13 Further combining graphene with gold improves the flow of charges and improves the biomolecular detection. Gold nanoparticle (GNP) is a good and well-established material and is generally accepted for the development of various sensors due to its positive features, such as compatibility with a wide range of sensing surfaces, easy dispersal in water, biological inertness, and ability to tailor production with similar and varied nanosizes. Moreover, gold nanoparticle-conjugated aptamers and antibodies showed higher stability of biomolecules and improved immobilization on the sensing system.14 In the present study, aptamers and antibodies were conjugated with GNP, and their interactions with HA of pdmH1N1 were thoroughly studied. Furthermore, the sandwich patterns of aptamer-HA-antibody and antibody-HA-aptamer on multiwalled carbon nanotube (MWCNTs)-modified IDE sensors were evaluated. The primary goal of this research is to generate a sensitive system by exploring the various possibilities using both aptamers and antibodies as the probe. Furthermore, fine-tuning was carried out employing the sandwich pattern. Using the combination of ideal probing surface and nanoparticle incorporation, generating a high-performance system is attempted towards the point-of-care system for pdmH1N1 surveillance.
Materials And Methods
Materials And Biological Reagents
Gold nanoparticle (GNP), 3-aminopropyl) triethoxysilane, 16-mercaptohexadecanoic acid (16-MHA) and ethanol were obtained from Sigma-Aldrich, USA. HA of A/California/07/2009 (H1N1) and A/Panama/2007/1999 (H3N2) and Anti-HA A/California/07/2009 antibodies were procured from Sino Biological (Beijing, China). Influenza B/Tokyo/53/99 was received from Prospec-Tany TechnoGene Ltd., Israel. RNase inhibitor was purchased from Epicentre Biotechnologies, USA. N-hydroxysuccinimide (NHS) and N-ethyl-N’-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) were procured from GE Healthcare (USA). All other reagents were used as recommended by the supplier.
Fabrication And Characterization Of Interdigitated Electrode (IDE)
The IDE sensing surface was fabricated as described previously.15 The Silver IDE electrode was deposited on the silicon wafer sample <100> using the traditional wet etching method. Positive photo resist (PR) was coated on the silicon wafer followed by soft baking for 90 sec. Ultraviolet (UV) light exposure was conducted for 10 sec to allow the pattern transfer from the IDE mask onto the surface of the samples. After that step, the development process was carried out for 15 sec by using an RD-6 developer. Then, the sample was baked at 110 °C to remove unwanted moisture and improve the adhesion between the silver and SiO2 layer. Finally, the unexposed area was removed using the silver etchant for 23 s and then cleaned with acetone.
The fabricated IDE sensing area was characterized by surface morphology analysis using scanning electron microscopy (SEM) on a bare silicon wafer with accelerated electrons at 20 kV. Furthermore, 3D-Nano profilometer-assisted high-power microscopy was supported for surface morphology observation of the dielectric gap by controlling PZT at scan rates of 13.45 and 78.67 for the upper and lower limits, respectively. The 5x magnification was used to obtain clear images with gap and electrode regions.
In Vitro Enzymatic Synthesis Of Anti-Influenza Aptamer And Complementary Sequences
Stable aptamers were prepared enzymatically in vitro using a Cellscript transcription kit (Epicentre Biotechnologies, USA), as described earlier.16–18 In brief, using the template DNA-oligonucleotide sequences (5ʹ-GGAGCTCAGCCTTCACTGC CAAAAAGTTAGGCCAGCAAATTGCGAGCTGATCCGGTGACTGGCTACAGGAGGCCTTGTCCACGGCCGTATTGGCACCACCGTCGGATCC-3ʹ), random regions and primer regions flanked at both the 5ʹ and 3ʹ ends and polymerase chain reaction were performed. The constant primer sequences at the 5′ and 3′ ends, in addition to the T7 promoter region at the 5ʹ forward primer region (5′-AGTAATACGACTCACTATAGGGGGAGCTCAGCCTTCACTGCCAAA-3′ and 5′-GGATCCGACGGTGGTGCCAAT-3′), were prepared for amplification. PrimeSTAR Max DNA Polymerase with dTTP, dATP, dCTP, and dGTP was used for the amplification. The obtained double-stranded DNAs were precipitated and further used to prepare the RNA aptamer by in vitro transcription (37°C 3 h). Upon completion of the reaction, it was stopped by adding 2 times the volume of urea buffer containing 50 mM EDTA, 7 M urea, and 90 mM Tris-borate with bromophenol blue (0.05%) and heated 2 min at 94°C before being loaded on a 10% polyacrylamide gel (with 7 M urea). The located RNA band was cut from the gel and ethanol precipitated using the crush and soak method, and the concentration was determined spectrophotometrically (at 260 nm). Similarly, the complementary aptamer sequences prepared (5ʹ-CCTCGAGTCGGAAGTGACGGTTTTTCAATCCGGTCGTTTAACGCTCGACTAGGCCACTGACCGATGTCCTCCGGAACAGGTGCCGGCATAACCGTGGTGGCAGCCTAGG-3ʹ) for the negative control.
