See Hansen and Møller (doi:10.1093/brain/awz318) for a scientific commentary on this article.
Using polygenic risk scores from a genome-wide association study in generalized and focal epilepsy, Leu et al. reveal a significantly higher genetic burden for epilepsy in multiple cohorts of people with epilepsy compared to population controls. Quantification of common variant burden may be valuable for epilepsy prognosis and treatment.
Keywords: epilepsy, genetics, genetic generalized epilepsy, common variant risk
Abstract
Rare genetic variants can cause epilepsy, and genetic testing has been widely adopted for severe, paediatric-onset epilepsies. The phenotypic consequences of common genetic risk burden for epilepsies and their potential future clinical applications have not yet been determined. Using polygenic risk scores (PRS) from a European-ancestry genome-wide association study in generalized and focal epilepsy, we quantified common genetic burden in patients with generalized epilepsy (GE-PRS) or focal epilepsy (FE-PRS) from two independent non-Finnish European cohorts (Epi25 Consortium, n = 5705; Cleveland Clinic Epilepsy Center, n = 620; both compared to 20 435 controls). One Finnish-ancestry population isolate (Finnish-ancestry Epi25, n = 449; compared to 1559 controls), two European-ancestry biobanks (UK Biobank, n = 383 656; Vanderbilt biorepository, n = 49 494), and one Japanese-ancestry biobank (BioBank Japan, n = 168 680) were used for additional replications. Across 8386 patients with epilepsy and 622 212 population controls, we found and replicated significantly higher GE-PRS in patients with generalized epilepsy of European-ancestry compared to patients with focal epilepsy (Epi25: P = 1.64×10−15; Cleveland: P = 2.85×10−4; Finnish-ancestry Epi25: P = 1.80×10−4) or population controls (Epi25: P = 2.35×10−70; Cleveland: P = 1.43×10−7; Finnish-ancestry Epi25: P = 3.11×10−4; UK Biobank and Vanderbilt biorepository meta-analysis: P = 7.99×10−4). FE-PRS were significantly higher in patients with focal epilepsy compared to controls in the non-Finnish, non-biobank cohorts (Epi25: P = 5.74×10−19; Cleveland: P = 1.69×10−6). European ancestry-derived PRS did not predict generalized epilepsy or focal epilepsy in Japanese-ancestry individuals. Finally, we observed a significant 4.6-fold and a 4.5-fold enrichment of patients with generalized epilepsy compared to controls in the top 0.5% highest GE-PRS of the two non-Finnish European cohorts (Epi25: P = 2.60×10−15; Cleveland: P = 1.39×10−2). We conclude that common variant risk associated with epilepsy is significantly enriched in multiple cohorts of patients with epilepsy compared to controls—in particular for generalized epilepsy. As sample sizes and PRS accuracy continue to increase with further common variant discovery, PRS could complement established clinical biomarkers and augment genetic testing for patient classification, comorbidity research, and potentially targeted treatment.
See Hansen and Møller (doi:10.1093/brain/awz318) for a scientific commentary on this article.
Introduction
Epilepsy is a common chronic neurological disorder, affecting approximately 1% of individuals (Ngugi et al., 2010). Lifetime prevalence is 8–10% for a seizure and 3–4% for epilepsy (Hesdorffer et al., 2011). The median incidence of epilepsy is 50 per 100 000 person-years (Ngugi et al., 2011). Individuals at high risk for recurrent seizures (epilepsy) benefit from early antiseizure drug treatment, compared to no treatment or delayed treatment (Kim et al., 2006). Predicting whether an individual will develop epilepsy after the first epileptic seizure is difficult (MacDonald et al., 2000; Bell et al., 2016), with recurrence risk varying from 27% to 71% (Hopkins et al., 1988; Berg and Shinnar, 1991; Kwan and Sander, 2004).
Epileptic seizures either have a generalized (involving both cerebral hemispheres) or a focal (originating from one cerebral hemisphere) onset (Scheffer et al., 2017). Generalized epilepsies account on average for 54%, focal epilepsies for 40%, and unclassifiable epilepsies for 7% of incident epilepsies in population-based studies of all ages (Banerjee et al., 2009). Distinguishing between the two types of epilepsy can be difficult: focal epilepsy can present with bilateral tonic-clonic seizures (secondary-generalization), patients with generalized epilepsy can have focal features on EEG (Japaridze et al., 2016), and some individuals have a mix of focal and generalized epilepsy (Scheffer et al., 2017). Since commonly used antiseizure drugs for focal epilepsy can be ineffective or exacerbate generalized epilepsies, differentiating between focal and generalized epilepsy is important (Japaridze et al., 2016). Hence, there is a clinical need for biomarkers that can help to distinguish individuals at high versus low risk to develop either focal or generalized epilepsy.
