Zhang, Dilliott et al. examine a unique family with early- and late-onset Alzheimer’s disease phenotypes, as well as disease-discordant monozygotic triplets. The triplets and the patient with early-onset disease are carriers of the APOE ε4-allele plus rare substitutions in other genes. Epigenetic analyses suggest accelerated ageing in the early-onset patient.
Keywords: Alzheimer's disease, monozygotic triplets, DNA methylation, ageing
Abstract
Age at onset of Alzheimer’s disease is highly variable, and its modifiers (genetic or environmental) could act through epigenetic changes, such as DNA methylation at CpG sites. DNA methylation is also linked to ageing—the strongest Alzheimer’s disease risk factor. DNA methylation age can be calculated using age-related CpGs and might reflect biological ageing. We conducted a clinical, genetic and epigenetic investigation of a unique Ashkenazi Jewish family with monozygotic triplets, two of whom developed Alzheimer’s disease at ages 73 and 76, while the third at age 85 has no cognitive complaints or deficits in daily activities. One of their offspring developed Alzheimer’s disease at age 50. Targeted sequencing of 80 genes associated with neurodegeneration revealed that the triplets and the affected offspring are heterozygous carriers of the risk APOE ε4 allele, as well as rare substitutions in APP (p.S198P), NOTCH3 (p.H1235L) and SORL1 (p.W1563C). In addition, we catalogued 52 possibly damaging rare variants detected by NeuroX array in affected individuals. Analysis of family members on a genome-wide DNA methylation chip revealed that the DNA methylation age of the triplets was 6–10 years younger than chronological age, while it was 9 years older in the offspring with early-onset Alzheimer’s disease, suggesting accelerated ageing.
Introduction
Alzheimer’s disease is the most common neurodegenerative disease with a brain pathology characterized by neuronal loss, inflammation, amyloid plaques (consisting of amyloid-β peptides encoded by APP), and tau inclusions (encoded by MAPT). Rare mutations in APP, PSEN1 and PSEN2 could cause autosomal dominant early-onset Alzheimer’s disease (<65 years); however, ∼50% of early-onset sporadic Alzheimer’s disease cases or families with a mix of early/late-onset cases are genetically unexplained. Common late-onset Alzheimer’s disease is associated with 28 well-confirmed Alzheimer’s disease loci, including the APOE ε4 allele with the largest risk (Ghani et al., 2018).
Alzheimer’s disease age at onset is highly variable (e.g. 39–85 years in APP carriers), suggesting the existence of genetic or environmental modifiers (Ghani et al., 2018), both of which could act through epigenetic changes, such as DNA methylation (DNAm) at CpG sites. There is not a strict dichotomy between the action of genetic and epigenetic factors (Zhang et al., 2016, 2018). For instance, the Exome Aggregation Consortium (ExAC) reported that CpGs are the most mutable sites in the genome (Lek et al., 2016).
DNAm is also linked to ageing—the strongest Alzheimer’s disease risk factor. The collective assessment of DNAm at 353 CpGs generates DNAm age, which is an accurate predictor of chronological age across multiple tissues, including blood and brain (Horvath, 2013). Deviation of DNAm age from chronological age (DNAm-age acceleration) may be linked to biological ageing via changes in gene expression, and was associated with several neurodegenerative diseases, including Parkinson’s disease (Horvath and Ritz, 2015; Picillo et al., 2018) and C9orf72-related disease (Zhang et al., 2017). Accelerated DNAm age was also linked to the degree of amyloid pathology or cognitive decline (Levine et al., 2015); and reported to be a significant predictor of dementia (Degerman et al., 2017).
Alzheimer’s disease concordance in monozygotic twins is 80%, suggesting high heritability (Martin et al., 1997). Monozygotic siblings provide the best opportunity to investigate risk/protective factors in disease development. Here we conducted clinical, genetic and epigenetic analyses of a unique Alzheimer’s disease family with monozygotic triplets.
