Abstract
Background
Studying expression of genes by direct sequencing and analysis of metatranscriptomes at a particular time and space can disclose structural and functional insights about microbial communities. The present study reports comparative analysis of metatranscriptome from two distinct soil ecosystems referred as M1 (agriculture soil) and O1 (organic soil).
Results
Analysis of sequencing reads revealed Proteobacteria as major dominant phyla in both soil types. The order of the top 3 abundant phyla in M1 sample was Proteobacteria > Ascomycota > Firmicutes, whereas in sample O1, the order was Proteobacteria > Cyanobacteria > Actinobacteria. Analysis of differentially expressed genes demonstrated high expression of transcripts related to copper-binding proteins, proteins involved in electron carrier activity, DNA integration, endonuclease activity, MFS transportation, and other uncharacterized proteins in M1 compared to O1. Of the particular interests, several transcripts related to nitrification, ammonification, stress response, and alternate carbon fixation pathways were highly expressed in M1. In-depth analysis of the sequencing data revealed that transcripts of archaeal origin had high expression in M1 compared to O1 indicating the active role of Archaea in metal- and pesticide-contaminated environment. In addition, transcripts encoding 4-hydroxyphenylpyruvate dioxygenase, glyoxalase/bleomycin resistance protein/dioxygenase, metapyrocatechase, and ring hydroxylating dioxygenases of aromatic hydrocarbon degradation pathways had high expression in M1. Altogether, this study provided important insights about the transcripts and pathways upregulating in the presence of pesticides and herbicides.
Conclusion
Altogether, this study claims a high expression of microbial transcripts in two ecosystems with a wide range of functions. It further provided clue about several molecular markers which could be a strong indicator of metal and pesticide contamination in soils. Interestingly, our study revealed that Archaea are playing a significant role in nitrification process as compared to bacteria in metal- and pesticide-contaminated soil. In particular, high expression of transcripts related to aromatic hydrocarbon degradation in M1 soil indicates their important role in biodegradation of pollutants, and therefore, further investigation is needed.
Keywords: Metatranscriptomics, Differential expression, Sequencing, Pollutants, Cypermethrin, Heavy metals
Backgrounds
Understanding microbial community structure and function of the soil ecosystem is vital to delineate ecological roles of the associated microbiome [1, 2]. Several studies have shown that more than 97% of microbes in different ecological habitats (soil, water, acid mine drainage, hot spring) cannot be cultured, and hence, their functional and ecological roles in various biochemical processes remained unexplored [3, 4]. Advances in next-generation sequencing have provided novel insights about structural and functional organization of bacterial genomes and about the key physiological processes and mechanisms bacteria employ to acclimatize under a set of environmental conditions [5–7]. Recently, genome sequencing and analysis of lignocelluloses degrading saprophytic fungi deciphered complete information about the enzymatic machinery these fungi have used to degrade lignocelluloses [8]. Though whole metagenome sequencing and assembly offers opportunity for researchers to profile gene diversity and function under normal conditions, it cannot predict gene functions and pathways which upregulate under particular environmental conditions [9]. In sharp contrast to this, metatranscriptome sequencing and analysis hold immense potential to unravel the functional role of diverse microbiota under various environmental conditions [10, 11]. Nonetheless, next-generation sequencing carried out from couple of complex microbial communities (marine, sediments, and soil) has successfully addressed the role of microbes in various ecological processes [12–17]. In agricultural lands where microbes interact within and between, various groups of organisms makes it very difficult to discover the role of pesticides and metals in shaping microbial community structure and function [18]. Furthermore, large variation in sorption, desorption, and degradation of pesticides has been reported in different soil types [19, 20]. Pesticide usage in agriculture increases the number of pesticides degrading bacteria or archaea in soil [21]. Interestingly, nitrification test as a pesticide side effect has been reported as the best way to depict the role of microorganisms in soil [22]. Studying expression of genes by direct sequencing and analysis of metatranscriptomes at a particular time and space can disclose structural and functional divergence of microbial communities [23].
Spraying of chemicals, e.g., pesticides, herbicides, and fertilizers, in agricultural lands of Punjab (India) is a regular practice [24]. We hypothesized that long usage of such chemicals may alter microbial community structure and function; thus, comparative metatranscriptome can allow researchers to investigate the ecological roles of microbial communities in such environments. Herein, we sequence, analyze, and compare the whole metatranscriptome of agricultural versus organic soil.
Methods
Site description and sampling
Soil samples were collected in duplicates from the agricultural field (M1) of Malwa region of Punjab, India (29° 30′ and 31° 10′ north latitudes and 73° 50′ and 76° 50′ east longitudes), and from the normal organic soil (O1) where no modern agricultural practices are carried out. Samples were collected at a depth of 0–10 cm in September 2014 (atmospheric temperature 28 °C in RNA later (Ambion) (2 ml RNA later added to 2 g of soil) and stored at − 80 °C. Agricultural soil was loamy and had pH of 7.5 to 8.0 whereas the organic soil was acidic and had pH of 6.5–7.0; it was deep brown and porous in nature. Soil samples were transported to the laboratory in dry ice and stored in − 80 °C freezer till its further use for extraction of RNA. The estimation of metals in soil was performed by inductively coupled plasma mass spectrometer (ICP-MS Agilent 7700), whereas the pesticides in the two soil types were estimated by gas chromatography-mass spectrometry (GC-MS, Agilent technologies) as reported in our previous study [25].
RNA extraction
Total RNA was extracted from 2 g soil in duplicates each from M1 and O1 using RNA PowerSoil® Total RNA isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA 92010, USA). Briefly, ~ 2 g soil was homogenized in a 15-ml tube containing silica carbide beads, lysis buffers, phenol to chloroform to isoamyl alcohol (pH 6.5–8), and IRS, to ensure complete lysis of all microorganisms and neutralization of RNases. Clear lysates were precipitated to concentrate the total nucleic acids and were re-suspended in a buffer optimized for binding to anion-exchange gravity flow columns. RNA was eluted using a high salt buffer and was precipitated to obtain the final pure RNA which was resuspended in RNase-free water. RNA from two extractions was pooled (both from M1 and O1) before cDNA preparation and sequencing.
cDNA library construction and Illumina sequencing
Total RNA was treated with DNase to remove DNA contamination. RNA quality was assessed using NANODROP LITE spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Double-stranded cDNA was generated from amplified RNA using the superscript IITM double-strand cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA), as per manufacturer’s instructions. Library preparation, processing, and sequencing were performed at the SciGenom Labs Private Limited, Kerala, India, using the Illumina HighSeq2500 with paired-end (PE) sequencing. Data was submitted with MG-RAST server [26]. MG-Rast ID of the data sets is available for O1 with accession number is mgm4653349.3; M1 dataset was provided a Gold ID 0eeec568676d676d343733323034392e33 and is under progress at present; however, it can be accessed upon request to the server.
FASTQ files quality checking
Raw sequences obtained after the sequencing of metatranscriptomes were analyzed using FastQC tool. Base distribution, base composition, and GC distribution of reads were calculated from the QC result. Based on results, we trimmed the sequence read wherever it was necessary in order to retain high-quality sequences for further analysis. Additionally, reads with more than 10% of “N”s, Illumina adapter contaminated reads, and low-quality reads were excluded from the analysis.
Adapter removal
Raw sample reads were initially pre-processed by removing adapter (Illumina) sequences. Adapter removal was carried out using the cutadapt tool (version 1.7.1) with default parameters.
Other non-coding RNA removals
The other non-coding reads such as tRNA and rRNA sequences were filtered via aligning the adapter filtered reads against reference tRNA sequences downloaded from Genomic tRNA database (GtRNA: http://gtrnadb.ucsc.edu/) using Bowtie-2 tool (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). rRNA sequences were removed using SortMeRNA (http://bioinfo.lifl.fr/RNA/sortmerna/).
Sequence clustering
The reads from each sample were clustered using UCLUST tool (http://www.drive5.com/usearch) with the cutoff value of > 90%. From clustered sequences, the representative sequencing reads were used as a query for functional annotation.
Taxonomy and functional annotation of sequencing reads
Read-based annotations of unique cDNA sequences were carried out against non-redundant sequence database using standalone BlastX program (http://www.ncbi.nlm.nih.gov/) with optimal e value of 10e−5. The best hits showing sequence similarity greater than 90 and lowest e value were retrieved. The predicted gene functions from each read were annotated using Gene Ontology (GO) and KEGG (Kyoto Encyclopedia of Genes and Genomes analysis). The GI (sequence accession number) of each functional hit was retrieved and queried against UniProt database (http://www.UniProt.org/). The transcript id and gene function is provided in the supplementary figures and tables (Additional files 1, 2, 3, 4 and 5).
Differential expression studies
The analysis of differentially expressed genes was carried out based on the read counts of all the samples using DESeq package (http://bioconductor.org/packages/release/bioc/html/ DESeq.html). Initially, the read counts of common transcripts from all samples were considered as an input table for DESeq. All the transcript frequencies were brought to the common scale to make them comparable by normalizing the read counts from each sample. Differential expression was carried out by negative binomial test, and as a result, the mean read count of sample combination, fold change, and p values were estimated. The p value between sample combinations shows the significance of differential expression.
