Abstract
Plant root system architecture in response to nitrate availability represents a notable example to study developmental plasticity, but the underlying mechanism remains largely unknown. Xyloglucan endotransglucosylases (XTHs) play a critical role in cell wall biosynthesis. Here we assessed the gene expression of XTH1-11 belonging to group I of XTHs in lateral root (LR) primordia and found that XTH9 was highly expressed. Correspondingly, an xth9 mutant displayed less LR, while overexpressing XTH9 presented more LR, suggesting the potential function of XTH9 in controlling LR development. XTH9 gene mutation obviously alters the properties of the cell wall. Furthermore, nitrogen signals stimulated the expression of XTH9 to promote LRs. Genetic analysis revealed that the function of XTH9 was dependent on auxin-mediated ARF7/19 and downstream AFB3 in response to nitrogen signals. In addition, we identified another transcription factor, OBP4, that was also induced by nitrogen treatment, but the induction was much slower than that of XTH9. In contrast to XTH9, overexpressing OBP4 caused fewer LRs while OBP4 knockdown with OBP4-RNAi or an artificial miRNA silenced amiOBP4 line produced more LR. We further found OBP4 bound to the promoter of XTH9 to suppress XTH9 expression. In agreement with this, both OBP4-RNAi and crossed OBP4-RNAi & 35S::XTH9 lines led to more LR, but OBP4-RNAi & xth9 produced less LR, similar to xth9. Based on these findings we propose a novel mechanism by which OBP4 antagonistically controls XTH9 expression and the OBP4-XTH9 module elaborately sustains LR development in response to nitrate treatment.
Author summary
Nitrate is not only a nutrient, but also a signal that controls downstream signaling genes at the whole-plant level. In plants, changes in root system architecture in response to nitrate availability represent a notable example of developmental plasticity in response to environmental stimuli. However, the molecular mechanisms underlying nitrate-associated modulation are largely unknown. Here, we identified a nitrogen-responsive signaling module that comprises both xyloglucan endotransglucosylase 9 (XTH9) and the Dof transcription factor OBP4 and controls lateral root (LR) development. We used root gravitropic bending assays to observe the gene expression of group 1 xyloglucan endotransglucosylases (XTHs) involved in LR primordia. The results showed that XTH9 expression patterns were changed and that xth9 knockout mutants displayed altered LR growth. XTH9 was expressed in the LRs and in response to nitrate treatment, and the xth9 mutants were defective in nitrate-promoted LR growth. Moreover, XTH9 overexpression increased LR length and increased tolerance to low-nitrate stress. We found that OBP4 could negatively regulate XTH9 and inhibited root growth. OBP4 and XTH9 worked downstream of ARF7/9. We conclude that OBP4 and XTH9 constitute a regulatory module which contributes to LR growth in response to different environmental nitrate concentration signals.
Introduction
In the biosphere, nitrate is the major form of nitrogen, and nitrate availability is important for plant development. Nitrate is not only a nutrient, but also a signal that controls downstream signaling genes at the whole-plant level [1, 2]. Previous genomic studies have shown that the nitrate response is comprehensive, but the molecular mechanisms of nitrate signal transduction and the downstream gene expression changes that lead to developmental responses, such as changes in root system architecture (RSA), are still unclear. In the last few years, several nitrate regulatory genes functioning in the primary nitrate response have been characterized. One key regulator is nitrate transporter 1.1 (NRT1.1), which functions not only as a dual-affinity nitrate transporter, but also as a nitrate sensor [3–5]. The CBL-interacting protein kinases, CIPK8 and CIPK23, are involved in the primary nitrate response [6–8]. Similarly, the MADS family NO3−-inducible transcription factor (TF) ANR1 and Lateral Organ Boundaries Domain (LBD) regulate lateral root (LR) growth in response to nitrate treatment [8, 9]. NIN-like protein 7 (NLP7) orchestrates the early response to nitrate in plants [10], and via a systems biology approach, TGACG SEQUENCE -SPECIFIC BINDING PROTEIN 1(TGA1), AUXIN SIGNALING F-BOX 3 (AFB3), ARABIDOPSIS NAC DOMAIN-CONTAINING PROTEIN 79 (NAC4), and OBP4 genes have been identified as nitrate regulators involved in nitrate signaling [2,11,12].
In Arabidopsis, RSA plays a role in nutrient and water absorption, which is important for plant growth and development. Thus, plants require modulation of RSA plasticity to respond to various dynamic environmental conditions. RSA is a plastic characteristic in many plant species, but the plasticity of the RSA phenotype is controlled via inherent genetic mechanisms. Lateral root primordia (LRP) are derived from many primary root pericycle cells. New LRs originate from LRP, followed by several rounds of cell division [13]. LRP play a critical role in the initial stages of LR initiation [14]. LRs serve as a fascinating model for determining how adjacent cells synchronize growth and development [15, 16]. The cell cycle inhibitor Kip-associated gene 2 exhibits important gene functions in the cycling of pericycle cells and blocks the G1-to-S phase transition [17]. In a more distal region, protoxylem pericycle cells proceed to the G2 phase and become competent for LR initiation [18, 19]. Furthermore, several important studies linking cell wall modifications and LR development have been published [20, 21], and several studies have also reported a role for xyloglucan endotransglucosylases (XTHs) during LR development [20, 22].
Auxin is an important hormone in LR development [23]. To emerge, the LRP must break through the overlying tissues (including endodermal, cortical, and epidermal cells). Development subsequently progresses via auxin signaling reprogramming of overlying tissues to undergo cell separation, promoting the emergence of the LRs [20, 24–27]. Auxin signaling during the formation of LRP is associated with protein degradation. First, auxin binds to AFB/TIR1 receptors and mediates their association with and the degradation of indoleacetic acid (IAA) proteins. Furthermore, auxin-induced Aux/IAA degradation enables auxin-response factors (ARFs) to transactivate the transcription of downstream genes [28]. Both Aux/IAA proteins and ARF transcriptional regulators are critical factors for sensing the concentration of auxin and subsequently translating that signal into the expression of downstream genes, further influencing the output of patterning in plant growth and development [29, 30].
XTHs play pivotal roles in the reconnection and splitting of xyloglucan crosslinks by catalyzing the molecular grafting of the xyloglucans to form a framework [31–33]. XTH also plays important roles in plant cell wall construction and disassembly [33]. XTHs compose a large gene family in multiple plant species. A total of 33 XTH genes have been identified in Arabidopsis, and one-third of them occur as clusters, potentially reflecting genomic duplication events [34]. Every member of the XTH gene family is likely controlled by specific cues that respond to the cell wall dynamics of certain tissues and/or cell types [35]. For example, AtXTH27 is crucial for tertiary vein development in rosette leaves and is also involved in cell wall modification of tracheary elements [36], and the function of AtXTH31 is important during cell elongation under Al stress because it modulates XTH activity [37]. Comprehensive analyses of XTH gene expression have revealed that many of these enzymes are expressed in the roots [38]. Despite these studies, little is known about the role of XTH genes in nitrate signaling with respect to the regulation of root system development.
In the present study, to determine whether XTH genes are responsible for LR development, we used a reverse genetics method to analyze the root development phenotypes of class 1 XTH genes. We subsequently identified an XTH9 mutation that causes a defect in LR development. Analysis of proXTH9 fused to GUS revealed that XTH9 is expressed in the LRs in response to nitrate treatment, and auxin regulates XTH9 expression in the roots via ARF7/19. Furthermore, we observed that the nitrate regulation of XTH9 occurs downstream of the AFB3-IAA14 signaling pathway. We also identified a Dof TF, OBP4, that negatively regulates XTH9. OBP4 was reported to be regulated by nitrate directly as a signal, and systems approaches showed that it was acting downstream of AFB3 [2, 12]. The OBP4 gene fine-tunes LR development in response to environmental nitrate availability. OBP4 and XTH9 constitute a regulatory module which controls LR growth in response to variation in nitrate concentrations. Thus, we deduce that the OBP4-XTH9 regulatory module has a specific function in LR development in response to nitrate signaling in Arabidopsis.
Results
Expression of group I XTH genes in LRP
The XTH/hydrolyase enzyme plays a role in the endo-transglucosylation and/or hydrolysis of xyloglucan molecules. In the Arabidopsis genome, 33 genes encoding XTH proteins have been isolated. Individual members of this gene family exhibit specific expression patterns, both temporally and spatially. Most of the XTH genes are functionally redundant, but some members have roles in specific aspects of plant growth and development. Among these members, 11 (XTH1-11) cluster into group I [38–40]. Because the expression patterns of many group I XTH genes occur in the roots, we initially investigated whether the expression of the cell wall modification-related XTH gene changed during LR development.
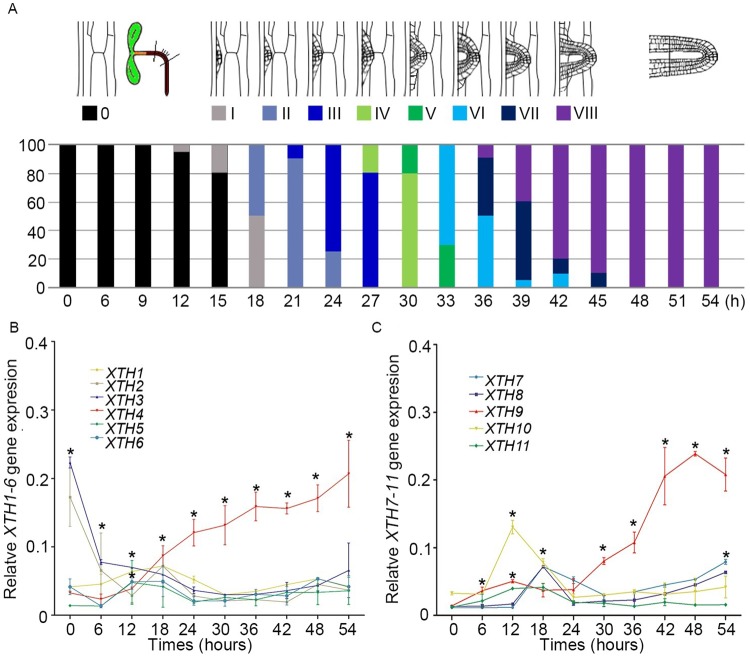
Both mechanical and/or gravitropic stimuli can facilitate the initiation of LRs [41–44]. LR initiation occurs in a highly synchronized manner on the outer surface of a bending root and can be measured with 90° gravitropic stimulus assays [27, 45]. We can classify this developmental progress into many different stages. In stage I, LRP was observed at 18 hours post-gravitropic induction (pgi), and subsequently, at approximately 6-hour intervals, each subsequent primordium stage was measured until the LR emerged at 42 hours pgi in stage VIII (Fig 1A). Because we measured the microdissected root bends every 6 hours pgi, we profiled XTH gene expression at all stages of LR development (Fig 1A). In this study, five-day-old Col-0 plants grown under normal conditions were used for gravity stimulus assays, and then the transcriptional profiles of the 11 XTHs were measured. We found increased expression levels of XTH4 and XTH9 during LR development. XTH10 gene expression was strongly increased during the first 12 hours after treatment and then sharply decreased (Fig 1B and 1C). The transcriptional level of other genes in group I did not present a dramatic change, suggesting the potential function of XTH4, XTH9, or XTH10 during LR initiation. To verify the accuracy of our experimental system, we further assessed the transcript level of the well-known LR regulator LBD29. Consistent with the previous study (S1 Fig) [46, 47], the transcriptional level of LBD29 was substantially upregulated at the indicated time points.
Fig 1. Transcriptomic analysis of class 1 XTH genes during lateral root (LR) initiation.
(A) After gravitropic stimulation to induce the synchronized initiation of lateral root primordia (LRP) at the surface of bending roots, LRP stages from I to VIII, in accordance with previous reports, were assessed every 6 hours for 0 to 54 hours after gravity stimulation and are represented here as a percentage of the total number of observed LRP at each time point. At least 70–80 LRP were observed at each time point. (B, C) All class 1 XTH gene expression patterns at each time point during LR initiation were measured every 6 hours from 6 to 54 hours pgi. Bending roots of a population of 5-day-old seedlings were microdissected at 10 time points, which were then used for RNA extraction (approximately 220 per time point for three independent replicates). Expression patterns of all class 1 XTH genes are shown. The error bars denote SDs (n = 4). Student’s t-test was applied. The asterisks (*) show statistically significant differences (p<0.05).
