Abstract
In Eukaryotes, tRNAs, 5S RNA and U6 RNA are transcribed by RNA polymerase (Pol) III. Human Pol III is composed of 17 subunits. Three specific Pol III subunits form a stable ternary subcomplex (RPC62-RPC39-RPC32α/β) being involved in pre-initiation complex formation. No paralogues for subunits of this subcomplex subunits have been found in Pols I or II, but hRPC62 was shown to be structurally related to the general Pol II transcription factor hTFIIEα. Here we show that these structural homologies extend to functional similarities. hRPC62 as well as hTFIIEα possess intrinsic ATP-dependent 3′-5′ DNA unwinding activity. The ATPase activities of both proteins are stimulated by single-stranded DNA. Moreover, the eWH domain of hTFIIEα can replace the first eWH (eWH1) domain of hRPC62 in ATPase and DNA unwinding assays. Our results identify intrinsic enzymatic activities in hRPC62 and hTFIIEα.
INTRODUCTION
Transcription in eukaryotes is carried out by three nuclear DNA-dependent RNA polymerases (Pol I, II, III) with specialized functions. Pol III transcribes tRNAs and 5S rRNA, which are both indispensable for translation. Furthermore, Pol III transcribes RNAs that are involved in the regulation of transcription (7SK RNA, Alu RNA), RNA processing (U6 RNA, RNase P RNA, RNase RMP RNA) and protein translocation (7SL RNA) (reviewed in (1)). Pol III is recruited to its cognate promoters by gene-specific assemblies of the general transcription factors (GTFs) TFIIIA, TFIIIB, TFIIIC and SNAPc/PTF (reviewed in (2,3)). Pol III transcription activity is correlated with cell growth and division (reviewed in (2)) and it is in particular deregulated during tumoral transformation (4).
The three eukaryotic RNA polymerases are composed of multiple subunits that have been conserved during evolution. Consequently, the three enzymes possess five identical subunits (plus another two that are identical in Pols I and III) and several other subunits that are highly related to each other (5). Interestingly, Pol III contains three or five additional subunits for which no paralogues are found in Pols I or II, respectively. Three specific Pol III subunits (in humans RPC62/POLR3C, RPC39/POLR3F, RPC32α/POLR3G) form a stable ternary subcomplex that can be dissociated from the other 14 subunits in yeast and human cell extracts (6,7). It was suggested that this ternary subcomplex contributes to pre-initiation complex formation (PIC) by interacting with the general transcription factors TFIIIB, TFIIIC and the enzymatic core of Pol III (8,9). In vertebrates, RPC32β/POLR3GL, a paralogue of RPC32/POLR3G is expressed in all cells. In contrast, RPC32α/POLR3G expression is restricted to undifferentiated cells and regulates embryonic stem cell and muscle differentiation (10–12).
Previously, we reported the crystal structure of hRPC62 (13). This protein is composed of two long α-helixes surrounded by four extended winged helix (eWH) domains. hRPC62 showed structural similarities with the N-terminal part of hTFIIEα (component of TFIIE, a general Pol II initiation factor). Comparable to other WH proteins, hRPC62 specifically binds to single-stranded but not double-stranded DNA (13). However, little is known about functions of hRPC62 and hTFIIEα during transcription.
Here, we report that hRPC62 possesses DNA-dependent ATPase and helicase activities. hRPC62 is able to unwind double-stranded (ds)DNA in an ATP-dependent manner, exclusively in the 3′ to 5′ direction with respect to the single-stranded (ss)DNA flanking the duplex. Unwinding activity is observed with highly purified recombinant human rhRPC62 and with affinity-purified human RNA polymerase III. Moreover, dsDNA unwinding and ATPase activities are likewise associated with hTFIIEα. We conclude that hRPC62 and the Pol II transcription factor hTFIIEα can be considered as novel human DNA-dependent ATPases and 3′-5′ DNA helicases. Collectively, our results point to an unexpected role for hRPC62 and hTFIIEα in DNA unwinding which may be implicated in promoting DNA strand separation during transcription and/or other yet to be discovered processes.
MATERIALS AND METHODS
Plasmids and recombinant protein purification
Plasmids encoding wild type human full-length or mutated hRPC62 proteins and hTFIIEα have been described previously in (13) and (14). The rhRPC62-eWH-TFIIEα expression constructs are schematically shown in Figure 7A and was described in Suppl. Supporting Appendix Materials and Methods. Point mutations of hRPC62 (R81A, R84A, R87A) were generated by PCR-mediated mutagenesis through the use of primer pairs: for hRPC62-R81A:
5′-GCCCAGTGCAGCGCCGTATTGCGAAT-3′,
5′-ATTCGCAATACGGCGCTGCACTGGGC-3′,
Figure 7.
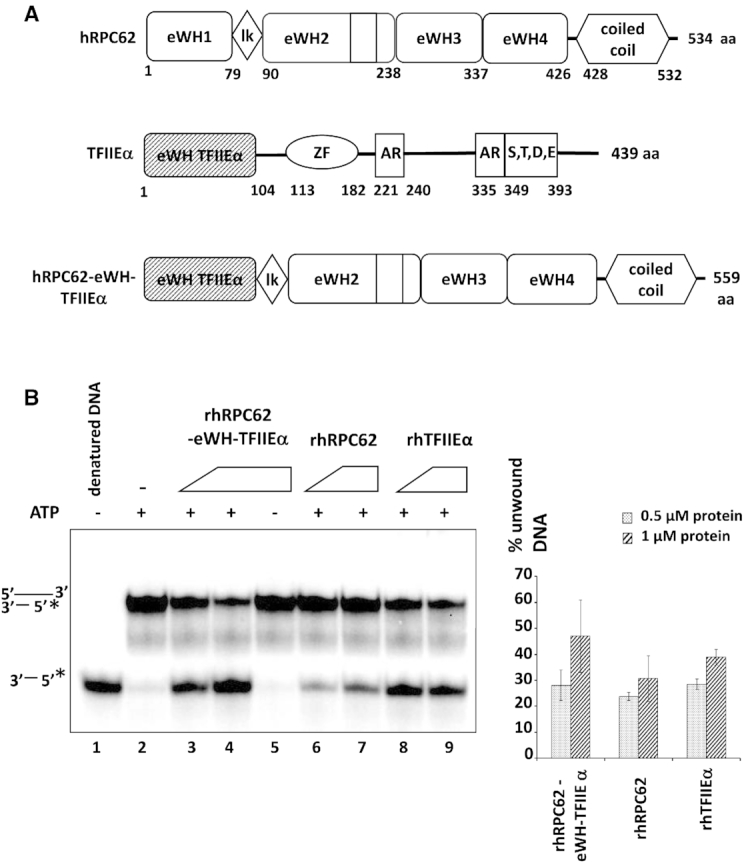
eWH domain of rhTFIIEα can replace eWH1 of rhRPC62. (A) Schematic representation of rhRPC62, rhTFIIEα and rhRPC62-eWH-TFIIEα. Amino acid positions that delimit individual domains are indicated below each scheme. eWH: extended winged helix domain; lk: linker; ZF: zinc finger; AR: Alanine-rich; S, T, D, E: a region that consists of serine (S), threonine (T), aspartic acid (D), and glutamic acid (E) residues. (B) The chimeric rhRPC62-eWH-TFIIEα expression vector was constructed, expressed and purified as described in Supporting Appendix Materials and Methods. Similar molar amounts of rhRPC62 (lanes 6 and 7), rhTFIIEα (lanes 8 and 9) and the chimeric rhRPC62-eWH-TFIIEα protein (lanes 3–5) were analyzed in helicase assays as described previously. Assays were carried out in the presence of 0.5 μM (lanes 3, 6 and 8) or 1 μM (lanes 4, 5, 7 and 9) protein. 4 mM ATP were included into reactions loaded onto lanes 2–4 and 6–9. Lane 1: no protein and denatured substrate DNA. Lane 2: substrate DNA without protein. The quantification of three independent experiments is shown in the right panel.
for hRPC62-R84A:
5′-TGCAGCCGGGTATTGGCTATGCTTAGATATCC-3′,
5′-GGATATCTAAGCATAGCCAATACCCGGCTGCA-3′,
for hRPC62-R87A:
5′- GTATTGCGAATGCTTGCATATCCCCGGTACATC-3′,
5′-GATGTACCGGGGATATGCAAGCATTCGCAATAC-3′
Other mutations of hRPC62 were described previously in Lefèvre et al. All recombinant proteins, are 6-His tagged and were expressed in Escherichia coli BL21 (DE3) Gold pLysS and purified as described (13). The purification scheme for the recombinant proteins is presented in Supplementary Figure S1A. Protein concentration was determined by Bradford assay (Bio-rad protein assay) and purified protein fractions were analyzed by SDS-PAGE and Coomassie Blue staining or by western blot (Supplementary Figure S1B).
Nucleic acid substrates
Oligonucleotides (Sigma Aldrich) were derived from the adenoviral VA1 sequence (Supplementary Figure S8), labeled at the 5′ end by phosphorylation with [γ-32P] ATP (Perkin Elmer). dsDNA substrates used in helicase assays were constructed by mixing labeled oligonucleotides with a 3-fold molar excess of the complementary unlabeled strand in 7 mM Tris–HCl pH 8.5, 50 mM NaCl, 7 mM MgCl2. Samples were heated to 95°C for 5 min followed by slow cooling to room temperature. Double-strand DNA substrates were then purified by native 12% PAGE.