Genetic factors can explain a substantial portion of cases of epilepsy, particularly severe epilepsy (EpiPM Consortium et al., 2015). For rare and early onset childhood epilepsies, >100 epilepsy-related genes have been discovered in recent years (Heyne et al., 2019). The identified genetic variants are rare and of large effect, ranging from large deletions that confer on average ∼7-fold risk for epilepsy (Pérez-Palma et al., 2017) to single, causative de novo variants in >33 genes (Heyne et al., 2018). These variants are diagnostically relevant and can influence patient management. For example, treatment with sodium channel blockers can exacerbate seizures in patients with Dravet syndrome or other early-onset epileptic syndromes caused by SCN1A mutations, whereas these drugs are beneficial in patients with gain-of-function variants in SCN2A (Guerrini et al., 1998; Löscher, 2009; Wolff et al., 2017). While rare variation of large effect has a clear impact in clinical practice for rare epilepsy syndromes (McTague et al., 2016), patients affected by common types of epilepsy rarely carry such variants and routine genetic testing is therefore not established for the common epilepsies.
Genome-wide association studies (GWAS) for common forms of epilepsy have identified common genetic risk variants for generalized epilepsy, focal epilepsy, and febrile seizures (Kasperavičiūtė et al., 2013; Feenstra et al., 2014; International League Against Epilepsy Consortium on Complex Epilepsies, 2014, 2018). Common genetic risk variants associated with a disease are usually of small effect size (1.33 median odds ratio) (Hindorff et al., 2009) and cannot individually quantify risk or to inform prognosis and treatment. However, polygenic risk scores (PRSs) that combine the effect sizes of thousands of variants into a single score can stratify affected and healthy individuals. For five common disorders, a recent study showed a 3- to 5-fold increased risk for patients with a high disease-specific PRS, similar to the range of risk conferred by rare monogenic variants, such as LDLR missense variants for coronary artery disease or rare BRCA variants in breast cancer (Khera et al., 2018). Based on these results, the authors proposed that PRS-based prediction may be reliable enough to consider their utility in clinical practice.
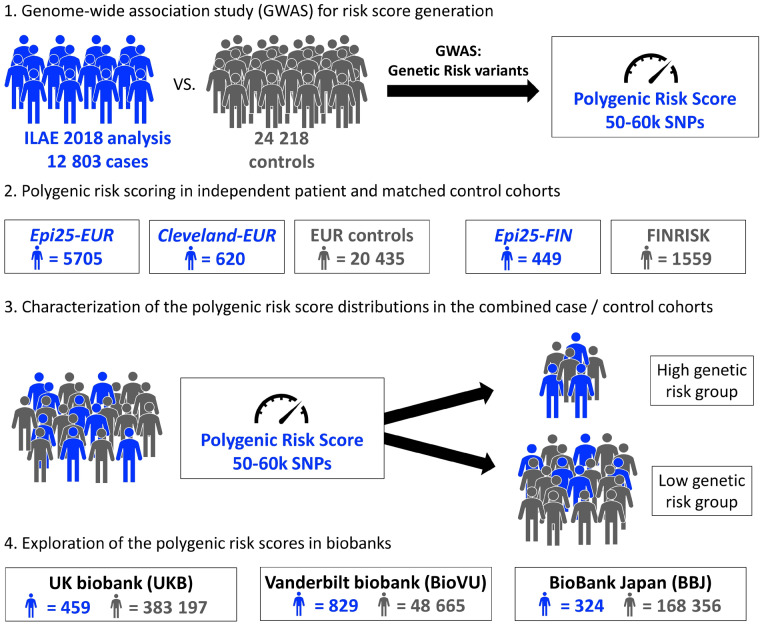
Genome-wide PRS based on thousands of common variants associated with epilepsy may help distinguish healthy individuals from those who develop epilepsy (Speed et al., 2014). However, no studies have directly investigated whether PRSs derived from well-phenotyped cohorts stratify patients in clinical practice or population-based cohort studies. Here, we calculate PRSs for the two main subtypes of epilepsy (generalized and focal) from the largest GWAS in epilepsy to date (International League Against Epilepsy Consortium on Complex Epilepsies, 2018) and (i) quantify the burden of PRSs derived from GWAS studies of well-phenotyped cohorts in patients with generalized or focal epilepsy; (ii) explore if PRS can differentiate patients with generalized from those with focal epilepsy; and (iii) explore if patients with generalized or focal epilepsy are enriched particularly in the upper extreme of the PRS burden distribution compared to controls. Our overall study design is presented in Fig. 1. Across two independent research cohorts, one clinically ascertained cohort, and three biobanks (repositories with clinical data and DNA samples available for research); data from 630 598 individuals were available for the PRS analyses.
Figure 1.
Study design. (1) PRSs for the two main classes of epilepsy (generalized and focal) were derived from the largest GWAS in epilepsy to date (International League Against Epilepsy Consortium on Complex Epilepsies, 2018). (2) PRS were calculated in patients with generalized or focal epilepsy and in population controls and (3) tested in their ability to identify significant differences of common variant burden among groups. (4) The UK and Vanderbilt biobanks were available to test the behaviour of the PRSs in individuals ascertained by ICD-10 codes for epilepsy, while the Biobank Japan was available to test the performance in a non-European population. SNP = single nucleotide polymorphism.