XTH9 mutation decreased LR density
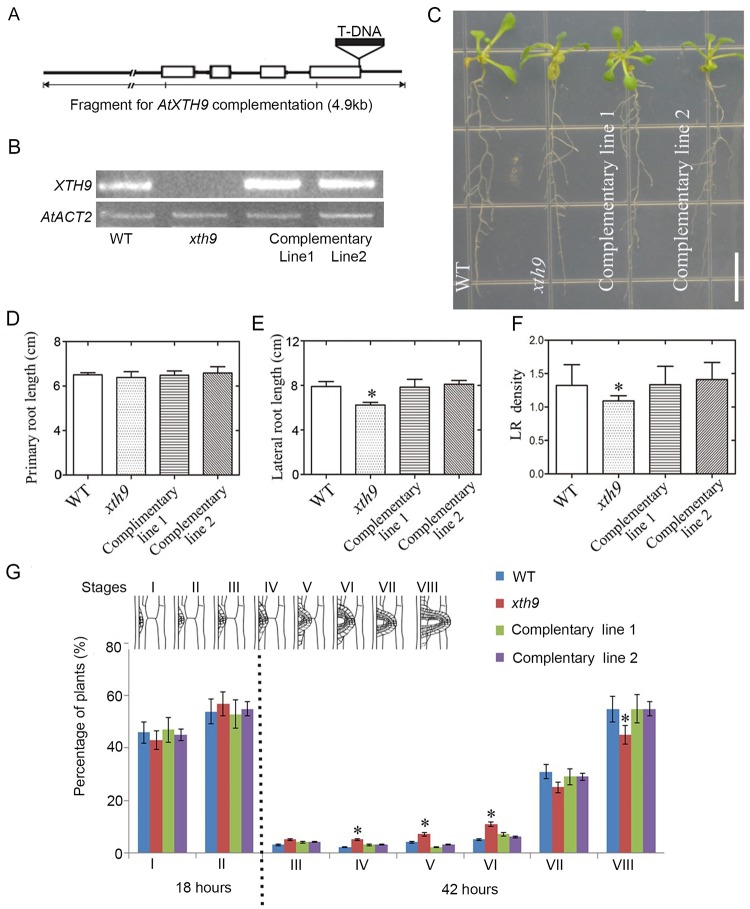
The expression patterns of XTH genes were altered during root bending under gravity stimuli; therefore, we sought to obtain further insight into the roles of the group I XTH family member, XTH1-11. We isolated and analyzed the phenotypes of homogeneous T-DNA insertion lines to observe the LR phenotypes of all 11 group I XTH genes (S2A Fig & S3 Table). Among these lines, the SALK_101024 line (named xth9) had a T-DNA insertion in the 3′ UTR of XTH9 (Fig 2A). After assessing the mRNA level of XTH9 in this mutant background, we determined that these plants represented a knockout line (Fig 2B). We also assessed other T-DNA insertion lines of XTH9 (SALK_063401, SALK_002571, and SALK_023274). These mutations were unable to downregulate XTH9 expression, which was likely due to all the T-DNA inserts detected in the promoter region of XTH9. After careful observation, we discovered that the xth9 mutant exhibited defective LR growth. However, no obvious root phenotype was observed in XTH4, XTH10 (S2B and S2C Fig) or the other group I XTH gene mutants. Therefore, we focused on XTH9. Under the experimental growth conditions, this gene mutation reduced LR development, but did not affect main root development (Fig 2C–2F). Furthermore, we used the full-length 4.9-kb genomic DNA of XTH9 to complement the mutant phenotype, and the results showed that the transgenic lines exhibited LR development that was similar to that of the wild-type (WT; Col-0) (Fig 2C). Next, the WT and mutant roots were exposed to a gravitropic stimulus, and the LRP were counted and staged at 18 and 42 hours pgi. The WT plants accumulated stage I and II LRP at 18 hours pgi and stage VII and VIII LRP at 42 hours pgi. However, compared with the WT, the xth9 mutant exhibited a higher percentage of stage III-VI LRP and a lower accumulation of stage VII-VIII LRP at 42 hours pgi (Fig 2G). The lateral root emergence (LRE) of the complemented plants was consistent with that of the WT plants (Fig 2G). Altogether, these results suggested that XTH9 positively regulates LR development; thus, we further examined the molecular mechanism underlying this phenotype.
Fig 2. XTH9 gene mutation affects lateral root (LR) development.
(A) The genomic organization of the XTH9 gene. The white boxes show the positions and sizes of the XTH9 gene exons. The black box indicates the structure of the T-DNA, the site of which is indicated by the triangle. The genomic sequences used to complement the xth9 mutation are underlined with a thick line. (B) XTH9 transcript levels in wild-type (WT) (Col-0) plants, the xth9 mutant, and xth9 complementary lines. The mRNA abundance of the XTH9 gene in the roots was measured using RT-PCR in various genotype backgrounds of Arabidopsis. (C) Root phenotypes of WT plants, the xth9 mutant, and complementary lines. The plants were grown vertically on media for 10 days. The white bar indicates 1 cm. (D) Primary root length and (E) total LR length. (F) LR density analysis of WT plants, the xth9 mutant, and xth9 complementary lines. The error bars denote SDs (n = 16–21). (G) Phenotypic analysis of lateral root emergence (LRE) was achieved by synchronizing LR formation with a gravity stimulus for 18 and 42 hours. Compared with the WT plants, the xth9 mutants showed delayed LRE. Mutant plants transformed with a 4.9-kb-long full-length XTH9 genomic fragment exhibited a WT LRE phenotype for LR induction. The data are shown as percentages, and the error bars represent SDs (n = 15–17). At least 80–100 total LR primordia were observed for each plant, and the asterisks (*) indicate statistically significant differences (p<0.05).
Since XTHs are involved in hemicellulose regulation, we tested the hemicellulose content of xth9 mutant. The total residual sugar in hemicellulose was reduced in xth9 mutant (S3A Fig). XTH can cleave or rejoin xyloglucan (XyG) chains, resulting in modified content or structure of the XyG, and XyG is shown to regulate the ability of cell wall extension in response to environmental changes. We analyzed the xyloglucan content in the root cell wall by enzyme digestion and MALDI-TOF analysis. The overall XyG repeat was obviously reduced in xth9 mutant, especially XXXG, XXFG, XLFG, and XXLG (S3B Fig). These results suggest that mutations in the XTH9 alter the properties of cell wall. The Group II XTH gene family has XET activities, we found that the XET activity of xth9 mutant was reduced under our growth conditions by using endogenous xyloglucan as the donor substrate (S3D Fig). These data indicate that the XET action in the roots requires XTH9.
XTH9 expression in the LRs and in response to nitrate treatment
Given the upregulation of XTH9 in the 90° gravitropic stimulus assay and the decreased LR growth in the xth9 mutant, RT-qPCR was used to examine the XTH9 mRNA levels in various plant organs, including shoots, flowers, roots, buds, and rosette leaves, as well as in seedlings. XTH9 was expressed in all the organs examined and relatively high expression levels were detected in seedlings, flowers, and roots (S3C Fig). Furthermore, we generated a proXTH9::GUS transgenic line driven by its 2.3-kb native promoter. We obtained 11 independent transgenic lines, and we selected three of them to observe GUS signals in the LRP. GUS signals were observed during LR initiation and elongation in all these lines. XTH9 expression was observed across the entire LR (Fig 3A–3H). This expression pattern suggests an important role for this gene in root development. To further identify XTH9 nitrate responses and understand their involvement in LR growth, we performed an XTH9 expression survey in Arabidopsis roots. First, we checked the expression of XTH9 in response to various abiotic stresses (mannitol, heat, and salt), nutrients (nitrogen and phosphorus), and hormones (ABA and GA). The results showed that XTH9 expression increased in response to nitrate treatments (S4 Fig). Therefore, we focused on XTH9-mediated LR formation and nitrate signaling. Nitrate significantly affects LR development [48], and we observed that nitrate treatment promoted XTH9 expression (Fig 3I–3M). Additional transcript profiling and reporter studies demonstrated that XTH9 is expressed in response to nitrate treatment. We concluded that the XTH9 gene regulates LR development in response to nitrate signals.
Fig 3. Observation of proXTH9::GUS transgenic lines during lateral root (LR) development.
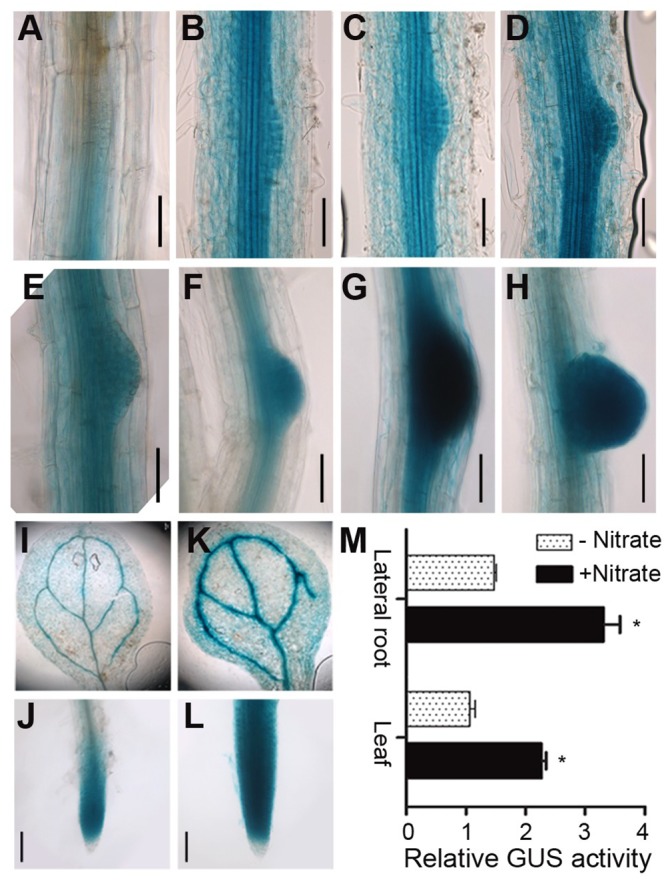
(A–H) GUS staining in proXTH9::GUS Arabidopsis roots at various developmental stages. The seedlings were incubated on media supplemented without (I, J) or with (K, L) 5 mM nitrate for 6 hours. Images of the leaves (I, K) and LRs (J, L) were obtained, and representative images are shown. (M) Relative GUS activity before and after nitrate treatment is shown. Leaf GUS activity in the absence of nitrate was used as a control. The asterisks (*) show statistically significant differences (p<0.05) and the error bars represent SDs (n = 4).
The xth9 mutant was defective in nitrate-promoted LR growth
Furthermore, datasets involving various concentrations of N-supplied roots were analyzed. We conducted RT-qPCR analysis of XTH9 in the roots of 12-day-old Col-0 Arabidopsis seedlings grown under various concentrations of nitrate (Fig 4).
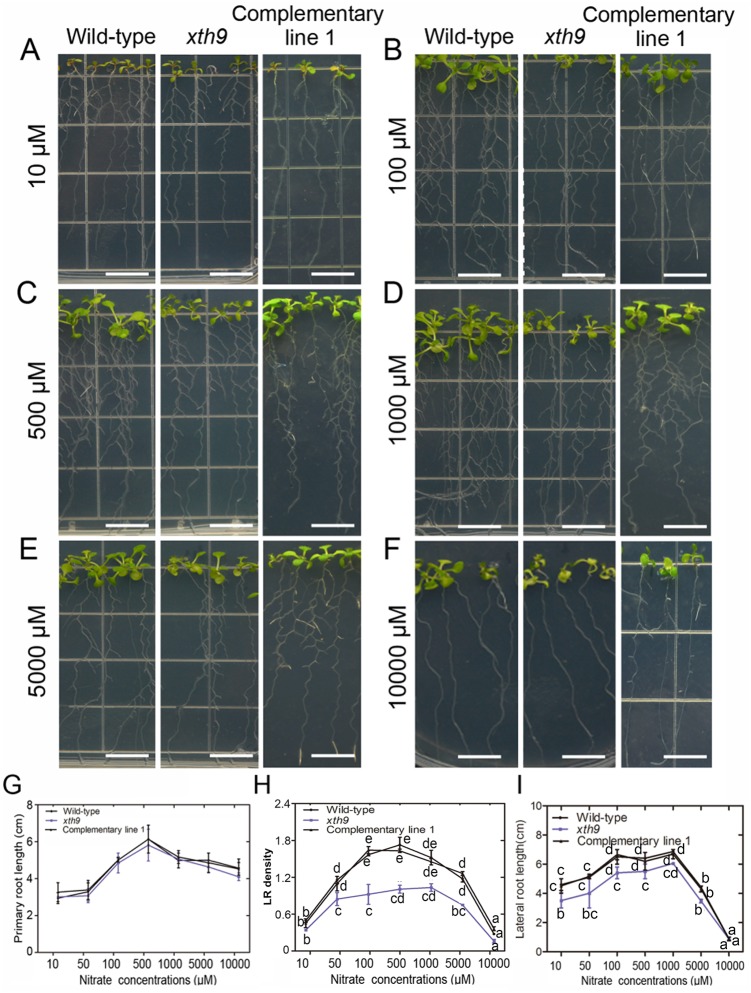
Fig 4. XTH9 gene mutation altered lateral root (LR) development in response to nitrate treatments.