Results
Soil characterization and estimation of pesticides and heavy metals
The soil surface was sandy loam in all samples, and the pH of soil for the organic and agriculture soil was 8 and 9.2 respectively, at the time of collection. Out of several pesticides analyzed, M1 soil revealed the presence of cypermethrin I and III in the range of 0.019 ± 0.001 ppb, whereas cypermethrin II and IV were found to be 0.018 ± 0.002 and 0.017 ± 0.002 ppb, respectively. Heavy metals such as nickel (47.7 ± 2.91 mg kg−1 and 58.7 ± 2.91 mg kg−1), mercury (23.8 ± 5.58 mg kg−1 and 28.8 ± 5.58 mg kg−1), selenium (21.1 ± 4.25 mg kg−1 and 21.1 ± 4.25 mg kg−1), and cadmium (3.1 ± 0.8 mg kg−1 and 5.1 ± 0.8 mg kg−1) are also reported in our previous studies [25]. No residues of pesticides and metals were detected in the O1 soil sample.
RNA extraction, quality checking and, Gene Ontology (GO)
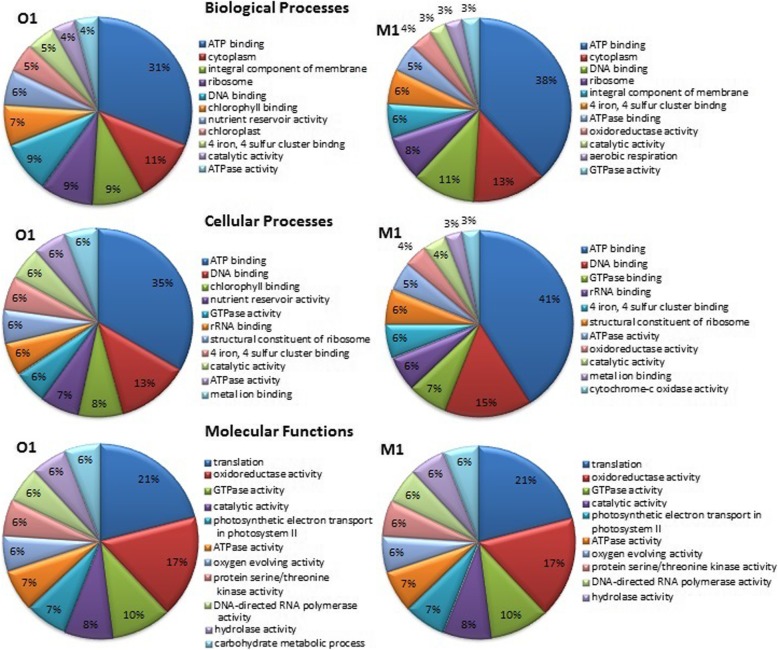
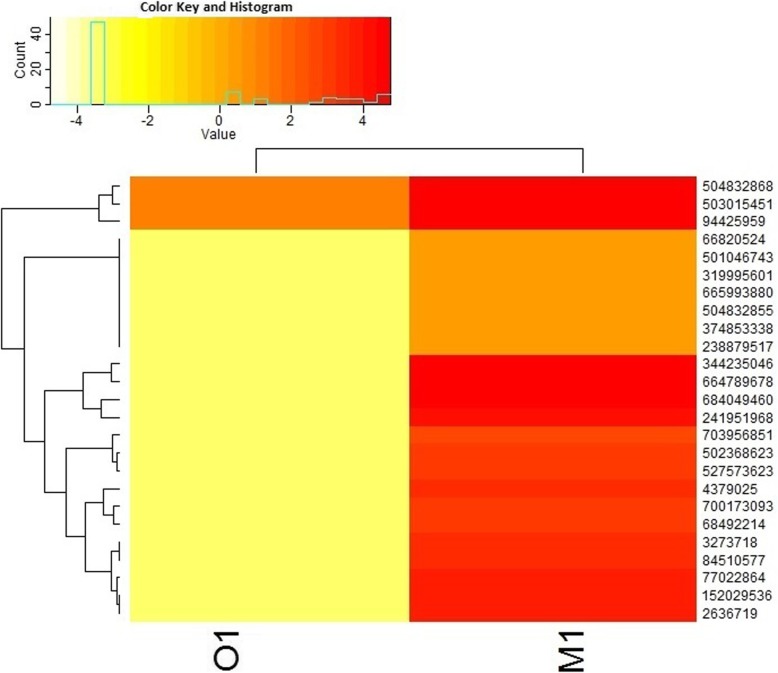
High-quality RNA was extracted from agricultural soil (M1) and organic soil (O1) and was quantified using Nanodrop. Paired-end Illumina cDNA libraries from two datasets yielded ~ 11.49 GBp and ~ 11.9 GBp data for the M1 and O1 samples, respectively (Table 1). A total of 114,908,828 and 119,019,498 raw data (R1 and R2) was sequenced for both M1 and O1respectively. The sequencing data showed 55% GC content and a Phred score ≥ 30. After removal of adapter and non-coding RNA, we assembled the metatranscriptome sequencing reads; however, very less number of reads could be assembled due to complexity and heterogeneity in the metatranscriptome datasets (data not shown). Alternatively, we used a read-based annotation after sequence clustering, as recommended previously [27, 28]. Clustering of sequences resulted in ~ 32,768 representative mRNA reads for M1 and 5611 for O1 respectively. The average length of the mRNA representative reads was > 100 and < 250 bp. Read-based annotation of clustered sequences were classified according to cellular (GO 00005575), molecular (GO 0003674), and biological (GO 0008150) processes [28]. Altogether, from GO assignments, it is evident that the two soil types share similar molecular function; however, they displayed a significant level of variations with respect to cellular and biological function. Percent distribution of transcripts assigned to biological, cellular, and molecular functions among the two datasets is shown in Fig. 1. Gene Ontology (GO) assignments and taxonomical classification are depicted in Additional file 3: Table S1 and Additional file 4: Table S2. The two soil systems however demonstrated great variations for processes involving energy generation, metabolism, transportation, and ribosomal activity. High ATPase and GTPase activity is directly related to energy, motility, host-pathogen interactions, and export of toxins and other wastes out of the cell and in multidrug resistance [29] The iron and sulfur binding metalloproteases play important roles in cellular and biological activities in both prokaryotes and eukaryotes [30]. Several transcripts playing crucial role in oxidation-reduction processes showed enhanced expression in M1. The top 25 transcripts from various organisms demonstrating abundant expression in M1 compared to O1 are depicted in Fig. 2 and Table 2. Other differentially expressed genes enlisted in Additional file 5: Table S3 were also analyzed to understand various metabolic processes taking place in soil.
Table 1.
Description of the raw reads, percent of GC content, and quality score
| Sample ID | Total reads (R1 and R2) | Sequence length (bp) | Total data (Gb) | Percent of GC content | Percent of data ≥ 30 Phred score |
|---|---|---|---|---|---|
| M1 | 114,908,828 | 100 × 2 | ~ 11.49 | 55 | ≥ 30 |
| O1 | 119,019,498 | 100 × 2 | ~ 11.9 | 55 | ≥ 30 |
Fig. 1.
Percent distribution of transcripts assigned to biological processes, molecular functions, and cellular components mapped using Gene Ontology
Fig. 2.
Heat map showing expression of top 25 expressed genes in M1 compared to O1, with p value ≤ 0.05. The details description of the gene IDs is shown in Table 2
Table 2.