DNA helicase assays
The helicase assay contained a 32P-labeled substrate in 10 μl helicase buffer (20 mM HEPES pH 7.9, 2 mM MgCl2, 65 mM KCl, 4 mM ATP, 0.2 mg/ml BSA, 3 mM dithiothreitol and 10% glycerol). Reactions were initiated by addition of the enzyme at the concentrations indicated in each figure, incubated for 30 min at 37°C and terminated by the addition of 5 μl stop buffer (0.1 mM Tris–HCl pH 7.5, 20 mM EDTA, 0.5% SDS, 0.1% NP40, 0.1% bromophenol blue, 0.1% xylene cyanol and 25% glycerol). Reaction products were resolved by native 12% PAGE. The percentage of unwound substrate (quantified using the ImageQuant software (GE Healthcare)) was determined as (% DNA unwound) = 100 × [ssDNA / total DNA], where total DNA comprises both ssDNA and the dsDNA substrate.
ATPase assays
ATPase activity was determined by using thin layer chromatography (TLC PEI Cellulose F, Merck). The reaction mixture (10 μl) contained 20 mM Hepes pH 7.9, 2 mM MgCl2, 65 mM KCl, 50 μM unlabeled ATP, 0.3 pmol [γ32P] ATP (3000 Ci/mmol Perkin Elmer), 0.2 mg/ml BSA, 1 mM dithiothreitol. Reactions were initiated by adding recombinant human RPC62, incubated at 37°C for 30 min, and stopped with 5 μl 20 mM EDTA. Finally, 1 μl of this mixture was spotted onto a pre-coated polyethyleneimine-cellulose plate and was separated during 1h with 2 M acetic acid and 0.5 M LiCl2. Phosphate release was visualized by phosphoimager and analyzed using ImageQuant software.
UV crosslinking
ATP binding of individual proteins was assayed by UV crosslinking with [α32P] ATP. Reaction mixtures (10 μl final) containing 1 μg of protein in 20 mM Hepes pH 7.9; 65 mM KCl; 0.5 mM DTT; 10% Glycerol and 20 μCi of [α32P] ATP 3000 Ci/mmol (Perkin Elmer) were first incubated 20 min at 37°C. Cross-linking was performed on ice using Stratalinker 1800 (Stratagene) with a total irradiation energy of 0.9 J/cm2. Then 4 mM of unlabeled ATP was added to the reaction mixtures. Samples were resolved by SDS-10% PAGE. The gel was stained with Coomassie Blue, dried and analysed by phosphoimager.
UV induced crosslinking with 4-thiouridine and crosslink identification
Purified recombinant rhRPC62 (3 μM) was incubated for 10 min on ice with 5x molar excess of 4-thiouridine (Jena Biosciences) in reaction mixture containing 20 mM Hepes pH 7.9, 60 mM KCl and a 5× molar excess of a single strand DNA oligonucleotide (1–37 f). Reaction mixtures were then subjected to 365 nm radiations for 3 min at 5 cm distance. Biochemical enrichment of protein-nucleotide heteroconjugates and data analysis was performed as described previously (15,16). The samples were digested with trypsin in 1:20 (w/w) ratio in the presence of 1 M urea. Non-cross-linked nucleotides were depleted by reverse phase C18 chromatography and peptide-nucleotide heteroconjugates were enriched by TiO2 affinity chromatography. Mass spectrometric analysis was performed on a Q Exactive HF (Thermo Scientific), coupled to Dionex UltiMate 3000 UHPLC system (Thermo Scientific), equipped with an in-house-packed C18 column (ReproSil-Pur 120 C18-AQ, 1.9 μm pore size, 75 μm inner diameter, 30 cm length, Dr Maisch GmbH). Chromatographic separation was performed with 43 min gradient starting at 8% and increasing to 45% B (mobile phase A – 0.1% FA (v/v); mobile phase B – 80% ACN, 0.08% FA (v/v), flow rate 300 nl/min). Acquisition was performed at 120 000 resolution for survey MS scans and 30 000 resolution for fragment MS/MS scans, dynamic exclusion was set to 9 s. The resulting files were analyzed with the RNPxlSearch pipeline of the OpenMS suite (2.4.0). Cross-link candidate spectra were manually validated and annotated.
RESULTS
Recombinant human rhRPC62 unwinds double-stranded DNA with a 3′ single-stranded DNA overhang
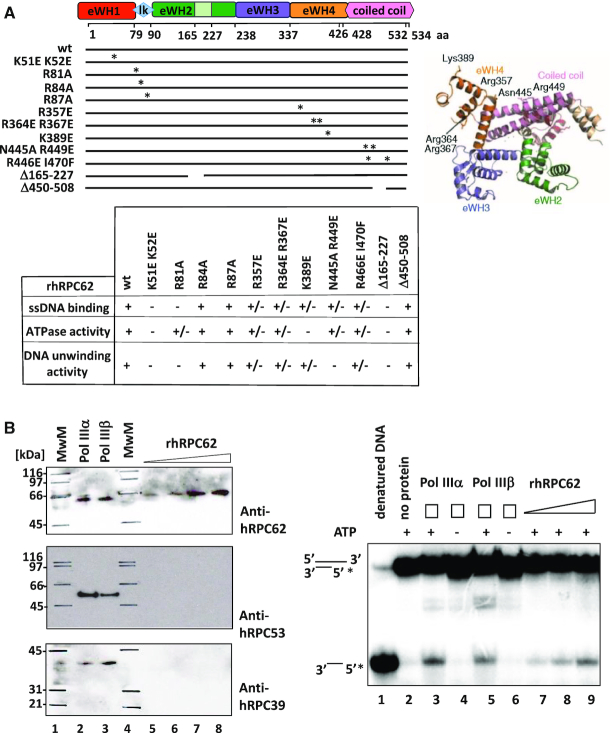
Several studies with yeast or human cells proposed the participation of RPC62, RPC39 and RPC32 in transcription initiation and open complex formation (6,17–19). In line with the idea of bridging DNA recognition and GTF interactions, it was shown that rhRPC62 binds to single-stranded DNA (13) and Saccharomyces cerevisiae RPC82 could be cross-linked near the transcription initiation site (at position –8/–7 and +11) (20,21). Therefore, we assumed that human RPC62 could participate in the unwinding of double-stranded dsDNA. We purified recombinant human 6-His-tagged-RPC62 (rhRPC62) to homogeneity (Supplementary Figure S1) and employed it together with a variety of several radiolabeled DNA substrates in helicase assays. These substrates contained 55-nucleotides of either 3′- or 5′-ssDNA overhang in addition to a 22-nucleotides stretch of dsDNA. As shown in Figure 1A, rhRPC62 efficiently separated dsDNA with a 3′-ssDNA overhang, but not the substrate containing a 5′-ssDNA overhang (compare lanes 3, 4 and 11, 12). Kinetic studies showed that rhRPC62 unwound the duplex DNA (22-bp dsDNA with a 55- nt 3′-overhang) in a time-dependent manner (Supplementary Figure S2A). This experiment allowed estimating an unwinding rate of about 0.3 nM 22-nt dsDNA. min−1. μM−1 rhRPC62. For comparison, human WRN and DHX9 helicases have unwinding rates of about 0.25 and 0.08 nM (17-mer). min−1. μM−1 enzyme, respectively (22). Similar helicase assays were also performed with purified recombinant human 6His-tagged-rhRPC39 protein. No DNA unwinding activity was detected, arguing for a specific enzymatic activity residing in rhRPC62. In addition, the possibility of contaminating bacterial helicases as a source of unwinding activity was furthermore ruled out by analyzing highly purified wild type rhRPC62 purifications used in these experiments by mass spectrometry. Around 90% of the peptides in the purified sample could be assigned to hRPC62. An analysis of the remaining peptides, representing bacterial contaminants in these purifications, did not reveal the presence of any protein that could be held responsible for the observed dsDNA unwinding activity (Supplementary Table S1). An analysis of hRPC62 mutants described below (Figure 3) confirmed that this enzymatic activity resides in wt hRPC62.
Figure 1.
