Abstract
Summary
Data visualization plays critical roles in proteomics studies, ranging from quality control of MS/MS data to validation of peptide identification results. Herein, we present PDV, an integrative proteomics data viewer that can be used to visualize a wide range of proteomics data, including database search results, de novo sequencing results, proteogenomics files, MS/MS data in mzML/mzXML format and data from public proteomics repositories. PDV is a lightweight visualization tool that enables intuitive and fast exploration of diverse, large-scale proteomics datasets on standard desktop computers in both graphical user interface and command line modes.
Availability and implementation
PDV software and the user manual are freely available at http://pdv.zhang-lab.org. The source code is available at https://github.com/wenbostar/PDV and is released under the GPL-3 license.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS) is the leading technique in proteomics, and it generates large volumes of data. Analyzing the raw data requires many different tools that produce additional data in various formats. These formats include open format MS/MS data (such as mzML and mzXML) converted from raw MS/MS data, protein identification results from database searching or de novo sequencing and proteomic data from public databases such as PRIDE (Jones, 2006) or spectral libraries such as PeptideAtlas (Desiere et al., 2006). Visualization at each stage of the analysis is critical in exploiting and understanding the information derived from these data (Oveland et al., 2015).
Currently, several tools are available to visualize proteomics data, such as TOPPView (Sturm and Kohlbacher, 2009), PRIDE Inspector (Wang et al., 2012), MS-Viewer (Baker and Chalkley, 2014) and BatMass (Avtonomov et al., 2016). TOPPView provides a graphical user interface (GUI) for visualizing MS/MS data. PRIDE Inspector is a Java-based tool for visualizing MS/MS data and protein identification results in PRIDE XML or mzIdentML formats. MS-Viewer is a web-based tool for visualizing protein identification results from database searching. However, MS-Viewer cannot handle de novo sequencing results and cannot visualize MS/MS data in mzML or mzXML format. BatMass is a recently published tool for visualizing MS/MS data in mzML or mzXML format. All of these tools are originally designed for the visualization of a specific data type. However, a proteomics study commonly requires visualizing data at different stages and in multiple formats, requiring researchers to use multiple tools. Furthermore, most tools are only available as a GUI and do not feature a batch mode. This makes the efficient generation of high quality figures for large scale datasets and implementation in other pipelines difficult.
Here, we propose PDV, a standalone software programmed in Java that can be used to visualize different kinds of proteomics data in both GUI and command line modes. The functions of PDV include visualization of open format MS/MS data, database search results, de novo sequencing results, proteogenomics files and data from public databases such as PRIDE and PeptideAtlas. In addition, PDV provides an interface for using proteoQC to generate various quality control figures and tables for assessing proteomic data quality.
2 Features and implementation
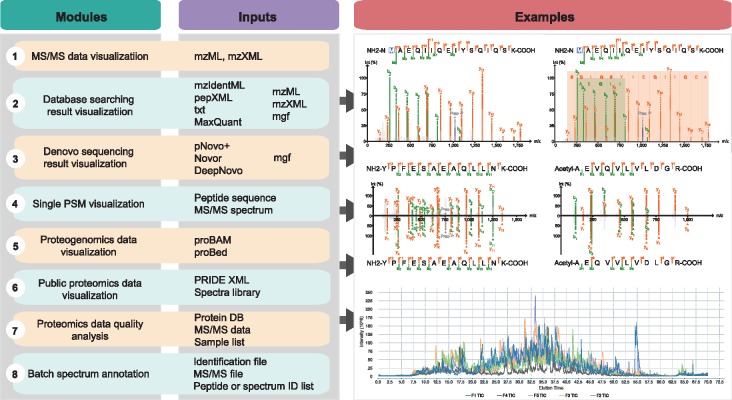
PDV is platform independent and written in Java 1.8. It can be run in GUI or command-line mode. The functions of PDV can be divided into the following modules as illustrated in Figure 1.
Fig. 1.
An overview of PDV
2.1 MS/MS data visualization module
PDV supports input of MS/MS data in mzML or mzXML format for visualization. It supports visualizing multiple files at once. PDV displays a table that includes meta information about the MS/MS data and a panel with the total ion current (TIC) chromatogram enabling the assessment of the chromatographic performance by visualizing the intensity distribution over the retention time (Supplementary Fig. S1). The MSDK library (https://github.com/msdk/msdk) is used to quickly extract TIC data from mzML or mzXML files.
2.2 Database searching result visualization module
PDV accepts identification result files in the mzIdentML standard format (Jones et al., 2012), the pepXML format, or a tab-delimited text format. The tab-delimited text file requires four columns: spectrum index, peptide sequence, charge state and modification information. In order to generate a spectrum annotation figure, an MS/MS data file in MGF, mzXML, or mzML format is also needed. The identification result from the widely used MaxQuant (Cox and Mann, 2008) software is also supported. In order to quickly visualize the results, multithreading technology and SQL database technology are used. In addition, if a user has an MS/MS spectrum from a synthetic peptide which is also identified by a spectrum in the input identification file, PDV provides a function to support direct visual comparison of the two spectra matching to the same peptide (Supplementary Fig. S2). The spectrum visualization and annotation in PDV is mainly based on the compomics-utilities library (Barsnes et al., 2011). Both MSDK (https://github.com/msdk/msdk) and MSFTBX (https://github.com/chhh/MSFTBX) libraries were used to access spectra data in mzML or mzXML file.