Patients and methods
Study cohorts
Patients of European ancestry with generalized epilepsy or focal epilepsy were recruited through the Epi25 project (http://epi-25.org/), an international multicentre epilepsy genetics research consortium [exploration cohorts for generalized (GE) and focal (FE) epilepsy: GE-Epi25-EUR and FE-Epi25-EUR, respectively] and from a single clinical centre, the Cleveland Clinic Epilepsy Center (replication cohorts for generalized and focal epilepsy: GE-Cleveland-EUR and FE-Cleveland-EUR, respectively). A Finnish-ancestry population isolate was recruited from the Epi25 project (GE-Epi25-FIN and FE-Epi25-FIN, respectively). Ancestry-matched population controls were recruited from several in-house projects, the Partners HealthCare Biobank (Karlson et al., 2016), and the FINRISK study (Borodulin et al., 2017). Three large-scale biobank repositories [UKB: UK Biobank (Sudlow et al., 2015); BioVU: Vanderbilt University biorepository (Roden et al., 2008); and BBJ: BioBank Japan (Nagai et al., 2017)] were used for additional explorations. All cohorts, totalling 630 598 individuals, are detailed in Table 1 and the Supplementary material.
Table 1.
Study cohorts after quality control
| Cohort name | Ascertainment type | Ethnicity | Generalized epilepsy (GE) | Focal epilepsy (FE) | Controls |
|---|---|---|---|---|---|
| Epi25-EUR | Research | EUR | 2256 | 3449 | 20 435 |
| Cleveland-EUR | Clinic | EUR | 85 | 535 | 20 435 |
| Epi25-FIN | Research | FIN | 112 | 337 | 1559 |
| UKB | Biobank | EUR | 246 | 213 | 383 197 |
| BioVU | Biobank | EUR | 293 | 536 | 48 665 |
| BBJ | Biobank | JPN | 219 | 105 | 168 356 |
Generalized and focal epilepsy were diagnosed in the Epi25-EUR, Cleveland-EUR, Epi25-FIN, and BBJ cohorts according to clinical criteria (clinical interview, neurological examination, EEG, imaging data). For the UK and BioVU biobanks, ICD-10 G40.3 codes were used to identify people with generalized epilepsy, and G40.0 to G40.2 codes to identify people with focal epilepsy.
BBJ = BioBank Japan; BioVU = Vanderbilt University biorepository; Cleveland-EUR = European-ancestry Cleveland Clinic Epilepsy Center cohort; Epi25-EUR = European-ancestry Epi25 cohort; Epi25-FIN = Finnish-ancestry Epi25 cohort; UKB = UK Biobank.
Polygenic risk scoring in the study cohorts
Single-nucleotide polymorphism (SNP) weights for PRS were derived from summary statistics of the ILAE Consortium on Complex Epilepsies GWAS for generalized and focal epilepsy (International League Against Epilepsy Consortium on Complex Epilepsies, 2018). SNP weights for negative control PRS were derived from the UKB GWAS for type 2 diabetes for all cohorts excluding the UKB, and from the DIAGRAM-type 2 diabetes GWAS (Scott et al., 2017) for the UKB. PRS for each individual were generated using the allelic scoring function, as implemented in PLINK v1.9 (Chang et al., 2015). Individual PRSs were calculated as the sum of weighted effect alleles divided by the number of SNPs in the analysis. We generated the PRSs at the P-value threshold 0.5, found to be the best predicting threshold in a random split (80% training, 20% validation) of our exploration cohort (Epi25-EUR, Supplementary material 4.8, Supplementary Tables 4 and 5). We excluded individuals if their data were included in the GWAS studies used for PRS development. Details of the method to detect overlapping individuals across cohorts and the SNP quality control applied are given in the Supplementary material.
Statistical analysis
We used logistic regression adjusted for sex and for the first four principal components of ancestry to determine the ability of PRS to stratify cases from controls. The proportion of phenotypic variance explained by PRS was calculated using Nagelkerke’s pseudo-R2, by comparing the full model of the logistic regression (PRS plus all covariates: sex and the first four principal components of ancestry) to the null model (covariates only). Following the example of Khera et al. (2018) we assessed the enrichment of the two epilepsy phenotypes (generalized epilepsy or focal epilepsy) in progressively more extreme tails of the PRS distribution (top 20%, 5%, 0.5%) against the remainder of the distribution in a logistic regression model predicting disease status, adjusted for sex and the first four principal components of ancestry. The threshold for statistical significance after Bonferroni correction was set to α = 1.67 × 10−2 (three tests per cohort). Fixed-effect meta-analysis, with adjustment for the effective sample size, was performed using METAL (Willer et al., 2010).
Data availability
The data that support the findings of this study are available from the Epi25 Consortium, upon reasonable request. The biobank data are available from the UKB, BioVU, and BBJ upon successful project application.