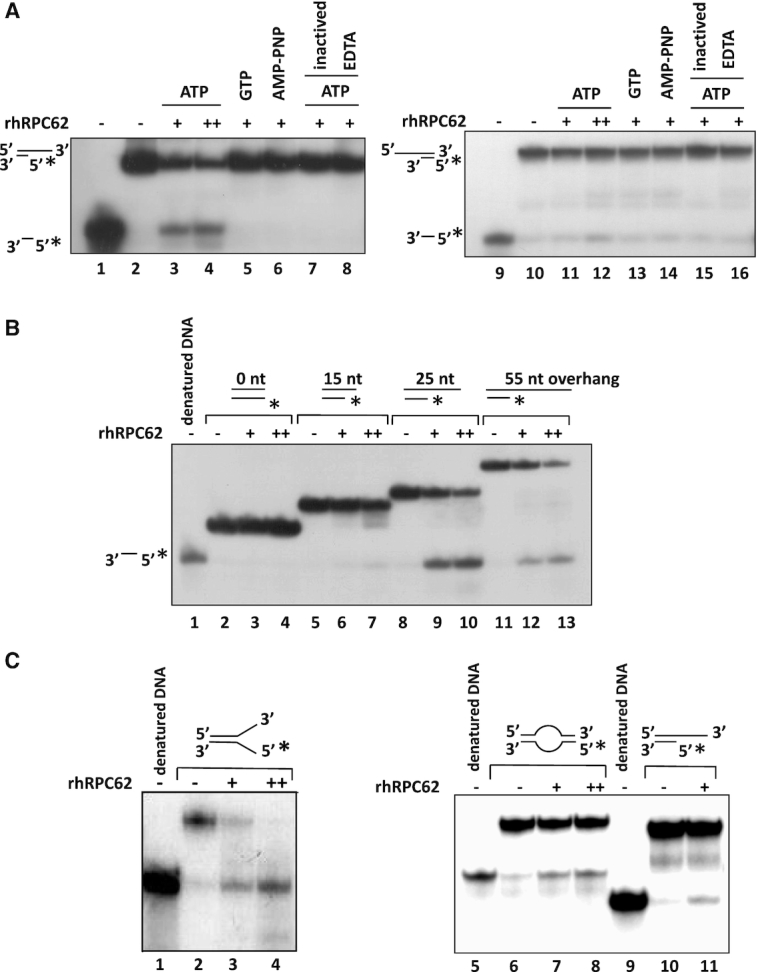
DNA unwinding activity of rhRPC62. Recombinant human rhRPC62 was produced and verified as described in Lefèvre et al. 2011. (A) rhRPC62 displays ATP-dependent 3′-5′ DNA unwinding activity. DNA helicase assays in the presence of DNA substrates contained 22-nt dsDNA in addition to 55-nt ssDNA overhangs. Lanes 1–8: 3′ overhang substrates. Lanes 9–16: 5′ overhang DNA. DNA structures are schematically shown to the left of the autoradiogram. * indicates 32P-labeled 5′-ends of oligonucleotides. 4 mM ATP, GTP or AMP-PNP were added to the reactions as appropriately indicated above the lanes. Lanes 1, 9: no protein and denatured DNA substrate. Lanes 2, 10: no protein. Lane 7, 15: heat denatured protein. (+) Lanes 3, 5, 6, 7, 8, 11, 13, 14, 15, 16: 1 μM rhRPC62. (++) Lanes 4, 12: 1.5 μM rhRPC62. (B) Helicase assays with DNA substrates containing 3′-ssDNA overhangs of different lengths. All substrates contained 22-nt dsDNA; individual substrates had either no overhang DNA (blunt-ended substrate DNA; lanes 2–4), or 3′-ssDNA overhangs of 15-nt (lanes 5–7), 25-nt (lanes 8–10) or 55-nt (lanes 11–13). The DNA sequences utilized for the construction of the substrates are shown in Supplementary Figure S8. Control reactions were incubated without rhRPC62 (lanes 2, 5, 8 and 11). The migration of denatured substrate DNA is shown in lane 1 and schematized to the left of the autoradiogram. The quantification of three independent experiments is presented in the Supplementary Figure S2C. (C) rhRPC62 unwinding activity on forked DNA substrates (left panel) and on a bubble DNA substrate (right panel). Oligonucleotide sequences employed for these reactions are given in Supplementary Figure S8. Left panel: The forked DNA substrates corresponded to 21-nt dsDNA with 26-nt 3′ ssDNA overhang. The bubble DNA substrate contained a bubble of 12 nucleotides. The amounts of hRPC62 are indicated: (−) no protein, (+) 0.5 μM, (++) 1 μM. Denatured substrate is shown in lanes 1, 5, 9. The amounts of hRPC62 are indicated: no protein (−; lanes 1, 2, 5, 6, 9, 10), 0.5 μM (+; lanes 3, 7, 11), 1 μM (++; lane 4, 8).
Figure 3.
Enzymatic activity of mutant rhRPC62 proteins and purified human Pol IIIα and Pol IIIβ. Recombinant human rhRPC62 (wt or mutant) was produced and verified as described in Lefèvre et al. 2011 (Supplementary Figure S1). (A) Schematic presentation of rhRPC62 domains; numbers of amino acids (aa) at the borders of individual domains are indicated. Point mutations are indicated by (*) and the positions of deletions are symbolized by discontinued lines. Similar molar amounts of recombinant wild type or mutant hRPC62 were analyzed for ssDNA-binding (as described in Lefèvre et al., 2011), for ATPase activity and for their ability to unwind a 22-nt dsDNA substrate with a 55-nt ssDNA 3′ overhang. Results are shown in the table. (+), (+/−) and (−) respectively, indicate full, defective and no DNA unwinding, ATPase and ssDNA binding activities as appropriately indicated to the left. The experimental data are presented in Supplementary Figures S3 and S4. (B) RNA polymerases III α (Pol IIIα) and III β (Pol IIIβ) were purified over M2 agarose as described in Haurie et al., 2010. Left panel: normalization of purified Pol III by western blot. 10 μl of immunopurified Pol IIIα (lane 2) or Pol IIIβ (lane 3) were probed with antibodies directed against hRPC62 (upper panel), hRPC53 (middle panel) or hRPC39 (lower panel). Signal intensities were compared to those of defined amounts of purified rhRPC62 (0.05, 0.1, 0.2, 0.3 μM) (lanes 5, 6, 7, 8 respectively). MwM: molecular weight marker proteins retraced with a marker pen (lanes 1, 4). The molecular masses are appropriately indicated to the left. Right panel: immunopurified human Pol IIIα and Pol IIIβ are able to separate 22-nt dsDNA with a 55-nt 3′ ssDNA overhang. 10 μl of Pol IIIα (lanes 3, 4), Pol IIIβ (lanes 5, 6) or 0.1, 0.2, 0.3 μM of rhRPC62 (lanes 7, 8, 9 respectively) were employed for helicase assays in the presence (+; lanes 2, 3, 5, 7–9) or absence (−; lanes 1, 4, 6) of 4 mM ATP. Lane 1: heat denatured DNA probe. Lane 2: control reaction without protein.
As shown in Figure 1A, substrate unwinding by rhRPC62 requires ATP (lanes 3, 4), which cannot be replaced by the nonhydrolyzable ATP analogue 5′-adenylyl-β,γ-imidodiphosphate (AMP-PNP) (lane 6) or GTP (lane 5). These results suggest that the DNA unwinding function of rhRPC62 is dependent on ATP hydrolysis. In addition, unwinding requires MgCl2, which could not be replaced by other cation salts like MnCl2 or CaCl2 (Supplementary Figure S2B compare lanes 3, 4 with 6, 7 or 9, 10 respectively). Moreover various DNA substrates showed that efficient dsDNA template unwinding (>35% unwinding of input DNA) by rhRPC62 required a 3′-overhang of at least 25-nt (Figure 1B, Supplementary Figure S2C).
In conclusion, these experiments show that rhRPC62 is able to unwind dsDNA substrates with 3′DNA overhangs in the presence of ATP and MgCl2.
RNA polymerase III transcription requires the formation and maintenance of the transcription bubble, which is characterized by about 22-nt melted DNA (20,23). We constructed several dsDNA templates that recapitulate certain features of or mimic the transcription bubble. First, we generated a dsDNA fork, consisting of 21-bp dsDNA and of two juxtaposed ssDNA tails of 26-nt. Figure 1C (left panel) shows that rhRPC62 unwound forked dsDNA as efficiently as 22-nt dsDNA with a 3′ 55-nt ssDNA overhang. Figure 1C (right panel) shows that rhRPC62 can also unwind DNA bubbles of 12 nucleotides at an efficiency close to that observed with DNA duplex substrates with 3′ overhang (unwinding rate > 30% with 0.5 μM rhRPC62). These analyses show the capacity of rhRPC62 to unwind DNA structures that mimic the transcription bubble.
rhRPC62 is a DNA-dependent ATPase
The results shown in Figure 1A suggested that DNA unwinding by rhRPC62 is dependent on an ATPase activity, similar to what was shown for other helicases (reviewed in (24)). Incubation of rhRPC62 with ATP induced the release of inorganic phosphate (Pi) indicating ATP hydrolysis (Figure 2A). ATP hydrolysis correlated to the amount of rhRPC62 in the reaction (lanes 4–5). rhRPC62 ATPase activity was barely detectable in the absence of DNA (Figure 2A, lane 3 and Figure 2B). 70% of ATP was hydrolyzed by 1 μM rhRPC62 in the presence of ssDNA and 35% in the presence of dsDNA (Figure 2A, lanes 4–7 and Figure 2B).
Figure 2.
