Abstract
Motivation
Comparison of RNA 3D structures can be used to infer functional relationship of RNA molecules. Most of the current RNA structure alignment programs are built on size-dependent scales, which complicate the interpretation of structure and functional relations. Meanwhile, the low speed prevents the programs from being applied to large-scale RNA structural database search.
Results
We developed an open-source algorithm, RNA-align, for RNA 3D structure alignment which has the structure similarity scaled by a size-independent and statistically interpretable scoring metric. Large-scale benchmark tests show that RNA-align significantly outperforms other state-of-the-art programs in both alignment accuracy and running speed. The major advantage of RNA-align lies at the quick convergence of the heuristic alignment iterations and the coarse-grained secondary structure assignment, both of which are crucial to the speed and accuracy of RNA structure alignments.
Availability and implementation
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The biological functions of non-coding RNAs depend on their tertiary structures. RNAs with similar fold may exert similar functions. For example, riboswitches performing similar function of transcription termination share structural similarity near the termination region (Gong et al., 2015). In light of the implication of structure to function, several RNA structure alignment programs have been developed to infer function similarity or to assess quality of predicted RNA structures. For instance, ARTS (Dror et al., 2006) and STAR3D (Ge and Zhang, 2015) identify seed matches of conserved stack regions, followed by the extension of the seed matches to global alignments. SARA (Capriotti and Marti-Renom, 2008) minimizes the root-mean-square deviation (RMSD) between unit vector representations of RNA structures. Rclick (Nguyen et al., 2017) matches RNA backbones represented as cliques. Despite the usefulness, these programs share two caveats. First, the scoring functions are usually built on base pairing scores and RMSD, both of which are dependent on the length of aligned RNAs. Such length dependency renders the absolute value of the structure similarity meaningless. RMalign (Jinfang et al., 2018) attempts to address this issue by a normalization factor derived from radius of gyration statistics, but the resulting scoring function is still slightly dependent on the RNA length. Second, the alignments in these programs are mainly derived from RNA secondary structure (SS) assignment based on full-atomic structure. Such reliance not only results in cumbersome dependences on third-party SS assignment packages, but also makes the alignment over-sensitive to atomic details and irregularities of the base pairs.
To address these issues, we developed RNA-align. To our knowledge, it is the first RNA alignment tool whose scoring function is truly independent of the length of RNA. RNA-align starts from initial alignments of both coarse-grain SS descriptors and gapless sliding, which ensure reasonable alignments can be generated for RNA structure pairs lacking significant similarity in base stacking pattern.
2 Materials and methods
As an extension of the TM-align program (Zhang and Skolnick, 2005), RNA-align measures structure similarity between RNA structure pairs by TM-scoreRNA, which is inspired by the standard TM-score for protein structure comparison (Zhang and Skolnick, 2004):
| (1) |
Here, is the length of the target RNA and the number of aligned nucleotides. is the distance between the ith aligned pair of nucleotides. By default, RNA-align is on C3’ atoms but the program provides an option for user to specify other backbone atoms (Supplementary Fig. S1A). is a scaling factor to ensure the score of random RNA pairs is independent of RNA length (Supplementary Material):
| (2) |
To construct structural alignments, RNA-align first creates three initial alignments by (i) gapless sliding of one sequence along another; (ii) dynamic programing (DP) alignment of SS (Supplementary Fig. S2); (ii) DP on a 50/50 combination of the SS score and the distance score of the first initial alignment. Starting from each initial alignment, iterative alignment refinement is performed. Each iteration derives a new rotation matrix and a translation vector to optimally superpose the previously aligned nucleotides of the two RNAs; a new alignment is then computed by DP using the distance score from the superposed structures. Here, ‘alignment’ refers to establishment of nucleotide equivalence between the two sequences, while ‘superposition’ to the rotation and translation operation to overlay two structures by maximizing the TM-scoreRNA. The alignment-superposition iterations are repeated until the alignment converges, where the alignment with the highest TM-scoreRNA is the final alignment.
3 Results
3.1 TM-scoreRNA is a size-independent metric to quantify RNA structure similarity
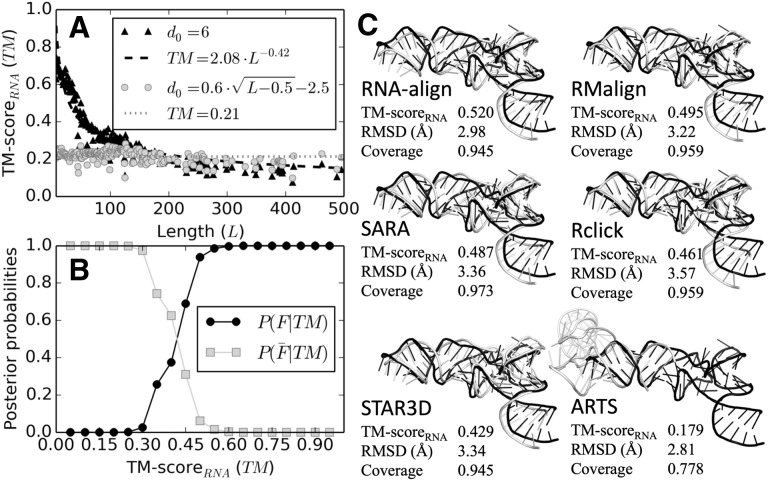
The parameters to calculate the scaling factor are derived from 6 571 496 random RNA pairs from the PDB. Average TM-scoreRNA has no dependence on the length of the RNA structures (Fig. 1A).