Materials and methods
Participants
A Canadian Alzheimer’s disease family of Ashkenazi Jewish origin (Fig. 1) was recruited from Baycrest Health Sciences (Toronto). The patients met the National Institute on Aging and Alzheimer’s Association criteria for probable Alzheimer’s disease (McKhann et al., 2011). We also used 192 neurologically normal Canadian controls (59% female; average age 74 ± 8 years) (Li et al., 2008), as well as whole-exome sequencing data from Washington Heights-Inwood Columbia Aging Project (WHICAP), including 1397 Alzheimer’s disease cases and 2198 controls (>65 years old) residing in northern Manhattan, NY (Tosto et al., 2019). Informed consent was obtained from each participant in accordance with the Research Ethics Boards.
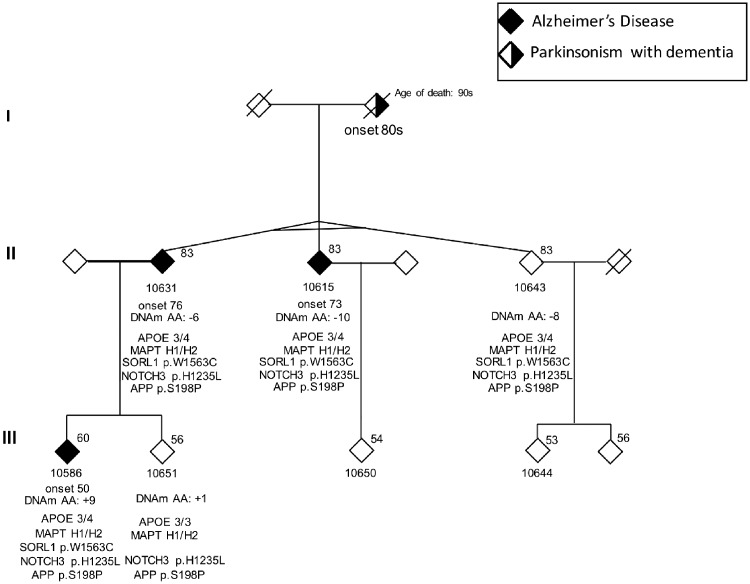
Figure 1.
Pedigree of the Ashkenazi family with monozygotic triplets. Gender is masked. Age at time of sample collection is shown above the symbol. Age of onset, DNAm-age acceleration (AA) and genetic findings by ONDRISeq are indicated below the symbol.
Genetic analyses
Blood genomic DNA was extracted using a QIAGEN kit. Genotyping was conducted for family members (the triplets, one affected and one unaffected offspring), as well as 192 Canadian controls. First, we used a next-generation sequencing panel developed by the Ontario Neurodegenerative Disease Research Initiative (ONDRISeq) that targets 80 genes associated with neurodegeneration (Farhan et al., 2016). ONDRISeq data were generated/processed as reported (Dilliott et al., 2018), and interrogated for copy number variants by VarSeq® (Golden Helix, Bozeman, MT, USA), which uses normalized depth of coverage analysis to identify large-scale deletions/duplications (Iacocca et al., 2017). ONDRISeq data of the 192 controls were provided to the algorithm, from which 49 with the lowest per cent difference in coverage data were selected as a reference. A coverage ratio ≤0.7 and a z-score of ≤−5.0 indicates heterozygous deletions, while a ratio ≥1.30 and z-score ≥5.0 suggests duplications. We also used the NeuroX array (Illumina), which includes the Exome BeadChip (∼240 000 variants) and ∼24 000 variants tailored to study neurodegenerative diseases (Ghani et al., 2015). APOE genotypes were based on rs429358 and rs7412 by ONDRISeq. MAPT H1/H2 haplotypes and C9orf72 repeat number were obtained as reported (Bruni et al., 2007; Zhang et al., 2017).