(A–F) Effects of various concentrations of nitrate (10, 50, 100, 500, 1000, 5000, and 10000 μM) on root growth. WT, xth9 mutant, and the complementary line 1 were grown vertically in media supplemented with various nitrate concentrations for 8 days. (G) Primary root length; (H) LR density; (I) Lateral root length were quantified. LR density was calculated by dividing the LR number per 1 cm of primary root. The error bars represent SDs (n = 22–31). Bars with different capital letters indicate significant differences among different concentrations of nitrate treatment in the given genotype by ANOVA analysis. The white bars indicate 1.2 cm.
The effects of nitrate on LR growth depended on the concentration. Moderate concentrations promoted LR growth, but low or high levels of nitrate inhibited LR growth [49]. We then investigated the root development of WT and xth9 mutant plants on media supplemented with various concentrations (10, 50, 100, 500, 1000, 5000, and 10000 μM) of nitrate. The results showed that, under N-deficient conditions (10 μM nitrate), both the WT and xth9 plants showed N-deficient phenotypes with strongly inhibited LR initiation. Additionally, the xth9 plants had fewer LRs than the WT plants (Fig 4A, 4H and 4I). When the nitrate concentration increased to 100–1000 μM, these moderate concentrations of nitrate substantially promoted LR growth in WT plants, but the xth9 plants were insensitive to these nitrate concentrations. Moreover, the LR density and total root length were also lower in the xth9 plants than in the WT control plants (Fig 4B–4D, 4H and 4I). High nitrate concentrations (5000–10000 μM) inhibited LR initiation in both the WT and xth9 mutant plants (Fig 4E–4I). In response to increasing nitrate concentrations, the LR density and total LR length curve for the xth9 mutant plants was lower than that for WT plants (Fig 4H and 4I). However, in the complementary transgenic lines, the phenotype recovered to that of the WT (S5 Fig). Therefore, in the xth9 mutant, nitrate-promoted LR growth was defective, but the main root elongation was not substantially affected (Fig 4G).
XTH9 gene overexpression increased LRs and enhanced tolerance to low-nitrate stress
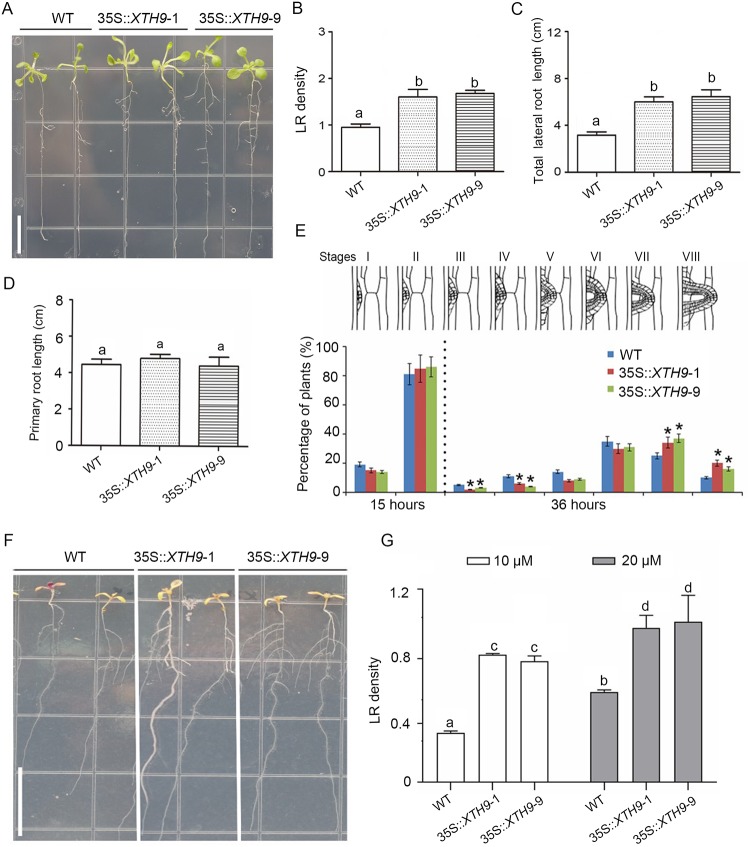
Due to the decreased LR growth in the xth9 mutant, we investigated the root system development in constitutive XTH9 overexpression lines. Nine of the 10 independent transgenic lines we obtained presented high XTH9 expression levels (S6 Fig), and the transgenic plants exhibited substantially increased LRs (Fig 5A–5D). The WT and transgenic roots were then exposed to a gravitropic stimulus, and LRP were counted and staged at 15 and 36 hours pgi (Fig 5E). The WT plants accumulated stage I and II LRP at 15 hours pgi and stage III to VIII LRP at 36 hours pgi. LR initiation and first divisions were not affected in the 35S::XTH9 plants, but showed an increased percentage of stage VII–VIII LRP at 36 hours pgi (Fig 5E). This result indicated that XTH9 overexpression affected lateral root emergence (LRE). Because the xth9 gene mutation decreased sensitivity to nitrate treatment, we investigated whether XTH9 overexpression increased low-nitrate stress tolerance. The results showed that, on 10 μM nitrate media, WT and transgenic plants showed an N-deficient phenotype, but the transgenic plants exhibited much better growth and a substantially increased LR system (Fig 5F and 5G). Therefore, we concluded that XTH9 overexpression improved low-nitrate tolerance.
Fig 5. Overexpression of the XTH9 gene increased lateral root (LR) development and improved low-nitrate tolerance.
(A) Root phenotype observation and analysis of (B) LR density, (C) total LR length, and (D) primary root length in WT and XTH9 overexpression lines. Bars with different capital letters indicate significant differences among different nitrate treatment in the given genotype by ANOVA analysis. The white bar indicates 1 cm. (E) Phenotypic analysis of lateral root emergence (LRE) was achieved by synchronizing LR formation with a gravity stimulus for 15 and 36 hours. Compared with the WT (Col-0) plants, the XTH9 overexpression plants showed increased LRE. The data are shown as percentages, and the error bars represent SDs (n = 19–22). The error bars denote SDs (n = 16–23). Approximately 90 total LRP were observed in each plant. (F) WT and transgenic plants were grown vertically in media supplemented with 10 μM nitrate for 8 days. (G) Average LR lengths were quantified. The error bars represent SDs (n = 20–30). Bars with different capital letters indicate significant differences among different concentrations of nitrate treatment in the given genotype by ANOVA analysis. The white bars indicate 1.2 cm.
Auxin regulates XTH9 expression in the roots via ARF7 and ARF19
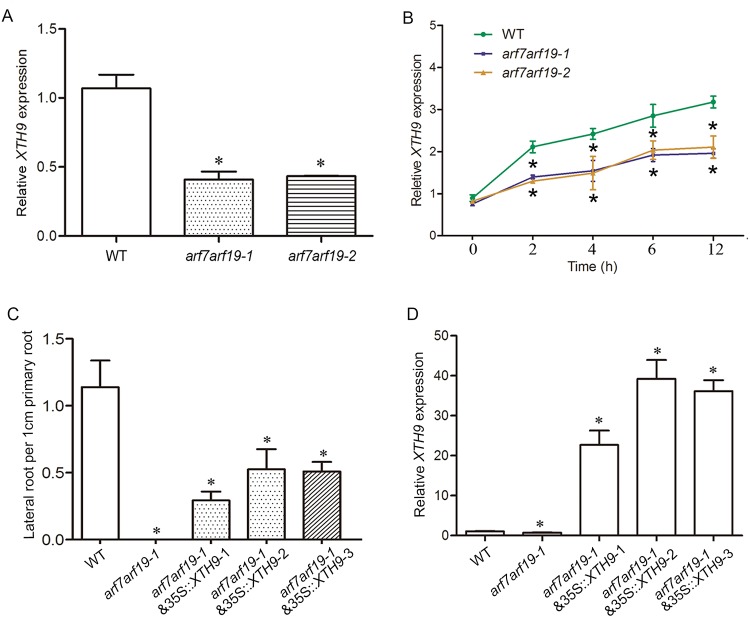
The products of ARF genes function as TFs that regulate downstream auxin-responsive genes. Among these genes, ARF7 and ARF19 play critical roles during LR development and show redundant functions. The arf7arf19 double mutant showed no LR initiation [50]; thus, we examined whether auxin regulates XTH9 expression through ARF7/19 in the roots. We initially investigated whether the regulatory mechanisms controlling auxin-promoted XTH9 expression depend on ARF. We observed that the XTH9 mRNA level was affected in the arf7arf19 double mutant background (Fig 6A), suggesting that XTH9 expression depends on the ARF TF [51–53]. Furthermore, transcript profiles of the roots of WT and arf7arf19 Arabidopsis mutants exposed to external auxin for varying lengths of time were examined by using RT-qPCR. Transcript profiling revealed that auxin-increased XTH9 mRNA expression was affected in the arf7arf19 mutant background (Fig 6B). Although XTH9 showed sustained auxin-dependent induction, a weakened auxin effect was detected in the arf7arf19 double mutants. Similarly, when the proXTH9::GUS reporter was expressed in arf7arf19, downregulated root GUS signaling was observed in arf7/19 & proXTH9::GUS plants [22] (S7 Fig). Thus, we concluded that auxin regulates XTH9 expression in an ARF7/19-dependent manner. To identify whether XTH9 acts downstream of ARFs to regulate LR development, we introduced 35S::XTH9 into arf7arf19 mutants to get 35S::XTH9 & arf7arf19 plants. As with 35S::XTH9, the 35S::XTH9 & arf7arf19 plant showed more LR development, indicating XTH9 is functional downstream of ARF7/ARF19 (Fig 6C and 6D).
Fig 6. Auxin regulates XTH9 expression in an ARF7/19-dependent manner.
(A) XTH9 expression level in the arf7arf19 double mutant background. RNA was isolated from 7-day-old Arabidopsis roots. (B) After treatment with 1 mM IAA for the indicated time, the wild-type (WT) and arf7arf19 double mutant roots were collected at the indicated time points. Values were determined from three independent plant roots. The auxin-dependent repression of XTH9 expression in the roots is ARF7/19 dependent. (C) Relative length of lateral root per 1 cm of primary root and (D) XTH9 expression levels in the WT, arf7arf19-1 and arf7arf19-1 & 35S::XTH9 transgenic plants. *indicates significant differences (p<0.05). The error bars represent SDs (n = 3).