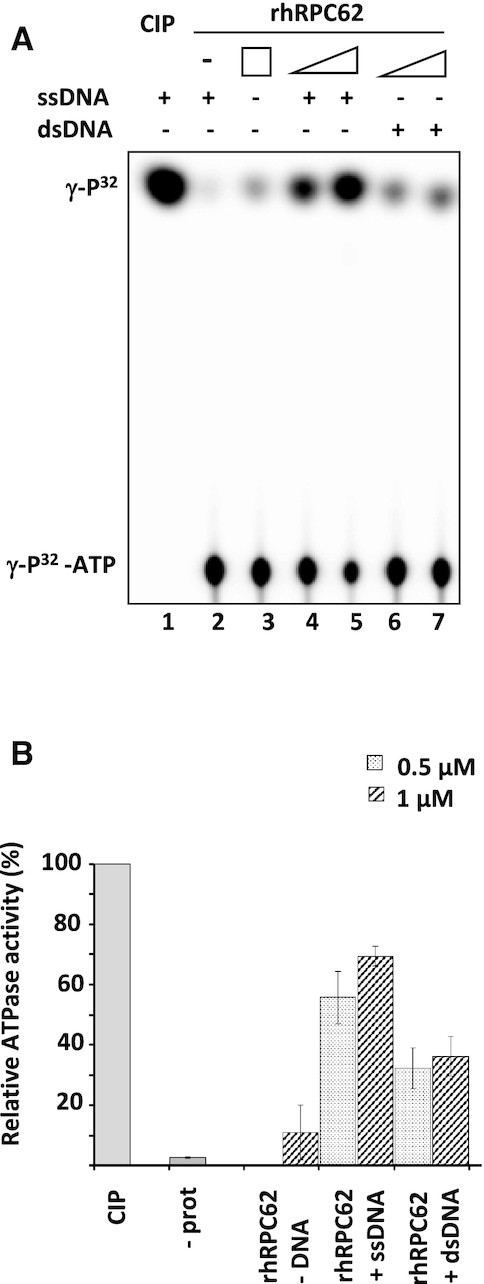
Characterization of rhRPC62 ATPase activity. (A) ATPase assays were conducted in presence of 0.5 μM of rhRPC62 (lanes 4, 6) or 1 μM of rhRPC62 (lanes 3, 5, 7). Lane 1: positive control with 1 unit of calf intestinal phosphatase (CIP); lane 2: negative control without protein. 2 μg of ssDNA were added to the reactions loaded in lanes 1, 2, 4 and 5. 2 μg of dsDNA were added to the reactions in lanes 6 and 7. (B) A quantification of three independent ATPase assays.  : 0.5 μM protein,
: 0.5 μM protein,  : 1 μM protein.
: 1 μM protein.
Mutagenesis of rhRPC62 confirms requirement of DNA binding for helicase and ATPase activities
The analysis of hRPC62 protein sequence did not reveal known helicase motifs. As described previously in Lefèvre et al., hRPC62 comprises two central α-helixes (residues 428–534) surrounded by four extended winged helix (eWH) domains (schematically shown in Figure 3A). In a previous study, mutants of conserved residues on the surface of eWH1, eWH4 and of the coiled-coil domain demonstrated that rhRPC62 eWH domains are required for ssDNA binding (13). All rhRPC62 mutant proteins were purified following the same experimental procedure, ensuring that wild type and mutant rhRPC62 were of comparable purity (Supplementary Figure S1).
First we analyzed the mutants previously characterized for their ability to bind DNA (R357E, R364 R367E, N455A N449E, R446E I470F, K389E, K51E K52E) in Lefèvre et al. (13). All mutants that could not bind to ssDNA (13) were also deficient in unwinding dsDNA with a 3′-ssDNA overhang and did not contain ATPase activity (Figure 3A, Supplementary Figures S3 and S4). These results underscore that ssDNA binding by rhRPC62 is required for helicase activity. In addition, these results support that the observed helicase activity resides within rhRPC62 and is not provided by bacterial proteins that might have co-purified with rhRPC62. To confirm this point we analyzed two truncated forms of rhRPC62 in DNA-binding and ATPase assays. One of them (rhRPC62 Δ164–227) is able to bind ssDNA and the other one (rhRPC62 Δ450–508) is not (13). Again, the inability of binding the ssDNA is correlated with lack of duplex DNA dissociation and of ATPase activity (Figure 3A, Supplementary Figures S3 and S4). Collectively, rhRPC62 possesses intrinsic helicase and ATPase activities that are dependent on single-stranded DNA binding.
RNA polymerase III purified from human cells contains intrinsic ATP dependent 3′-5′ DNA unwinding activity
We next wanted to determine whether human RNA polymerase III likewise contains helicase activity. Since hRPC62 is a constituent of the two isoforms of human RNA polymerase III (Pol IIIα and Pol IIIβ) containing either RPC32α or its paralogue RPC32β, we purified both enzymes from HeLa cell nuclear extracts by immunoaffinity chromatography as described previously by Haurie et al. (10). The purified samples were analyzed by western blot for the presence of several Pol III subunits (hRPC62, hRPC53 and hRPC39). Roughly similar amounts of the three subunits were detected in both purified enzymes (Figure 3B, left panel, lanes 2, 3). Since hRPC53 is not part of the ternary complex and present in equimolar amounts relative to RPC62 and RPC39 in purified Pol III, it can be assumed that the purification contains entire Pol III. We estimated the amounts of hRPC62 in each purified Pol III sample by comparing them with defined quantities of rhRPC62 (Figure 3B, left panel, lanes 5 to 8). Pol IIIα and Pol IIIβ both contained ∼0.2–0.3 μM of hRPC62 (Figure 3B, left panel, compare lanes 2–3 to 7–8). Equivalent amounts of each immunopurified Pol IIIα and Pol IIIβ were analyzed for their ability to unwind dsDNA with 3′-overhangs and compared to the activity of the same molar amounts of rhRPC62. Both Pol III isoforms were able to unwind dsDNA (Figure 3B, right panel, lanes 3, 5) in an ATP-dependent manner (compare lanes 3 and 5 to 4 and 6, respectively). Strand separation activities of the two isoforms of Pol III are comparable to that residing in similar molarities of rhRPC62 (lanes 7–9). These results support the notion that hRPC62 contains a DNA unwinding activity and reveal a novel enzymatic function for Pol III. In addition, they indicate that this activity is not altered by the absence or presence of the other 16 Pol III subunits.
Determination of rhRPC62 residues that are in contact with ATP
After having demonstrated that human RPC62 contains DNA unwinding and ATPase activities, we wished to determine whether RPC62 is able to bind ATP and which amino acids are involved in this process. We incubated rhRPC62 with [α32P] ATP and exposed the reaction to UV light. Wild type rhRPC62 and a positive control, the kinase domain of hRIO2 protein were efficiently crosslinked to ATP (Figure 4A, lanes 7–8). The specificity of ATP binding and crosslinking was verified employing the same molar amounts of rhTBP in control reactions (lane 9). Recombinant proteins are shown in lanes 2–4. These data show that rhRPC62 is able to bind ATP.
Figure 4.
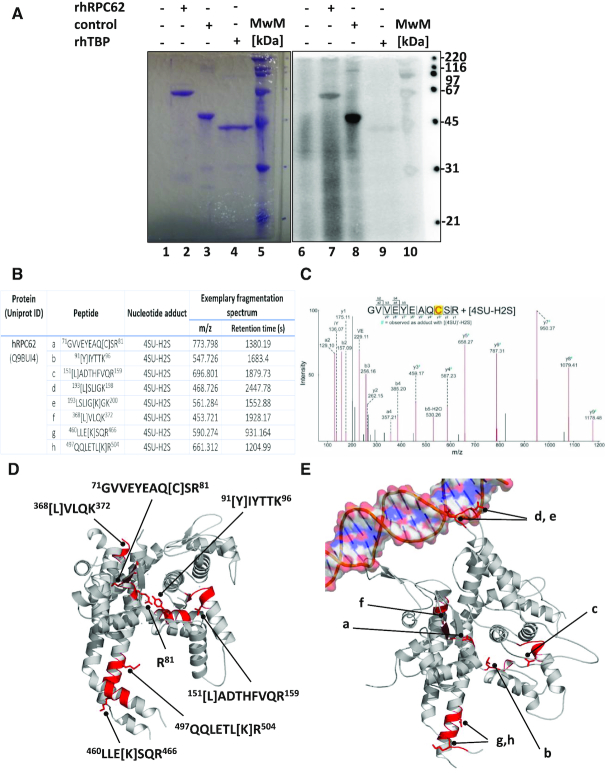
hRPC62 crosslinking with 4-thiouridine. (A) rhRPC62 is able to crosslink ATP. 1 μM purified rhRPC62 (lanes 2 and 7), positive control (fragment of the kinase Rio protein) (lanes 3 and 8) or rhTPB (lanes 4 and 9), were UV-irradiated in the presence of [α32P]-ATP as described in Materials and Methods. Reaction products were separated by SDS 10% PAGE, Coomassie Blue-stained (left panel) and analyzed by phosphoimaging (right panel). MwM: molecular weight marker proteins (lanes 5, 10) and their molecular masses are appropriately indicated to the right. Lanes 1 and 6: Control reactions without protein. (B) Overview of identified protein-nucleotide cross-links of hRPC62 with 4-thiouridine. The cross-linked amino acid is enclosed in square brackets. (4SU-H2S–4-thiouridine monophosphate with net loss of H2S; m/z – experimentally observed precursor mass to charge ratio). (C) MS/MS fragment spectrum of hRPC62 peptide 71GVVEYEAQCSR81 cross-linked to 4-thiouridine. The a-, b-, y-immonium and internal ion fragments with their corresponding m/z values are annotated in the spectrum. Ion mass shifts with nucleotide fragment adducts are indicated with a superscript – #. The prominent shifted y-ion series confidently localize the cross-linked amino acid at the 79Cys residue. (4SU-H2S-4-thiouridine monophosphate with net loss of H2S; (4SU)′-H2S-4-thiouracil with net loss of H2S). (D) Peptides identified by mass-spectrometry and amino acid R81 are displayed as red areas on the hRPC62 structure. The cross-linked residues (square brackets) are shown as stick representations. (E) The corresponding peptides that were cross-linked on hRPC62 are displayed on RPC82 taken from the complete RNA polymerase III pre-initiation complex structure (25). The incoming double stranded DNA is also displayed to show the proximity of unresolved structural elements of hRPC62 that are visible in RPC82.