Detailed description of the top 25 high expressed genes in M1 compared to O1 (p value ≤ 0.05)
| Transcript_ID | UNIPROT ID | Protein names | Gene names | Organism | Gene length |
|---|---|---|---|---|---|
| 504832868 | K0IHK8 | Monooxygenase, subunit C | amoC1 Ngar_c25120 | Nitrososphaera gargensis (strain Ga9.2) | 184 |
| 503015451 | D8P7X5 | Uncharacterized protein | NIDE3937 | Candidatus Nitrospira defluvii | 1645 |
| 94425959 | Q1MYB0 | Uncharacterized protein | RED65_03040 | Bermanella marisrubri | 119 |
| 66820524 | Q553U6 | Putative actin-22 | act22 DDB_G0275023 | Dictyostelium discoideum (slime mold) | 376 |
| 501046743 | A7HGP7 | 10 kDa chaperonin (GroES protein) | groS groES Anae109_3714 | Anaeromyxobacter sp. (strain Fw109-5) | 97 |
| 319995601 | E8MYN6 | Cell division protein FtsZ | ftsZ ANT_23460 | Anaerolinea thermophila | 387 |
| 665993880 | A0A075MRB7 | Cytochrome b subunit of the bc complex | NTE_01577 | Candidatus Nitrososphaera evergladensis SR1 | 546 |
| 504832855 | K0ILC3 | Methylmalonyl-CoA mutase, small subunit | Ngar_c24990 | Nitrososphaera gargensis (strain Ga9.2) | 141 |
| 374853338 | H5SJB3 | MFS transporter | HGMM_F35G12C20 HGMM_F51D07C02 | Uncultured Acetothermia bacterium | 412 |
| 238879517 | C4YNC2 | Phosphoenolpyruvate carboxykinase | CAWG_01381 | Candida albicans (strain WO-1) (yeast) | 553 |
| 344235046 | G3IQD4 | Putative uncharacterized protein | I79_026257 | Cricetulus griseus (Chinese hamster) (Cricetulus barabensis griseus) | 60 |
| 664789678 | A0A075KDX2 | Uncharacterized protein | UFO1_2808 | Pelosinus sp. UFO1 | 128 |
| 684049460 | A0A094MWR7 | Uncharacterized protein | Thpro_01067 | Thiobacillus prosperus | 71 |
| 241951968 | B9WC89 | d-arabinose dehydrogenase | CD36_22310 | Candida dubliniensis (strain CD36/ATCC MYA-) | 326 |
| 703956851 | A0A0A3BUY2 | Uncharacterized protein | MEO_05149 | Candida albicans P94015 | 1541 |
| 502368623 | C6E7X5 | Multicopper oxidase type 2 | GM21_0071 | Geobacter sp. (strain M21) | 1299 |
| 527573623 | S8JMM6 | Endonuclease | SAG0065_03165 | Streptococcus agalactiae CCUG 37742 | 1275 |
| 4379025 | O95662 | KpnI repetitive sequence | Homo sapiens (human) | 194 | |
| 700173093 | A0A0A0JA99 | Uncharacterized protein | N798_05235 | Knoellia flava TL1 | 1571 |
| 68492214 | Q59K39 | Retrotransposon Tca2 | POL98 | Candida albicans | 1566 |
| 3273718 | O74209 | Pol polyprotein (fragment) | pol | Candida albicans (yeast) | 1576 |
| 84510577 | A3V3H5 | Uncharacterized protein | SKA53_01511 | Loktanella vestfoldensis SKA53 | 91 |
| 77022864 | Q3MP87 | Putative uncharacterized protein CaJ7.0300 | CaJ7.0300 CaO19.6451 | Candida albicans (yeast) | 1566 |
| 152029536 | A7HF08 | Multicopper oxidase type 2 | Anae109_3108 | Anaeromyxobacter sp. (strain Fw109-5) | 1430 |
| 2636719 | O13308 | POL protein (fragment) | Candida albicans (yeast) | 1576 |
Phylogeny analysis
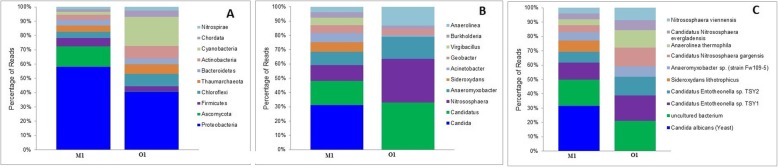
Taxonomic analysis (Additional file 3: Table S1) of the annotated reads, as presented in Fig. 3, revealed Proteobactreria as major phylum in the two soil types, the order of top 3 abundant phyla in the M1 was Proteobacteria > Ascomycota > Firmicutes, whereas sample O1 demonstrated the following order Proteobacteria>Cyanobacteria>Actinobacteria (Fig. 3a). Proteobacteria encompass enormous diversity in terms of physiology, morphology, and metabolism and is considered as a key player in carbon and nitrogen cycle. Deltaproteobacteria and Alphaproteobacteria showed abundant presence in M1, whereas Lilliopsida and Deltaproteobacteria were more in O1. Another important phylum identified in this M1 soil sample belongs to Thaumarchaeota. At the genus level, ~ 1312 transcripts were assigned to the M1 sample, and the most dominant genus identified was Candida (30%) followed by Candidatus and Nitrosophaera in M1 (Fig. 3b). At the species level, Candida albicans (30%) and Sideroxydans lithotrophicus (8%) revealed their unique occurrence in M1. Interestingly, Anaeromyxobacter sp. Fw109-5, Candidatus Nitrososphaera gargensis, N. Viennensis, Anaerolinea thermophila, Candidatus Entotheonella sp. TSY1, and Candidatus Entotheonella sp. TSY2 were less represented in the M1 sample as compared to O1 (Fig. 3c).
Fig. 3.
Percent distribution of transcripts representing various taxonomical groups at phylum, genus, and species level in M1 and O1. a Phylum. b Genus. c Species
Comparing nitrification process in agriculture (M1) and organic soil (O1)
Nitrification is used to monitor the side effects of contaminants on microbial community structure, function, and soil quality. KEGG (Kyoto Encyclopedia of Genes and Genomes analysis) analysis of nitrification pathway revealed that several transcripts encoding genes of nitrification were highly expressed in M1 that mainly include amoC from AOA (ammonia-oxidizing archaea) (amoC1, C2 = 4.3, 4.17-fold in N. gargensis, amoC3 = 1.46 in Nitrososphaera viennensis EN76, NTE_00725 = 3, NTE_01100 = 1.44-fold in Candidatus Nitrososphaera evergladensis SR1), indicating active contribution of AOA in nitrification process in M1 soil. Furthermore, we observed a high expression of putative NxrA2 and NxrB1/B2, i.e., nitrite oxidoreductase (NXR), in the M1 soil sample (Additional file 4: Table S4) from C. Nitrospira defluvii and a beta subunit of nitrite oxidoreductase from Nitrospira moscoviensis, a key enzyme of NO2 oxidation in NOB (nitrate-oxidizing bacteria). In addition, copper-containing nitrite reductases (NirK) from uncultured bacterium also had high expression in the M1 soil. Interestingly, a membrane-bound dissimilatory nitrate reductase (~ 2.7-fold) Tbd_1403 of T. denitrificans was also highly expressed in agriculture soil M1. High expression of the small subunit of methylmalonyl-CoA mutase (EC 5.4.99.2) that convert malonyl-CoA to succinyl-CoA as the main product of an alternative carbon fixation pathway was also observed in M1.
Analysis of stress-related transcripts
Several chaperones demonstrated high expression (~ 1.6–3-fold increase, Fig. 4 and Additional file 3: Table S2) that include DnaK (HSP70) (heat shock 70 kDa protein) with transcript ID 570743717, 10 kDa chaperonin (GroES protein) transcript ID 644122643 (Protein Cn10), ATP-dependent chaperone ClpB transcript ID 503058777, 60 kDa chaperonin (GroEL protein) transcript ID 751261482, and ATPase with chaperone activity and a copper chaperones (CCH-related protein) with transcript ID_11542043. Additionally, several proteases also showed high expression in the polluted soil sample that includes Lon protease (EC 3.4.21.53) (ATP-dependent protease La) of Candidatus Entotheonella sp., serine protease of Nitrosococcus oceani, ATP-dependent zinc metalloprotease FtsH (EC 3.4.24.-) ftsH from Nitrosospira, ATP-dependent zinc metalloprotease FtsH (EC 3.4.24.-) ftsH from Caldilinea aerophila, carboxyl-terminal protease (EC 3.4.21.102) of Niastella koreensis, ATP-dependent zinc metalloprotease FtsH (EC 3.4.24.-) ftsH of Anaerolinea thermophila, and Lon protease (EC 3.4.21.53) (ATP-dependent protease La) of Anaeromyxobacter sp. Abundant expression of transcripts encoding endonuclease (transcript ID_527573623) from Streptococcus agalactiae indicates heavy metal contamination in the M1 soil sample, in which both Ni and Cd potentially affect the activity of LINE-1.
Fig. 4.
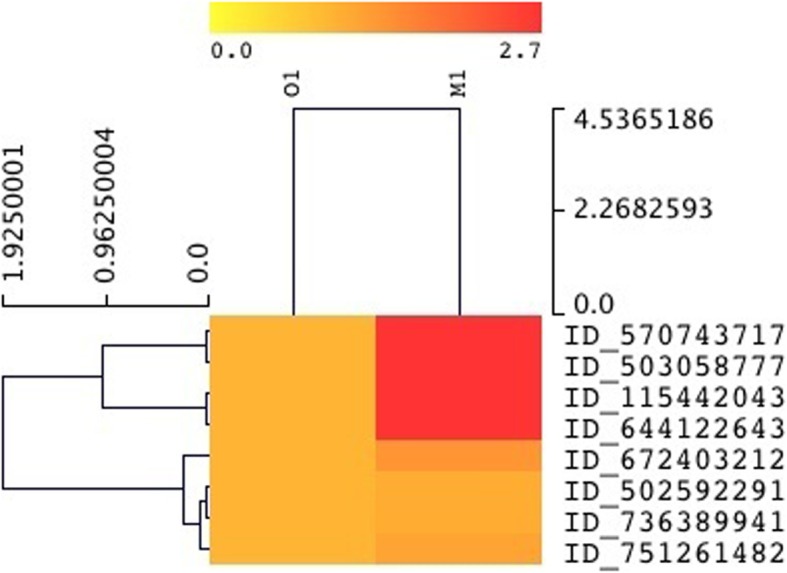
Heat map showing high expression of heat shock proteins in the M1 soil, refer to Additional file 4: Table S2 for details
Aromatic and hydrocarbon metabolism
Transcripts associated with the metabolism of aromatic hydrocarbon demonstrated their high expression in cypermethrin- and metal-contaminated M1 soil (Fig. 5 and Table 3) compared to O1. Increased expression of transcripts related to aromatic metabolism was widespread and associated with both central and peripheral aromatic metabolic pathways. Interestingly, high expression of transcripts that include 4-hydroxyphenylpyruvate dioxygenase (HPPD), glyoxalase/bleomycin resistance protein/dioxygenase, metapyrocatechase, and some uncharacterized protein share similarity with vicinal oxygen chelate superfamily (VOC family). Another transcript-encoding 2-nitropropane dioxygenase [EC 1.13.12.16] also showed high expression in M1 sample; this enzyme catalyzes oxidation of nitroalkanes into their corresponding carbonyl product. Nitronate monooxygenase (NMO), formerly referred as 2-nitropropane dioxygenase (NPD) (EC 1.13.11.32), is an FMN-dependent enzyme that uses molecular oxygen to oxidize (anionic) alkyl nitronates. Of the particular interests, we observed high expression of transcripts encoding ring hydroxylating dioxygenases (EC 1.14.12.12) from Novosphingobium aromaticivorans (strain DSM 12444/F199) and intradiol ring-cleavage dioxygenase from Novosphingobium resinovorum.