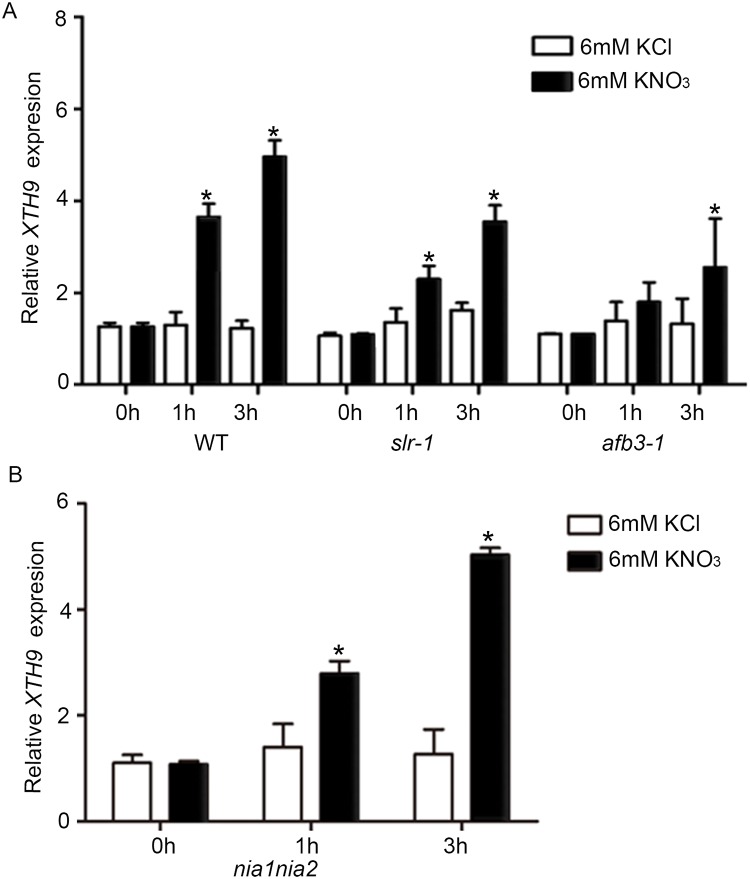
Nitrate regulation of XTH9 acts downstream of the AFB3-IAA14 signaling pathway
Previous studies involving systems approaches have revealed that the nitrate-regulated miR393/AFB3 module controls RSA [1, 2]. Because the cell wall-related gene XTH9 is predicted to be involved in auxin and nitrate signaling, XTH9 represents a likely downstream candidate of AFB3 for mediating the effects of nitrate on LR development. Therefore, we assessed XTH9 expression levels over time in response to nitrate treatment in WT, afb3-1, and slr-1 (a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene gain-of-function of IAA14) mutants using RT-qPCR. Compared with the WT plants, the mutant plants exhibited delayed transient responses to nitrate after treatment. The results showed that AFB3-IAA14 (SLR) function is required for the nitrate-mediated regulation of XTH9. The nitrate responses of XTH9 were compromised in the afb3-1 and slr-1 mutant backgrounds (Fig 7A). Previous studies have shown that AFB3 is induced by nitrate in a nitrate reductase-null nia1nia2 double mutant. Given that XTH9 function depends on AFB3, we assessed XTH9 expression levels in the nia1nia2 background in response to nitrate treatment. The results showed that these genes were still regulated by nitrate in the NR-null mutant, suggesting that the genes respond to nitrate, but not to N metabolites (Fig 7B), which is consistent with the hypothesis that the XTH9 gene is regulated by nitrate signaling. In addition, the LR phenotype in nitrate-treated WT, afb3-1, and slr-1 are shown in S8 Fig.
Fig 7. The nitrate response of XTH9 is altered in afb3-1, slr-1 mutant background and XTH9 is still regulated by nitrate in NR-null nia1nia2 mutant.
(A) WT (Col-0) and afb3-1 and slr-1 mutant plants were grown on media supplemented with ammonium succinate for one week and subsequently treated with 6 mM KNO3 or 6 mM KCl for 1–3 hours. The XTH9 gene expression level in the roots was measured via RT-qPCR. (B) Nitrate reductase-null plants (nia1nia2) were grown in media supplemented with ammonium succinate for one week and then treated with 6 mM KNO3 or 6 mM KCl for 1–3 hours. The RNA level of the XTH9 gene was measured via RT-qPCR. KCl treatment results are shown with white bars, and the KNO3 treatment results are shown with black bars. *indicates significant differences (p<0.05). The error bars represent SDs (n = 18–24).
The Dof TF OBP4 negatively regulates XTH9 in the roots
After assessing the XTH9 promoter, we identified many Dof-binding motifs (Fig 8A). To independently assess the role of other regulatory regions, we used an XTH9 promoter-deletion approach (S9A Fig). The approximately 2.3-kb-long XTH9 promoter was truncated at four different positions (Δ1, Δ2, Δ3, and Δ4) to generate increasingly shorter promoter fragment lengths, after which these constructs were subsequently transformed into the xth9 mutant background. Multiple transgenic lines expressing each proXTH9::XTH9 promoter deletion were initially scored for complementation of the xth9 LR phenotype. All Δ1 and Δ2 promoter deletion lines fully complemented the xth9 LR defect (S9B Fig). This result demonstrates that the 1100-bp promoter sequence of XTH9 is sufficient to promote LR emergence. In contrast, all Δ3 promoter deletion lines partially complemented the xth9 LR phenotype, whereas no complementation was observed for any of the Δ4 promoter deletion lines (S9B Fig).
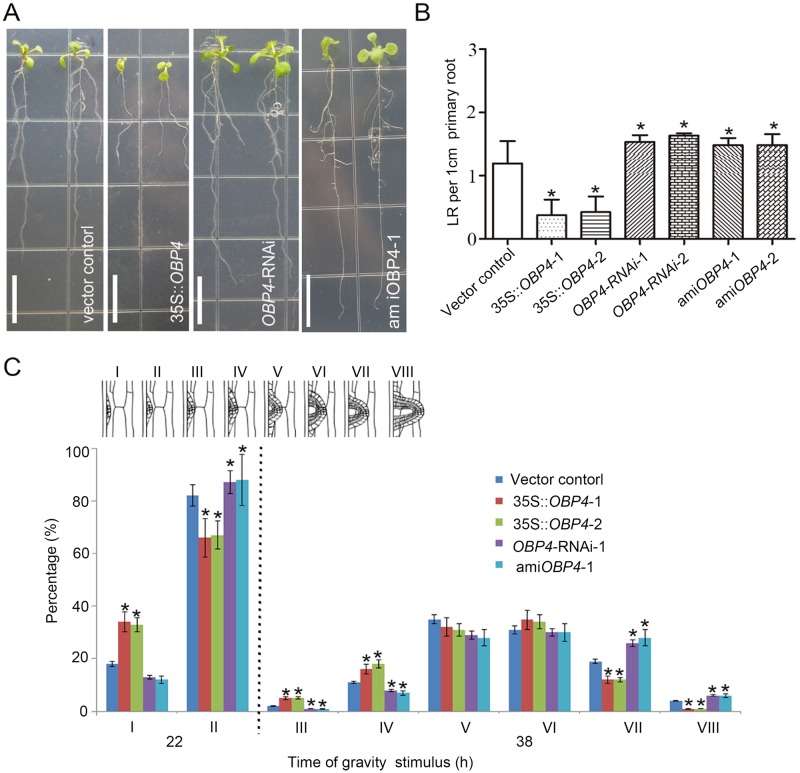
Fig 8. OBP4 regulates plant root system architecture.
(A) Phenotypic observation and (B) analysis of the root system in constitutive overexpression and knockdown OBP4 transgenic plants. The white bar indicates 1.2 cm in length. (C) Phenotypic analysis of lateral root emergence (LRE) was performed by synchronizing lateral root (LR) formation with a gravity stimulus for 22 and 38 hours. Compared with that in the control plants, the LRE in the OBP4 overexpression plants and OBP4 knockdown plants was delayed and promoted, respectively. The data are shown as percentages and the error bars represent SDs (n = 8–12). Approximately 24–36 lateral root primordia (LRP) were observed in 35S::OBP4 plants. Approximately 100 LRP were observed in OBP4 knockdown transgenic plant. *indicates significant differences (p<0.05).
Based on our previous results, the XTH9 expression level was downregulated in the Dof family TF OBP4 overexpression transgenic lines, OBP4 codes for a TF that negatively targets XTH9 [54]. The OBP4 expression pattern in the roots is similar to that of XTH9 in both the LR and vascular tissue [55], and OBP4 negatively regulated XTH9 expression in the roots (S9C Fig). Similar to XTH9, OBP4 also responds to nitrogen signals downstream of AFB3 [2, 12]. We performed ChIP analysis to test the relative enrichment of OBP4 in the selected fragments. An antibody directed against HA was used to precipitate OBP4::HA and its crosslinked DNA, which was isolated from pER8::OBP4::HA roots after estradiol induction. These results showed that OBP4 binded to XTH9 gene promoter regions. Based on this information, we get the conclusion that OBP4 negatively targets XTH9 to participate in nitrogen signal regulation.
OBP4 negatively regulates plant root growth
To directly examine the functional importance of OBP4 in XTH9 nitrate-inducible expression, we identified mutants carrying a T-DNA insertion, namely, lines SALK_116433, SALK_118463, WiscDsLoxHs071, and CS69190. These mutations were unable to downregulate OBP4 expression, most likely because all the T-DNA inserts were detected in the promoter region of OBP4. Studies have shown that overexpression of OBP4 under a constitutive promoter resulted in dwarf plants with shorter primary roots and fewer LRs (Fig 8A and S10A Fig) [54, 55]. Furthermore, we generated OBP4 knockdown lines by an RNA interference method (OBP4-RNAi) and artificial miRNA-mediated RNA silencing method (amiOBP4). Both OBP4-RNAi and amiOBP4 transgenic plants significantly reduced the OBP4 transcript level (S10B and S10C Fig). To validate the interference specificity, we quantified the levels of OBP1, the closest homolog of OBP4. OBP1 transcripts were unaltered in OBP4-RNAi and amiOBP4 seedlings (S10D Fig). In contrast to the 35S::OBP4 plants, phenotype analysis showed that OBP4-RNAi and amiOBP4 seedlings developed longer primary roots and more LRs than controls (Fig 8A and 8B). These results indicate that OBP4 acts as a repressor of root growth. For 20 hours after gravity stimulation, the vector control root bends contained primarily stage I and stage II LRP, whereas, by 38 hours, many primordia were close to emergence (stage VI or VII). In the case of the 35S::OBP4 plants, 20 hours after gravity stimulus, the transgenic plant roots proportionately displayed more stage I primordia than did the vector control roots. Thirty-eight hours after gravity stimulus, 35S::OBP4 transgenic plants had a higher proportion of stage II to IV primordial, but the OBP4-RNAi and amiOBP4 plants had a higher proportion of stage VII and VIII primordial (Fig 8C). These results were consistent with the hypothesis that OBP4 functions in the regulatory pathway of LR emergence and indicate that OBP4 negatively regulates LR growth.
XTH9 functions downstream of OBP4 in control of LR growth in response to variation of nitrate concentrations
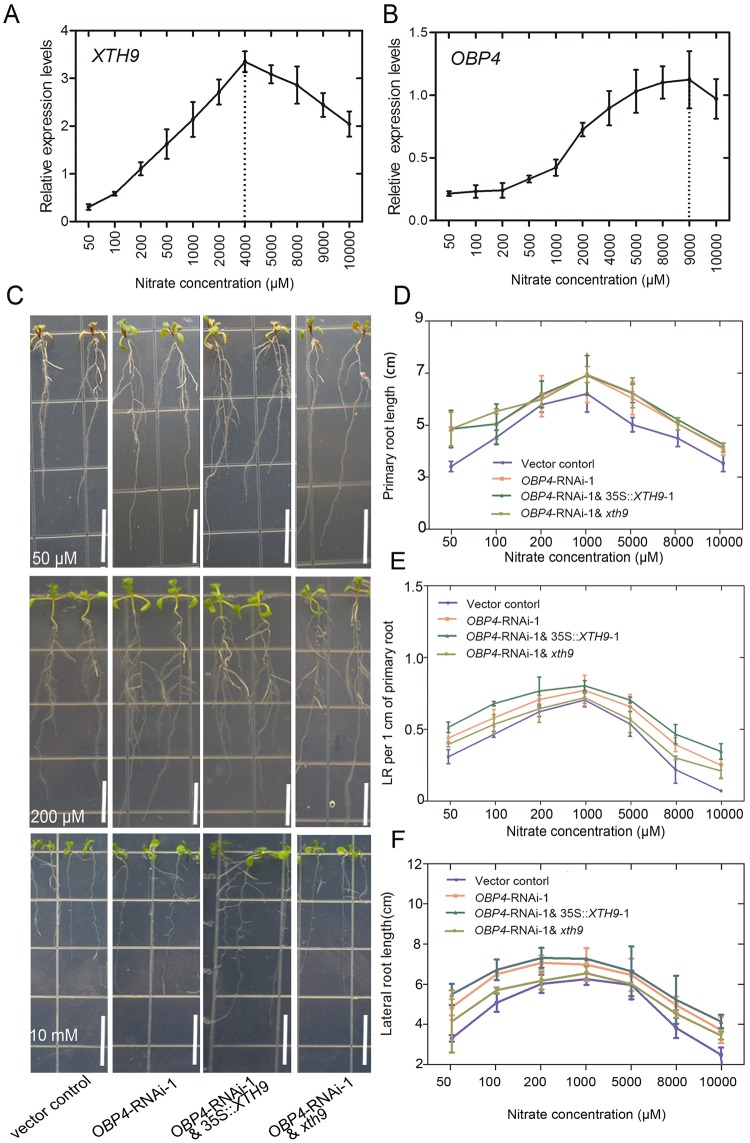
We tested how OBP4 and XTH9 regulated LR growth at nitrate concentrations ranging from 50 to 10000 μM (Fig 9A and 9B). First, we found that the expression of XTH9 increased rapidly at low concentrations. The induction concentration of maximal XTH9 expression was detected at 4000 μM. Higher nitrate concentrations inhibited XTH9 expression (Fig 9A). However, the OBP4 expression level was not increased under low nitrate concentrations of 50–1000 μM. Starting from nitrate induction with 1000 μM, the expression of OBP4 increased rapidly. When the expression of OBP4 reached the highest level, the nitrate-induced concentration was 8000–9000 μM (Fig 9B). The induction expression pattern indicates that OBP4 may have a special role under high nitrate conditions.