After demonstrating that hRPC62 can bind and hydrolyse ATP, we wanted to determine the potential ATP binding site by identifying the domains of hRPPC62 that can be crosslinked to ATP using mass spectrometry. However, ATP exhibits very low photoreactivity, and while we were able to visualize its ability to bind hRPC62 thanks to radioactive labeling, we were not able to clearly identify the regions crosslinked to ATP by mass spectrometry. In order to overcome this problem, we substituted ATP with a modified nucleotide: 4-thiouridine (4-thioUTP), a uridine analog in which the keto oxygen at position 4 of the pyrimidine ring is replaced by a sulfur atom. 4-thioUTP has the advantage of being much more photo reactive than unmodified nucleotides allowing to significantly increase the efficiency of UV crosslinking. We assumed that an ATP binding site should exhibit high affinity towards nucleotides and we have good chances to crosslink a highly reactive nucleotide close to this site. As summarized in figure 4B, we identified eight major peptides efficiently crosslinked to 4-thioUTP. This confirms the ability of rhRPC62 to bind directly a nucleotide. The first two peptides (a: 71GVVEYEAQCSR81 and b: 91YIYTTK96) are located in the region between eWH1 and 2 (Figure 4D). One peptide (c: 151LADTHFVQR159) corresponds to the region of eWH2 preceding the insertion loop of eWH2. Two peptides (d: 193LSLIGK198 and e: 193LSLIGKGK200) are localized in the unstructured insertion loop of eWH2. This region is not visible in figure 4D. One peptide (f: 368LVLQK372) is located in eWH4 and two others (g:460LLEKSQR466 and h: 497QQLETLKR504) in the coiled coil domain (Figure 4D). In order to understand the relevance of the determined crosslinks, we used the published structure of the RNA polymerase III open complex (25) and localized the corresponding peptides on the S. cerevisiae homolog of hRPC62 (Figure 4E). This structure shows that the insertion loop of eWH2 enters in direct interaction with the DNA template. This may explain the crosslinks of the region between residues 193 and 200 (peptides d and e). In the initially transcribing complex structure of S. cerevisiae Pol III, we can see the eWH4 of RPC82 oriented towards the DNA template, suggesting that this domain exhibits an affinity towards nucleic acids, possibly explaining the crosslinks localized in eWH4 of hRPC62. On the other hand, peptides a, b and c do not enter in close proximity with the DNA template or the nascent RNA but their localization on the hRPC62 structure shows that they surround a pocket like area inside the protein. This suggests that they might be implicated in the interaction with a nucleotide and potentially ATP. The last two identified peptides (g and h) are located on the bottom of the coiled coil domain and oriented towards the surface of the RNA polymerase. This suggests that the observed crosslinks with these two peptides reflect an affinity of this region towards nucleotides but is not likely to correspond to an ATP binding site.
The peptide 71 GVVEYEAQCSR 81attracted our attention since Lee et al. (26) reported the crystal Structures of the Bacillus stearothermophilus CCA-adding Enzyme and its complexes with ATP or CTP. In this study, the authors showed a direct interaction of ATP with a motif RxxRxxR. This motif resembles the sequence R81xxR84xxR87 found in hRPC62 in the area where ATP could be coordinated. Therefore, we completed the DNA-binding, helicase and ATPase analyses with three additional point mutants (R81A, R84A, R87A) of hRPC62 positioned in the linker between the first two eWH domains (Supplementary Figure S5). As with the previous mutations, we confirm the perfect correlation between the lack of ssDNA binding (Supplementary Figure S5A), ATPase (Supplementary Figure S5B) and DNA unwinding activities (Supplementary Figure S5C). It is noteworthy that the R81A mutation most dramatically affects DNA binding and enzymatic activities.
Mutation of the R81 homologous residue in Saccharomyces cerevisiae RPC82 strongly impairs transcription in vivo
In order to assess whether mutations of residues involved in DNA binding of hRPC62 were impacting on transcriptional activity in vivo, we resorted to the yeast S. cerevisiae (Sc). It was published that ScRPC82 is essential for life in S. cerevisiae. However, a mutant rpc82-6 was isolated and showed reduced growth rate (27).
We sequenced this mutant and determined that it corresponds to the mutation G126E. Interestingly, G126 in ScRPC82 is found at the same structural position as R81 in human RPC62 (Supplementary Figure S6A and according to HHPRED alignment). We first established haploid strains containing the deleted rpc82Δ:::HIS3 gene rescued by either the WT (RPC82) or the mutant allele (rpc82-6). These strains were tested at permissive (24°C) or restrictive (37°C) temperatures and confirmed the growth defect at restrictive temperature (Supplementary Figure S6B). Then we established a cell line expressing TAP-tagged RPC82 and we purified ScPol III from this cell line demonstrating that this mutation does not affect the assembly of Pol III since all subunits are present in the mutant and the wt enzyme (compare lanes 1 and 2 in Supplementary Figure S6C). As shown in Supplementary Figure S6D, the mutant enzyme exhibited significantly reduced transcription of several pre-tRNA genes and also of the 5S gene. Due to difficulties in growth, it was not possible to purify sufficient amounts of mutant Pol III for conducting helicase and ATPase assays.
Collectively, these results indicate that mutation of G126 in ScRPC82, being homologous to the R81 residue in human RPC62 leads to reduced transcription activity of mutant Pol III. These results hint to the importance of this amino acid for normal Pol III activity and may be consistent with a role of helicase and ATPase activities in Pol III transcription efficiency.
hTFIIEα exhibits functional similarities with hRPC62
hRPC62 eWH domains are structurally related to the N-terminus of hTFIIEα (13). To investigate whether these structural homologies extend to functional similarities, we purified recombinant human 6-His-tagged-TFIIEα (rhTFIIEα; Figure 5A) for subsequent DNA-binding, ATPase and DNA helicase assays. Comparable to rhRPC62, rhTFIIEα hydrolyzed ATP in a DNA-dependent manner (Figure 5B, compare lanes 3 and 5; quantification shown in the graph to the right) and it bound to ssDNA (Figure 5C). Moreover and also comparable to rhRPC62, rhTFIIEα contained 3′-5′ DNA unwinding activity requiring a 3′-ssDNA overhang (Figure 5D lanes 3, 4). Like rhRPC62, rhTFIIEα was unable to unwind blunt-ended dsDNA substrates or with a 55-nt 5′-ssDNA overhang (Figure 5D lanes 6–7, 9–10). rhTFIIEα DNA unwinding activity also depended on divalent cations similar to what we showed for rhRPC62 (Supplementary Figure S7A). Unlike rhRPC62, TFIIEα does not necessarily require Mg2+, but was also is able to dissociate duplex DNA in the presence of Mn2+ cations (Supplementary Figure S7A). Kinetic studies allowed an estimation of the unwinding rate of 22-nt dsDNA by rhTFIIEα at about 0.43 nM hybride (22-mer). min−1. μM−1 protein (Figure 6A and Supplementary Figure S7B). Like rhRPC62, rhTFIIEα unwound various dsDNA substrates such as forked DNA and DNA bubble structures (Figure 6B and 6C). Collectively, these data show that hRPC62 is not only structurally but also functionally related to hTFIIEα.
Figure 5.