Conjugation Of GNP With Aptamer Or Antibody
A thiol (SH)-linker with six carbon spacers on the aptamer was used to conjugate GNP and aptamer. Various concentrations of aptamer (30, 60, 125, 250, 500 and 1000 nM) were mixed independently with 10 µl of GNP and kept for 30 min at room temperature (RT). Centrifuged at a speed of 10,000 × g for 10 min to separate the unbound aptamers. Then, aptamer-GNP conjugates were washed with distilled water and kept at 4°C for further use. The adsorption of GNP on the aptamer was analyzed by a UV-visible spectrophotometer.
To conjugate the anti-HA antibody on the surface of GNP, briefly, 1 mL of GNP was added with 2 mM 16-MHA and stirred at RT for 15 min. Upon activation by EDC and NHS, different concentrations of anti-HA (30, 60, 125, 250 and 500 nM) with 15 µl were added independently to the above solution and kept at RT for 2 h with continuous stirring. The antibody-conjugated GNP (anti-HA-GNP) was separated by centrifugation, washed three times with distilled water, and kept at 4°C until further analysis. The adsorption of GNP with the anti-HA was analyzed by UV-visible spectrophotometer.
GNP-Aptamer And GNP-Antibody Immobilization On MWCN-Modified IDE Surface
To prepare the probe-modified IDE surface, the first surface was modified by a commercially obtained multiwalled carbon nanotube (MWCNTs). To that end, 1 g of acid-cleaned MWCN was diluted in 2% of APTES (in 30% ethanol), dropped onto the active IDE surface and kept for 2 h at RT. After 2 h, the surface was washed with 30% ethanol followed by water, and then the prepared GNP-aptamer or GNP-antibody conjugated probe was dropped onto the MWCN-modified surface. The remaining sensing surface was blocked by 1 M ethanolamine to avoid the nonspecific adsorption of HA protein.
Comparison Of HA Detection With GNP-Aptamer And GNP-Antibody Modified IDE Surfaces
HA was detected and compared on both GNP-aptamer and GNP-antibody modified MWCN surfaces. HA concentration with 100 pM was dropped onto these surfaces after the blocking step was performed with ethanolamine. The current changes before and after (30 min) the addition of HA was measured to find the interaction of HA with aptamer or antibody. Based on this detection, further concentration-dependent analysis was performed.
Determination Of Limit Of Detection Of HA On GNP-Aptamer And GNP-Antibody Modified IDE Surfaces
After confirming the binding of HA with aptamer and antibody, the detection limit of HA with aptamer and antibody was analyzed by titrating different HA concentrations on both surfaces. The HA was diluted from the lower femtomolar (10 fM) until the higher concentration (100 pM) and dropped onto both GNP-aptamer- and GNP-antibody-modified surfaces. For each experiment, 10 min incubation was employed to interact HA with aptamer or antibody, and then the surface was washed thoroughly with 1X HEPES buffer (10 mM, pH 7.4) to remove the unbound HA. Before and after HA interaction with aptamer or antibody, the current change was recorded, and the difference in changes was drawn by the linear graph to find the limit of detection of HA with aptamer and antibody. The limit of detection (LOD) was considered to be the lowest concentration of an analyte (from the calibration line at low concentrations) against the background signal (S/N = 3:1); in other words, LOD = standard deviation of the baseline (σ) + 3σ.
Discrimination Of Influenza Viral Types: Specificity Analysis
The higher-performing surface from the above analysis was used for the high-performance analysis by determining the specificity. HAs from influenza A/Panama/2007/1999 (H3N2) and influenza B/Tokyo/53/99 were used to compare the specificity of the anti-HA aptamer against pdmH1N1 (A/California/07/2009). A similar concentration (100 pM) was used based on the above experiments. To that end, after aptamer-GNP was immobilized onto the IDE surface, these two proteins (HA) were dropped independently, instead of interacting with HA of pdmH1N1, and the changes in the current were observed. Another control experiment was carried out with the complementary aptamer for pdmH1N1. In this experiment, the complementary GNP was immobilized on the sensing surface, the HA was dropped onto the surface, and the changes in the current were monitored. The same concentration of HA (pdmH1N1) was used for specificity analysis against the complementary sequences of the aptamer desired. Furthermore, support was rendered by the reproducibility analysis with different molecular assemblies.
Sandwich Pattern Of Aptamer-HA-Antibody And Antibody-HA-Aptamer
To improve the detection of HA with aptamer and antibody, sandwich patterns with aptamer and antibody were carried out with the lower level of HA protein detected. The optimized minimum detection level of HA with aptamer and antibody was used to compare the efficiency of sandwich. Two types of sandwich, aptamer-HA-antibody and antibody-HA-aptamer, were performed and compared. In the case of the aptamer-HA antibody, initially, the optimized concentration of aptamer-GNP was immobilized on the MWCN-modified IDE surface, and then the remaining surfaces were blocked by 1 M ethanolamine. Then, 10 fM of HA was dropped on the surface, and the optimized antibody-GNP concentration was interacted. In other cases of antibody-HA-aptamer, the optimized concentration of antibody-GNP was immobilized on the MWCN-modified IDE surfaces, and then the remaining surfaces were blocked by 1 M ethanolamine. Then, 1 pM of HA was interacted on the surface followed by the addition of the optimized aptamer-GNP. The subsequent changes were observed before and after the immobilization of molecules.