Results
Higher PRS burden in patients with epilepsy compared to controls
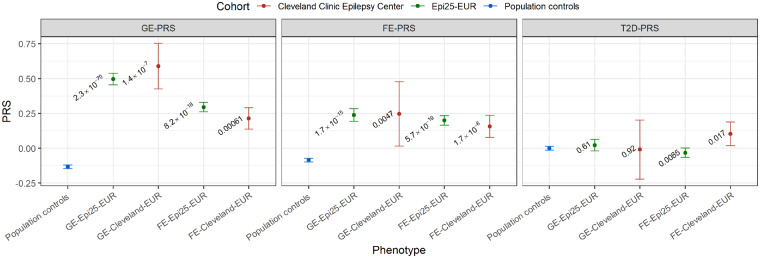
To determine if common variants associated with epilepsy are enriched in independent cohorts of patients with generalized epilepsy or focal epilepsy compared to population controls, we conducted a PRS analysis in two independent epilepsy cohorts of European ancestry. We found that in the GE-Epi25-EUR cohort, genome-wide polygenic risk scores for generalized epilepsy (GE-PRS) were significantly higher in patients with generalized epilepsy (n = 2256 cases) than in population controls (n = 20 435 controls; P = 2.35 × 10−70; Fig. 2 and Supplementary Table 1). GE-PRS explained 2.8% of the total phenotypic variance (composed of genetic, environmental, and genetic-environmental interaction variances) among the case and control group of the GE-Epi25-EUR cohort (Supplementary Table 1). This observation was replicated in the clinical GE-Cleveland-EUR cohort (P = 1.43×10−7; n = 85 cases; 2.6% phenotypic variance explained). In the FE-Epi25-EUR cohort, the genome-wide polygenic risk for focal epilepsy (FE-PRS) was significantly higher in patients with focal epilepsy (n = 3449 cases) than in population controls (P = 5.74×10−19), with 0.6% of the phenotypic variance explained. This observation was replicated in the clinical FE-Cleveland-EUR cohort (n = 535; P = 1.69×10−6; 0.5% phenotypic variance explained). As expected, PRSs for type 2 diabetes (negative control) were not significantly higher in patients with generalized epilepsy or focal epilepsy than in population controls (Fig. 2 and Supplementary Table 1).
Figure 2.
Genome-wide polygenic risk for generalized epilepsy or focal epilepsy in the exploration and replication cohorts. Shown are the means of the standardized GE-, FE-, and type 2 diabetes-PRS with 95% confidence intervals for the European-ancestry population controls (highlighted in blue; n = 20 435), the European-ancestry generalized epilepsy and focal epilepsy Epi25 exploration cohorts (highlighted in green; GE-Epi25-EUR, n = 2256; FE-Epi25-EUR, n = 3449), and the European-ancestry generalized epilepsy and focal epilepsy Cleveland Clinic replication cohorts (highlighted in red; GE-Cleveland-EUR, n = 85; FE-Cleveland-EUR, n = 535). The P-values for the differences between cases and population controls are given as numbers. The threshold for statistical significance after Bonferroni correction was set to α = 1.67 × 10−2 (three tests per cohort).
To test the utility of PRSs across different populations, we investigated the power of the PRS derived from the European population in the isolated Finnish population. The GE-PRS was significantly higher in patients with generalized epilepsy (n = 112 cases) than in the population controls (n = 1559 controls; P = 3.11×10−4; Supplementary Fig. 2 and Supplementary Table 2). However, the PRSs explained less phenotypic variance than generalized epilepsy cohorts of European ancestry (2% phenotypic variance explained). The FE-PRS were not significantly different between Finnish patients with focal epilepsy (n = 337 cases) and controls (P = 0.55).
Higher PRS burden in generalized compared to focal epilepsy
To determine if common variants associated with generalized epilepsy are enriched in patients with generalized epilepsy compared to patients with focal epilepsy, we regressed the GE-PRS against the diagnosis of generalized epilepsy or focal epilepsy. In the Epi25-EUR cohort, GE-PRSs were significantly higher in patients with generalized epilepsy than in those with focal epilepsy (P = 1.64×10−15), explaining 1.7% of the phenotypic variance (Supplementary Table 1). This observation was replicated in the Cleveland-EUR cohort (P = 2.85×10−4, 3.9% phenotypic variance explained) and the Epi25-FIN cohort (P = 1.80×10−4, 4.6% phenotypic variance explained, Supplementary Table 2). Overall, the PRS had the most predictive power in the corresponding epilepsy phenotype group: GE-status was best predicted by GE-PRS (P = 2.35×10−70, 2.8% phenotypic variance explained in Epi25-EUR) over FE-PRS (P = 1.71×10−15, 0.6% phenotypic variance explained) and FE-status was best predicted by FE-PRS (P = 5.74×10−19, 0.6% phenotypic variance explained in Epi25-EUR) over GE-PRS (P = 8.21×10−18, 0.5% phenotypic variance explained, Supplementary Table 1).