Fig 9. Relative expression of XTH9 and OBP4 genes in response to nitrate treatments and OBP4-XTH9 in control of LR growth in response to variation of nitrate concentrations.
Wild-type (WT) (Col-0) plants were grown on media supplemented with ammonium succinate for one week and subsequently treated with various concentrations of nitrate for 2 hours, and the relative (A) XTH9 and (B) OBP4 gene expression levels were assessed. (C) Phenotypic observations and analysis of (D) the primary roots, (E) the lateral root (LR) density, (F) the LR length of 4-day-old vector control, OBP4-RNAi, OBP4-RNAi-1 & 35S::XTH9-1 and OBP4-RNAi-1 & xth9 transgenic plants in response to various concentrations of nitrate (50, 100, 200, 1000, 5000, 8000, and 10000 μM) treatment for five days. LR density was calculated by dividing the LR number per 1 cm of primary root. *indicates significant differences (p<0.05). The error bars show SDs (n = 20–32). The white bars indicate 1.2 cm.
In addition, we observed an altered nitrate response of OBP4 in afb3-1 and slr-1 mutant backgrounds (S11 Fig). We further aimed to identify whether OBP4 acted upstream of XTH9 in controlling root development in response to nitrate. Because it is difficult to screen hygromycin-positive plants and 35S::OBP4 plants are pollen sterile, we crossed the OBP4-inhibited transgenic plants with xth9 mutants and 35S::XTH9 plants to investigate how the coordination between OBP4 and XTH9 can regulate LR development under nitrate signaling. Phenotype analyses in vector control, OBP4-RNAi, OBP4-RNAi & 35S::XTH9, and OBP4-RNAi & xth9 were performed (Fig 9C). The results showed that OBP4-RNAi had more LRs and longer primary roots than the vector control (Fig 9D). OBP4-RNAi & 35S::XTH9 further increased the LRs, and OBP4-RNAi & xth9 decreased LRs more than OBP4-RNAi plants (Fig 9E and 9F). However, the lengths of primary roots in OBP4-RNAi, OBP4-RNAi & 35S::XTH9, and OBP4-RNAi & xth9 plants were almost identical, but longer than the vector control (Fig 9D). The results showed clearly that OBP4 negatively regulates root growth, while XTH9 functions downstream of OBP4 and promotes LR growth.
We also treated the vector control, OBP4-RNAi, OBP4-RNAi & 35S::XTH9, and OBP4-RNAi & xth9 plants with different concentrations of nitrate. We observed that the growth of LRs was nitrate-dependent (Fig 9C). In the lower concentration range (50–1000 μM), the expression of the XTH9 gene upregulated rapidly and the expression of the OBP4 gene upregulated slowly with an increase of nitrate concentration. XTH9 promoted the growth of LRs induced by low nitrate concentration (Fig 9A). It was also observed that XTH9 played an important role in promoting LR growth in low concentrations. However, the expression of OBP4 was increased at higher concentrations (Fig 9B). OBP4 plays an important role in restraining the growth of LRs. Therefore, the growth of LRs in vector control plants decreased most rapidly with an increase of nitrate at a range of high concentrations (Fig 9E and 9F). Altogether, our current model demonstrated that under low nitrate conditions, XTH9 mainly worked downstream of ARF7/9 to promote LR growth. However, under high nitrate conditions, OBP4 antagonistically regulated XTH9 at the late stage of nitrogen stimulation, which inhibited XTH9 expression level to fine-tune LR maintenance. Therefore, in response to signaling at various concentrations of nitrate the OBP4-XTH9 regulatory module elaborately controls LR growth in Arabidopsis (Fig 10).
Fig 10. The OBP4-XTH9 module regulates lateral root (LR) development in response to nitrate signaling.
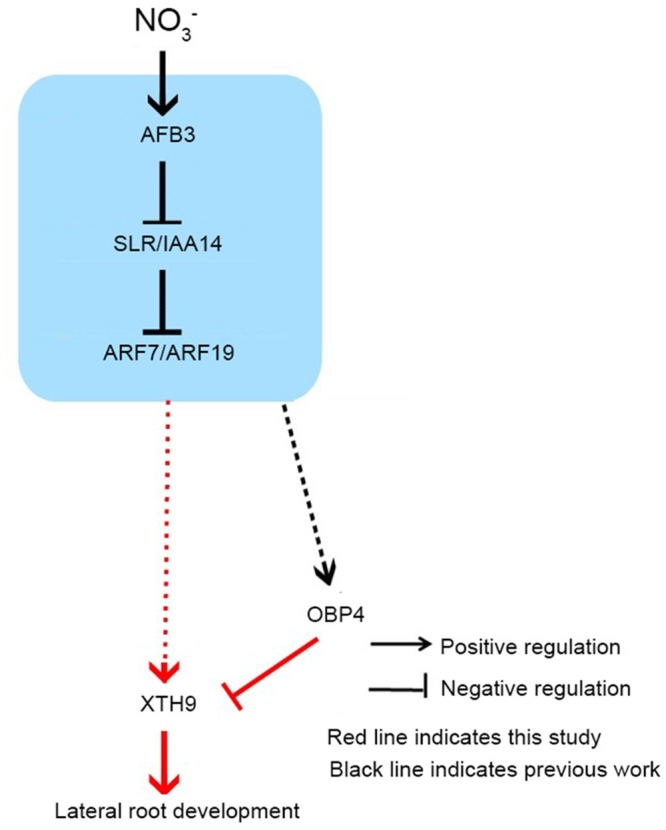
Temporal control of the lateral root emergence (LRE) gene regulatory network involving OBP4 and XTH9. Auxin is perceived by AFB3 and triggers the degradation of IAA14, which releases ARF7/19. Auxin regulates XTH9 expression in an ARF7/19-dependent manner. XTH9 is also negatively regulated by OBP4, which fine-tunes the XTH9 expression level in response to environmental nitrate availability. This synergistic regulation of OBP4-XTH9 has a specific function in lateral root (LR) development in response to nitrate signaling in Arabidopsis.
Discussion
In the Arabidopsis genome, 33 genes encoding XTH proteins have been identified. Individual members of this gene family exhibit specific expression patterns, temporally, and spatially [38]. Here, we isolated 11 of these genes, namely, XTH1–11, which are clustered into group I (S12 Fig). First, we determined whether these genes are involved in primary and/or lateral root development. The results from 90° gravitropic stimulus assays revealed that LR initiation occurs in a highly synchronized manner at the outer surface of bending roots. We profiled the class 1 XTH genes at all stages of LR development, and the results showed that XTH9 mRNA levels were highly upregulated during LR development (Fig 1B and 1C). We subsequently isolated mutants of each XTH gene family member. We carefully observed the effects of XTH9 on LR development and compared it with that in the WT plants. The density of emerged LRs at stages VII and VIII decreased in the xth9 mutant plants (Fig 2G). Although xth9 mutant showed decreased LR growth under all nitrate concentrations, with the increase of nitrogen concentration, the curve curvature of lateral root density of mutant was significantly lower than that of control group. And in the XTH9 over-expressing lines, the variation range of lateral roots treated with exogenous nitrogen was smaller than that of the wild type. So, we got the conclusion that XTH9 played an important role in mediating plant lateral root development in response to exogenous nitrogen signals. Furthermore, we generated transgenic proXTH9::GUS lines and observed GUS signals during the formation of LRP and during LR initiation and elongation (Fig 3A–3H). The results of transcript profiling and reporter studies showed that XTH9 was expressed in response to nitrate treatment (Fig 3I–3M). In the xth9 mutant, the sensitivity to nitrate-promoted LR growth was defective, but the main root elongation was not affected (Fig 4G). Moreover, in response to increasing nitrate concentrations, the average LR length curve for the xth9 mutant plants was lower than that for Col-0 plants (Fig 4H). In addition, XTH9 gene overexpression increased in lateral roots and enhanced tolerance to low nitrate stress (Fig 5).
Previous systems approaches have revealed that the nitrate-regulated miR393/AFB3 module controls RSA [2]. To identify whether the nitrogen-responsive cell wall-related XTH9 gene controls root development downstream of AFB3, we determined the expression levels of XTH9 over time in response to nitrate treatment in WT and afb3-1 and slr-1 mutants. The nitrate response of XTH9 was clearly compromised in the mutant background (Fig 7A). We also observed that XTH9 was still regulated by nitrate in the NR-null mutant (Fig 7B), suggesting that this gene responds to nitrate signaling, but not to N metabolites. The nitrate-specific induction of AFB3 in the roots might control a specific component of Aux/IAA and ARF TFs that facilitate LR growth. In Arabidopsis, LR development depends on multiple Aux/IAA-ARF modules to generate new LRs. The IAA14 (SLR)-ARF7 and ARF19 modules regulate the posterior asymmetric cell division of founder cells in preparation of LR initiation. We initially investigated whether these ARF-dependent regulatory mechanisms control auxin-promoted XTH9 expression. We observed that the XTH9 mRNA level was clearly downregulated in the arf7arf19 mutant background (Fig 6A). Furthermore, the 35S::XTH9 & arf7arf19 plant showed more LR development than the arf7arf19 plant, indicating XTH9 is functional downstream of ARF7/ARF19 (Fig 6C and 6D). In summary, the present study identified the XTH9 components that are downstream of the AFB3-dependent network and that regulate root system growth in response to nitrate.
To independently assess the role of other regulatory regions, we used an XTH9 promoter-deletion approach. The 2.3-kb-long XTH9 promoter was truncated at four different positions to generate increasingly shorter promoter lengths, and the resulting constructs were subsequently transformed into the xth9 mutant background. All Δ1 and Δ2 promoter deletion lines fully complemented the xth9 LR defect (S9A and S9B Fig). This result demonstrates that the 1100-bp upstream promoter sequence of XTH9 is sufficient to drive nitrate-inducible expression and promote LR emergence. Therefore, we concluded that the Dof-binding motifs of the XTH9 gene promoter are important for gene activity. In addition, OBP4 negatively regulates XTH9 expression in the roots (S9C Fig). The OBP4 expression pattern in the roots is similar to that of XTH9 in both LRs and vascular tissue [54, 55]. OBP4 is a promising candidate as an intermediary transcriptional regulator of XTH9. Therefore, we subsequently used constitutive OBP4 overexpression and OBP4 knockdown lines. In the LR system, 35S::OBP4 inhibited LR development, while the OBP4::RNAi and pER8::amiOBP4 plants promoted LR development (Fig 8A and 8B). Furthermore, by treating the vector control, OBP4-RNAi, OBP4-RNAi & 35S::XTH9 and OBP4-RNAi & xth9 with various concentrations of nitrate, we found that OBP4-RNAi plants increased LR growth under high nitrate conditions (Fig 9). We concluded that OBP4 is involved in the nitrate signaling pathway.
Here, we identified a nitrogen-responsive signaling module comprising the cell wall-related gene, XTH9, and the Dof TF, OBP4, downstream of AFB3. This module plays an important role in controlling root development. We proposed that the OBP4 TF is involved in the regulation of LRE and the LR elongation stage. However, the major role of XTH9 seems to be associated with LR elongation. Although OBP4 can negatively regulate XTH9 expression level, it may associate with other downstream target genes involved in the nitrate signaling pathway. Moreover, phenotype analyses in vector control, OBP4-RNAi, OBP4-RNAi & 35S::XTH9, and OBP4-RNAi & xth9 transgenic plants showed that OBP4 and XTH9 respond to changes in nitrate concentration in plant LR growth (Fig 9). Altogether, we propose that under low nitrate conditions, XTH9 mainly works downstream of ARF7/9 to promote LR growth, while under high nitrate conditions, OBP4 negatively regulates XTH9, which inhibits XTH9 expression level and results in the further inhibition of LR growth. Therefore, OBP4 and XTH9 function as a regulatory module in response to various concentrations of nitrate signaling to control LR growth in Arabidopsis (Fig 10).