Results And Discussion
Current influenza detection methods, including immunochromatography, are gaining popularity for the rapid diagnosis of influenza viruses and discriminating influenza viral types A and B. In contrast, doctors are learning that immunochromatography testing is not adequately sensitive and is especially not ideal for detecting the early stages of influenza infection, where a lower count of virus is present in the nasopharyngeal tract.19 Currently available conventional tests are not able to recognize the influenza viruses until the onset of fever after viral infection. In this research, two different probes (aptamer and antibody) were tested and further expanded with sandwich patterns of these probes to detect HA of influenza virus H1N1 (A/California/07/2009) at its lower count, leading to the generation of an earlier-stage detection system.
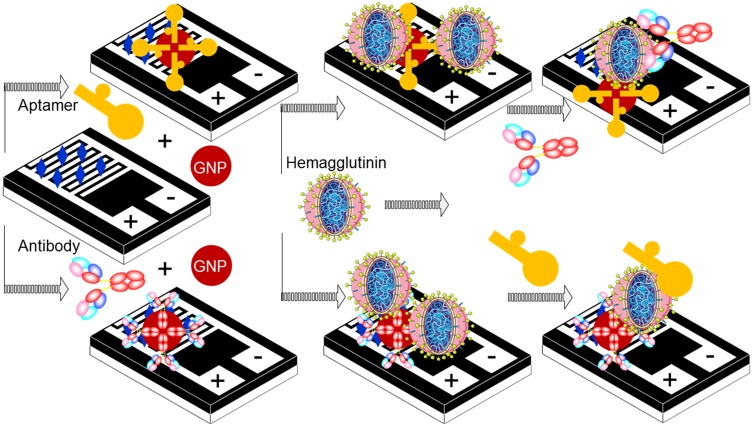
It is well-known that aptamers and antibodies have unique positive characteristics in biosensor development and are also able to interact at different binding sites of the target molecule. This study revealed options for creating a sandwich pattern with aptamers and antibodies to improve the diagnosis of influenza viruses and discrimination. The main purpose of this study was to increase the sensitivity and selectivity of influenza detection. The sensitivity of this system mainly depends on the selection of the capture and the detection molecules. This approach requires careful consideration to determine these molecules; generally, in the case of antibodies, to reach the equilibrium constant, a longer period of time is required due to the different binding affinity with larger sizes and a slower diffusing rate in the solution. However, aptamers are smaller in size compared to antibodies and exhibit superior characteristics. Generation of a detection system with a combination of aptamers and antibodies is useful, as it can retain the above advantages. The main goal of this study is to capture the advantages of both aptamers and antibodies for the detection of HA from the influenza virus pdmH1N1, thereby creating different detection systems. Figure 1 shows the schematic representation of different approaches with the aptamer and antibody to be used as the probes on the IDE-sensing surface.
Figure 1.
Schematic representation for the detection of HA protein on the MWCN-IDE surface. Different pattern detections with aptamer-conjugated GNP and antibody-conjugated GNP are shown.
Preliminary Confirmations On Molecules And Sensing System
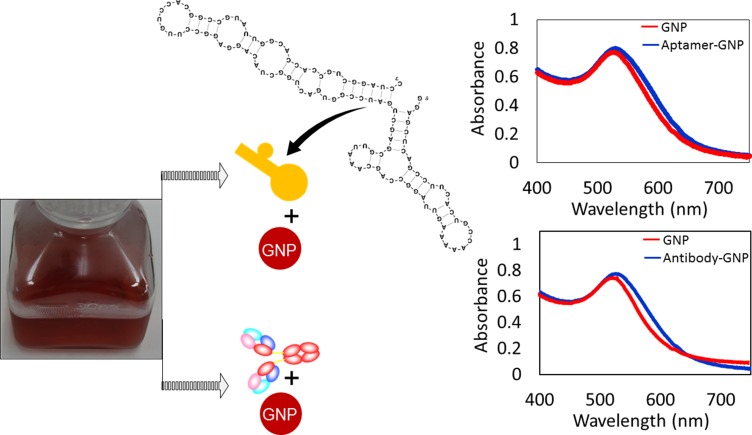
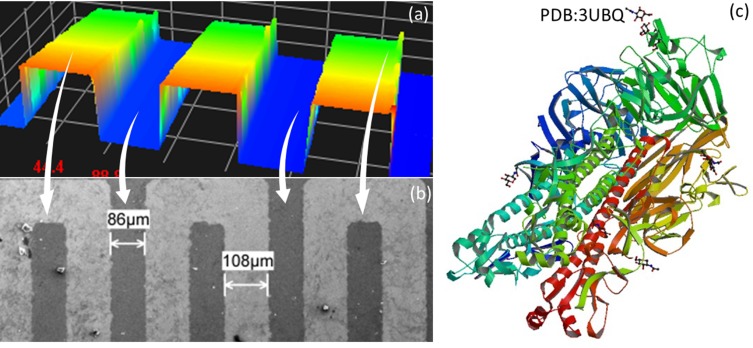
Before detecting HA on IDE, the complex formation of GNPs with aptamers and antibodies was confirmed by UV-visible spectrophotometer analysis. As shown in Figure 2, the peak maximum of GNP was confirmed at 520 nM, and the shift was found after complex formation with aptamer and antibody. These changes differ slightly between the aptamer and antibody attachment on the GNP. The prediction of aptamer secondary structure was analyzed by mfold online software-based folding. The stability of the prepared is sufficient to withstand both in vitro and in vivo systems because it was modified by 2ʹ-fluoro incorporation. These results confirmed the suitability and complex formation of the probe with the GNP to be immobilized on the MWCN-IDE surfaces. The prepared sensing surface was analyzed by high-resolution microscopies, and as shown in Figure 3A and B by 3D nanoprofiler and SEM, the surface was intact with the proper finger and gap regions. With these clearances, the detection of HA from the influenza pdmH1N1 was detected by the aptamer generated. The 3D-crystal structure of HA was analyzed from the data bank (PDB accession: 3UBQ), as displayed in Figure 3C with the exposed ribbon and helices.20
Figure 2.