2.3 De novo sequencing result visualization module
De novo sequencing is a popular technique in proteomics for identifying peptides from tandem mass spectra without relying on a protein sequence database. PDV can import the identification results from PepNovo (Frank and Pevzner, 2005), pNovo+ (Chi et al., 2013), Novor (Ma, 2015) and DeepNovo (Tran et al., 2017). The basic module of this function is developed based on DeNovoGUI (Muth et al., 2014). The result visualization panel includes a table presenting the result for each spectrum and a panel to present the spectrum annotation figure (Supplementary Fig. S3).
2.4 Single PSM visualization module
In order to visualize a single PSM, PDV provides a separate panel allowing users to input a peptide sequence and an MS/MS spectrum in the MGF format (Supplementary Fig. S4). In this panel, users can manually set modifications that occurred in the peptide sequence. All modifications from Unimod are available (Supplementary Fig. S5). In addition, the user can tune the spectrum annotation by changing the peptide spectrum matching parameters.
2.5 Proteogenomics data visualization module
Two new standard formats, proBAM and proBed, were recently released to facilitate the integration of genomics, transcriptomics, and protoemics data in proteogenomics studies (Menschaert et al., 2018). PDV supports input of proBAM and proBed files thanks to the Htsjdk library (https://github.com/samtools/htsjdk) (Supplementary Fig. S6).
2.6 Public proteomics data visualization module
Proteomics data from the PRIDE repository in the PRIDE XML format and PeptideAtlas in sptxt format are supported. The visualization panel is similar to the database searching visualization panel (Supplementary Fig. S7).
2.7 Data quality analysis module
PDV provides an interface for using proteoQC (bioconductor.org/packages/proteoQC) for proteomics data quality assessment (Supplementary Fig. S8). It accepts MS/MS data (MGF, mzML or mzXML) and a protein database and generates an HTML-based report that includes identification-free (ID-free) metrics and identification-based (ID-based) metrics within a single experiment, as well as across multiple experiments.
2.8 Batch spectrum annotation module
PDV provides a command line module to produce figures of annotated spectra or TIC in batch mode. It can be used to generate figures according to a list of peptide sequences or a list of spectrum indexes. This function is especially useful when requiring the generation of a large number of high quality figures for publication (Wang et al., 2018).
3 Results and discussion
We have developed a fast and easy-to-use software named PDV for visualization of different kinds of proteomics data. The PDV GUI enables users lacking programing experience to visualize proteomics data interactively while the command line interface allows users to generate annotated spectra or TIC figures in batch mode for large-scale datasets. Furthermore, the use of multi-threading and SQL database technologies in PDV enables efficient processing of large datasets. For example, loading a 2 GB mzIdentML file takes less than 15 s and loading 20 raw files (20 GB) takes about 30 s with a Windows computer (Intel Core i3-7100, 8 GB of RAM and 256 GB of SanDisk X400 SSD). We anticipate that researchers from the proteomics community will benefit from PDV for the interpretation and validation of proteomics data.
Supplementary Material
Acknowledgements
The authors thank Dr. Harald Barsnes for his helpful suggestions for PDV development. They also thank Dr. Sara Savage for proof-reading the manuscript.
Funding
This work was supported by grant U24CA210954 from the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC), by grant CPRIT RR160027 from the Cancer Prevention and Research Institutes of Texas, by funding from the McNair Medical Institute at The Robert and Janice McNair Foundation and by funding from the National Key R&D Program of China (2017YFC0908400).
Conflict of Interest: none declared.
References
- Avtonomov D.M. et al. (2016) BatMass: a Java software platform for LC-MS data visualization in proteomics and metabolomics. J. Proteome Res., 15, 2500–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P.R., Chalkley R.J. (2014) MS-viewer: a web-based spectral viewer for proteomics results. Mol. Cell Proteomics, 13, 1392–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsnes H. et al. (2011) compomics-utilities: an open-source Java library for computational proteomics. BMC Bioinformatics, 12, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H. et al. (2013) pNovo+: de novo peptide sequencing using complementary HCD and ETD tandem mass spectra. J. Proteome Res., 12, 615–625. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol., 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Desiere F. et al. (2006) The PeptideAtlas project. Nucleic Acids Res., 34, D655–D658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank A., Pevzner P. (2005) PepNovo: de Novo peptide sequencing via probabilistic network modeling. Anal. Chem., 77, 964–973. [DOI] [PubMed] [Google Scholar]
- Jones A.R. et al. (2012) The mzIdentML data standard for mass spectrometry-based proteomics results. Mol. Cell Proteomics, 11, M111 014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. (2006) PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucleic Acids Res., 34, D659–D663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B. (2015) Novor: real-time peptide de novo sequencing software. J. Am. Soc. Mass Spectrom, 26, 1885–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menschaert G. et al. (2018) The proBAM and proBed standard formats: enabling a seamless integration of genomics and proteomics data. Genome Biol., 19, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth T. et al. (2014) DeNovoGUI: an open source graphical user interface for de novo sequencing of tandem mass spectra. J. Proteome Res., 13, 1143–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oveland E. et al. (2015) Viewing the proteome: how to visualize proteomics data? Proteomics, 15, 1341–1355. [DOI] [PubMed] [Google Scholar]
- Sturm M., Kohlbacher O. (2009) TOPPView: an open-source viewer for mass spectrometry data. J. Proteome Res., 8, 3760–3763. [DOI] [PubMed] [Google Scholar]
- Tran N.H. et al. (2017) De novo peptide sequencing by deep learning. Proc. Natl. Acad. Sci., 114, 8247–8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. et al. (2012) PRIDE Inspector: a tool to visualize and validate MS proteomics data. Nat. Biotechnol., 30, 135–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. (2018) Detection of proteome diversity resulted from alternative splicing is limited by trypsin cleavage specificity. Mol. Cell Proteomics, 17, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.