In fact, we propose that both the expression of XTH9 and OBP4 are regulated by nitrogen signals through ARF7/19, but their functions are antagonistic: XTH9 positively controls LR development while OBP4 negatively controls LR development via suppressing XTH9. OBP4 is a safeguard mechanism that prevents the over-accumulation of XTH9 in response to nitrogen signals. It is possible that XTH9 is strongly induced for LR establishment at the beginning of nitrogen stimulation, once XTH9 reaches a saturated level; the plant initiates a feedback mechanism to suppress XTH9 expression. It is also reasonable to assume that plants do not need too much XTH9 once adequate levels are reached in response to nitrogen signals, thus the plant initiates the expression of OBP4 to suppress the transcription of XTH9 during the later stage. As a result, the plant fine-tunes the number of LR by an elaborate OBP4-XTH9 regulatory module, including OBP4, which antagonistically suppresses the expression of XTH9 to coordinate the activity of XTH9 and LR architecture. A mechanism in plants to regulate LR growth in response to different concentrations of nitrate in the surrounding environment is beneficial in the effective utilization of nitrogen by plants.
Materials and methods
Material and plant growth conditions
The Col-0 ecotype of Arabidopsis thaliana (L.) Heynh was used in all experiments. The germinated seeds were transferred to media in growth chambers at 22±2°C under a 16-hour/8-hour (day/night) photoperiod, and the seeds were surface sterilized [56]. The seeds were then subjected to synchronized germination at 4°C in the dark for 3 days, after which they were subsequently transplanted to Murashige and Skoog (MS) media under a 16-hour/8-hour (day/night) photoperiod. The arf7/arf19-1 and arf7/arf19-2 mutants [51], xth9 mutants (SALK_101024, SALK_063401, SALK_023274, SALK_002571), nia1nia2 double mutant (CS2356) and obp4 (SALK_116433, SALK_118463, WiscDsLoxHs071, CS69190) mutants were obtained from the Arabidopsis Biological Resource Center.
Plasmid construction and transgenic plant generation
XTH9 cDNA was cloned from Arabidopsis thaliana. The XTH9 gene was then subcloned into a pHB vector to generate 35S::XTH9 transgenic lines. The 2.3-kb-long XTH9 promoter was fused with the XTH9 coding DNA sequence (CDS) and then subcloned into a pCAMBIA1300 vector. Generation of transgenic xth9 mutants expressing the XTH9 chimeric gene was carried out as previously described [57, 58]. For inducible and constitutive OBP4 overexpression, the OBP4 ORF was cloned into the binary vector pER8 and pHB. For inducible and constitutive knockdown of OBP4, about 250 bp target sequence was cloned from genome. This sequence was introduced into the pCAMBIA1301 vector in the sense and antisense orientation. Next, the entire fragment was removed by digesting the pCAMBIA1301-based construct, and the fragment was inserted into pHB vector. We obtained several OBP4 knockdown transgenic lines, the highly downregulated lines pHB::OBP4::RNAi were used for our experiments. For artificial microRNA (amiRNA) mediated gene silencing of OBP4. We used the WMD3 online tool (http://wmd3.weigelworld.org) to the design of amiRNA specific to OBP4 coding region, the artificial miRNA vector pRS300 was requested from the Weigel lab. The final fragment was inserted into pHB vector. We obtained several transgenic lines, the highly OBP4 downregulated lines pHB::amiOBP4 was used for our experiments. Col-0 plants were transformed by the floral-dip method. The positive transgenic plants were screened by 50 mg/L Hygromycin (Roche, USA), and RT-qPCR was performed to measure the gene expression levels in the transgenic plants; the primers used are listed in S1 Table in Supporting Information.
Cloning of full promoters and promoter deletions
Point-mutated promoters were cloned back into pCAMBIA1300-XTH9-GUS and sequenced to ensure that no other mutations were generated during PCR. For the PCR-generated promoter deletions, a combination of primers for Δ1, Δ2, Δ3, and Δ4 was used. The purified PCR products were subsequently subcloned into the pCAMBIA1300 vector.
Expression analysis via RT-qPCR
Total RNA was isolated using an RNeasy Plant Mini Kit (Qiagen, USA) and stored at -80°C. One gram of total RNA was used for cDNA synthesis via Novo Script First-Strand cDNA Synthesis SuperMix E041 (Novoprotein, China). The experiments were conducted using Hieff qPCR SYBR Green Master Mix (Yeasen, China) according to the manufacturer’s instructions. The data were analyzed by using LightCycler 96 analysis software 1.1 (ΔΔCT method). The AtACT2 gene was used as an internal control. The RT-qPCR primers used are listed in S2 Table, and the assays were performed in triplicate.
Nitrate treatment assays
Approximately 1000 seedlings were hydroponically planted on nitrogen-free basal salt media (M531, Phytotechnology Laboratories) that were supplemented with various concentrations of nitrate (10, 50, 500, 1000, 5000 and 10000 μM) and 1% sucrose. The seedlings were then subjected to a 16-hour/8-hour (day/night) photoperiod at 22°C.
Histochemical analysis and microscopy
With respect to the histochemical analysis of GUS activity, Arabidopsis seedlings were incubated at 37°C in reaction buffer (100 mM sodium phosphate buffer [pH 7.0], 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, 0.1% [vol/vol] Triton X-100 and 0.1% [wt/vol] sodium lauroyl sarcosine) plus 1 mM X-Glu. GUS staining and clearing was performed as previously described [54]. The seedlings were cleared and imaged using differential interference contrast (DIC) optics via a Leica microscope, and at least 12 plants were used for each treatment. The activity of β-glucuronidase (GUS) in intact plant tissue was measured using 4-methylumbelliferyl β-D-glucuronide (4-MUG) as a substrate according to the methods of [59].
Analysis of root architecture traits
Initiating and emerging LRs (stages I, II, III, IV, V, VI, VII and VII) [22] were counted using DIC optics via a Leica microscope. For root measurements, the plants were scanned using a Nikon photo scanner, and the roots were measured using ImageJ software. The data were subsequently analyzed using Graph Pad Prism 5 software.
XET activity assay
The XET activity of XTH was determined according to a method reported by Vissenberg [60]. The roots were cultured in a 5 mM XGO-SR mixture (XLLG-SR.XXLGSR.XXXG-SR) dissolved in 25 mm MES buffer at a pH of 5.5 for 1 hour and then rinsed with ethanol: formic acid: water (15:1:4, v/v/v) for 10 minutes to remove the remaining unreacted residue XGO-SR. The XGO-SR was further incubated overnight in 5% formic acid to remove exogenous, non-wall-bound XGO-SR [61]. Samples were mounted on glass slides and inspected under a confocal microscope using excitation light of 540 nm.
Root cell wall extraction
Extraction of root crude cell wall materials were performed according to Zhu [37] with minor modifications. Roots were ground with a mortar and pestle in liquid nitrogen and then homogenized with 75% ethanol for 20 min in an ice-cold water bath. The sample was then centrifuged at 8000 rpm for 10 min, and the supernatant was removed. The pellets were homogenized and washed with acetone, methanol:chloroform at a ratio of 1:1, and methanol, respectively, for 20 min each, with each supernatant being removed after centrifugation between the washes. The remaining the cell wall material was dried and stored at 4°C for further use.
Determination of total sugar residues
The content of total sugar residues in the hemicellulosic fractions was determined by the phenol-sulfuric acid method and expressed as Glc equivalents. Briefly, 200 mL of hemicellulose extracts was incubated with 1 mL of 98% H2SO4 and 10 mL of 80% phenol at room temperature for 15 min and then incubated at 100°Cfor 15 min. After cooling, the absorbance at 490 nm was measured spectrophotometrically.
MALDI-TOF mass spectrometry analysis of XyG oligosaccharides
Arabidopsis roots were preserved in 100% ethanol. At 37°C overnight, 1 unit of xyloglucanase (Megazyme, Brae, Ireland) was treated with 50 mM sodium acetate buffer (pH 5.0) to remove ethanol and rehydrated to produce xylooligosaccharide. MALDI-TOF mass spectrometry of XyG oligosaccharides was recorded with an Applied Biosystems using super-DHB (Sigma-Aldrich, USA) as a matrix [62, 63].
Chromatin immunoprecipitation (ChIP) assay
The ChIP assays were performed as described previously with some changes [54]. In brief, 5 g of 8-day-old pER8::OBP4-HA transgenic seedlings were extracted and immersed in 1% formaldehyde to cross-link DNA with DNA-binding proteins. Next, the chromatin pellets were extracted and sheared by sonication. Anti-HA antibody was used to immunoprecipitate the DNA-OBP4 protein complexes. DNA was released by protease K and purified for PCR analysis. The enrichment of DNA fragments was checked by qRT-PCR. Primers are indicated in S2 Table.
Root phenotyping
The seedlings were grown on vertical plates subjected to 90° gravity stimulation for 54 hours [45]. In addition, 10-day-old seedlings were grown vertically and harvested to analyze the developmental stages of LRP. In this phenotypic study, the total number and stages of LRP were counted and determined according to the methods of Malamy and Benfey [64, 65]. The root length was measured using ImageJ software.
Statistical analysis and multiple alignments
Statistics were performed with Graphpad 8.0, by using Student’s t-test or ANOVA analyses. Differences were considered significant when p<0.05. Multiple alignments of the predicted amino acid sequences and phylogenetic analyses were performed using DNAMAN 6.0 and MEGA 4.1 software, respectively [66].
Accession numbers
The Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: XTH9, At4g03210; ARF7, At5g20730; ARF19, At1g19220; OBP4, At5g60850; NIA1, At1g7760; NIA2, At1g37130; IAA14, At4g14550; and AFB3, At1g12820.
Supporting information
LBD29 expression pattern at each time point during LR initiation every 6 hours from 6 to 54 hours pgi. The bending roots of a population of 5-d-old seedlings were microdissected at each of the 10 time points and used for RNA extraction (approximately 200 per time point). The error bars show the SDs (n = 3).
(TIF)
(A) RT-PCR analysis of XTH genes expression level in WT and T-DNA insertion mutants. Root phenotypes of WT plants and the xth4, xth10 mutants. The plants were grown vertically on media for 10 days. (B) Primary root. (C) LR density analysis of WT plants and the mutant lines. The error bars denote the SDs. Actin2 was used as internal control.
(TIF)
(A) Total sugar residues in extractable hemicellulose of Col-0 and xth9 mutant. Cell wall material from roots was fractionated into different polysaccharide classes. Data are means ± SD. n = 3 (B) Cell wall material was extracted from Col-0 and xth9 mutant roots and digested with XEG. The oligosaccharides obtained were analyzed by MALDI-TOF MS. Data are means ± SD; n = 2. The asterisk shows a significant difference between xth9 and Col-0 at p < 0.05 by Student’s t test. (C) Analysis of the XTH9 gene expression patterns in seedling, root, shoot, rosette leaf, flower, and bud tissues. The error bars show the SDs (n = 6). The asterisk (*) shows a significant difference at p<0.05 by Student’s t test. (D) XET activity action expressed as fluorescence relative to untreated wild type. Roots were subjected to cytochemical assays of XET action for 1 h. Data are means SD (n = 3). (*) indicate significant differences at p < 0.05 by Student’s t test.
(TIF)
Ten-day-old wild-type plants grown on half-strength MS-agar plates were treated with 200 mM mannitol, heat (30°C), 100 mM NaCl, 500 μM KNO3, 1 mM KH2PO4 and plant growth hormones (1 μM ABA and 20 μM GA) for 3 and 8 hours. For drought treatment, the plants were transferred to dry 3M paper for 3 and 8 hours. RT-qPCR was used to check XTH9 expression levels at various time points. The error bars show the SDs (n = 3).
(TIF)
LR density (number of LRs per 1 cm of primary root length) in WT and complimentary lines grown in media supplemented with various concentrations of nitrate. The error bars show the SDs (n = 3).
(TIF)
XTH9 expression level in the 2-week-old 35S::XTH9 transgenic plant leaves. *indicates significant differences (p<0.05). The error bars show the SDs (n = 3).
(TIF)
*indicates significant differences (p<0.05), and the error bars show the SD (n = 3).