UV-vis spectroscopy confirmation for aptamer-GNP and antibody-GNP conjugates. The secondary structure folding pattern for the aptamer is also shown.
Figure 3.
Surface and molecular analysis. (A) 3D-profiler image of the IDE sensing surface; (B) SEM image of the IDE sensing surface; (C) crystal structure of HA (PDB Accession: 3UBQ).20
Optimum Level Of Aptamer-GNP On The MWCN Surface
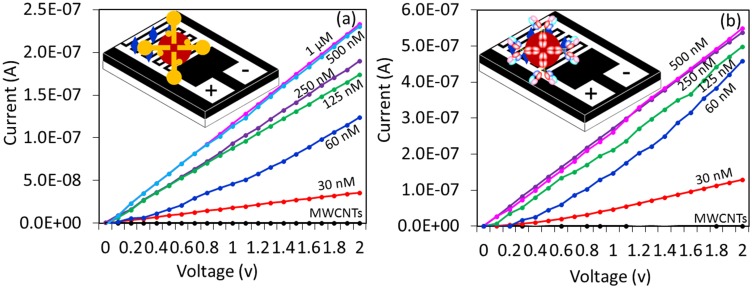
Figure 4A displays the optimum level of the GNP-aptamer complex needed to cover the surface of the MWCN-IDE. The aim of this experiment is to immobilize the maximum number of aptamers on the surface, which may lead to capturing the higher HA. As shown in Figure 4A, aptamer-GNP with a range of 30 nM to 1 µM was dropped independently onto the MWCN-IDE surface. MWCN immobilization on the amine surface shows a current response of 1.68 E−10. After attaching 30 nM of aptamer-GNP, the intensity of current was increased to 3.58 E−2. This immobilization occurred due to the reaction of amine in the APTES with GNP. The GNP-conjugated aptamer is stabilized with anionic citric ions and can bind to the cationic APTES by electrostatic interactions. This binding is observed because the positively charged amine groups in APTES also attract the negatively charged GNP.21,22 Furthermore, with increasing aptamer concentration, the current intensity was also increased. At 500 nM and 1 µM, aptamer-GNP shows the same levels of current, indicating the attainment of the saturation level. This result indicated that 500 nM of aptamer-GNP is the optimum concentration to cover the complete surface of the MWCN-IDE surface.
Figure 4.
Optimal aptamer-GNP and antibody-GNP attachments. (A) Determination by aptamer-GNP. Different concentrations of aptamer-GNP (30 nM to 1 µM) were tested on the amine-modified MWCN surfaces. 500 nM of aptamer reached saturation. (B) Determination by antibody-GNP. Different concentrations (30 nM to 500 nM) were tested on the amine-modified MWCN surfaces. The antibody (250 nM) reached saturation. Figure inset shows the schematic.
Optimum Level Of Antibody-GNP On The MWCN Surface
Similar to aptamer, antibody-GNP saturation on the MWCN surface was determined by dropping different concentrations of antibody-GNP conjugates onto the amine-modified MWCN surfaces. The interactions occurred in two ways: the amine group of the APTES could bind to the COOH group of the antibody, and the positively charged amine groups in APTES attracted the negatively charged GNP.23 As shown in Figure 4B, the antibody-GNP with a concentration of 30 nM to 500 nM was dropped independently onto the MWCN-IDE surfaces, and the changes in current were determined to identify the saturated concentration of antibody. With increasing antibody-GNP concentrations, the current levels also gradually increased concomitantly. Different concentrations with 30, 60, 120, 250, and 500 of antibody-GNP show current levels of 1.29, 4.57, 4.99, 5.38 and 5.42 E−07, respectively. Moreover, it was found that 250 and 500 nM shows a similar response to current levels, indicating saturation. This result made it possible to choose 250 nM as the level of saturation, and this level was used for further analysis to interact HA of the influenza virus pdmH1N1.
Comparative Analysis On Detection Of HA Using Aptamer-GNP And Antibody-GNP Complexes
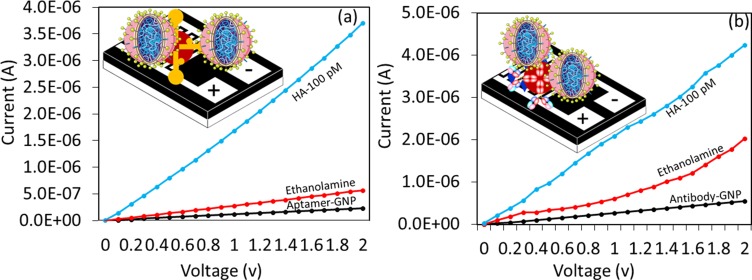
For this analysis, HA was detected on the optimized level of aptamer-GNP and antibody-GNP immobilized surfaces. A similar concentration at 100 pM of HA was tested on both of these surfaces after the surface was blocked by ethanolamine. As shown in Figure 5A, the black line is the aptamer-GNP immobilization, and after adding the ethanolamine, the current level was increased from 2.33 E−07 to 5.68 E−07. Then, 100 pM of HA (blue line) was interacted, and the current increased from 5.68 E−07 to 3.71 E−06. This result clearly shows the interaction of aptamer with HA was significant. The difference in current changes was noticed as 3.15 E−06. A similar concentration (100 pM) of HA interacted on the antibody-GNP-modified surfaces. As shown in Figure 5B, the current change was determined to be from 2.03 E−06 to 4.24 E−06. The difference in current was found to be 2.21E−06. By analyzing these two HA detection strategies, aptamer-GNP shows higher current changes compared with antibody-GNP with the same concentration of HA. This finding might be due to the strong binding affinity of HA with aptamer and the higher number of aptamers bound on the single GNP compared with antibody due to the smaller aptamer.