Minor allele frequencies (MAF) were acquired from gnomAD, including the Ashkenazi Jewish cohort (http://gnomad.broadinstitute.org/), ExAC (http://exac.broadinstitute.org/), and the Healthy Exome (HEX) database (https://www.alzforum.org/exomes/hex) containing ∼500 neuropathologically normal autopsy cases (age >60). The variant filtering process included two steps. First, both the ONDRISeq (Table 2) and NeuroX results were filtered for rare variants with MAF <0.005 (ExAC-ALL). Second, NeuroX variants that were predicted to be damaging by both SIFT and PolyPhen-2 were selected for Supplementary Table 3, which includes allele frequencies for the above mentioned datasets.
Table 2.
Genetic analysis of the family using the ONDRISeq panel identified three heterozygous missense variants that are rare in ExAC-ALL database (MAF <0.005)
| Gene | Variation (transcript) | Predicted effects | SNP | ExAC MAF | gnomAD MAF | HEX MAF | SIFT prediction (score) | PolyPhen-2 prediction (score) | Phenotype associated with the gene | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | American | NFE | Ashkenazi Jewish | ||||||||
| SORL1 | c.G4689C (NM_003105) | p.W1563C | rs138580875 | 0.0005 | 0.0005 | 0.0007 | 0.0005 | 0 | D (0.003) | D (1) | AD |
| NOTCH3 | c.A3704T (NM_000435) | p.H1235L | rs55882518 | 0.004 | 0.003 | 0.007 | 0.02 | 0.01 | D (0.03) | B (0) | CADASIL |
| APP | c.T592C (NM_000484) | p.S198P | rs145081708 | 0.0005 | 0.0007 | 0.0007 | 0.01 | 0.001 | D (0.009) | D (0.999) | AD |
B = benign; CADASIL = cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy; D = probably damaging or deleterious; ExAC = Exome Aggregation Consortium; HEX = Healthy Exome database; nFE = non-Finnish European; WHICAP = Washington Heights-Inwood Columbia Aging Project.
Epigenetic analyses
DNAm analysis was conducted as reported (Zhang et al., 2017). Briefly, DNA was bisulfite converted using the Imprint DNA modification kit (Sigma), and assessed on genome-wide Infinium MethylationEPIC chip (Illumina) at the Centre for Applied Genomics (Toronto). Raw DNAm data was processed using GenomeStudio (Illumina). DNAm-age calculator tool (https://dnamage.genetics.ucla.edu) analysed 334 age-related CpGs, which includes 90% of the CpGs from the discontinued 450K BeadChip and has a similar capability to estimate DNAm-age (Logue et al., 2017).
Data availability
The data that support the findings of this study are available on request from the corresponding authors (E.R., M.F.). The data are not publicly available because of information that could compromise the privacy of the research participants.
Results
Clinical findings
The family structure is presented in Fig. 1. The triplet’s mother (deceased at age 97) had a history of parkinsonism with dementia. Current age of the triplets is 85. Two of them (Patients 10615 and 10631) were diagnosed with late-onset Alzheimer’s disease in their mid-seventies, while triplet Subject 10643 still has no cognitive complaints or deficits in daily living activities, and functions normally based on self/family-reporting. The offspring (Patient 10586) of triplet Subject 10631 developed early-onset Alzheimer’s disease at age 50. Imaging results are presented in Supplementary Figs 1–3. Detailed clinical findings are available in Supplementary Table 1, and performance profiles on the Toronto Cognitive Assessment (TorCA) (Freedman et al., 2018) are presented in Supplementary Fig. 4.