(TIF)
(A) Observations of wild-type, afb3-1 and slr-1 plant root development in response of 500 mM nitrate treatment for 2 day. The bar indicates 1 cm. (B) Analysis of LR density (LR per 1 centimeter of primary root). *indicates significant differences (p<0.05), and the error bars show the SD (n = 3).
(TIF)
(A) The PLACEcare online tool was used to search for motifs. Many motifs including GATA-box-binding elements, the W-box elements and Dof TF-binding elements were found in the XTH9 promoter. Representation of the XTH9 full promoter from -2314 bp to the start codon (ATG). Promoter deletions (named Δ1, Δ2, Δ3 and Δ4) were generated and cloned upstream of XTH9::GUS. Dof-binding motifs are indicated as vertical solid lines. The sequence areas used for the ChIP experiment are marked in the gene promoters (from a to f, top panel). The number of Dof-binding motifs is indicated for each promoter deletion below the panel. (B) LR density measurements (number of LR per 1 cm of primary root length). The error bars represent the SDs. (n≥10). (C) Relative XTH9 activity in pER8 vector transgenic lines and inducible OBP4 expression and RNAi lines before and after 20 μM estradiol induction for 2 days. *indicates significant differences (p<0.05), and the error bars show the SD, n = 3. (D) 8-day-old pER8::OBP4::HA transgenic plants grown on MS-agar plates were used for ChIP assays. The enrichment shown was calculated as the DNA level of each fragment in the β-estradiol-treated sample divided by that in the DMSO-treated sample. Anti-HA antibody was used to precipitate OBP4-HA. Three measurements were averaged for individual assays. Bars indicate the SD. The values in Col-0 plants were set to 1 after normalization to ACT2 for qPCR analysis. Asterisks indicate significant differences, p< 0.05.
(TIF)
(A-C) Relative OBP4 expression levels in the 2-week-old 35S::XTH9, RNAi-OBP4 and amiOBP4 transgenic plants leaves. (D) OBP1 expression levels in the vector control and RNAi-OBP4-1 and amiOBP4-1 transgenic plants. *indicates significant differences (p<0.05). The error bars show the SDs (n = 3).
(TIF)
WT (Col-0), afb3-1 and slr-1 mutant plants were grown in media supplemented with ammonium succinate for one week and subsequently treated with 5 mM KNO3 or 5 mM KCl for 1–3 hours. The OBP4 gene expression level in plant roots was measured via RT-qPCR. The KCl treatment results are shown with white bars, and the KNO3 treatment results are shown with black bars. *indicates significant differences (p<0.05), and the error bars show the SDs (n = 12–18).
(TIF)
(A) Alignment and (B) Phylogram of Arabidopsis class 1 XTH family proteins. Multiple sequence alignment of the predicted amino acid sequence and phylogenetic analysis were performed via DNAMAN 6.0 and MEGA 4.1 software.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Dr. Jiying Xu from Suzhou University for providing the afb3-1 mutant and slr-1 gain-of-function mutant.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China (Grant No. 31500236) to PPX, the National Natural Science Foundation of China (Grant Nos. U1738107, 31600684 and 31570859) to WMC, the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant Nos. XDA04020202-15 and XDA04020415) to WMC, and the China Manned Space Flight Technology Project to WMC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Vidal EA, Tamayo KP Gutierrez RA (2010). Gene networks for nitrogen sensing, signaling, and response in Arabidopsis thaliana. Wiley Interdiscip Rev Syst Biol Med 2, 683–93. 10.1002/wsbm.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vidal EA, Moyano TC, Riveras E, Contreras-Lopez O, and Gutierrez RA (2013). Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. P Natl Acad Sci USA 110:12840–12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leran S, Munos S, Brachet C, Tillard P, Gojon A, Lacombe B (2013). Arabidopsis NRT1.1 is a bidirectional transporter involved in root-to-shoot nitrate translocation. Mol Plant 6, 1984–7. 10.1093/mp/sst068 [DOI] [PubMed] [Google Scholar]
- 4.Warzybok A, Migock M (2012). The function of nitrate transporters NRT1 in plants. Postepy Biochem 58, 61–8. [PubMed] [Google Scholar]
- 5.Parker JL, Newstead S (2014). Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 507, 68–72. 10.1038/nature13116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang C, Zhang W, Li Z, Zhen Li, Bi YJ, Crawford NM, Wang Y (2018). FIP1 Plays an Important Role in Nitrate Signaling and Regulates CIPK8 and CIPK23 Expression in Arabidopsis. Front Plant Sci 9, 593 10.3389/fpls.2018.00593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krouk G, Crawford NM, Coruzzi GM, Tsay YF (2010). Nitrate signaling: adaptation to fluctuating environments. Curr Opin Plant Biol 13, 266–73. 10.1016/j.pbi.2009.12.003 [DOI] [PubMed] [Google Scholar]
- 8.Gan Y, Filleur S, Rahman A, Gotensparre S, Forde BG (2005). Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 222, 730–42. 10.1007/s00425-005-0020-3 [DOI] [PubMed] [Google Scholar]
- 9.Feng Z, Zhu J, Du X, Cui X (2012). Effects of three auxin-inducible LBD members on lateral root formation in Arabidopsis thaliana. Planta 236, 1227–37. 10.1007/s00425-012-1673-3 [DOI] [PubMed] [Google Scholar]
- 10.Zhao L, Zhang W, Yang Y, Zehui Li, Na Li, Qi SD, Crawford NM, Wang Y (2018). The Arabidopsis NLP7 gene regulates nitrate signaling via NRT1.1-dependent pathway in the presence of ammonium. Sci Rep 8, 1487 10.1038/s41598-018-20038-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarez J. M., Riveras E., Vidal E. A., Gras D. E., ntrerasLópez O, & Tamayo K. P., et al. (2015). Systems approach identifies tga1 and tga4 transcription factors as important regulatory components of the nitrate response of arabidopsis thaliana roots. Plant Journal for Cell & Molecular Biology, 80(1), 1–13. [DOI] [PubMed] [Google Scholar]
- 12.Vidal E.A., Araus V., Lu C., Parry G., Green P.J., Coruzzi G.M., and Gutierrez R.A. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. P Natl Acad Sci USA 107:4477–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatfield SP, Capron R, Severino A, Penttila PA, Alfred S, Nahal H, Trobacher C, Raizada MN, Provart NJ (2014). Incipient stem cell niche conversion in tissue culture: using a systems approach to probe early events in WUSCHEL-dependent conversion of lateral root primordial into shoot meristems. Plant J 77:665–666. [DOI] [PubMed] [Google Scholar]
- 14.Dubrovsky JG, Rost TL, Colon-Carmona A, Doerner P (2001). Early primordium morphogenesis during lateral root initiation in Arabidopsis thaliana. Planta 214:30–36. [DOI] [PubMed] [Google Scholar]
- 15.Kircher S, Schopfer P (2016). Priming and positioning of lateral roots in Arabidopsis. An approach for an integrating concept. J Exp Bot 67:1411–1420. 10.1093/jxb/erv541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vilches-Barro A, Maizel A (2015). Talking through walls: mechanisms of lateral root emergence in Arabidopsis thaliana. Curr Opin Plant Biol 23:31–38. 10.1016/j.pbi.2014.10.005 [DOI] [PubMed] [Google Scholar]
- 17.Malamy JE (2005). Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 28:67–77. [DOI] [PubMed] [Google Scholar]
- 18.Himanen K, Boucheron E, Vanneste S, Engler JD, Inze D, Beeckman T (2002). Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14:2339–2351. 10.1105/tpc.004960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanz L, Dewitte W, Forzani C, Patell F, Nieuwland J, Wen B, Quelhas P, De Jager S, Titmus C Campilho A, Ren Hong, Estelle Mark, Wang Hong, Murray James AH (2011). The Arabidopsis D-Type Cyclin CYCD2;1 and the Inhibitor ICK2/KRP2 Modulate Auxin-Induced Lateral Root Formation. Plant Cell 23:641–660. 10.1105/tpc.110.080002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis DR, Olex AL, Lundy SR, Turkett WH, Fetrow JS, Muday GK (2013). A kinetic analysis of the auxin transcriptome reveals cell wall remodeling proteins that modulate lateral root development in Arabidopsis. The Plant cell 25:3329–3346 10.1105/tpc.113.114868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucas M, Kenobi K, von Wangenheim D, Vobeta U, Swarup K, De Smet I, Van Damme D, Lawrence T, Peret B, Moscardi E, Barbeau D, Godin C, Salt D, Guyomarc’h S, Stelzer EHK, Maizel A, Laplaze L, Bennett MJ (2013). Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proceedings of the National Academy of Sciences of the United States of America 110:5229–5234. 10.1073/pnas.1210807110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swarup K, Benkova E, Swarup R, Casimiro I, Peret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, Levesque MP, Carrier D, James N, Calvo V, Ljung K, Kramer E, Roberts R, Graham N, Marillonnet S, Patel K, Jones JDG, Taylor CG, Schachtman DP, May S, Sandberg G, Benfey P, Friml J, Kerr I, Beeckman T, Laplaze L, Bennett MJ (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10:946–954. 10.1038/ncb1754 [DOI] [PubMed] [Google Scholar]
- 23.Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L (2013). Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci 18:455–463. [DOI] [PubMed] [Google Scholar]
- 24.De Smet I, Tetsumura T, De Rybel B, Frey NFD, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, Inzé D, Bennett MJ, Beeckman T (2007). Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134:681–690. 10.1242/dev.02753 [DOI] [PubMed] [Google Scholar]
- 25.Farquharson KL (2010). Gibberellin-Auxin Crosstalk Modulates Lateral Root Formation. Plant Cell 22:540–540. 10.1105/tpc.110.220313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukaki H, Okushima Y, Tasaka M (2007). Auxin-mediated lateral root formation in higher plants. Int Rev Cytol 256:111–137. 10.1016/S0074-7696(07)56004-3 [DOI] [PubMed] [Google Scholar]
- 27.Peret B, Li GW, Zhao J, Band LR, Voss U, Postaire O, Luu DT, Da Ines O, Casimiro I, Lucas M, Wells DM, Lazzerini L, Nacry P, King JR, Jensen OE, Schäffner AR, Maurel C, Bennett MJ (2012). Auxin regulates aquaporin function to facilitate lateral root emergence. Nature cell biology 14 (10):991 10.1038/ncb2573 [DOI] [PubMed] [Google Scholar]
- 28.Nibau C, Gibbs DJ, Coates JC (2008). Branching out in new directions: the control of root architecture by lateral root formation. New Phytologist 179:595–614. 10.1111/j.1469-8137.2008.02472.x [DOI] [PubMed] [Google Scholar]
- 29.Arase F, Nishitani H, Egusa M, Nishimoto N, Sakurai S, Sakamoto N, Kaminaka H (2012). IAA8 Involved in Lateral Root Formation Interacts with the TIR1 Auxin Receptor and ARF Transcription Factors in Arabidopsis. Plos One 7(8):e43414 10.1371/journal.pone.0043414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H (2012). Multiple AUX/IAA-ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos T R Soc B 367:1461–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asada K, Okazawa K, Nakagawa T, Sato Y, Kato I, Tomita E, Nishitani K (1993). Molecular-Cloning of Endo-Xyloglucan Transferase (Ext), a New Transferase Involved in Cell-Wall Construction. Faseb J 7:A1299–A1299. [Google Scholar]
- 32.Nishitani K, Tominaga R (1992). Endoxyloglucan Transferase, a Novel Class of Glycosyltransferase That Catalyzes Transfer of a Segment of Xyloglucan Molecule to Another Xyloglucan Molecule. J Biol Chem 267:21058–21064. [PubMed] [Google Scholar]
- 33.Rose JKC, Braam J, Fry SC, Nishitani K (2002). The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant and Cell Physiology 43:1421–1435. 10.1093/pcp/pcf171 [DOI] [PubMed] [Google Scholar]
- 34.Yokoyama R, Nishitani K (2001). A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant and Cell Physiology 42:1025–1033. 10.1093/pcp/pce154 [DOI] [PubMed] [Google Scholar]
- 35.Osato Y, Yokoyama R, Nishitani K (2006). A principal role for AtXTH18 in Arabidopsis thaliana root growth: a functional analysis using RNAi plants. Journal of plant research 119:153–162. 10.1007/s10265-006-0262-6 [DOI] [PubMed] [Google Scholar]
- 36.Matsui A, Yokoyama R, Seki M, Ito T, Shinozaki K, Takahashi T, Komeda Y, Nishitani K (2005). AtXTH27 plays an essential role in cell wall modification during the development of tracheary elements. Plant J 42:525–534. 10.1111/j.1365-313X.2005.02395.x [DOI] [PubMed] [Google Scholar]