Enrichment of patients with epilepsy in the highest PRS burden percentile
To explore if the GE- and FE-PRS enrichment in patients with epilepsy is due to a few patients with a very high burden or due to many with a slightly elevated burden, we characterized the epilepsy PRS distribution in the European-ancestry cohorts. Patients with epilepsy and population control subjects were ranked according to their PRSs and tested for enrichment of patients with generalized epilepsy or focal epilepsy in the extreme tails of the PRS distribution. Strikingly, in the combined GE-Epi25-EUR and control cohorts, we observed a significant 4.63-fold enrichment of patients with generalized epilepsy in the group with the highest GE-PRS (top 0.5%, P = 2.60×10−15; involving 2.39% of the GE-Epi25-EUR cohort; Table 2). This observation was replicated in the clinical GE-Cleveland-EUR cohort (4.47-fold enrichment; P = 1.39×10−2; 3.53% of the GE-Cleveland-EUR cohort). In the FE-Epi25-EUR cohort, we observed a significant 2-fold enrichment of patients with focal epilepsy in the group with the highest FE-PRS (top 0.5%, P = 5.57×10−4; 1.16% of the FE-Epi25-EUR cohort; Table 2). This observation was not replicated in the smaller clinical FE-Cleveland-EUR cohort (P = 0.22). All patients with top 0.5% highest PRS were found in the top decile of the GE- and FE-PRS distributions of the Epi25-EUR cohort (Supplementary Figs 3 and 4). Measures of the diagnostic accuracy of the PRS are given in Supplementary Table 8.
Table 2.
Enrichment of patients with epilepsy in the extreme tails of the PRS distribution
| Reference group | OR | 95% CI | P-value | Cases/controls upper PRS% | Cases/controls lower PRS% | Sensitivity | Specificity | |
|---|---|---|---|---|---|---|---|---|
| GE–PRS / GE-Epi25 | ||||||||
| Top 20% of distribution | Remaining 80% | 2.04 | 1.86–2.25 | 4.61 × 10−48 | 887/3652 | 1369/16 783 | 0.393 | 0.821 |
| Top 5% of distribution | Remaining 95% | 2.39 | 2.06–2.76 | 4.39 × 10−32 | 305/830 | 1951/19 605 | 0.135 | 0.959 |
| Top 0.5% of distribution | Remaining 99.5% | 4.63 | 3.16–6.76 | 2.60 × 10−15 | 54/60 | 2202/20 375 | 0.024 | 0.997 |
| GE–PRS / GE-Cleveland | ||||||||
| Top 20% of distribution | Remaining 80% | 2.09 | 1.33–3.28 | 1.31 × 10−3 | 35/4070 | 50/16 365 | 0.412 | 0.801 |
| Top 5% of distribution | Remaining 95% | 2.02 | 1.00–3.72 | 3.44 × 10−2 | 11/1016 | 74/19 419 | 0.129 | 0.950 |
| Top 0.5% of distribution | Remaining 99.5% | 4.47 | 1.07–12.6 | 1.39 × 10−2 | 3/100 | 82/20 335 | 0.035 | 0.995 |
| FE–PRS / FE-Epi25 | ||||||||
| Top 20% of distribution | Remaining 80% | 1.35 | 1.24–1.48 | 2.32 × 10−11 | 992/3785 | 2457/16 650 | 0.288 | 0.815 |
| Top 5% of distribution | Remaining 95% | 1.44 | 1.25–1.66 | 7.74 × 10−7 | 292/903 | 3157/19 532 | 0.085 | 0.956 |
| Top 0.5% of distribution | Remaining 99.5% | 2.00 | 1.34–2.95 | 5.57 × 10−4 | 40/80 | 3409/20 355 | 0.012 | 0.996 |
| FE–PRS / FE-Cleveland | ||||||||
| Top 20% of distribution | Remaining 80% | 1.49 | 1.22–1.82 | 1.04 × 10−4 | 148/4047 | 387/16 388 | 0.277 | 0.802 |
| Top 5% of distribution | Remaining 95% | 1.55 | 1.09–2.14 | 1.17 × 10−2 | 40/1009 | 495/19 426 | 0.075 | 0.951 |
| Top 0.5% of distribution | Remaining 99.5% | 1.83 | 0.62–4.32 | 0.22 | 5/100 | 530/20 335 | 0.009 | 0.995 |
Odds ratios (OR) and P-values were calculated by comparing those within the top 0.5, 5, and 20% of the PRS distribution, to the remainder of the PRS distribution in a logistic regression model adjusted for sex and the first four principal components of ancestry. The threshold for statistical significance after Bonferroni correction was set to α = 1.67 x 10−2 (three tests per cohort). CI = confidence interval; EUR = European.