Briefly, the progressive memory problems of Patient 10615 started at age 73. CT brain scan was normal at age 75, but a single photon emission computed tomography (SPECT) scan showed mild symmetrical decreased perfusion in the posterior parietal lobes compatible with Alzheimer’s disease. At that time, Mini-Mental State Examination (MMSE) score was 23/30 and score on the Behavioural Neurology Assessment – Short Form (Darvesh et al., 2005) was 70/114 (cut-off for dementia is 82). At age 85, the MMSE score was 26/30 and TorCA total score was 202 (impaired <257). The progressive memory problems of Patient 10631 (monozygotic sibling of Patient 10615) started at age 76. SPECT scan at age 80 showed decreased bilateral parietotemporal lobe perfusion. MMSE score was 23/30 and TorCA score was 219. At age 85, MMSE was 21/30 and TorCA total score dropped to 165. For both siblings, insight, ability to show empathy and sympathy were reduced. Their medical history is also remarkable for hypertension and long-standing obsessive-compulsive behaviour.
Triplet Subject 10643 has remained free from dementia, despite some deficits on cognitive testing at age 85 (MMSE = 22/30). TorCA scores were obtained on two occasions (10.5 weeks apart) because of poor sleep before the first assessment. Both times TorCA total scores were impaired (179 and 198). Other medical history includes hypertension and sarcoma at age 60. There were no features of obsessive-compulsive disorder.
The progressive memory problems of Patient 10586 (offspring of monozygotic triplet Patient 10631) started at age 50. Brain MRI at age 52 showed a few small foci of non-specific subcortical white matter of uncertain significance (mainly in the left frontal lobe); and a SPECT scan showed moderate decreased perfusion within the left parietotemporal region consistent with Alzheimer’s disease. At age 53, global hypoperfusion was slightly more prominent in the left frontal, temporal and parietal lobes. The score on the Behavioural Neurology Assessment–Short Form was 64/114; and MMSE score was 24/30, declining to 9/30 by age 55 and 4/30 by age 57. Behavioural symptoms (e.g. verbal aggression and resistance to care) started at age 59. Other medical history included Crohn’s disease (inactive for years).
Genetic findings
The monozygosity of the triplets was confirmed by the identical ONDRISeq and NeuroX genotypes. The triplets and Alzheimer’s disease-affected offspring are carriers of normal C9orf72 alleles (≤8 repeats), but heterozygous for the risk APOE ε4 allele and MAPT H1 haplotype (Table 1 and Fig. 1). None of the 80 genes included on ONDRISeq had deletions/duplications, but we detected three heterozygous substitutions in NOTCH3 (p.H1235L), APP (p.S198P) and SORL1 (p.W1563C) with MAF<0.005 in ExAC-ALL (Table 2). The investigation of these variants in the WHICAP dataset did not reveal association with Alzheimer’s disease (Supplementary Table 2). Although the WHICAP dataset could be under power to study very rare variants, considerations below also argue against the pathogenic nature of the substitutions in NOTCH3 and APP, whereas the significance of SORL1 p.W1563C in Alzheimer’s disease could not be excluded.
Table 1.
Genotyping results of C9orf72, MAPT and APOE in the family members
| DNA # | Status | Age of onset, years | C9orf72 genotype | MAPT haplotype | APOE genotype |
|---|---|---|---|---|---|
| 10631 | Alzheimer’s disease | 76 | 2/8 | H1/H2 | 4/3 |
| 10615 | Alzheimer’s disease | 73 | 2/8 | H1/H2 | 4/3 |
| 10643 | Unaffected | NA | 2/8 | H1/H2 | 4/3 |
| 10586 | Alzheimer’s disease | 50 | 2/2 | H1/H2 | 4/3 |
| 10651 | Unaffected | NA | 2/8 | H1/H2 | 3/3 |
NA = not applicable.