- 37.Zhu XF, Shi YZ, Lei GJ, Fry SC, Zhang BC, Zhou YH, Braam J, Jiang T, Xu XY, Mao CZ, Pan YJ, Yang JL, Wu P, Zheng SJ (2012). XTH31, Encoding an in Vitro XEH/XET-Active Enzyme, Regulates Aluminum Sensitivity by Modulating in Vivo XET Action, Cell Wall Xyloglucan Content, and Aluminum Binding Capacity in Arabidopsis. Plant Cell 24:4731–4747. 10.1105/tpc.112.106039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Becnel J, Natarajan M, Kipp A, Braam J. (2006). Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol Biol 61:451–467. 10.1007/s11103-006-0021-z [DOI] [PubMed] [Google Scholar]
- 39.Kaewthai N, Gendre D, Eklof JM, Ibatullin FM, Ezcurra I, Bhalerao RP, Brumer H (2013). Group III-A XTH Genes of Arabidopsis Encode Predominant Xyloglucan Endohydrolases That Are Dispensable for Normal Growth. Plant Physiol 161:440–454. 10.1104/pp.112.207308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okamoto S, Tomita E, Nishitani K (1997). Expression pattern of endoxyloglucan transferase (EXGT-A1) in Arabidopsis thaliana, and its implications for cell wall construction. Plant Physiol 114:23–23. [Google Scholar]
- 41.Parizot B, De Rybel B, Beeckman T (2010). VisuaLRTC: a new view on lateral root initiation by combining specific transcriptome data sets. Plant physiology 153:34–40. 10.1104/pp.109.148676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ditengou FA, Tealea WD, Kochersperger P, Flittner KA, Kneuper I, van der Graaff E, Nziengui H, Pinosa F, Li XG., Nitschke R, Laux T, Palme K (2008). Mechanical induction of lateral root initiation in Arabidopsis thaliana. P Natl Acad Sci USA 105:18818–18823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malamy JE, Ryan KS (2001). Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol 127:899–909. [PMC free article] [PubMed] [Google Scholar]
- 44.Wightman F, Thimann KV (1976). Hormonal-Regulation of Lateral Root Initiation in Pisum-Sativum. Plant Physiol 57:52–52. [Google Scholar]
- 45.Voss U, Wilson MH, Kenobi K, Gould PD, Robertson FC, Peer WA, Lucas M, Swarup K, Casimiro I, Holman TJ, Wells DM, Péret BJ, Goh T, Fukaki H, Charlie Hodgman T., Laplaze L, Halliday KJ, Ljung K, Murphy AS, Hall AJ, Webb AR, Bennett MJ (2015). The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana. Nat Commun 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perrine-Walker FM, Jublanc E (2014). The localization of auxin transporters PIN3 and LAX3 during lateral root development in Arabidopsis thaliana. Biol Plantarum 58:778–782. [Google Scholar]
- 47.Porco S, Larrieu A, Du YJ, Gaudinier A, Goh T, Swarup K, Swarup R, Kuempers B, Bishopp A, Lavenus J, Casimiro I, Hill K, Benkova E, Fukaki H, Brady SM, Scheres B, Péret B, Bennett MJ (2016). Lateral root emergence in Arabidopsis is dependent on transcription factor LBD29 regulation of auxin influx carrier LAX3. Development 143:3340–3349. 10.1242/dev.136283 [DOI] [PubMed] [Google Scholar]
- 48.Lavenus J, Goh T, Guyomarc’h S, Hill K, Lucas M, Voss U, Kenobi K, Wilson MH, Farcot E, Hagen G, Guilfoyle TJ, Fukaki H, Laplaze L, Bennett MJ (2015). Inference of the Arabidopsis Lateral Root Gene Regulatory Network Suggests a Bifurcation Mechanism That Defines Primordia Flanking and Central Zones. Plant Cell 27:1368–1388. 10.1105/tpc.114.132993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patterson K, Cakmak T, Cooper A, Lager I, Rasmusson AG, Escobar MA (2010). Distinct signaling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant Cell Environ 33:1486–1501. 10.1111/j.1365-3040.2010.02158.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okushima Y, Fukaki H, Theologis A, Tasaka M (2005a). Analysis of ARF7- and ARF19-regulated genes in Arabidopsis lateral root formation. Plant and Cell Physiology 46:S197–S197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ito J, Fukaki H, Onoda M, Li L, Li CY, Tasaka M, Furutani M (2016). Auxin-dependent compositional change in Mediator in ARF7-and ARF19-mediated transcription. P Natl Acad Sci USA 113:6562–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19:118–130. 10.1105/tpc.106.047761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Alison Smith, Yu G, Theologis A (2005b). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17:444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu PP, Chen HY, Ying L, Cai WM (2016). AtDOF5.4/OBP4, a DOF Transcription Factor Gene that Negatively Regulates Cell Cycle Progression and Cell Expansion in Arabidopsis thaliana. Sci Rep-Uk 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramirez-Parra E, Perianez-Rodriguez J, Navarro-Neila S, Gude I, Moreno-Risueno MA, Del Pozo JC (2017). The transcription factor OBP4 controls root growth and promotes callus formation. The New phytologist 213:1787–1801. 10.1111/nph.14315 [DOI] [PubMed] [Google Scholar]
- 56.Clough SJ, and Bent AF (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- 57.Xu PP, Cai WM (2017). Functional characterization of the BnNCED3 gene in Brassica napus. Plant Sci 256:16–24. 10.1016/j.plantsci.2016.11.012 [DOI] [PubMed] [Google Scholar]
- 58.Zeng Y, Raimondi N, Kermode AR (2003). Role of an ABI3 homologue in dormancy maintenance of yellow-cedar seeds and in the activation of storage protein and Em gene promoters. Plant molecular biology 51:39–49. 10.1023/a:1020762304937 [DOI] [PubMed] [Google Scholar]
- 59.Murashige T, Skoog F (1962). A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol Plantarum 15:473–497. [Google Scholar]
- 60.Vissenberg K., & Fry S. C. (2000). In vivo colocalization of xyloglucan endotransglycosylase activity and its donor substrate in the elongation zone of arabidopsis roots. Plant Cell, 12(7), 1229–1237. 10.1105/tpc.12.7.1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ (1992) Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem J 282 (3):821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cavalier D. M., Olivier L., Lutz N., Kazuchika Y., Antje R., & Glenn F., et al. (2008). Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell, 20(6), 1519–1537. 10.1105/tpc.108.059873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rocha J., Félix Cicéron, Sanctis D. D., Lelimousin M., & Breton C. (2016). Structure of Arabidopsis thaliana fut1 reveals a variant of the gt-b class fold and provides insight into xyloglucan fucosylation. The Plant Cell, 28 (10), 2352 10.1105/tpc.16.00519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malamy JE, Benfey PN (1997a). Lateral root formation in Arabidopsis thaliana. Plant Physiol 114:277–277. [Google Scholar]
- 65.Malamy JE, Benfey PN (1997b). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124:33–44. [DOI] [PubMed] [Google Scholar]
- 66.Hall BG (2013). Building Phylogenetic Trees from Molecular Data with MEGA. Molecular biology and evolution 30:1229–1235. 10.1093/molbev/mst012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LBD29 expression pattern at each time point during LR initiation every 6 hours from 6 to 54 hours pgi. The bending roots of a population of 5-d-old seedlings were microdissected at each of the 10 time points and used for RNA extraction (approximately 200 per time point). The error bars show the SDs (n = 3).
(TIF)
(A) RT-PCR analysis of XTH genes expression level in WT and T-DNA insertion mutants. Root phenotypes of WT plants and the xth4, xth10 mutants. The plants were grown vertically on media for 10 days. (B) Primary root. (C) LR density analysis of WT plants and the mutant lines. The error bars denote the SDs. Actin2 was used as internal control.
(TIF)
(A) Total sugar residues in extractable hemicellulose of Col-0 and xth9 mutant. Cell wall material from roots was fractionated into different polysaccharide classes. Data are means ± SD. n = 3 (B) Cell wall material was extracted from Col-0 and xth9 mutant roots and digested with XEG. The oligosaccharides obtained were analyzed by MALDI-TOF MS. Data are means ± SD; n = 2. The asterisk shows a significant difference between xth9 and Col-0 at p < 0.05 by Student’s t test. (C) Analysis of the XTH9 gene expression patterns in seedling, root, shoot, rosette leaf, flower, and bud tissues. The error bars show the SDs (n = 6). The asterisk (*) shows a significant difference at p<0.05 by Student’s t test. (D) XET activity action expressed as fluorescence relative to untreated wild type. Roots were subjected to cytochemical assays of XET action for 1 h. Data are means SD (n = 3). (*) indicate significant differences at p < 0.05 by Student’s t test.
(TIF)
Ten-day-old wild-type plants grown on half-strength MS-agar plates were treated with 200 mM mannitol, heat (30°C), 100 mM NaCl, 500 μM KNO3, 1 mM KH2PO4 and plant growth hormones (1 μM ABA and 20 μM GA) for 3 and 8 hours. For drought treatment, the plants were transferred to dry 3M paper for 3 and 8 hours. RT-qPCR was used to check XTH9 expression levels at various time points. The error bars show the SDs (n = 3).
(TIF)
LR density (number of LRs per 1 cm of primary root length) in WT and complimentary lines grown in media supplemented with various concentrations of nitrate. The error bars show the SDs (n = 3).
(TIF)
XTH9 expression level in the 2-week-old 35S::XTH9 transgenic plant leaves. *indicates significant differences (p<0.05). The error bars show the SDs (n = 3).
(TIF)
*indicates significant differences (p<0.05), and the error bars show the SD (n = 3).
(TIF)
(A) Observations of wild-type, afb3-1 and slr-1 plant root development in response of 500 mM nitrate treatment for 2 day. The bar indicates 1 cm. (B) Analysis of LR density (LR per 1 centimeter of primary root). *indicates significant differences (p<0.05), and the error bars show the SD (n = 3).
(TIF)
(A) The PLACEcare online tool was used to search for motifs. Many motifs including GATA-box-binding elements, the W-box elements and Dof TF-binding elements were found in the XTH9 promoter. Representation of the XTH9 full promoter from -2314 bp to the start codon (ATG). Promoter deletions (named Δ1, Δ2, Δ3 and Δ4) were generated and cloned upstream of XTH9::GUS. Dof-binding motifs are indicated as vertical solid lines. The sequence areas used for the ChIP experiment are marked in the gene promoters (from a to f, top panel). The number of Dof-binding motifs is indicated for each promoter deletion below the panel. (B) LR density measurements (number of LR per 1 cm of primary root length). The error bars represent the SDs. (n≥10). (C) Relative XTH9 activity in pER8 vector transgenic lines and inducible OBP4 expression and RNAi lines before and after 20 μM estradiol induction for 2 days. *indicates significant differences (p<0.05), and the error bars show the SD, n = 3. (D) 8-day-old pER8::OBP4::HA transgenic plants grown on MS-agar plates were used for ChIP assays. The enrichment shown was calculated as the DNA level of each fragment in the β-estradiol-treated sample divided by that in the DMSO-treated sample. Anti-HA antibody was used to precipitate OBP4-HA. Three measurements were averaged for individual assays. Bars indicate the SD. The values in Col-0 plants were set to 1 after normalization to ACT2 for qPCR analysis. Asterisks indicate significant differences, p< 0.05.
(TIF)
(A-C) Relative OBP4 expression levels in the 2-week-old 35S::XTH9, RNAi-OBP4 and amiOBP4 transgenic plants leaves. (D) OBP1 expression levels in the vector control and RNAi-OBP4-1 and amiOBP4-1 transgenic plants. *indicates significant differences (p<0.05). The error bars show the SDs (n = 3).
(TIF)
WT (Col-0), afb3-1 and slr-1 mutant plants were grown in media supplemented with ammonium succinate for one week and subsequently treated with 5 mM KNO3 or 5 mM KCl for 1–3 hours. The OBP4 gene expression level in plant roots was measured via RT-qPCR. The KCl treatment results are shown with white bars, and the KNO3 treatment results are shown with black bars. *indicates significant differences (p<0.05), and the error bars show the SDs (n = 12–18).
(TIF)
(A) Alignment and (B) Phylogram of Arabidopsis class 1 XTH family proteins. Multiple sequence alignment of the predicted amino acid sequence and phylogenetic analysis were performed via DNAMAN 6.0 and MEGA 4.1 software.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.