Fig. 1.
(A) TM-scoreRNA versus length, calculated on 6 571 496 RNA pairs with sequence identity <0.4. A constant d0 results in raw TM-scoreRNA (black triangles) with a power law dependence on length (black dash line for fitted curve), while a length-normalized d0 results in length-independent TM-scoreRNA (grey circles). The average TM-scoreRNA with normalized d0 is 0.21 (grey dotted line). (B) Posterior probabilities of an RNA pair with a given TM-scoreRNA belonging to the same (black) or different (grey) Rfam families. (C) 3D structure alignments for two tRNAs (Rfam family RF00005) with PDB IDs 4kzz Chain-j (grey) and 5mrc Chain-bb (black). TM-scoreRNA and Coverage (number of aligned nucleotides divided by length) are normalized by length of 5mrc Chain-bb
To investigate the ability of TM-scoreRNA in RNA family assignment, we tested RNA-align on 463 203 RNA pairs from Rfam (Kalvari et al., 2018). Given a TM-scoreRNA, the posterior probability (Supplementary Material) of an RNA pair belonging to the same Rfam Family follows a sigmoid curve with a sharp transition at TM-scoreRNA = 0.45 (Fig. 1B). A similar transition was also observed in protein structure alignment but at the value of TM-score = 0.5 (Xu and Zhang, 2010).
3.2 Fast and accurate alignment by RNA-align enabled by coarse-grain SS assignment
RNA-align is tested on all-to-all alignment of 687 RNA structures, in control with five state-of-the-art RNA structure alignment programs (ARTS, Rclick, RMalign, SARA and STAR3D). Although RNA-align generate results for all 235 641 RNA pairs, SARA, STAR3D, ARTS and Rclick failed to generate an alignment for 10.2, 60.0, 65.5 and 0.006% of the pairs (Supplementary Table S1). For the common subset of 76 067 RNA pairs with results from all programs, RNA-align achieves an average TM-scoreRNA 7.0% higher than that from the best control algorithm (RMalign), while being 38.3 times faster than the latter. Figure 1C illustrates an example to align two tRNAs, where the RNA-align alignment has the highest TM-scoreRNA (0.520) among all alignments.
In addition to the iterative refinement procedure, the high accuracy of RNA-align can be partially attributed to the SS assignment. Supplementary Figure S3 shows a case study where the use of SS results in completely different alignments from that without SS: TM-scoreRNA of the former is 168% higher than the latter. Here, RNA-align uses a simple strategy to deduce SS from C3’ atom distances and base types (Supplementary Fig. S1). The SS assignment takes negligible time but has an appreciable accuracy of 87.6% on 4950 RNA structures from PDB, including those with pseudoknots (Supplementary Fig. S4).
3.3 The webserver interface of RNA-align
The RNA-align webserver, source code and dataset are freely available at https://zhanglab.ccmb.med.umich.edu/RNA-align/. The only required server inputs are two RNA structures in PDB or PDBx/mmCIF format. The output webpage contains the nucleotide alignment, TM-scoreRNA, and an interactive applet to visualize the superposed structures. Additionally, an option is provided to allow alignments sticking to the original nucleotide index to facilitate direct superposition of two structures when needed. Since DNA has similar local structure elements, the program can also align DNA structures.
Supplementary Material
Acknowledgements
We thank Dr Wei Zheng and Dr Xiaoqiong Wei for discussions.
Funding
The work was supported in part by the National Natural Science Foundation of China [31600592], the National Institutes of Health [GM083107, GM116960 and AI134678], the National Science Foundation [DBI1564756] and the Extreme Science and Engineering Discovery Environment (XSEDE).
Conflict of Interest: none declared.
References
- Capriotti E., Marti-Renom M.A. (2008) RNA structure alignment by a unit-vector approach. Bioinformatics, 24, I112–I118. [DOI] [PubMed] [Google Scholar]
- Dror O. et al. (2006) The ARTS web server for aligning RNA tertiary structures. Nucleic Acids Res., 34, W412–W415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge P., Zhang S.J. (2015) STAR3D: a stack-based RNA 3D structural alignment tool. Nucleic Acids Res., 43, e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S. et al. (2015) The regulation mechanism of yitJ and metF riboswitches. J. Chem. Phys., 143, 045103. [DOI] [PubMed] [Google Scholar]
- Jinfang Z. et al. (2018) RMalign: an RNA structural alignment tool based on a novel scoring function RMscore. BMC genomics, 20, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I. et al. (2018) Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res., 46, D335–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M.N. et al. (2017) Topology independent comparison of RNA 3D structures using the CLICK algorithm. Nucleic Acids Res., 45, e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.R., Zhang Y. (2010) How significant is a protein structure similarity with TM-score=0.5? Bioinformatics, 26, 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Skolnick J. (2004) Scoring function for automated assessment of protein structure template quality. Proteins, 57, 702–710. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Skolnick J. (2005) TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res., 33, 2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.