The p.H1235L substitution in exon 22 of NOTCH3 is deleterious by SIFT, but benign by PolyPhen-2; and is not rare in the Ashkenazi Jewish population (gnomAD MAF = 0.02; 10 368 chromosomes) (Table 2). It is mapped outside the mutation hotspot for NOTCH3-related dementia caused by the loss/gain of cysteine residues encoded by exons 2–5 or 7–11 (Joutel et al., 2004). Moreover, NOTCH3 mutations are associated with diffuse white matter abnormalities (absent on the neuroimages of the family members). Similarly, the p.S198P in exon 5 of APP is mapped outside the Alzheimer’s disease mutation hot-spot (exons 16–17 encoding amyloid-β domain) (Ghani et al., 2018). Although p.S198P is predicted to be damaging by SIFT and PolyPhen-2, it is not very rare in the Ashkenazi Jewish population (gnomAD MAF = 0.01) (Table 2). One of the WHICAP controls is homozygous for p.S198P, arguing against its pathogenic nature (Supplementary Table 2).
In contrast, the p.W1563C substitution in the Fibronectin type-III domain of SORL1 is very rare in the Ashkenazi Jewish population (gnomAD MAF = 0.0005) and elderly controls (one Canadian, one WHICAP, and none in the HEX database). It is predicted to be damaging by SIFT, PolyPhen-2 (Table 2), and strongly damaging by the Combined Annotation Dependent Depletion score (>30) (Kircher et al., 2014).
NeuroX confirmed the substitutions in SORL1, NOTCH3 and APP. In addition, we catalogued 52 possibly damaging rare variants (five truncating variants) detected by NeuroX (24 in the triplets and 28 in the offspring with early-onset Alzheimer’s disease) to be followed-up in large Alzheimer’s disease datasets (Supplementary Table 3).
Epigenetic findings
In agreement with our prior study of monozygotic twins, demonstrating that many DNAm changes are genetically controlled (Zhang et al., 2016), the genome-wide blood DNAm profiles of the monozygotic triplets are much more similar than between first-degree relatives, who share 50% of their genetic material (Supplementary Fig. 5). Four CpGs in KCNS2, CADM1, RAB3IL1 and MATN2 had DNAm difference >30% between the triplets affected by Alzheimer’s disease in their seventies and the triplet without dementia at age 85 (Supplementary Table 4), warranting further investigation of these CpGs in Alzheimer’s disease.
The discordance in age at onset between the triplets could not be attributed to DNAm-age acceleration (their DNAm age was 6–10 years younger than chronological age) (Fig. 1). In contrast, the DNAm age in the offspring with early-onset Alzheimer’s disease (Patient 10586) was 9 years older than chronological age, suggesting accelerated ageing, which might be driven by five age-related CpGs that showed a >20% difference in DNAm levels between Patient 10586 and the triplets with late-onset Alzheimer’s disease (Table 3). Notably, the DNAm age of the unaffected sibling Subject 10651 of Alzheimer’s disease Patient 10586, was only 1 year older than the chronological age.
Table 3.
The five age-related CpG sites
| CpG ID | Gene symbol | Beta 10631 | Beta 10615 | Beta 10643 | Beta 10586 | Delta beta |
|---|---|---|---|---|---|---|
| cg00945507 | SEC61G | 0.27 | 0.28 | 0.33 | 0.69 | 0.39 |
| cg12768605 | LYPD5 | 0.32 | 0.32 | 0.32 | 0.52 | 0.20 |
| cg13854874 | CHAF1B | 0.24 | 0.23 | 0.20 | 0.49 | 0.27 |
| cg22901840 | DIRAS3 | 0.70 | 0.74 | 0.68 | 0.48 | -0.23 |
| cg07770222 | C8orf31 | 0.60 | 0.55 | 0.59 | 0.81 | 0.23 |
Presented are the age-related CpG sites that showed a >20% difference in DNAm levels between the patient with early-onset Alzheimer’s disease (Patient 10586) and the triplets (Patients 10631, 10615 and 10643).
Discussion
We report a family with monozygotic triplets, two of whom were diagnosed in their seventies with late-onset slow progressing Alzheimer’s disease (mild dementia 9 and 12 years post-onset). In contrast, an offspring of one of the affected triplets has early-onset Alzheimer’s disease, which progressed to severe dementia within 5 years. The unaffected triplet was impaired on cognitive testing, but is independent in daily activities at age 85 and thus did not meet criteria for dementia or mild cognitive impairment.