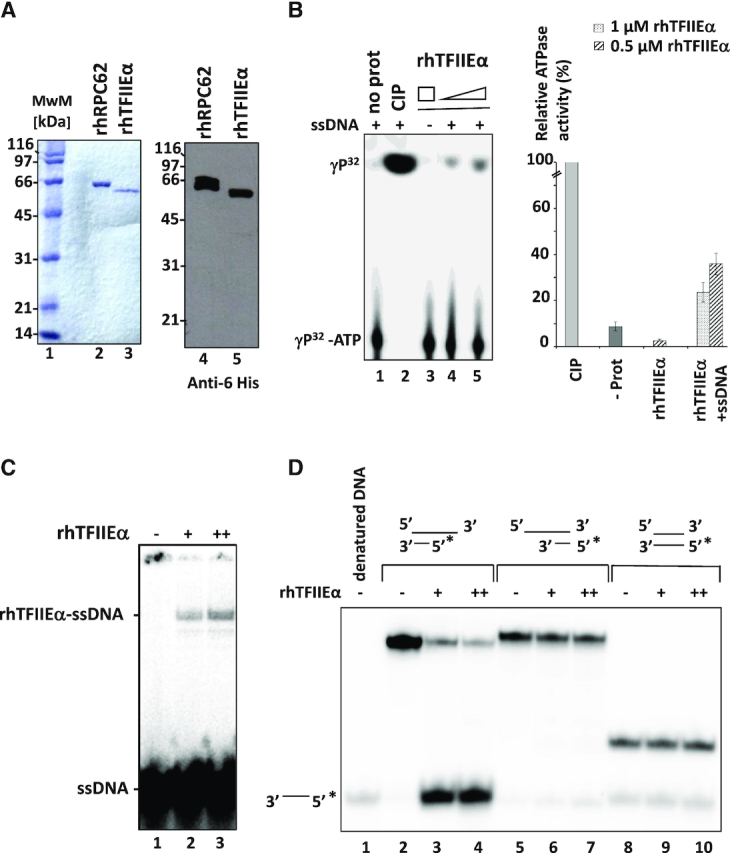
Characterization of rhTFIIEα ssDNA-binding, ATPase and helicase activities. (A) Purified recombinant 6-His-tagged hRPC62 and rhTFIIEα. 1 μg in left panel and 0.1 μg in right panel were separated by SDS 10% PAGE and stained with Coomassie Brilliant Blue (left panel) or processed by western blot (right panel). Anti-6 histidine antibodies were employed for the blot. MwM: molecular weight marker proteins; the molecular masses of MwM are indicated to the left of the gel and autoradiogram. (B) ATPase Assays were carried out in the presence (lanes 1, 2, 4, 5) or absence (lane 3) of 2 μg ssDNA with 0.5 μM (lane 4) or 1 μM (lanes 3 and 5) of rhTFIIEα. Lane 1: negative control without protein. Lane 2: positive control with 1 unit of calf intestinal phosphatase. Quantification of relative ATPase activities is represented to the right. The average of three independent experiments is shown.  : 0.5 μM protein,
: 0.5 μM protein,  : 1 μM protein with or without 2 μg of ssDNA (as indicated). (C) ssDNA-binding properties of recombinant hTFIIEα. EMSA was carried out as described in Lefèvre et al., 2011 with radiolabelled oligonucleotide (1–77) f, shown in Supplementary Figure S8. Lane 1: (−) no protein; lane 2: (+) 0.5 μM rhTFIIEα; lane 3: (++) 1 μM rhTFIIEα. Positions of ssDNA and the rhTFIIEα-ssDNA complex are presented to the left of the autoradiogram. (D) Recombinant TFIIEα unwinds 22-nt dsDNA with a 55-nt ssDNA 3′-overhang. The following DNA substrates were analyzed in helicase assays: 22-nt dsDNA with 55-nt 3′ ssDNA overhang (lanes 2–4), 22-nt dsDNA with 55-nt 5′ ssDNA overhang (lanes 5–7) and 22-nt blunt-ended dsDNA (lanes 8–10). Reactions were carried out in the presence of 0.5 μM (+; lanes 3, 6 and 9) or 1 μM (++; lanes 4, 7 and 10) of rhTFIIEα. Lanes 2, 5, 8: Control reactions without protein. Lane 1: Denatured DNA. (*) indicates the position of the radiolabeled single-stranded nucleotide.
: 1 μM protein with or without 2 μg of ssDNA (as indicated). (C) ssDNA-binding properties of recombinant hTFIIEα. EMSA was carried out as described in Lefèvre et al., 2011 with radiolabelled oligonucleotide (1–77) f, shown in Supplementary Figure S8. Lane 1: (−) no protein; lane 2: (+) 0.5 μM rhTFIIEα; lane 3: (++) 1 μM rhTFIIEα. Positions of ssDNA and the rhTFIIEα-ssDNA complex are presented to the left of the autoradiogram. (D) Recombinant TFIIEα unwinds 22-nt dsDNA with a 55-nt ssDNA 3′-overhang. The following DNA substrates were analyzed in helicase assays: 22-nt dsDNA with 55-nt 3′ ssDNA overhang (lanes 2–4), 22-nt dsDNA with 55-nt 5′ ssDNA overhang (lanes 5–7) and 22-nt blunt-ended dsDNA (lanes 8–10). Reactions were carried out in the presence of 0.5 μM (+; lanes 3, 6 and 9) or 1 μM (++; lanes 4, 7 and 10) of rhTFIIEα. Lanes 2, 5, 8: Control reactions without protein. Lane 1: Denatured DNA. (*) indicates the position of the radiolabeled single-stranded nucleotide.
Figure 6.
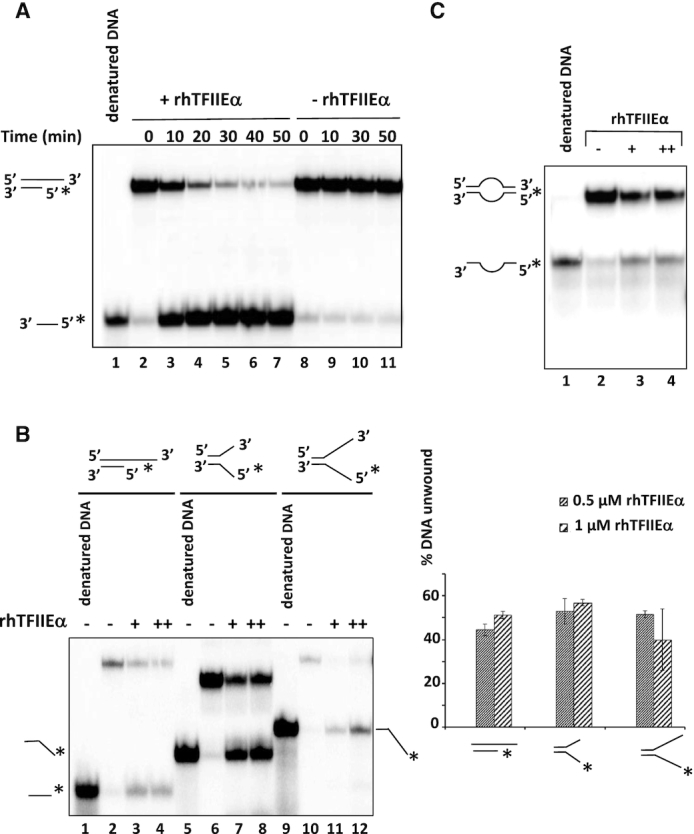
Characterization of rhTFIIEα DNA unwinding activities. (A) Kinetic analysis of DNA unwinding by 1 μM rhTFIIEα. Reactions were performed as described in Materials and Methods employing a 22-nt dsDNA with a 55-nt 3′ overhang ssDNA as substrate. Reaction products were separated by 12% PAGE and quantified with a phosphoimager. The plot (presented in Supplementary Figure S7B) shows percent unwinding against time. (B) TFIIEα unwinding activity on forked DNA substrates. A substrate of 22-nt dsDNA and 55-nt ssDNA 3′ overhang was employed in lanes 1–4. Forked DNA constructs contained in addition to 22-nt dsDNA 5′ and 3′-ssDNA overhangs of 15-nt (lanes 5–8) or 25-nt (lanes 9–12). No protein was added to the reactions loaded in lanes 1, 2, 5, 6, 9 and 10 (indicated by a (−) above the lanes). Molar amounts of rhTFIIEα that were added to reactions are indicated by (+) for 0.5 μM (lanes 3, 7 and 11) and (++) for 1 μM protein (lanes 4, 8 and 12). Denatured DNA substrates were loaded onto lanes 1, 5 and 9. The migration of unwound ssDNA of 22 and 37-nt is shown to the left, that of 47-nt to the right. Substrate DNA structures are schematized on top of the autoradiogram. (*) indicates the position of the labelled nucleotide. The quantification of three independent experiments is shown in the right panel. (C) TFIIEα unwinding activity on bubbled DNA substrate. Denatured substrate is shown in lane 1 and the corresponding radiolabelled (*) fragments are schematically shown to the left. The amounts of TFIIEα are indicated: no protein (−; lanes 2), 0.5 μM (+; lanes 3), 1 μM (++; lane 4). Oligonucleotide sequences employed in this experiment are provided in Supplementary Figure S8.
eWH1 domain of rhRPC62 can be replaced by the eWH domain of rhTFIIEα
eWH1 of hRPC62 is indispensable for ssDNA-binding, helicase and ATPase activities of this protein, since the K51E K52E mutant of eWH1 has lost these three activities (13) (Figure 3A, Supplementary Figures S3 and S4). Given that eWH1 domain of hRPC62 exhibits the highest structural similarity to the eWH of hTFIIEα (13), we fused the eWH domain (amino acids 1–104) of rhTFIIEα to amino acids 80–534 of rhRPC62 (Figure 7A), expressed the fusion protein (rhRPC62-eWH-TFIIEα) as described previously. DNA unwinding by rhRPC62-eWH-TFIIEα was as efficient as that of similar molar amounts of rhRPC62 or of rhTFIIEα (Figure 7B, compare lanes 3–4 with 6–7 and 8–9) and it required ATP (Figure 7B compare lanes 3–4 to 5). These results demonstrated that the eWH domain of rhTFIIEα is able to functionally replace the eWH1 domain of rhRPC62, suggesting that their structure, but not their primary amino acid sequence is the crucial determinant for DNA unwinding.
DISCUSSION
Here, we present evidence in support of DNA-dependent helicase and ATPase activities for hRPC62 and its structural homologue, the general Pol II transcription factor hTFIIEα.
rhRPC62 unwinds DNA in the presence of ATP
Our results reveal that rhRPC62 is able to unwind dsDNA with a 3′-ssDNA overhang in an ATP-dependent manner (Figure 1). All rhRPC62 mutants without ssDNA-binding activity did not show unwinding and DNA-dependent ATPase activities, indicating that these activities are interconnected. The dependence of helicase and ATPase activities on rhRPC62 DNA-binding indicate that these enzymatic activities are intrinsic to rhRPC62 and not an artifact of co-purifying bacterial helicases, which should be expected to function even in the absence of rhRPC62 DNA binding. In addition, about 90% of the peptides identified in our mass spectrometry analysis corresponded to RPC62. The strongest contaminants summed up to less than 0.1% of the peptides indicating that they were ∼1000-fold less represented in the sample compared to RPC62. Thus a potential contaminating helicase would be expected to possess a calculated unwinding rate of more than 0.3 μM hybrid (22-mer. min−1. μM−1) which is not imaginable under normal conditions.