Figure 5.
Comparative analysis. Detection of 100 pM of HA by (A) aptamer-GNP; (B) antibody-GNP. Figure inset shows the schematic.
Sensitivity Of HA By Aptamer-GNP And Antibody-GNP
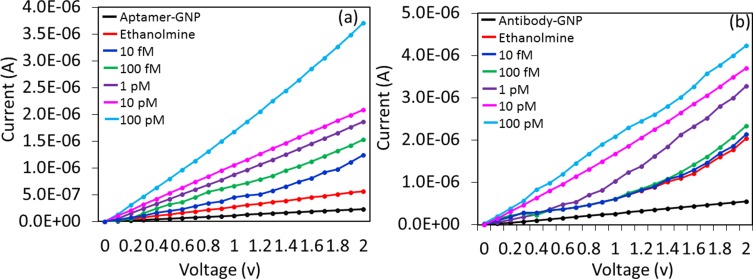
It was proven that aptamer-GNP shows higher changes in current for a similar concentration of HA. To expand these results further, the limit of detection of HA was analyzed on both aptamer-GNP- and antibody-GNP-modified MWCN surfaces for comparison. For this study, HA was titrated from the lower femtomolar to the higher picomolar concentrations (10 fM to 100 pM) and interacted on aptamer-GNP- and antibody-GNP-modified surfaces. Figure 6A and B shows the results obtained upon binding of HA with aptamer and antibody. It was observed that aptamer HA was detected from 10 fM. At 10 fM of HA, and the response shows a difference of 6.8 E−07 from the ethanolamine surface to HA interacted surface. Moreover, with concentrations increasing further to 100 fM, 1 pM, 10 pM, and 100 pM, the current differences also increased gradually as 8.7E−07, 1.47 E−06, 2.34 E−06, and 2.68 E−05, respectively (Figure 6A). At 10 fM of HA, a clear difference in current and increased current levels gradually increased with increasing concentrations of up to 100 pM. These results concluded that the sensitivity was 10 fM when using aptamer-GNP. A similar experiment was carried out with an antibody-GNP-immobilized surface with concentrations from 10 fM to 100 pM of HA. These molecules were dropped onto the sensing surface individually, and the current changes were noted. As shown in Figure 6B, ethanolamine (red line) shows the current change of 2.03 E−06. After the addition of 10 fM of HA, there was not a clear difference in current (dark blue line). At a concentration of 100 fM, the current showed a slight change of 2.33 E−06 and then increased further with the concentration of HA from 1 pM, 10 pM, and 100 pM, indicating the current levels as 3.28 E−06, 3.71 E−06, and 4.24 E−06, respectively. From 1 pM of HA, the change in current was clear. The sensitivity of HA with antibody-GNP was found to be 1 pM, which was 100 times lower than the sensitivity with aptamer-GNP. This finding might be attributable to the higher binding affinity of aptamer with HA, with more aptamers being bound on the GNP surface, leading to the lower level of detection. The sensitivity level shown in this study is ~7-fold higher than the previous study, which was carried out by surface plasmon resonance analysis.7
Figure 6.
Sensitivity analysis. Detected on (A) aptamer-GNP (B) antibody-GNP. HA concentrations from 10 fM to 100 pM were tested individually on both aptamer- and antibody-modified surfaces.
High-Analytical Performance On Sensing HA By GNP-Aptamer
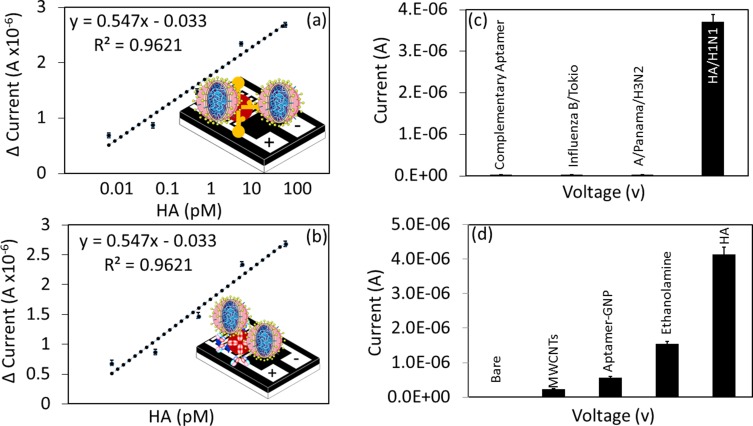
To evaluate the limit of detection, a linear regression analysis was performed by plotting the current levels from different concentrations of HA interaction with aptamers or antibodies. A linear graph was plotted using the response obtained from 0.01 to 100 pM. Based on 3σ, we could estimate the limit of detection to be 10 fM for aptamer, and it was 1 pM for antibody, as indicated by the sensitivity analysis (Figure 7A and B). Table 1 summarizes the current detection systems and compared them with the reported data.