The APOE ε4 allele may explain the late-, but not the early-onset Alzheimer’s disease in the family. Also, the pathogenic role of SORL1 p.W1563C cannot be excluded. Notably, APOE and SORL1 interact in the amyloid-β pathway, and rare SORL1 variants (p.N674S) could increase penetrance of Alzheimer’s disease in APOE ɛ4 carriers (Louwersheimer et al., 2017). Multiple evidence support SORL1 as an Alzheimer’s disease gene, and truncating SORL1 variants were even suggested for consideration in clinical practice, similar to PSEN1, PSEN2 and APP mutations (Rogaeva et al., 2007; Holstege et al., 2017). However, it is challenging to determine penetrance of rare substitutions, and their pathogenic impact remain unclear without functional investigations. A recent study sequenced SORL1 in 1895 cases and 3206 controls, and proposed that the pathogenicity of the 181 detected variants could be classified based on their predicted damaging effect and MAF in databases, which placed p.W1563C among the variants with uncertain significance (Holstege et al., 2017).
Our study has limitations. A single, although unique, family has limited statistical power in the context of genome-wide data. Furthermore, we did not search for ultra-rare mutations that may have occurred after the blastocyst split (Weber-Lehmann et al., 2014); and de novo mutations have been reported to contribute to the discordance between monozygotic twins for some neurological disorders, such as schizophrenia (Tang et al., 2017). Both limitations will be resolved by analysing this family together with others in our ongoing whole-genome sequencing Alzheimer’s disease project. We also could not exclude Alzheimer’s disease-associated genetic variability in brain, although a recent study of brain samples reported that somatic variants in Alzheimer’s disease genes is not a common Alzheimer’s disease cause (Nicolas et al., 2018). Finally, it would be important to estimate the polygenic risk score for individuals affected by late- versus early-onset Alzheimer’s disease. However, association studies related to polygenic risk/hazard scores are currently done only in large European or North American Alzheimer’s disease datasets (Leonenko et al., 2019) and their utility is not yet clear for individual application in specific ethnic groups with a unique genetic makeup (e.g. Ashkenazi Jewish). Notably, of the 54 rare possibly damaging variants detected by NeuroX in our family (ExAC-ALL MAF <0.005), only 14 are also rare in the Ashkenazi Jewish population (Supplementary Table 3).
Environmental/ageing factors triggering epigenetic changes may also affect disease manifestation. It was reported that a cluster of genes with DNAm changes may influence biological networks, but DNAm at specific CpG sites may not be sufficient for modifying phenotypes in monozygotic twins discordant for major depression (Malki et al., 2016) or amyotrophic lateral sclerosis (Zhang et al., 2016). In the current study, DNAm age was similar between the monozygotic triplets (6–10 years younger than chronological age regardless of Alzheimer’s disease onset/status). In contrast to the parental generation with late-onset Alzheimer’s disease, DNAm-age was accelerated by 9 years in the offspring with early-onset Alzheimer’s disease. This is in line with previous findings that DNAm age of the prefrontal cortex is associated with amyloid load and cognitive function (Levine et al., 2015). A longitudinal study revealed that individuals at age 70–80 were epigenetically younger (by 2–3 years) than their chronological age compared to baseline at 55–65 age (Degerman et al., 2017). However, this systematic deviation did not affect the conclusion that accelerated epigenetic age at the age of 55–80 years may increase risk of dementia. In the current study, correcting for the possible error would bring the DNAm-age of the monozygotic triplets to 3–7 years younger than chronological age and would still be noticeably different from the early-onset patient whose DNAm age was 69 at age 60. Future case-control Alzheimer’s disease studies are required to assess if DNAm-age acceleration is associated with age at onset.