Fig. 5.
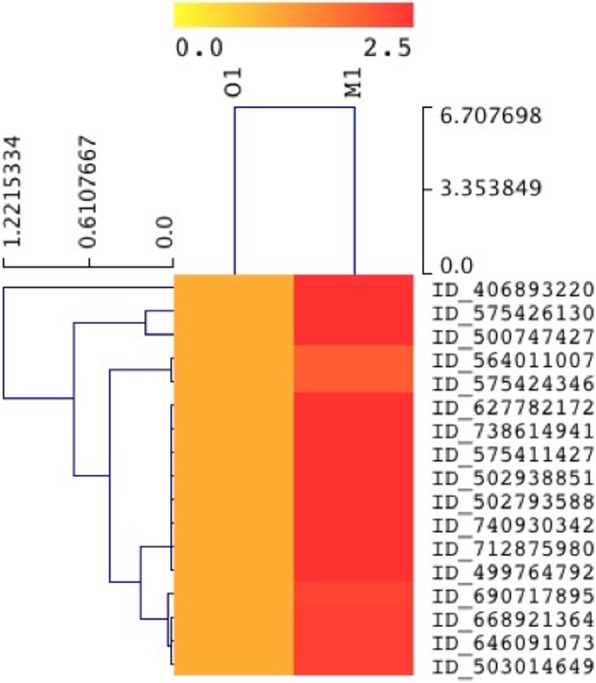
Heat map showing expression of genes involved in aromatic compound degradation in M1 compared to O1 with p value ≤ 0.05. The detailed description of the genes is provided in Table 3
Table 3.
Transcripts showing high expression related to aromatic hydrocarbon degradation in M1 compared to O1 (p value ≤ 0.05)
| Transcript_ID | UniProt ID | Protein names | Gene names | Organism | Gene length |
|---|---|---|---|---|---|
| 627782172 | A0A023XH36 | Metapyrocatechase | BJS_04241 | Bradyrhizobium japonicum SEMIA 5079 | 326 |
| 668921364 | A0A085DUZ9 | Protocatechuate 3,4-dioxygenase | DK37_06335 | Halomonas sp. SUBG004 | 196 |
| 738614941 | A0A031K144 | Ring hydroxylating dioxygenase, alpha subunit | BV97_01833 | Novosphingobium resinovorum | 425 |
| 575411427 | W4LH05 | Uncharacterized protein | ETSY1_22950 | Candidatus Entotheonella sp. TSY1 | 171 |
| 575426130 | W4MGP2 | Uncharacterized protein | ETSY2_00190 | Candidatus Entotheonella sp. TSY2 | 169 |
| 564011007 | W9BZ28 | 2-Nitropropane dioxygenase | Q27BB25_02350 | Blastomonas sp. CACIA14H2 | 467 |
| 575424346 | W4MC15 | Uncharacterized protein | ETSY2_08610 | Candidatus Entotheonella sp. TSY2 | 380 |
| 502938851 | D6XZD0 | 2-Nitropropane dioxygenase NPD | Bsel_2926 | Bacillus selenitireducens | 318 |
| 502793588 | D5CM51 | 2-Nitropropane dioxygenase NPD | Slit_0424 | Sideroxydans lithotrophicus (strain ES-1) | 416 |
| 646091073 | A0A069IMT4 | 4-Hydroxyphenylpyruvate dioxygenase | ER13_02875 | Brevundimonas sp. EAKA | 373 |
| 500747427 | A7HAK0 | Glyoxalase/bleomycin resistance protein/dioxygenase | Anae109_1540 | Anaeromyxobacter sp. (strain Fw109-5) | 121 |
| 740930342 | A0A063B6R9 | Glyoxalase/bleomycin resistance protein/dioxygenase | LIG30_4306 | Burkholderia sp. lig30 | 183 |
| 406893220 | K1Z0Y1 | Glyoxalase/bleomycin resistance protein/dioxygenase | ACD_75C00771G0006 | Uncultured bacterium | 157 |
| 690717895 | A0A098SBR0 | Homogentisate 1,2-dioxygenase | IX84_02245 | Phaeodactylibacter xiamenensis | 390 |
| 712875980 | A0A0A4BJC1 | Indoleamine 2,3-dioxygenase | MG7_04021 | Candida albicans P34048 | 440 |
| 499764792 | Q2G757 | Intradiol ring-cleavage dioxygenase | Saro_1876 | Novosphingobium aromaticivorans (strain DSM 12444/F199) | 313 |
| 503014649 | D8PHR7 | Putative homogentisate 1,2-dioxygenase (EC 1.13.11.5) | NIDE3110 | Candidatus Nitrospira defluvii | 371 |
Discussion
Investigation of transcripts encoding functional protein by direct sequencing and analysis at a particular time and space can disclose structural and functional features of microbial communities. In this study, analysis of metatranscriptome samples from two distinct soils revealed divergences in functional features of microbial communities. Interestingly, during phylogeny analysis, we observed an abundant presence of archaea especially Thaumarchaeota, a phylum which is widely distributed in extreme, non-extreme [31], terrestrial [32], and metal-contaminated soil [33]; these observations primarily indicate important consequences of archaea, especially towards degradation of the pesticides, ammonia oxidation [34], nitrate leaching in greenhouse gas production, and soil subsidence [35]. Additionally, the abundance of Candida albicans and Sideroxydans lithotrophicus in the M1 soil sample suggests their high tolerance to metals as also reported previously [36]. It was interesting to note that despite of low abundance of archaea in the M1 soil, it has shown high expression of genes related to several pathways, thus indicating archaeal and better adaptation and active contribution in pesticide- and metal-contaminated environment, e.g., several transcripts related to nitrification process, which is used to monitor side effects of pesticide contamination [37], showed high expression in M1. A highly expressed amoC transcript identified in the M1 soil (belongs to group I.1.a of archaea as depicted in Additional file 1: Figure S1, which harbor an additional copy of amoC gene) is well correlated previously with high metal resistance [38–40]. Additionally, we observed high expression of putative NxrA2 and NxrB1/B2, i.e., nitrite oxidoreductase (NXR) (Additional file 4: Table S4), from C. Nitrospira defluvii and β subunit of nitrite oxidoreductase from Nitrospira moscoviensis, key enzymes of NO2 oxidation in NOB indicating active participation of these organisms in nitrification in M1 soil. On further analysis, we found that these genes have shared similarity with NxrA2 and NxrB of N. moscoviensis and with other known NO2−/NO3−-binding molybdoenzymes, such as NXR of Nitrobacter or bacterial nitrate reductases (NARs) which shuttles two electrons per oxidized NO2 into the electron transport chain [41]. Another transcript (NirK) from uncultured bacterium also had high expression in M1; this transcript has shared maximum homology (> 70%) with NirK of Chloroflexi thus indicating denitrification through AOB (ammonia-oxidizing bacteria) [42] that forms NO from NO2 [43]. Besides nirK homologs, no other genes typically contributing to the denitrification process showed high expression in the M1 soil. We therefore propose that high expression of transcripts nirK along with amoC can be a useful molecular marker to monitor soil ammonia oxidation in agricultural soil contaminated with pesticides and metals. Additionally, high expression (~ 2.7-fold) of a membrane-bound dissimilatory nitrate reductase Tbd_1403 of T. denitrificans in agriculture soil indicates nitrate-dependent oxidation of metals with high reduction potentials and can also be used as molecular marker to study metal resistance pathways among denitrifying bacteria showing high metal tolerance U(IV) and Fe(II) [44–46].