Figure 7.
High-performance analysis. (A) Linear graph shows the dose-dependent binding of HA with aptamer-GNP. (B) Linear graph shows the dose-dependent binding of HA with antibody-GNP. (C) Specificity analysis. Complementary aptamer sequences of HA and other influenza strains were used. The current changes were noticed with only HA of pdmH1N1. (D) Reproducibility analysis. Molecules attached to different surfaces are shown with multiple experimental setups.
Table 1.
Influenza Detection By Various Methods And Comparison
| Target | Detection Molecule | Detection Method | Limit Of Detection | Reference |
|---|---|---|---|---|
| Influenza A pdmh1N1 | Aptamer and antibody | Volumetric assay | 10 fM | This work |
| Influenza M1 protein | Antibody | Electrochemical sensor | 1 fg/mL | 28 |
| H3N2 virus | Aptamer | RAMAN scattering | 1X10−4 HA units/probe | 29 |
| Influenza A H1N1 | Aptamer | Quantum dot | 138 pg/mL | 30 |
| Influenza A H1N1 | Sialic acid receptors | Electrical detection | 1X10−4 HAU | 31 |
| Influenza virus | Antibody | Capacitive Immunosensor | 0.25 pg/mL | 32 |
| Influenza virus | Antibody | Impedance immunosensor | 10 pg/mL | 33 |
| Avian Influenza virus | Complementary for HA strands | Luminescence sensor | 7 pM | 34 |
| H1N1 | Micro RNA | Non-PCR MARS assay | 1 nM | 35 |
| Influenza A H1 pdm | DNA | RT-PCR | 7.5 pg/mL | 36 |
| Influenza virus H5N1 | Antibody | Waveguide mode sensor | 1 nM | 19 |
Furthermore, to determine the specific interaction of HA with GNP-aptamer on the MWCN-IDE surface, control experiments were performed with the complementary sequences of aptamer [against HA of A/California/07/2009 (pdmH1N1)] and with two different influenza strains, influenza A/Panama/2007/1999 (H3N2) and influenza B/Tokyo/53/99. The values obtained were compared with the specific interaction by HA of influenza A/California/07/2009 against the specific aptamer. It was found that HA of pdmH1N1 could specifically interact only with the aptamer-GNP. Other molecules did not show significant current changes with aptamer-GNP, indicating the specific detection of HA by its aptamer (Figure 7C). Due to the specific binding region of HA with the aptamer, it could easily discriminate the other closely related strains. Previously, Lakshmipriya et al.24 generated an aptamer against influenza/B/Tokyo that could differentiate other closely related influenza strains. This study also validated the reproducibility of current levels with different preparations of bare, MWCN-, aptamer-GNP-, ethanolamine- and HA-modified surfaces (Figure 7D). The analysis clearly resulted in higher reproducibility rates on the MWCN-IDE sensing surface with minimal error range.
Specific Detection Of HA On Sandwich Patterns: Signal Enhancement
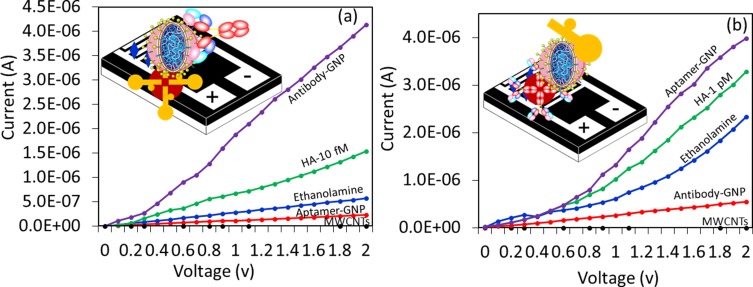
The sandwich pattern is a specific method of detection and has been widely appreciated in the field of sensors. In general, sandwich patterns with mono- and polyclonal antibodies were used to improve the performance of detection.25–27 Both types of antibodies were used either as capture or detection probes. Since monoclonal antibodies have a higher specificity with a single epitope, they have been used as the detection probe predominantly to detect and quantify minor differences with antigen binding. Polyclonal antibodies have been used as capture antibodies to pull down possible antigen molecules at a higher rate. Highly sensitive molecules were chosen as the capture probe to improve the limit of detection. In this study, comparative analysis was performed with the sandwich pattern of aptamer-HA-antibody and antibody-HA-aptamer. To that end, a lower level of HA was used to evaluate the efficiency of the sandwich pattern. In the case of the aptamer-HA-antibody, 10 fM of HA was used for detection, while 1 pM was used in the case of the antibody-HA-aptamer. In both cases, the current response was clearly increased compared to the single probe system, revealing a possible improvement in the sensitivity of detection with the sandwich pattern (Figure 8A and B).
Figure 8.
Sandwich assay. (A) aptamer-HA-antibody. 500 nM of aptamer-GNP was immobilized on the MWCN surfaces, and then 10 fM of HA was tested. A 250 nM antibody was used to make the sandwich. (B) Antibody-HA-aptamer. Approximately 250 nM of antibody-GNP was immobilized on the MWCN surfaces, and 1 pM of HA interacted. 500 nM of aptamer was used to make the sandwich pattern.