Supplementary Material
Acknowledgements
We gratefully acknowledge Mindy Halper for assisting with cognitive assessment and data collection relating to the triplets, as well as Fidelma Serediuk and Gary Gallagher for facilitating collection of blood for genetic testing.
Funding
This work was in part supported by the Canadian Consortium on Neurodegeneration in Aging (E.R., M.Z.), Ontario Neurodegenerative Disease Research Initiative (A.A.D., J.R., R.H., M.F., P.S.G.H., E.R.), the Alzheimer Society of London and Middlesex (A.A.D.), the Saul A. Silverman Family Foundation as a Canada International Scientific Exchange Program and Morris Kerzner Memorial Fund (M.F.), the Shanghai Pujiang Program 19PJ1410300 (M.Z.), National Institutes of Health RF1AG054080 (C.R., E.R.) and R21AG054832 (G.T.).
Competing interests
The authors report no competing interests.
Glossary
Abbreviations
- DNAm =
DNA methylation
- MAF =
minor allele frequency
- TorCA =
Toronto Cognitive Assessment
References
- Bruni AC, Momeni P, Bernardi L, Tomaino C, Frangipane F, Elder J, et al. Heterogeneity within a large kindred with frontotemporal dementia: a novel progranulin mutation. Neurology 2007; 69: 140–7. [DOI] [PubMed] [Google Scholar]
- Degerman S, Josefsson M, Nordin Adolfsson A, Wennstedt S, Landfors M, Haider Z, et al. Maintained memory in aging is associated with young epigenetic age. Neurobiol Aging 2017; 55: 167–71. [DOI] [PubMed] [Google Scholar]
- Dilliott AA, Farhan SMK, Ghani M, Sato C, Liang E, Zhang M, et al. Targeted next-generation sequencing and bioinformatics pipeline to evaluate genetic determinants of constitutional disease. J Vis Exp: JoVE 2018; 134: e57266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan SMK, Dilliott AA, Ghani M, Sato C, Liang E, Zhang M, et al. The ONDRISeq panel: custom-designed next-generation sequencing of genes related to neurodegeneration. NPJ Genom Med 2016; 1: 16032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman M, Leach L, Carmela Tartaglia M, Stokes KA, Goldberg Y, Spring R, et al. The Toronto cognitive assessment (TorCA): normative data and validation to detect amnestic mild cognitive impairment. Alzheimers Res Ther 2018; 10: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvesh S, Leach L, Black SE, Kaplan E, Freedman M. The behavioural neurology assessment. Can J Neurol Sci 2005; 32: 167–77. [DOI] [PubMed] [Google Scholar]
- Ghani M, Lang AE, Zinman L, Nacmias B, Sorbi S, Bessi V, et al. Mutation analysis of patients with neurodegenerative disorders using NeuroX array. Neurobiol Aging 2015; 36: 545e9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghani M, Reitz C, George-Hyslop P, Rogaeva E. Genetic complexity of early-onset Alzheimer’s disease In: G D, S E, editors. Neurodegenerative diseases: Cham: Springer; 2018. p. 29–50. [Google Scholar]
- Holstege H, van der Lee SJ, Hulsman M, Wong TH, van Rooij JG, Weiss M, et al. Characterization of pathogenic SORL1 genetic variants for association with Alzheimer's disease: a clinical interpretation strategy. Eur J Hum Genet 2017; 25: 973–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol 2013; 14: R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Ritz BR. Increased epigenetic age and granulocyte counts in the blood of Parkinson's disease patients. Aging 2015; 7: 1130–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacocca MA, Wang J, Dron JS, Robinson JF, McIntyre AD, Cao H, et al. Use of next-generation sequencing to detect LDLR gene copy number variation in familial hypercholesterolemia. J Lipid Res 2017; 58: 2202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joutel A, Monet M, Domenga V, Riant F, Tournier-Lasserve E. Pathogenic mutations associated with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy differently affect Jagged1 binding and Notch3 activity via the RBP/JK signaling Pathway. Am J Hum Genet 2004; 74: 338–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014; 46: 310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonenko G, Sims R, Shoai M, Frizzati A, Bossu P, Spalletta G, et al. Polygenic risk and hazard scores for Alzheimer's disease prediction. Ann Clin Transl Neurol 2019; 6: 456–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Lu AT, Bennett DA, Horvath S. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer's disease related cognitive functioning. Aging 2015; 7: 1198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, et al. Candidate single-nucleotide polymorphisms from a genome wide association study of Alzheimer disease. Arch Neurol 2008; 65: 45–53. [DOI] [PubMed] [Google Scholar]
- Logue MW, Smith AK, Wolf EJ, Maniates H, Stone A, Schichman SA, et al. The correlation of methylation levels measured using Illumina 450K and EPIC BeadChips in blood samples. Epigenomics 2017; 9: 1363–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwersheimer E, Cohn-Hokke PE, Pijnenburg YA, Weiss MM, Sistermans EA, Rozemuller AJ, et al. Rare genetic variant in SORL1 May increase penetrance of Alzheimer's disease in a family with several generations of APOE-varepsilon4 Homozygosity. J Alzheimer's Dis: JAD 2017; 56: 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malki K, Koritskaya E, Harris F, Bryson K, Herbster M, Tosto MG. Epigenetic differences in monozygotic twins discordant for major depressive disorder. Transl Psychiatry 2016; 6: e839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N, Boomsma D, Machin G. A twin-pronged attack on complex traits. Nat Genet 1997; 17: 387–92. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR Jr, Kawas CH, et al. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 2011; 7: 263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas G, Acuna-Hidalgo R, Keogh MJ, Quenez O, Steehouwer M, Lelieveld S, et al. Somatic variants in autosomal dominant genes are a rare cause of sporadic Alzheimer's disease. Alzheimers Dement 2018; 14: 1632–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picillo M, Lizarraga KJ, Friesen EL, Chau H, Zhang M, Sato C, et al. Parkinsonism due to A53E alpha-synuclein gene mutation: clinical, genetic, epigenetic, and biochemical features. Mov Disord 2018; 33: 1950–55. [DOI] [PubMed] [Google Scholar]
- Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet 2007; 39: 168–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Fan Y, Li H, Xiang Q, Zhang DF, Li Z, et al. Whole-genome sequencing of monozygotic twins discordant for schizophrenia indicates multiple genetic risk factors for schizophrenia. J Genet Genomics 2017; 44: 295–306. [DOI] [PubMed] [Google Scholar]
- Tosto G, Vardarajan B, Sariya S, Brickman AM, Andrews H, Manly JJ, et al. Association of variants in PINX1 and TREM2 With late-onset Alzheimer disease. JAMA Neurol 2019; 76: 942–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Lehmann J, Schilling E, Gradl G, Richter DC, Wiehler J, Rolf B. Finding the needle in the haystack: differentiating "identical" twins in paternity testing and forensics by ultra-deep next generation sequencing. Forensic Sci Int Genet 2014; 9: 42–6. [DOI] [PubMed] [Google Scholar]
- Zhang M, Ferrari R, Tartaglia MC, Keith J, Surace EI, Wolf U, et al. A C6orf10/LOC101929163 locus is associated with age of onset in C9orf72 carriers. Brain 2018; 141: 2895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Tartaglia MC, Moreno D, Sato C, McKeever P, Weichert A, et al. DNA methylation age-acceleration is associated with disease duration and age at onset in C9orf72 patients. Acta Neuropathol 2017; 134: 271–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Xi Z, Ghani M, Jia P, Pal M, Werynska K, et al. Genetic and epigenetic study of ALS-discordant identical twins with double mutations in SOD1 and ARHGEF28. J Neurol Neurosurg Psychiatry 2016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding authors (E.R., M.F.). The data are not publicly available because of information that could compromise the privacy of the research participants.