In addition to high expression of transcripts related with nitrification and ammonia oxidation from Thaumarchaeal viz N. gargensis, we observed high expression of PCK1 in archaea; PCK1 regulates 3-hydroxypropionate/4-hydroxybutyrate carbon fixation pathway as reported previously [47–50]. In particular, high expression of PCK1 gene from C. albicans that encodes protein of gluconeogenesis pathway indicate soil environment where carbon availability might be changing continuously enabling C. albicans to switch its metabolism by regulating PCK1, as also reported previously in metal-contaminated soil [51, 52]. Another transcript that demonstrated high expression in M1 belongs to a putative NADP+-specific d-arabinose dehydrogenase from C. dubliniensis sharing > 95% identity with C. albicans. The exact mechanism of action of d-arabinose dehydrogenase is unknown; however, its activity is shown to be inhibited by metals presence including Hg2+ [53]; thus, it may be hypothesized that high expression of d-arabinose dehydrogenase is indicating oxidative stress response of C. albicans in agriculture soil.
Microbes demonstrate multiple adaptations to survive under osmotic, oxidative, heavy metals, elevated temperatures, high salinity, and other stress conditions, one of the strategies that microbes employ to cope up these stresses is accumulation of compatible solutes and small soluble organic molecules [54]. In order to investigate the transcripts showing high expression in metal and pesticide stress conditions, we compared and analyze the transcripts associated with stress from two soil types; interestingly, high expression of UDP-glucose 6-dehydrogenase (EC 1.1.1.22) (udg_NIDE4145; ~ 2.7-fold) and pyruvate to ferredoxin oxidoreductase δ subunit (EC 1.2.7.1) (porD_NIDE0972; ~ 1.7-fold) indicate active synthesis of glycogen in Ca. N. defluvii in response to stress [38]. These solutes can either be transported into the cell or synthesized de novo. Indeed, glycogen deposits have been observed under electron microscopy in Nitrospira cells under stress conditions [55]. To mitigate oxidative damage, in contrast to most of the aerobic bacteria, N. defluvii lacks SOD and catalase, and its genome lacks superoxide dismutase (SOD) either. High expression of bacterioferritin transcripts in Ca. N. defluvii indicates ROS (reactive oxygen species) detoxification in M1 soil; it binds with free iron thus reduces the risk of ROS generation. Additionally, transcript ID_664789678 (Table 2) with unknown function from Pelosinus species also showed high expression in the M1 soil sample compared to O1. Previously, this organism has shown its presence in various sites that include Melton Branch Watershed, Oak Ridge, TN, USA ([56] and uranium-contaminated field site near Oak Ridge [57]. During its growth, it uses Fe(III) [58] and U(VI) [59]. As most of the proteins from Pelosinus are not characterized [60], we hypothesized that high expression of one of its transcripts in the M1 soil can be correlated with the presence of heavy metals in M1 compared to the O1 soil. Additionally, high expression of cell division-related protein FtsZ from Anaerolinea thermophila indicates active peptidoglycan biosynthesis in the M1 soil. Furthermore, a multicopper oxidase (transcript ID_ 502368623, 700173093, 152029536) from Geobacter sp., Knoellia sp., and Anaeromyxobacter sp. also showed high expression in the M1 soil. Catalytic sites of these proteins contain copper-binding sites and are implicated in bacterial copper resistance, oxidation of phenolic compounds, and in detoxification of Mn2+ [61]. Additionally, high expression of putative actin 22 in M1 indicates the formation of actin rods which was previously reported in Dictyostelium during the formation of spores under stress conditions such as heat shock and osmotic pressure as well as non-nutrient conditions [62, 63]. Many of these chaperones were of Proteobacterial origin and of high expression. These proteases were also of proteobacterial origin, and their role has not yet established in the metal- and pesticide-contaminated agriculture soil; however, we propose that their high expression can be correlated with high turnover in Proteobacteria which mostly depends on ATP-dependent proteases that are recruited by the cells in the cytosol (Lon, ClpAP, ClpXP, HslVU) and are well reported in cypermethrin- and cadmium-contaminated soil [64–67] or are associated with the inner membrane (FtsH) [68]. These enzymes in adverse conditions not only help in degradation of misfolded or abnormal proteins but also degrade unstable proteins, like σ32 heat shock factor. Another transcript Tca2 which is widely distributed in C. albicans, related with Ty1/copia-type retrotransposon [69], and is predominantly present in the M1 soil (transcript ID_68492214, 77022864, 703956851, 3273718) support the fact that Tca2 transpositional activity is favored at high-temperature stress and reveal a close relationship between the Tca2 expression and virulence in C. albicans endonuclease activity in vitro since it also utilizes Mg2+ as a cofactor for enzymatic activity [70]. It further raises the possibility of co-selection of antibiotic and heavy metal resistance dissemination through mobile genetic elements [71] observed in other transcripts in the current study. High expression of MFS (transcript id_374853338) in uncultured Acetothermia bacterium showing similarity with oxalate/formate antiporter of proteobacteria, i.e., Nitrosomonas sp., point out that it may be participating in exchange of external divalent oxalate with the intracellular monovalent formate [72, 73] to generate a proton-motive force that supports membrane functions, including ATP synthesis, accumulation of growth substrates, and extrusion of waste products [72–75].
Furthermore, the analysis of metabolic pathways associated with aromatic hydrocarbon metabolism revealed high expression of glyoxalase/bleomycin resistance protein/dioxygenase, metapyrocatechase, and some uncharacterized protein sharing similarity with vicinal oxygen chelate superfamily (VOC family) that is known to harbor a β-α-β motif in cypermethrin- and metal-contaminated soil (Fig. 5 and Table 3); similar observations were also reported in previous studies [76, 77]. This β-α-β motif provides a metal coordination environment for these enzymes and helps them in catalyzing different enzymatic reactions including isomerizations in glyoxalase I and epimerization in methylmalonyl-CoA epimerase, in oxidative cleavage of C-C bonds in extradiol dioxygenase and nucleophilic substitutions in fosfomycin/bleymucin resistance proteins [78]. Phylogenetically, these transcripts showed their origin from Geobacter, strains viz. G. metallireducens, G. pickeringii, and Geobacter uraniireducens (Additional file 2: Figure S2), which are often the dominant members of subsurface sediments, dwell under metal-reducing conditions [79], retrieve energy through dissimilatory reduction of metals ions, and efficiently oxidize monoaromatic compounds such as toluene and phenol [80, 81]. We have also observed high expression of transcripts encoding enzyme 2-nitropropane dioxygenase [EC 1.13.12.16], an enzyme catalyzing oxidation of nitroalkanes into their corresponding carbonyl product. Nitronate monooxygenase (NMO), formerly referred as 2-nitropropane dioxygenase (NPD) (EC 1.13.11.32) which oxidizes (anionic) alkyl nitronates, upregulation of NPD gene in the agriculture soil M1, indicates high nitronate contamination in cypermethrin- and metal-contaminated soil. Similar observations were reported previously where authors have shown upregulation of NPD in Sideroxydans lithotrophicus (strain ES-1) in iron-contaminated groundwater [82] indicating its stimulatory effect with Fe2+ in the agricultural soil [83–85]. Of the particular interests, high expression of transcripts encoding ring hydroxylating dioxygenases (EC 1.14.12.12) from Novosphingobium aromaticivorans (strain DSM 12444/F199) and intradiol ring-cleavage dioxygenase from Novosphingobium resinovorum indicate active participation of these organisms in the degradation process of polycyclic aromatic hydrocarbon that includes naphthalene and ethylbenzene [86–90].
Conclusion
In summary, a comparative metatranscriptomic study from agriculture and organic soil types illuminated structural and functional variations. The study further concluded that high expression of microbial transcripts in two soil types is associated with wide range of functions. It has also provided clue about several molecular markers which could be a strong indicator of metal and pesticide contamination in soils. Interestingly, our study revealed that archaea are playing a significant role in nitrification process compared to bacteria in metal- and pesticide-contaminated soil. In particular, high expression of transcripts related to aromatic hydrocarbon degradation provided clue about degradation potential associated in polluted soil communities.