Conclusion
The current study demonstrated possible ways to improve the detection of the pandemic influenza virus (pdmH1N1) by multiwalled carbon nanotube (MWCNT)-modified interdigitated electrode surface (IDE). The two predominant probes, aptamers and antibodies, were generated against hemagglutinin of pdmH1N1 and compared to the detection levels. The utilization of aptamer as the probe has shown 100-fold higher sensitivity than the antibody-based detection and 7-fold higher sensitivity than the detection shown with the surface plasmon resonance. Furthermore, a higher specificity and reproducibility were clearly demonstrated by discriminating different types and sub-types of influenza viral strains. In addition, a sandwich aptamer-HA antibody pattern improved the detection of HA on the MWCN-IDE sensing surface. The current study, employing nanocomposite-based modification on the dielectric sensing surface, clearly demonstrated a suitable sensing system for influenza surveillance and establishes a foundation for detecting and discriminating other medically important biomarkers. Furthermore, the demonstrated system can operate with both alternating current (AC) and direct current (DC), demonstrating the portability of this system. With this portability, the system will be ideal for point-of-care detection and influenza surveillance.
Disclosure
The authors have no conflicts of interest to declare in this work.
References
- 1.Kumar PKR. Monitoring intact viruses using aptamers. Biosensors. 2016;6(3):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar PKR. Systematic screening of viral entry inhibitors using surface plasmon resonance. Rev med virol. 2017;29(August):1–12. doi: 10.1152/ajplegacy.1975.229.3.754 [DOI] [PubMed] [Google Scholar]
- 3.Subbarao K, Katz J. Avian influenza viruses infecting humans. Cell Mol Life Sci. 2000;57(12):1770–1784. doi: 10.1007/PL00000657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matrosovich MN, Krauss S, Webster RG. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology. 2001;281(2):156–162. doi: 10.1006/viro.2000.0799 [DOI] [PubMed] [Google Scholar]
- 5.Skehel JJ, Wiley DC. Influenza haemagglutinin. Vaccine. 2002;20:S51–4. doi: 10.1016/S0264-410X(02)00131-7. [DOI] [PubMed] [Google Scholar]
- 6.Liao HY, Hsu CH, Wang SC, et al. Differential receptor binding affinities of influenza hemagglutinins on glycan arrays. J Am Chem Soc. 2010;132(42):14849–14856. doi: 10.1021/ja104657b [DOI] [PubMed] [Google Scholar]
- 7.Gopinath SCB, Kumar PKR. Aptamers that bind to the hemagglutinin of the recent pandemic influenza virus H1N1 and efficiently inhibit agglutination. Acta Biomater. 2013;9(11):8932–8941. doi: 10.1016/j.actbio.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 8.Nitsche A, Kurth A, Dunkhorst A, et al. One-step selection of Vaccinia virus-binding DNA aptamers by MonoLEX. BMC Biotechnol. 2007;7:48. doi: 10.1186/1472-6750-7-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruno J. Predicting the uncertain future of aptamer-based diagnostics and therapeutics. Molecules. 2015;20(4):6866–6887. doi: 10.3390/molecules20046866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao L, Huang Y, Dong Y, et al. Aptamers and aptasensors for highly specific recognition and sensitive detection of marine biotoxins: recent advances and perspectives. Toxins (Basel). 2018;10(11):427. doi: 10.3390/toxins10010035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang Y, Feng KJ, Chen JW, Jiang JH, Shen GL, Yu RQ. Electrochemical detection of thrombin by sandwich approach using antibody and aptamer. Bioelectrochemistry. 2008;73(1):76–81. doi: 10.1016/j.bioelechem.2008.04.024 [DOI] [PubMed] [Google Scholar]
- 12.Du F, Zhu L, Dai L. Carbon nanotube-based electrochemical biosensors. Biosens Based Nanomater Nanodevices. 2017;17(1):7–14. [Google Scholar]
- 13.Vashist SK, Zheng D, Al-Rubeaan K, Luong JHT, Sheu FS. Advances in carbon nanotube based electrochemical sensors for bioanalytical applications. Biotechnol Adv. 2011;29(2):169–188. doi: 10.1016/j.biotechadv.2010.10.002 [DOI] [PubMed] [Google Scholar]
- 14.Gopinath SCB, Perumal V, Kumaresan R, et al. Nanogapped impedimetric immunosensor for the detection of 16Â kDa heat shock protein against Mycobacterium tuberculosis. Microchim Acta. 2016;183(10):2697–2703. [Google Scholar]
- 15.Letchumanan I, Md Arshad MK, Balakrishnan SR, Gopinath SCB. Gold-nanorod enhances dielectric voltammetry detection of c-reactive protein: a predictive strategy for cardiac failure. Biosens Bioelectron. 2019;130(October 2018):40–47. doi: 10.1016/j.bios.2019.01.042. [DOI] [PubMed] [Google Scholar]
- 16.Gopinath SCB, Misono TS, Kawasaki K, et al. An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits haemagglutinin-mediated membrane fusion Printed in Great Britain. J Gener Virol. 2007;87(3):479–487. [DOI] [PubMed] [Google Scholar]