Epilepsy PRSs have limited value in biobank-derived cohorts based on ICD-10 codes
To evaluate whether common epilepsy variants identified in well-phenotyped patients are enriched in patient cohorts ascertained from patient registries and biobanks, we extended our GE- and FE-PRS analyses to include three biobank-derived cohorts. In the UKB and BioVU biobanks, diagnosis was ascertained by ICD-10 codes for epilepsy and by a standardized questionnaire for the attending physicians in the BBJ biobank. Fixed-effect meta-analysis of the two European biobanks (UKB and BioVU), adjusted for the effective sample size (Willer et al., 2010), revealed significantly higher GE-PRS in individuals coded as having generalized epilepsy than in population controls (P = 7.99×10−4, 539 patients with generalized epilepsy against 431 862 population controls). However, the PRS explained only very little of the phenotypic variance in each biobank (UKB: 0.12% variance explained; BioVU: 0.19%). In the BBJ, the GE-PRS were not significantly different between Japanese-ancestry patients with generalized epilepsy and controls (P = 0.33, 219 patients with generalized epilepsy against 168 356 controls). The FE-PRS were not significantly different between individuals coded as having focal epilepsy and controls in any of the three studied biobanks (UKB: P = 0.44; BioVU: P = 0.23; BBJ: P = 0.29). The PRSs for type 2 diabetes (negative control) were not significantly different in patients with generalized epilepsy or focal epilepsy than in population controls. The results in the three biobanks are detailed in Supplementary Table 3.
Discussion
We identified a significantly higher genetic burden for epilepsy, as quantified by PRS, in independent cohorts of patients with epilepsy as compared to population controls. While modest effect sizes preclude risk prediction based on single common genetic variants, PRSs that combine thousands of variants show predictive ability across a range of complex traits and diseases, including neuropsychiatric disorders (Khera et al., 2018). In the setting of a collaborative epilepsy genetics community, we demonstrate that available datasets have reached an adequate size to address the role of common genetic variants, each with small effect sizes, in large populations of patients with epilepsy. In line with previous studies showing significant differences between the genetic architectures of generalized epilepsy and focal epilepsy (Speed et al., 2014; International League Against Epilepsy Consortium on Complex Epilepsies, 2018), we also show that patients with generalized epilepsy have a significantly higher burden of common risk variants associated with generalized epilepsy than patients with focal epilepsy. The PRSs perform similarly in a multicentre research cohort and in an unselected—although much smaller—cohort ascertained through routine clinical practice in one single hospital. In contrast, significant, but small differences of PRS burden in large-scale biobanks provide evidence that ICD-10 epilepsy codes are not the best substitutes for precise clinical classifications by experts, despite our efforts to identify patients with generalized or focal epilepsy using stringent ICD code filtering. In line with recent evidence, we observe that PRSs derived from a European cohort have lower power when applied to populations of different genetic architecture, as observed in the cohorts of Finnish and Japanese ancestry (Martin et al., 2017).
By evaluating the PRS distribution, we identify patients with an effect size of polygenic variants at group level that is comparable to those in other studies with rare variants of large effect. Among the group of patients with high GE-PRS (top 0.5% of cases and controls with the highest scores), we observe an enrichment of patients with generalized epilepsy similar to that seen among carriers of established genetic risk factors, such as copy number variations and de novo variants: the largest copy number variation burden study in epilepsy to date showed a 7.45-fold enrichment of patients with generalized epilepsy (2.78% of all patients with generalized epilepsy) among all hotspot copy number variations carriers (cases/controls) (Pérez-Palma et al., 2017). The largest de novo variant study in neurodevelopmental disorders with epilepsy showed a 4.6-fold excess of de novo variants in known genes associated with developmental and epileptic encephalopathies in neurodevelopmental disorders with epilepsy when compared to those without epilepsy (Heyne et al., 2018). In this study, we identify a 4.63-fold enrichment of patients with generalized epilepsy in the top 0.5% highest GE-PRS in the Epi25 exploration cohort (2.39% of patients with generalized epilepsy) and a 4.47-fold enrichment of patients with generalized epilepsy in the top 0.5% highest GE-PRS in the clinical replication cohort (3.53% of patients with generalized epilepsy).
PRSs could have clinical implications for epilepsies because of their predictive power. Treatment with antiepileptic drugs after the first seizure has been debated, and the decision to start pharmacological treatment is usually based on relative risks, benefits, and lifestyle factors. After the first seizure, ∼50% of individuals go on to have a second seizure within 3–5 years, with most recurrences occurring within the first year (Kho et al., 2006; Wiebe et al., 2008). Several factors can increase the risk of seizure recurrence, including abnormal results on neurological examination, brain imaging, or EEG, a family history of epilepsy, or a personal history of remote symptomatic seizures (Wiebe et al., 2008). Patients at high risk for recurrence have been shown to benefit from immediate antiepileptic drug treatment after a first seizure compared to no treatment or delayed treatment (Kim et al., 2006). For an individual with new-onset epilepsy, it is also critical to differentiate between a focal versus generalized epilepsy to inform the selection of the first-line antiseizure drug (Perucca et al., 1998; Goldenberg, 2010). Differential diagnosis is especially challenging for focal epilepsy patients with secondary generalization or for those not found to have a relevant lesion on magnetic resonance imaging scans. A PRS indicating that a person is carrying an excessive amount of common risk variants for epilepsy or for generalized versus focal epilepsy could provide useful information for clinicians in deciding when to begin, and what type of treatment should be provided.