Supplementary information
Additional file 1: Fig. S1. Phylogenetic analysis of upregulated amoC genes obtained using neighbor-joining clustering method having bootstrap values out of 1000 replicates using MEGA 7.0 using UniProt ID with their species and gene name (GN). K0IHK8_ Nitrososphaera gargensis (strain Ga9.2)_amoC1, A0A075MNL6_ Candidatus Nitrososphaera evergladensis SR1_NTE00725, A0A060HQE0_ Nitrososphaera viennensis EN76_amoC3, A0A075MPN8_ Candidatus Nitrososphaera evergladensis SR1_ NTE_01100. It clearly indicates that thaumarchaea species harbor additional copy of amoC that could be used as a molecular marker in the detection of particular archeal community present in cypermerthrin and metal co-contaminated agriculture soils i.e. M1 soil sample. (JPG 82 kb)
Additional file 2: Fig. S2. Phylogenetic analysis of VOC superfamily transcript obtained in M1 soil sample using neighbor-joining clustering method having bootstrap values out of 1000 replicates using MEGA 7.0 using UniProt ID with their species (K1Z0Y1_uncultured bacterium, A0A063B6R9_ Burkholderia sp. lig30,A7HAK0_ Anaeromyxobacter sp. (strain Fw109-5), A0A069IMT4_ Brevundimonas sp. EAKA, A0A023XH36_ Bradyrhizobium japonicum SEMIA 5079) that clearly indicates the evolution of VOC superfamily mostely from Geobacter sp., Nitrospirae sp. Deulphobulbacae sp, actinobacteria sp., candidatus Entothella sp. present dominantly in M soil sample. (JPG 122 kb)
Additional file 3: Table S1. The transcript id, gene function, gene ontology (biological,molecular and cellular function) obtained M1 functional hits after quarried against UniPrt database. (XLS 5019 kb)
Additional file 4: Table S2. The transcript id, gene function, gene ontology (biological, molecular and cellular function) obtained O1 functional hits after quarried against UniProt database. (XLS 1457 kb)
Additional file 5: Table S3. List of Differentially Expressed genes in M1 and O1sample. (DOC 45 kb)
Acknowledgements
Not applicable
Ethical approval and consent to participate
Not applicable
Authors’ contributions
PKS designed, analyzed, wrote, proofread, and drafted the manuscript. VS helped in the analysis and writing of the manuscript. SS did the annotation, assembly, and expression analysis. GB analyzed the results. KS helped in the analysis, writing, and drafting of the manuscript. RK performed and contributed in analyzing and writing of the manuscript. Further, all authors have read and agreed for its submission and publication in Journal of Genetic Engineering and Biotechnology.
Funding
This research work was funded by the Science and Engineering Research Board (SERB) New Delhi, project file number: SB/YS/LS-63/2013 under fast track scheme for the young scientists. Dr. PKS would like to thank SERB for this financial support.
Availability of data and materials
The metatranscriptome data for M1 and O1 is submitted with MG-RAST under the accession number mgm4653349.3, whereas M1 dataset was provided a Gold ID 0eeec568676d676d343733323034392e33.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pushpender Kumar Sharma, Email: pushpg_78@rediffmai.com, Email: pushpg_78@rediffmail.com, Email: psnp7819@gmail.com.
Vinay Sharma, Email: vinay_mnd@yahoo.com.
Shailesh Sharma, Email: shailesh.sharma@nabi.res.in.
Garima Bhatia, Email: garibhat@gmail.com.
Kashmir Singh, Email: kashmir123@gmail.com.
Rohit Sharma, Email: rohitsharma20@yahoo.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s43141-019-0006-3.
References
- 1.Rappe MS, Vergin K, Giovannoni SJ. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol Ecol. 2000;33:219–232. doi: 10.1016/S0168-6496(00)00064-7. [DOI] [PubMed] [Google Scholar]
- 2.Bailly JL, Fraissinet-Tachet L, Verner MC, Debaud J, Lemaire M, Micheline W, Louvel S, Marmeisse R. Soil eukaryotic functional diversity, a metatranscriptomic approach. ISME J. 2007;1:632–642. doi: 10.1038/ismej.2007.68. [DOI] [PubMed] [Google Scholar]
- 3.Prosser J, Bohannan B, Curtis T, Ellis R, Firestone M, Freckleton R. The role of ecological theory in microbial ecology. Nat Rev Microbiol. 2007;5:384–392. doi: 10.1038/nrmicro1643. [DOI] [PubMed] [Google Scholar]
- 4.Fierer N. Microbial biogeography: patterns in microbial diversity across space and time. In: Zengler K, editor. in Accessing Uncultivated Microorganisms: From the Environment to Organisms and Genomes and Back. Washington, DC: ASM Press; 2008. pp. 95–115. [Google Scholar]
- 5.Gifford SM, Sharma S, Rinta-Kanto JM, Moran MA. Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J. 2011;5:461–472. doi: 10.1038/ismej.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tyson GW, Lo I, Baker BJ, Allen EE, Hugenholtz P, Baker BJ. Genome-directed isolation of the key nitrogen fixer Leptospirillum ferrodiazotrophum sp nov. from an acidophilic microbial community. Appl Environ Microbiol. 2005;71:6319–6324. doi: 10.1128/AEM.71.10.6319-6324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW. Deciphering the evolution and metabolism of an Anammox bacterium from a community genome. Nature. 2006;440:790–794. doi: 10.1038/nature04647. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert JA, Field D, Huang Y, Edwards R, Li WZ, Gilna P, Joint I. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One. 2003;3:e3042. doi: 10.1371/journal.pone.0003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tringe SG, Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 10.DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert JA, Field D, Huang Y, Edwards R, Li W, Gilna P. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One. 2008;3:e3042. doi: 10.1371/journal.pone.0003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, DeLong EF. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci U S A. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poretsky RS, Hewson I, Sun SL, Allen AE, Zehr JP, Moran MA. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol. 2009;11:358–1375. doi: 10.1111/j.1462-2920.2008.01863.x. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Hu M, Huang L, Hua Z, Kuang J, Li S, Shu W. Comparative metagenomic and metatranscriptomic analyses of microbial communities in acid mine drainage. ISME J. 2015;9:1579–1592. doi: 10.1038/ismej.2014.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damon C, Lehembre F, Oger-Desfeux C, Luis P, Ranger J. Metatranscriptomics reveals the diversity of genes expressed by eukaryotes in forest soils. PLoS One. 2012;7:e28967. doi: 10.1371/journal.pone.0028967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menezes A, Clipson N, Doyle E. Comparative metatranscriptomics reveals widespread community responses during phenanthrene degradation in soil. Environ Microbiol. 2012;14:2577–2588. doi: 10.1111/j.1462-2920.2012.02781.x. [DOI] [PubMed] [Google Scholar]
- 17.Tveit A, Urich TT. Svenning MM (2010) Metatranscriptomic analysis of arctic peat soil microbiota. Appl Environ Microbiol. 2014;80:5761–5772. doi: 10.1128/AEM.01030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker SS Buried treasure: soil biodiversity and conservation. Biodivers Conserv 2010;19(17):3743–3756
- 19.Wakelin SA, Chu G, Lardner R, Liang Y, McLaughlin M. A single application of Cu to field soil has long-term effects on bacterial community structure, diversity, and soil processes. Pedobiologia. 2010;53:149–158. doi: 10.1016/j.pedobi.2009.09.002. [DOI] [Google Scholar]
- 20.Bælum J, Jacobsen CS. TaqMan probe-based real-time PCR assay for detection and discrimination of class I, II, and III tfdA genes in soils treated with phenoxy acid herbicides. Appl Environ Microbiol. 2009;75:2969–2972. doi: 10.1128/AEM.02051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Floch C, Chevremont AC, Joanico K, Capowiez Y, Criquet S. Indicators of pesticide contamination: soil enzyme compared to functional diversity of bacterial communities via BiologW ecoplates. Eur J Soil Biol. 2011;47:256–263. doi: 10.1016/j.ejsobi.2011.05.007. [DOI] [Google Scholar]
- 22.Bælum J, Nicolaisen MH, Holben WE, Strobel BW, Sørensen J, Jacobsen CS. Direct analysis of tfdA gene expression by indigenous bacteria in phenoxy acid amended agricultural soil. ISME J. 2008;2:677–687. doi: 10.1038/ismej.2008.21. [DOI] [PubMed] [Google Scholar]
- 23.Todaka N, Moriya S, Saita K, Hondo T, Kiuchi I, Takasu H. Environmental cDNA analysis of the genes involved in lignocellulose digestion in the symbiotic protist community of Reticulitermes Speratus. FEMS Microbiol Ecol. 2007;59:592–599. doi: 10.1111/j.1574-6941.2006.00237.x. [DOI] [PubMed] [Google Scholar]
- 24.Mittal S, Kaur G, Vishwakarma GS. Effects of environmental pesticides on the health of rural communities in the Malwa region of Punjab (India): a review. Hum Ecol Risk Assess. 2014;20:366–387. doi: 10.1080/10807039.2013.788972. [DOI] [Google Scholar]