- 17.Gopinath SCB, Hayashi K, Kumar PKR. Aptamer that binds to the gD protein of herpes simplex Virus 1 and efficiently inhibits viral entry. J Virol. 2012;86:6732–6744. doi: 10.1128/JVI.00377-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gopinath SCB, Misono TS, Kawasaki K, et al. An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits haemagglutinin-mediated membrane fusion. J Gen Virol. 2006;87(2006):479–487. doi: 10.1099/vir.0.81508-0 [DOI] [PubMed] [Google Scholar]
- 19.Gopinath SCB, Awazu K, Fujimaki M. Detection of influenza viruses by a waveguide-mode sensor. Anal Methods. 2010;2(12):1880. [Google Scholar]
- 20.Xu R, McBride R, Nycholat CM, Paulson JC, Wilson IA. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J Virol. 2012;86(2):982–990. doi: 10.1128/JVI.06322-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams S, Davies P, Bowen J, Allender C. Controlling the nanoscale patterning of AuNPs on silicon surfaces. Nanomaterials. 2013;3(1):192–203. doi: 10.3390/nano3010192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castillo F, Perez E, de la Rosa E. Adsorption of gold nanoparticles on silicon substrate and their application in Surface Enhancement Raman Scattering. Rev Mex Fis. 2011;57(2):61–65. [Google Scholar]
- 23.Vashist SK, Marion Schneider E, Lam E, Hrapovic S, Luong JHT. One-step antibody immobilization-based rapid and highly-sensitive sandwich ELISA procedure for potential in vitro diagnostics. Sci Rep. 2014;4:4407. doi: 10.1038/srep04407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lakshmipriya T, Fujimaki M, Gopinath SCB, Awazu K. Generation of anti-influenza aptamers using the systematic evolution of ligands by exponential enrichment for sensing applications. Langmuir. 2013;29:15107–15115. doi: 10.1021/la4027283 [DOI] [PubMed] [Google Scholar]
- 25.Lakshmipriya T, Horiguchi Y, Nagasaki Y. Co-immobilized poly (ethylene glycol)-block-polyamines promote sensitivity and restrict biofouling on gold sensor surface for detecting factor IX in human plasma. Analyst. 2014;139(16):3977–3985. doi: 10.1039/c4an00168k. [DOI] [PubMed] [Google Scholar]
- 26.Lakshmipriya T, Fujimaki M, Gopinath SCB, Awazu K, Horiguchi Y, Nagasaki Y. A high-performance waveguide-mode biosensor for detection of factor IX using PEG-based blocking agents to suppress non-specific binding and improve sensitivity. Analyst. 2013;138:2863–2870. doi: 10.1039/c3an00298e. [DOI] [PubMed] [Google Scholar]
- 27.Vashist SK. Graphene-based immunoassay for human lipocalin-2. Anal Biochem. 2014;446:96–101. doi: 10.1016/j.ab.2013.10.022. [DOI] [PubMed] [Google Scholar]
- 28.Nidzworski D, Siuzdak K, Niedziałkowski P, et al. A rapid-response ultrasensitive biosensor for influenza virus detection using antibody modified boron-doped diamond. Sci Rep. 2017;7:15707. doi: 10.1038/s41598-017-15806-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kukushkin VI, Ivanov NM, Novoseltseva AA, et al. Highly sensitive detection of influenza virus with SERS aptasensor. PLoS One. 2019;14(4):e0216247. doi: 10.1371/journal.pone.0216247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee N, Wang C, Park J. User-friendly point-of-care detection of influenza A (H1N1) virus using light guide in three-dimensional photonic crystal. RSC Adv. 2018;8:22991–22997. doi: 10.1039/C8RA02596G [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horiguchi Y, Goda T, Matsumoto A, Takeuchi H, Yamaoka S, Miyahara Y. Direct and label-free influenza virus detection based on multisite binding to sialic acid receptors. Biosens Bioelectron. 2017;92:234–240. doi: 10.1016/j.bios.2017.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng C, Cui H, Wu J, Eda S. A PCR-free point-of-care capacitive immunoassay for influenza A virus. Microchim Acta. 2017;184:1649–1657. doi: 10.1007/s00604-017-2140-4 [DOI] [Google Scholar]
- 33.Nidzworski D, Pranszke P, Grudniewska M, Krol E, Gromadzka B. Universal biosensor for detection of influenza virus. Biosens Bioelectron. 2014;59:239–242. doi: 10.1016/j.bios.2014.03.050 [DOI] [PubMed] [Google Scholar]
- 34.Ye WW, Tsang MK, Liu X, Yang M, Hao J. Upconversion luminescence resonance energy transfer (LRET)-based biosensor for rapid and ultrasensitive detection of avian influenza virus H7 subtype. Small. 2014;10(12):2390–2397. doi: 10.1002/smll.201303766 [DOI] [PubMed] [Google Scholar]
- 35.Loo JFC, Wang SS, Peng F, et al. A non-PCR SPR platform using RNase H to detect MicroRNA 29a-3p from throat swabs of human subjects with influenza A virus H1N1 infection. Analyst. 2015;140(13):4566–4575. doi: 10.1039/c5an00679a [DOI] [PubMed] [Google Scholar]
- 36.Yamanaka K, Saito M, Kondoh K, et al. Rapid detection for primary screening of influenza A virus: microfluidic RT-PCR chip and electrochemical DNA sensor. Analyst. 2011;136:2064–2068. doi: 10.1039/c1an15066a [DOI] [PubMed] [Google Scholar]