Helicases use the energy of ATP hydrolysis for unwinding, remodeling and translocating nucleic acids or nucleic acid-protein complexes (reviewed in (24)). According to this definition, rhRPC62 can be regarded as a 3′-5′ helicase, which furthermore exhibits ATPase activity that is strongly stimulated by ssDNA.
Helicase activity in rhTFIIE
Mammalian TFIIE is a conserved heterodimer of TFIIEα and TFIIEβ corresponding to S. cerevisiae TFIIE subunits Tfa1p and Tfa2p, respectively (28). Tfa1p N-terminal half contains an eWH domain and shows strong structural similarity to archeal TFE (29). It was shown that the N-terminal part of P. furiosus TFE (containing the eWH motif) binds to ssDNA in the promoter at the upstream end of the transcription bubble (30) and that S. cerevisiae Tfa1p is positioned to DNA sequences overlapping the transcription bubble (31). These reports are in agreement with our results showing that recombinant human rhTFIIEα binds to ssDNA in EMSA (Figure 5C).
The importance of the N-terminal half of TFIIEα/Tfa1p for yeast cell growth (28) and for basal in vitro transcription with human cell extracts was demonstrated (32). Here we identified and characterized ATPase and helicase activities of rhTFIIEα. Characteristic functions (DNA template dependence, polarity, Mg2+- and ATP-dependence, rate of DNA unwinding) are similar to those of rhRPC62. Remarkably, the N-terminally located eWH of hTFIIEα can replace the first eWH domain of hRPC62 (Figure 7) in a fusion protein that conserves functional helicase activity. This result was very surprising, since the primary amino acid sequences of the eWH domains of hTFIIEα and of hRPC62 are only poorly related. Therefore, it is likely that the eWH structure and not the overall amino acid composition of the two domains were crucial for ATPase and helicase functions. Nevertheless, we cannot exclude that one or several conserved amino acids in the two eWH domains play essential roles in DNA-binding, helicase and/or ATPase function.
hRPC62 does not contain classical helicase motifs
We were neither able to identify classical ATPase or helicase domain in hRPC62, nor in hTFIIEα. In addition, we were not yet able to selectively inactivate the ATPase function without affecting the ability of hRPC62 to bind DNA. However, we were able to identify amino acids in human RPC62 that are in contact with 4-thioUTP (Figure 4). One of the residues in the domain contacting 4-thioUTP, R81, is essential for DNA binding, ATPase and helicase activities. Furthermore, mutation of the corresponding residue (G126) its mutation in the orthologous RPC82 subunit results in impaired transcription activity. These results are in agreement with a role for G126/R81 in conferring high transcriptional efficiency to Pol III. It was shown that nucleotide binding specificity in tRNA nucleotidyltransferase is mainly determined by motifs including the RxxRxxR sequence (26). This triad of amino acids was proposed to mediate base stacking interactions for incoming nucleotides. This sequence resembles closely to motif VI in superfamily 2 helicases, a motif that participates in coordinate binding and hydrolysis of triphosphate nucleotides (for review (24)).
Interestingly, previous studies showed that the WH domain of several RecQ proteins plays key roles in their helicase function. Indeed, mutants located in the WH domain of human RecQ1 affected DNA unwinding revealing that a prominent β-hairpin within the WH domain was required for the helicase activity (33–35). In addition, structural studies of Hel 308 and WRN helicases showed that the β-hairpin (or β-wing) of their respective WH domains plays a key role in ATPase and helicase functions (36). Another example of the direct implication of a WH domain in the DNA strand dissociation was reported for the Werner helicase. The co-crystal structure of the RecQ C-terminal (RQC) domain of WRN (including a Zn2+ binding domain and a winged helix structure) bound to dsDNA led Kitano et al. to propose that this domain acted like a ‘knife’ for base-pair separation (33). Taking into account that the WH domain is important for helicase function of several human RecQ-family proteins and it seems reasonable to propose that the eWH domains of hRPC62 and hTFIIEα are implicated in their helicase function. Interestingly, electron microscopy reconstructions of Pol II complexes progressing from closed to open promoter complexes, show a direct role of TFIIE and more precisely, of its eWH domain in promoter opening (37,38). The authors describe a repositioning of the E-wing contained in the eWH motif of TFIIEα from position -7 in closed initiation complex to the upstream edge of the DNA bubble. The presented structure strongly resembles the β-wing of RecQ helicases, suggesting a similar mechanism of action. Similarly, recent EM reconstructions of yeast Pol III preinitiation complexes allowed a fine dissection of structural changes promoting DNA melting and the transition from closed to open Pol III complexes (25,39). The described structures indicate a direct role of the ‘cleft loop’ of C82 (yeast homolog of hRPC62) in promoter opening similarly to what has been described for TFIIEα.
Putative role of hRPC62 and TFIIEα enzymatic functions in transcription initiation
Up to now, Pol II is the only enzyme that was shown to require ATP-dependent helicase activity for transcription (reviewed in (40)). In vitro transcription by Pol III does not require ATP hydrolysis, since it can be carried out in the presence of AMP-PNP (App(NH)p) instead of ATP (41,42). Therefore, the ATP-dependent DNA unwinding activity of hRPC62 appears not to be essential for in vitro transcription. However, we cannot exclude that the helicase activity may improve or stimulate transcription by Pol III in vivo. This assumption is supported by the results that we obtained in the yeast S. cerevisiae (Supplementary Figure S6).
TFIIE was suggested to contribute to TFIIH recruitment which in turn is directly implicated in promoter clearance via its XPB 3′-5′ helicase activity-containing subunit. However, XPB-independent promoter opening was recently reported (43) being in agreement with helicase-independent transcription or with the possibility that this activity resides in another protein involved in Pol II transcription. Moreover, other studies suggested roles for TFIIE which could be in accordance with the here described helicase activity of TFIIEα. Transcription from a supercoiled promoter DNA is significantly stimulated by TFIIE in the absence of TFIIH (44). In addition, TFIIE is not required for transcription directed by a promoter that has been melted from position –8 to +2 relative to the transcription start site (44). These results and other studies support the hypothesis that, depending on DNA topology, TFIIE may be required for transcription initiation, at least in vitro, by contributing to the opening of dsDNA. This hypothesis could be in agreement with a helicase activity of TFIIEα. In summary, our results identify hRPC62 and hTFIIEα as ATP-dependent DNA helicases. Unraveling specific roles of these enzymatic activities in transcription or other cellular processes will be subject for future research.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to A. Poterszman for providing the plasmid encoding hTFIIEα and R. G. Roeder for providing the antibodies directed against hRPC62, hRPC39 and hRPC53 (6,10). We thank Michel Werner for the kind gift of plasmids pRS314-RPC82 and pRS314-rpc82-6. We would like to thank Stéphanie Durrieu-Gaillard for technical support.
Notes
Present address: Leyla El Ayoubi, Max Planck Institute for Biophysical Chemistry, Department of Cellular Biochemitry, Am Faßberg 11, 37077 Göttingen, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Bordeaux, INSERM, CNRS; La Ligue Nationale Contre le Cancer (The French National Cancer League - Equipe labellisée and comité des Landes to M.T. and H.D.-O.); French National Cancer Institute INCa (to M.T.), L.E.A. was supported by PhD fellowships from the Ligue Nationale Contre le Cancer (The French National Cancer League and comité de Gironde et Dordogne) and Fondation ARC pour la Recherche Contre le Cancer. The work of H.U. and A.C. was supported by the Deutsche Forschungsgemeinschaft (DFG SFB860). Funding for open access charge: INSERM, CNRS, INCa, Fondation ARC and Ligue Contre le Cancer.
Conflict of interest statement. None declared.