- 25.Sharma PK, Sharma R. Metatranscriptome sequencing and analysis of agriculture soil provided significant insights about the microbial community structure and function. Ecological Genetics and Genomics. 2017;6:9–15. doi: 10.1016/j.egg.2017.10.001. [DOI] [Google Scholar]
- 26.Meyer F, Paarmann D, D'Souza M, Olson R, Glass E, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, et al. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mavromatis K, Land ML, Brettin TS, Quest DJ, Copeland A, et al. The fast changing landscape of sequencing technologies and their impact on microbial genome assemblies and annotation. PLoS One. 2012;7:e48837. doi: 10.1371/journal.pone.0048837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas T, Gilbert J, Meyer F. Metagenomics – a guide from sampling to data analysis. Microb Inform Exp. 2012;2:3. doi: 10.1186/2042-5783-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pang Q, Gao L, Hu W, An Y, Deng H, Zhang Y. De novo transcriptome analysis provides insights into immune related genes and the RIG-I-like receptor signaling pathway in the freshwater planarian (Dugesia japonica) PLoS One. 2016;11:e0151597. doi: 10.1371/journal.pone.0151597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lubelski J, Konings WN. Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol Mol Biol Rev. 2007;71:463–476. doi: 10.1128/MMBR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson KA, Bhushan S, Stahl A, Hallberg BM, Frohn A, Glaser E, Eneqvist T. The closed structure of presequence protease PreP forms a unique 10 000 A° 3 chamber for proteolysis. EMBO J. 2006;25:1977–1986. doi: 10.1038/sj.emboj.7601080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papa S, Bartoli G, Pellegrino A, Fioretto A. Microbial activities and trace element contents in an urban soil. Environ Monit Assess. 2010;165:193–203. doi: 10.1007/s10661-009-0938-1. [DOI] [PubMed] [Google Scholar]
- 33.Zhang P, Shao M. Spatial variability and stocks of soil organic carbon in the Gobi Desert of Northwestern China. PLoS One. 2014;165:e93584. doi: 10.1371/journal.pone.0093584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stefanowicz AM, et al. Metals affect soil bacterial and fungal functional diversity differently. Environ Toxicol Chem. 2008;27:591–598. doi: 10.1897/07-288.1. [DOI] [PubMed] [Google Scholar]
- 35.Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol. 2006;72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellis RJ, Morgan P, Weightman AJ, Fry JC. Cultivation-dependent and -independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Appl Environ Microbiol. 2003;69:3223–3230. doi: 10.1128/AEM.69.6.3223-3230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kowalchuk GA, Stephen JA. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol. 2001;55:485–529. doi: 10.1146/annurev.micro.55.1.485. [DOI] [PubMed] [Google Scholar]
- 38.Zhalnina K, Dias R, Leonard MT, Dorr de Quadros P, FAO C, Drew JC. Genome sequence of Candidatus Nitrososphaera Evergladensis from Group I.1b enriched from everglades soil reveals novel genomic features of the ammonia-oxidizing archaea. PLoS One. 2014;9:e101648. doi: 10.1371/journal.pone.0101648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin W, Amin SA, Lundeen RA, Heal KR, Martens-Habbena W, Turkarslan S, Urakawa H, Costa KC, Hendrickson EL, Wang T, Beck DA, Tiquia-Arashiro SM, Taub F, Holmes AD, Vajrala N, Berube PM, Lowe TM, Moffett JW, Devol AH, Baliga NS, Arp DJ, Sayavedra-Soto LA, Hackett M, Armbrust EV, Ingalls AE, Stahl DA. Stress response of a marine ammonia-oxidizing archaeon informs physiological status of environmental populations. ISME J. 2018;12:508–519. doi: 10.1038/ismej.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rastogi G. Microbial diversity in uranium mining-impacted soils as revealed by high-density 16S microarray and clone library. Microb Ecol. 2010;59:94–108. doi: 10.1007/s00248-009-9598-5. [DOI] [PubMed] [Google Scholar]
- 41.Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, Rattei T, Damsté JS, Spieck E, Le Paslier D, Daims HA. Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci U S A. 2010;107:13479–13484. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arp DJ, Chain PSG, Klotz MG. The impact of genome analyses on our understanding of ammonia-oxidizing bacteria. Annu Rev Microbiol. 2007;61:503–528. doi: 10.1146/annurev.micro.61.080706.093449. [DOI] [PubMed] [Google Scholar]
- 43.Starkenburg SR, Arp DJ, Bottomley PJ. Expression of a putative nitrite reductase and the reversible inhibition of nitrite-dependent respiretion by nitric oxide in Nitrobacter winogradskyi Nb-255. Environ Microbiol. 2008;10:3036–3042. doi: 10.1111/j.1462-2920.2008.01763.x. [DOI] [PubMed] [Google Scholar]
- 44.Beller HR, Chain PSG, Letain TE, Chakicherla A, Larimer FW, Richardson PM, Coleman MA, Wood AP, Kelly DP (2006) The genome sequence of the obligately chemolithoautotrophic, facultatively anaerobic bacterium Thiobacillus denitrificans J Bacteriol 188:1473–1488 [DOI] [PMC free article] [PubMed]
- 45.Sorokin DY, Tourova TP, Antipov AN, Muyzer G, Kuenen JG. Anaerobic growth of the haloalkaliphilic denitrifying sulfur-oxidizing bacterium Thoialkali vibrio thiocyanodenitrificans sp. nov. with thiocyanate. Microbiology. 2004;150:2435–2442. doi: 10.1099/mic.0.27015-0. [DOI] [PubMed] [Google Scholar]
- 46.Berg IA. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol. 2011;77:1925–1936. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318:1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- 48.Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hügler M. Autotrophic carbon fixation in archaea. Nat Rev Microbiol. 2010;8:447–460. doi: 10.1038/nrmicro2365. [DOI] [PubMed] [Google Scholar]
- 49.Berg IA, Ramos-Vera WH, Petri A, Huber H, Fuchs G. Study of the distribution of autotrophic CO2 fixation cycles in Crenarchaeota. Microbiology. 2010;156:256–269. doi: 10.1099/mic.0.034298-0. [DOI] [PubMed] [Google Scholar]
- 50.Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci U S A. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandai D, Zhikang Y, Selway L, Stead D, Walker J, Leach MD, Bohovych I, Ene IV, Kastora S, Budge S, Munro CA, Odds FC, Gow NAR, Brown AJP The evolutionary rewiring of ubiquitination targets has reprogrammed the regulation of carbon assimilation in the pathogenic yeast Candida albicans. mBio 2012;3:e00495–e00412 [DOI] [PMC free article] [PubMed]
- 52.Kim ST, Huh WK, Kim JY, Hwang SW, Kang SO. D-arabinose dehydrogenase and biosynthesis of erythroascorbic acid in Candida albicans. Biochim Biophys Acta. 1996;1297:1–8. doi: 10.1016/0167-4838(96)00077-5. [DOI] [PubMed] [Google Scholar]
- 53.Amako K, Fujita K, Shimohata TA, Hasegawa E, Kishimoto R, Goda K. NAD+-specific D-arabinose dehydrogenase and its contribution to erythroascorbic acid production in Saccharomyces cerevisiae. FEBS Lett. 2006;580:6428–6434. doi: 10.1016/j.febslet.2006.10.058. [DOI] [PubMed] [Google Scholar]
- 54.Watson SW, Bock E, Valois FW, Waterbury JB, Schlosser U. Nitrospira marina gen. nov. sp. nov.: a chemolithotrophic nitrite-oxidizing bacterium. Arch Microbiol. 1986;144:1–7. doi: 10.1007/BF00454947. [DOI] [Google Scholar]
- 55.Hansel CM, Fendorf S, Jardine PM, Francis CA. Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl Environ Microbiol. 2008;74:1620–1633. doi: 10.1128/AEM.01787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gihring TM. A limited microbial consortium is responsible for extended bioreduction of uranium in a contaminated aquifer. Appl Environ Microbiol. 2011;77:5955–5965. doi: 10.1128/AEM.00220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shelobolina ES, Nevin KP, Blakeney-Hayward JD, Johnsen CV, Plaia TW, Krader P, Woodard T, Holmes DE, Vanpraagh CG, Lovley DR. Geobacter pickeringii sp. nov., Geobacter argillaceus sp. nov. and Pelosinus fermentans gen. nov., sp. nov., isolated from subsurface kaolin lenses. Int J Syst Evol Microbiol. 2007;57:126–135. doi: 10.1099/ijs.0.64221-0. [DOI] [PubMed] [Google Scholar]
- 58.Ray AE. Evidence for multiple modes of uranium immobilization by an anaerobic bacterium. Geochim Cosmochim Acta. 2011;75:2684–2695. doi: 10.1016/j.gca.2011.02.040. [DOI] [Google Scholar]
- 59.Brown SD, Utturkar SM, Magnuson TS, Ray AE, Poole FL, Lancaster WA, Thorgersen MP, Adams MW, Elias DA. Complete genome sequence of Pelosinus sp. strain UFO1 assembled using single-molecule real-time DNA sequencing technology. Genome Announc. 2014;2:e00881–e00814. doi: 10.1128/genomeA.00881-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garrity GE, Holt JG. Bergey’s Manual of Systematic Bacteriology. 2001. Phylum BVI. Phylum BVI Chloroflexi phy nov; pp. 427–446. [Google Scholar]