Future research should determine if and how well PRS can improve existing prediction models when combined with other factors, including established genetic risk factors of individually larger effect. Although our study represents the first of its kind in epilepsy, it needs to be replicated in a prospective setting. The predictive power of the PRS is determined by the genetic homogeneity of the GWAS from which the PRS is generated and that of the cohort to which it is applied. For epilepsy, a strong Eurocentric bias in the only available large scale GWAS is impeding PRS prediction in non-European individuals. Possible approaches to improve the predictive power in the non-European population, such as including the target population in the training data (Márquez-Luna et al., 2017), have been explored, but to realize the full potential of the PRS, greater population diversity must be prioritized in future GWAS studies (Martin et al., 2019). It is possible that PRSs for focal epilepsies currently lack power because this group is genetically and phenotypically more heterogeneous than the group of generalized epilepsy (Speed et al., 2014; International League Against Epilepsy Consortium on Complex Epilepsies, 2018). The clinical value of the PRS will also be limited by the low prevalence rates of epilepsy, leading to high negative predictive values, but low positive predictive values. To facilitate the implementation of PRS into clinical practice, additional research with larger, better-differentiated cohorts from different populations with well-characterized epilepsy phenotypes will be needed.
In summary, common polygenic variant burden for epilepsy can be measured and is differently distributed among patients with epilepsy and controls as well as between the two main epilepsy phenotypes (i.e. generalized and focal). PRS for epilepsies can provide physicians with an estimate of an individual’s overall genetic risk for epilepsy that could aid in early diagnosis and targeted treatment in the future. In addition, a combination of rare and common variants that may predispose an individual to develop epilepsy provides a chance for more informative prediction tools that may lead to a paradigm shift from current practice in rare disorder genetics (presence or absence of a Mendelian, high-risk variant) to a liability threshold model that assumes for each individual a continuous liability composed of rare and common genetic risk variants.
Funding
This work is part of the Centers for Common Disease Genomics (CCDG) program, funded by the National Human Genome Research Institute (NHGRI) and the National Heart, Lung, and Blood Institute (NHLBI). CCDG-funded Epi25 research activities at the Broad Institute, including genomic data generation in the Broad Genomics Platform, are supported by NHGRI grant UM1 HG008895 (PIs: Eric Lander, Stacey Gabriel, Mark Daly, Sekar Kathiresan). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Competing interests
The authors report no competing interests.
Supplementary Material
Glossary
Abbreviations
- FE =
focal epilepsy
- GE =
generalized epilepsy
- GWAS =
genome-wide association study
- PRS =
polygenic risk score
References
- Banerjee PN, Filippi D, Allen Hauser W. The descriptive epidemiology of epilepsy-a review. Epilepsy Res 2009; 85: 31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell GS, Neligan A, Giavasi C, Keezer MR, Novy J, Peacock JL, et al. Outcome of seizures in the general population after 25 years: a prospective follow-up, observational cohort study. J Neurol Neurosurg Psychiatry 2016; 87: 843–50. [DOI] [PubMed] [Google Scholar]
- Berg AT, Shinnar S. The risk of seizure recurrence following a first unprovoked seizure: a quantitative review. Neurology 1991; 41: 965–72. [DOI] [PubMed] [Google Scholar]
- Borodulin K, Tolonen H, Jousilahti P, Jula A, Juolevi A, Koskinen S, et al. Cohort profile: the National FINRISK study. Int J Epidemiol 2017. [DOI] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015; 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EpiPM ConsortiumBerkovic SF, Scheffer IE, Petrou S, Delanty N, Dixon-Salazar TJ, et al. A roadmap for precision medicine in the epilepsies. Lancet Neurol 2015; 14: 1219–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenstra B, Pasternak B, Geller F, Carstensen L, Wang T, Huang F, et al. Common variants associated with general and MMR vaccine-related febrile seizures. Nat Genet 2014; 46: 1274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg MM. Overview of drugs used for epilepsy and seizures: etiology, diagnosis, and treatment. P T 2010; 35: 392–415. [PMC free article] [PubMed] [Google Scholar]
- Guerrini R, Dravet C, Genton P, Belmonte A, Kaminska A, Dulac O. Lamotrigine and seizure aggravation in severe myoclonic epilepsy. Epilepsia 1998; 39: 508–12. [DOI] [PubMed] [Google Scholar]
- Hesdorffer DC, Logroscino G, Benn EKT, Katri N, Cascino G, Hauser WA. Estimating risk for developing epilepsy: a population-based study in Rochester, Minnesota. Neurology 2011; 76: 23–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyne HO, Artomov M, Battke F, Bianchini C, Smith DR, Liebmann N, et al. Targeted gene sequencing in 6994 individuals with neurodevelopmental disorder with epilepsy. Genet Med 2019; doi: 10.1038/s41436-019-0531-0. [DOI] [PubMed] [Google Scholar]