REFERENCES
- 1. Dieci G., Fiorino G., Castelnuovo M., Teichmann M., Pagano A.. The expanding RNA polymerase III transcriptome. Trends Genet. 2007; 23:614–622. [DOI] [PubMed] [Google Scholar]
- 2. Dumay-Odelot H., Durrieu-Gaillard S., Da Silva D., Roeder R.G., Teichmann M.. Cell growth- and differentiation-dependent regulation of RNA polymerase III transcription. Cell Cycle. 2010; 9:3687–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dumay-Odelot H., Durrieu-Gaillard S., El Ayoubi L., Parrot C., Teichmann M.. Contributions of in vitro transcription to the understanding of human RNA polymerase III transcription. Transcription. 2014; 5:e27526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Durrieu-Gaillard S., Dumay-Odelot H., Boldina G., Tourasse N.J., Allard D., Andre F., Macari F., Choquet A., Lagarde P., Drutel G. et al.. Regulation of RNA polymerase III transcription during transformation of human IMR90 fibroblasts with defined genetic elements. Cell Cycle. 2018; 17:605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vannini A., Cramer P.. Conservation between the RNA Polymerase I, II, and III Transcription initiation machineries. Mol. Cell. 2012; 45:439–446. [DOI] [PubMed] [Google Scholar]
- 6. Wang Z., Roeder R.G.. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997; 11:1315–1326. [DOI] [PubMed] [Google Scholar]
- 7. Werner M., Hermann-Le Denmat S., Treich I., Sentenac A., Thuriaux P.. Effect of mutations in a zinc-binding domain of yeast RNA polymerase C (III) on enzyme function and subunit association. Mol. Cell Biol. 1992; 12:1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hsieh Y.J., Kundu T.K., Wang Z., Kovelman R., Roeder R.G.. The TFIIIC90 subunit of TFIIIC interacts with multiple components of the RNA polymerase III machinery and contains a histone-specific acetyltransferase activity. Mol. Cell Biol. 1999; 19:7697–7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Werner M., Chaussivert N., Willis I.M., Sentenac A.. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J. Biol. Chem. 1993; 268:20721–20724. [PubMed] [Google Scholar]
- 10. Haurie V., Durrieu-Gaillard S., Dumay-Odelot H., Da Silva D., Rey C., Prochazkova M., Roeder R.G., Besser D., Teichmann M.. Two isoforms of human RNA polymerase III with specific functions in cell growth and transformation. PNAS. 2010; 107:4176–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wong R.C., Pollan S., Fong H., Ibrahim A., Smith E.L., Ho M., Laslett A.L., Donovan P.J.. A novel role for an RNA polymerase III subunit POLR3G in regulating pluripotency in human embryonic stem cells. Stem Cells. 2011; 29:1517–1527. [DOI] [PubMed] [Google Scholar]
- 12. McQueen C., Hughes G.L., Pownall M.E.. Skeletal muscle differentiation drives a dramatic downregulation of RNA polymerase III activity and differential expression of Polr3g isoforms. Dev Biol. 2019; 454:74–84. [DOI] [PubMed] [Google Scholar]
- 13. Lefèvre S., Dumay-Odelot H., El-Ayoubi L., Budd A., Legrand P., Pinaud N., Teichmann M., Fribourg S.. Structure-function analysis of hRPC62 provides insights into RNA polymerase III transcription initiation. Nat. Struct. Mol. Biol. 2011; 18:352–358. [DOI] [PubMed] [Google Scholar]
- 14. Jawhari A., Uhring M., De Carlo S., Crucifix C., Tocchini-Valentini G., Moras D., Schultz P., Poterszman A.. Structure and oligomeric state of human transcription factor TFIIE. EMBO Rep. 2006; 7:500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kramer K., Sachsenberg T., Beckmann B.M., Qamar S., Boon K.L., Hentze M.W., Kohlbacher O., Urlaub H.. Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nat. Methods. 2014; 11:1064–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sharma K., Hrle A., Kramer K., Sachsenberg T., Staals R.H., Randau L., Marchfelder A., van der Oost J., Kohlbacher O., Conti E. et al.. Analysis of protein-RNA interactions in CRISPR proteins and effector complexes by UV-induced cross-linking and mass spectrometry. Methods. 2015; 89:138–148. [DOI] [PubMed] [Google Scholar]
- 17. Brun I., Sentenac A., Werner M.. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 1997; 16:5730–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Flores A., Briand J.F., Gadal O., Andrau J.C., Rubbi L., Van Mullem V., Boschiero C., Goussot M., Marck C., Carles C. et al.. A protein-protein interaction map of yeast RNA polymerase III. PNAS. 1999; 96:7815–7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thuillier V., Stettler S., Sentenac A., Thuriaux P., Werner M.. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J. 1995; 14:351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bartholomew B., Durkovich D., Kassavetis G.A., Geiduschek E.P.. Orientation and topography of RNA polymerase III in transcription complexes. Mol. Cell Biol. 1993; 13:942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kassavetis G.A., Han S., Naji S., Geiduschek E.P.. The role of transcription initiation factor IIIB subunits in promoter opening probed by photochemical cross-linking. J. Biol. Chem. 2003; 278:17912–17917. [DOI] [PubMed] [Google Scholar]
- 22. Chakraborty P., Grosse F.. WRN helicase unwinds Okazaki fragment-like hybrids in a reaction stimulated by the human DHX9 helicase. Nucleic Acids Res. 2010; 38:4722–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Braun B.R., Bartholomew B., Kassavetis G.A., Geiduschek E.P.. Topography of transcription factor complexes on the Saccharomyces cerevisiae 5 S RNA gene. J. Mol. Biol. 1992; 228:1063–1077. [DOI] [PubMed] [Google Scholar]
- 24. Fairman-Williams M.E., Guenther U.P., Jankowsky E.. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010; 20:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vorlander M.K., Khatter H., Wetzel R., Hagen W.J.H., Muller C.W.. Molecular mechanism of promoter opening by RNA polymerase III. Nature. 2018; 553:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li F., Xiong Y., Wang J., Cho H.D., Tomita K., Weiner A.M., Steitz T.A.. Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and its complexes with ATP or CTP. Cell. 2002; 111:815–824. [DOI] [PubMed] [Google Scholar]
- 27. Chiannilkulchai N., Stalder R., Riva M., Carles C., Werner M., Sentenac A.. RPC82 encodes the highly conserved, third-largest subunit of RNA polymerase C (III) from Saccharomyces cerevisiae. Mol. Cell Biol. 1992; 12:4433–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuldell N.H., Buratowski S.. Genetic analysis of the large subunit of yeast transcription factor IIE reveals two regions with distinct functions. Mol. Cell Biol. 1997; 17:5288–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meinhart A., Blobel J., Cramer P.. An extended winged helix domain in general transcription factor E/IIE alpha. J. Biol. Chem. 2003; 278:48267–48274. [DOI] [PubMed] [Google Scholar]
- 30. Grunberg S., Bartlett M.S., Naji S., Thomm M.. Transcription factor E is a part of transcription elongation complexes. J. Biol. Chem. 2007; 282:35482–35490. [DOI] [PubMed] [Google Scholar]
- 31. Miller G., Hahn S.. A DNA-tethered cleavage probe reveals the path for promoter DNA in the yeast preinitiation complex. Nat. Struct. Mol. Biol. 2006; 13:603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ohkuma Y., Hashimoto S., Wang C.K., Horikoshi M., Roeder R.G.. Analysis of the role of TFIIE in basal transcription and TFIIH-mediated carboxy-terminal domain phosphorylation through structure-function studies of TFIIE-alpha. Mol. Cell Biol. 1995; 15:4856–4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lucic B., Zhang Y., King O., Mendoza-Maldonado R., Berti M., Niesen F.H., Burgess-Brown N.A., Pike A.C., Cooper C.D., Gileadi O. et al.. A prominent beta-hairpin structure in the winged-helix domain of RECQ1 is required for DNA unwinding and oligomer formation. Nucleic Acids Res. 2011; 39:1703–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pike A.C., Shrestha B., Popuri V., Burgess-Brown N., Muzzolini L., Costantini S., Vindigni A., Gileadi O.. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. PNAS. 2009; 106:1039–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kitano K., Kim S.-Y., Hakoshima T.. Structural basis for DNA Strand separation by the unconventional Winged-Helix Domain of RecQ Helicase WRN. Structure. 2010; 18:177–187. [DOI] [PubMed] [Google Scholar]
- 36. Woodman I.L., Bolt E.L.. Winged helix domains with unknown function in Hel308 and related helicases. Biochem. Soc. Trans. 2011; 39:140–144. [DOI] [PubMed] [Google Scholar]
- 37. Plaschka C., Hantsche M., Dienemann C., Burzinski C., Plitzko J., Cramer P.. Transcription initiation complex structures elucidate DNA opening. Nature. 2016; 533:353–358. [DOI] [PubMed] [Google Scholar]
- 38. Nogales E., Louder R.K., He Y.. Cryo-EM in the study of challenging systems: the human transcription pre-initiation complex. Curr. Opin. Struct. Biol. 2016; 40:120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Abascal-Palacios G., Ramsay E.P., Beuron F., Morris E., Vannini A.. Structural basis of RNA polymerase III transcription initiation. Nature. 2018; 553:301–306. [DOI] [PubMed] [Google Scholar]
- 40. Grunberg S., Hahn S.. Structural insights into transcription initiation by RNA polymerase II. Trends Biochem. Sci. 2013; 38:603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bunick D., Zandomeni R., Ackerman S., Weinmann R.. Mechanism of RNA polymerase II–specific initiation of transcription in vitro: ATP requirement and uncapped runoff transcripts. Cell. 1982; 29:877–886. [DOI] [PubMed] [Google Scholar]
- 42. Carey M.F., Gerrard S.P., Cozzarelli N.R.. Analysis of RNA polymerase III transcription complexes by gel filtration. J. Biol. Chem. 1986; 261:4309–4317. [PubMed] [Google Scholar]
- 43. Alekseev S., Nagy Z., Sandoz J., Weiss A., Egly J.M., Le May N., Coin F.. Transcription without XPB Establishes a Unified Helicase-Independent mechanism of promoter opening in Eukaryotic gene expression. Mol. Cell. 2017; 65:504–514. [DOI] [PubMed] [Google Scholar]
- 44. Holstege F.C., Tantin D., Carey M., van der Vliet P.C., Timmers H.T.. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 1995; 14:810–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.