- 61.Singh V, Mani I, Chaudhary DK. Analysis of the multicopper oxidase gene regulatory network of Aeromonas hydrophila. Syst Synth Biol. 2012;6:51–59. doi: 10.1007/s11693-012-9096-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sameshima M, Kishi Y, Osumi M, Minamikawa-Tachino R, Mahadeo D, Cotter DA. The formation of actin rods composed of actin tubules in Dictyostelium discoideum spores. J Struct Biol. 2001;136:7–19. doi: 10.1006/jsbi.2001.4424. [DOI] [PubMed] [Google Scholar]
- 63.Pankaj Negi G, Gangola S, Khati P, Kumar G, Srivastava A, Sharma A. Differential expression and characterization of cypermethrin-degrading potential proteins in Bacillus thuringiensis strain, SG4. 3. Biotech. 2016;6:225. doi: 10.1007/s13205-016-0541-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang A, Crowley DE. Global gene expression responses to cadmium toxicity in Escherichia coli. J Bacteriol. 2005;187:3259–3266. doi: 10.1128/JB.187.9.3259-3266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seiler C, Berendonk TU. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front Microbiol. 2012;3:399. doi: 10.3389/fmicb.2012.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harrison JJ, Ceri H, Turner RJ. Multi metal resistance and tolerance in microbial biofilms. Nat Rev Microbiol. 2007;5:928–938. doi: 10.1038/nrmicro1774. [DOI] [PubMed] [Google Scholar]
- 67.Kanemori M, Yanagi H, Yura T. The ATP-dependent HslVU/ClpQY protease participates in turnover of cell division inhibitor SulA in Escherichia coli. J Bacteriol. 1999;181:3674–3680. doi: 10.1128/jb.181.12.3674-3680.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holton NJ, Goodwin TJ, Butler MI, Poulter RT. An active retrotransposon in Candida albicans. Nucleic Acids Res. 2001;29:4014–4024. doi: 10.1093/nar/29.19.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahmad I, Smith M, Meyers J, Russell B, Dorrah K, DeFreece C. Effect of the heavy metals cobalt and cadmium on LINE1 endonuclease activity. FASEB J. 2016;30:1. doi: 10.1096/fj.16-0101ufm. [DOI] [PubMed] [Google Scholar]
- 70.Smith M, Ahmad I, Meyers J, Russell B, Dorrah K, DeFreece C. The effect of the heavy metal nickel on the LINE1 endonuclease. The FASEB J. 2016;30:1. doi: 10.1096/fj.16-0101ufm. [DOI] [PubMed] [Google Scholar]
- 71.Rojo-Bezares B, Azcona-Gutiérrez JM, Martin C, Jareño MS, Torres C, Sáenz Y. Streptococcus agalactiae from pregnant women: antibiotic and heavy-metal resistance mechanisms and molecular typing. Epidemiol Infect. 2016;144:3205–3214. doi: 10.1017/S0950268816001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anantharam V, Allison MJ, Maloney PC. Oxalate:formate exchange. The basis for energy coupling in Oxalobacter J Biol Chem. 1989;264:7244–7250. [PubMed] [Google Scholar]
- 73.Ruan ZS, Anantharam V, Crawford IT, Ambudkar SV, Rhee SY, Allison MJ, Maloney PC. Measurement of the substrate dissociation constant of a solubilized membrane carrier. J Biol Chem. 1992;267:10537–10543. [PubMed] [Google Scholar]
- 74.Abe K, Hayashi H, Maloney PC. Exchange of aspartate and alanine. Mechanism for development of a proton-motive force in bacteria J Biol Chem. 1996;271:3079–3084. doi: 10.1074/jbc.271.6.3079. [DOI] [PubMed] [Google Scholar]
- 75.Lolkema JS, Poolman B, Konings WN. Bacterial solute uptake and efflux systems. Curr Opin Microbiol. 1998;1:248–253. doi: 10.1016/S1369-5274(98)80018-0. [DOI] [PubMed] [Google Scholar]
- 76.Akbar S, Sultan S, Kertesz M. Determination of cypermethrin degradation potential of soil bacteria along with plant growth-promoting characteristics. Curr Microbiol. 2015;70:75–84. doi: 10.1007/s00284-014-0684-7. [DOI] [PubMed] [Google Scholar]
- 77.Tallur PN, Megadi VB, Ninnekar HZ. Biodegradation of cypermethrin by Micrococcussp. strain CPN 1. Biodegradation. 2008;19:77–82. doi: 10.1007/s10532-007-9116-8. [DOI] [PubMed] [Google Scholar]
- 78.He P, Moran GR. Structural and mechanistic comparisons of the metal-binding members of the vicinal oxygen chelate (VOC) superfamily. J Inorg Biochem. 2011;105:1259–1272. doi: 10.1016/j.jinorgbio.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 79.Lovley DR, Holmes DE, Nevin KP. Dissimilatory Fe (III) and Mn (IV) reduction. Adv Microb Physiol. 2004;49:219–286. doi: 10.1016/S0065-2911(04)49005-5. [DOI] [PubMed] [Google Scholar]
- 80.Lovley DR, Phillips EJ. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microbiol. 1988;54:1472–1480. doi: 10.1128/aem.54.6.1472-1480.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lovley DR, Giovannoni SJ, White DC, Champine JE, Phillips EJ, Gorby YA, Goodwin S. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch Microbiol. 1993;159:336–344. doi: 10.1007/BF00290916. [DOI] [PubMed] [Google Scholar]
- 82.Lucas S, Copeland A, Lapidus A, Glavina del Rio T, Dalin E, Tice H, Bruce D, Goodwin L, Pitluck S, Sims D, Brettin T, Detter JC, Han C, Larimer F, Land M, Hauser L, Kyrpides N, Ovchinnikova G, Stolz J. Nucleotide sequence [large scale genomic DNA] 2009. [Google Scholar]
- 83.Chen J, Xu QX, Su Y, Shi ZQ, Han F. Phytoremediation of organic polluted soil. J Bioremed Biodegr. 2013;4:e132. [Google Scholar]
- 84.Abed RMM, Dobretsov S, Sudesh K. Applications of cyanobacteria in biotechnology. J Appl Microbiol. 2009;106:1–12. doi: 10.1111/j.1365-2672.2008.03918.x. [DOI] [PubMed] [Google Scholar]
- 85.Rastogi RP, Sinha RP. Biotechnological and industrial significance of cyanobacterial secondary metabolites. Biotechnol Adv. 2009;27:521–553. doi: 10.1016/j.biotechadv.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 86.Basta T, Keck A, Klein J, Stolz A. Detection and characterization of conjugative degradative plasmids in xenobiotics degrading Sphingomonas strains. J Bacteriol. 2004;186:3862–3872. doi: 10.1128/JB.186.12.3862-3872.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Basta T, Burger S, Stolz A. Structural and replicative € diversity of large plasmids from polycyclic aromatic compounds and xenobiotics degrading Sphingomonas strains. Microbiol. 2005;151:2025–2037. doi: 10.1099/mic.0.27965-0. [DOI] [PubMed] [Google Scholar]
- 88.Ogram A, Duan YP, Trabue SL, Feng X, Castro H, Ou LT. Carbofuran degradation mediated by three related plasmid systems. FEMS Microbiol Ecol. 2000;32:197–203. doi: 10.1111/j.1574-6941.2000.tb00712.x. [DOI] [PubMed] [Google Scholar]
- 89.Romine MF, Stillwell LC, Wong KK, Thurston SJ, Sisk EC, Sensen C, Gaasterland T, Fredrickson JK, Saffer JD. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J Bacteriol. 1999;181:1585–1602. doi: 10.1128/jb.181.5.1585-1602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fuchs G, Boll M, Heider J. Microbial degradation of aromatic compounds—from one strategy to four. Nat Rev Microbiol. 2011;9:803–816. doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Phylogenetic analysis of upregulated amoC genes obtained using neighbor-joining clustering method having bootstrap values out of 1000 replicates using MEGA 7.0 using UniProt ID with their species and gene name (GN). K0IHK8_ Nitrososphaera gargensis (strain Ga9.2)_amoC1, A0A075MNL6_ Candidatus Nitrososphaera evergladensis SR1_NTE00725, A0A060HQE0_ Nitrososphaera viennensis EN76_amoC3, A0A075MPN8_ Candidatus Nitrososphaera evergladensis SR1_ NTE_01100. It clearly indicates that thaumarchaea species harbor additional copy of amoC that could be used as a molecular marker in the detection of particular archeal community present in cypermerthrin and metal co-contaminated agriculture soils i.e. M1 soil sample. (JPG 82 kb)
Additional file 2: Fig. S2. Phylogenetic analysis of VOC superfamily transcript obtained in M1 soil sample using neighbor-joining clustering method having bootstrap values out of 1000 replicates using MEGA 7.0 using UniProt ID with their species (K1Z0Y1_uncultured bacterium, A0A063B6R9_ Burkholderia sp. lig30,A7HAK0_ Anaeromyxobacter sp. (strain Fw109-5), A0A069IMT4_ Brevundimonas sp. EAKA, A0A023XH36_ Bradyrhizobium japonicum SEMIA 5079) that clearly indicates the evolution of VOC superfamily mostely from Geobacter sp., Nitrospirae sp. Deulphobulbacae sp, actinobacteria sp., candidatus Entothella sp. present dominantly in M soil sample. (JPG 122 kb)
Additional file 3: Table S1. The transcript id, gene function, gene ontology (biological,molecular and cellular function) obtained M1 functional hits after quarried against UniPrt database. (XLS 5019 kb)
Additional file 4: Table S2. The transcript id, gene function, gene ontology (biological, molecular and cellular function) obtained O1 functional hits after quarried against UniProt database. (XLS 1457 kb)
Additional file 5: Table S3. List of Differentially Expressed genes in M1 and O1sample. (DOC 45 kb)
Data Availability Statement
The metatranscriptome data for M1 and O1 is submitted with MG-RAST under the accession number mgm4653349.3, whereas M1 dataset was provided a Gold ID 0eeec568676d676d343733323034392e33.