- Heyne HO, Singh T, Stamberger H, Abou Jamra R, Caglayan H, Craiu D, et al. De novo variants in neurodevelopmental disorders with epilepsy. Nat Genet 2018; 50: 1048–53. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 2009; 106: 9362–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins A, Garman A, Clarke C. The first seizure in adult life. Value of clinical features, electroencephalography, and computerised tomographic scanning in prediction of seizure recurrence. Lancet 1988; 1: 721–6. [DOI] [PubMed] [Google Scholar]
- International League Against Epilepsy Consortium on Complex Epilepsies. Genetic determinants of common epilepsies: a meta-analysis of genome-wide association studies. Lancet Neurol 2014; 13: 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International League Against Epilepsy Consortium on Complex Epilepsies. Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat Commun 2018; 9: 5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japaridze G, Kasradze S, Lomidze G, Zhizhiashvili L, Kvernadze D, Geladze K, et al. Focal EEG features and therapeutic response in patients with juvenile absence and myoclonic epilepsy. Clin Neurophysiol 2016; 127: 1182–7. [DOI] [PubMed] [Google Scholar]
- Karlson EW, Boutin NT, Hoffnagle AG, Allen NL. Building the Partners HealthCare Biobank at partners personalized medicine: informed consent, return of research results, recruitment lessons and operational considerations. J Pers Med 2016; 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasperavičiūtė D, Catarino CB, Matarin M, Leu C, Novy J, Tostevin A, et al. Epilepsy, hippocampal sclerosis and febrile seizures linked by common genetic variation around SCN1A. Brain 2013; 136: 3140–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018; 50: 1219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho LK, Lawn ND, Dunne JW, Linto J. First seizure presentation: do multiple seizures within 24 hours predict recurrence? Neurology 2006; 67: 1047–9. [DOI] [PubMed] [Google Scholar]
- Kim LG, Johnson TL, Marson AG, Chadwick DW; MRC MESS Study Group. Prediction of risk of seizure recurrence after a single seizure and early epilepsy: further results from the MESS trial. Lancet Neurol 2006; 5: 317–22. [DOI] [PubMed] [Google Scholar]
- Kwan P, Sander JW. The natural history of epilepsy: an epidemiological view. J Neurol Neurosurg Psychiatry 2004; 75: 1376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löscher W. Preclinical assessment of proconvulsant drug activity and its relevance for predicting adverse events in humans. Eur J Pharmacol 2009; 610: 1–11. [DOI] [PubMed] [Google Scholar]
- MacDonald BK, Johnson AL, Goodridge DM, Cockerell OC, Sander JW, Shorvon SD. Factors predicting prognosis of epilepsy after presentation with seizures. Ann Neurol 2000; 48: 833–41. [PubMed] [Google Scholar]
- Márquez-Luna C, Loh P-R; South Asian Type 2 Diabetes (SAT2D) Consortium; SIGMA Type 2 Diabetes Consortium, Price AL. Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genet Epidemiol 2017; 41: 811–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, et al. Human Demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet 2017; 100: 635–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet 2019; 51: 584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTague A, Howell KB, Cross JH, Kurian MA, Scheffer IE. The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol 2016; 15: 304–16. [DOI] [PubMed] [Google Scholar]
- Nagai A, Hirata M, Kamatani Y, Muto K, Matsuda K, Kiyohara Y, et al. Overview of the BioBank Japan Project: study design and profile. J Epidemiol 2017; 27: S2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngugi AK, Bottomley C, Kleinschmidt I, Sander JW, Newton CR. Estimation of the burden of active and life-time epilepsy: a meta-analytic approach. Epilepsia 2010; 51: 883–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngugi AK, Kariuki SM, Bottomley C, Kleinschmidt I, Sander JW, Newton CR. Incidence of epilepsy: a systematic review and meta-analysis. Neurology 2011; 77: 1005–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Palma E, Helbig I, Klein KM, Anttila V, Horn H, Reinthaler EM, et al. Heterogeneous contribution of microdeletions in the development of common generalised and focal epilepsies. J Med Genet 2017; 54: 598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perucca E, Gram L, Avanzini G, Dulac O. Antiepileptic drugs as a cause of worsening seizures. Epilepsia 1998; 39: 5–17. [DOI] [PubMed] [Google Scholar]
- Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008; 84: 362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer IE, Berkovic S, Capovilla G, Connolly MB, French J, Guilhoto L, et al. ILAE classification of the epilepsies: position paper of the ILAE Commission for Classification and Terminology. Epilepsia 2017; 58: 512–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RA, Scott LJ, Mägi R, Marullo L, Gaulton KJ, Kaakinen M, et al. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes 2017; 66: 2888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed D, O’Brien TJ, Palotie A, Shkura K, Marson AG, Balding DJ, et al. Describing the genetic architecture of epilepsy through heritability analysis. Brain 2014; 137: 2680–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015; 12: e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebe S, Téllez-Zenteno JF, Shapiro M. An evidence-based approach to the first seizure. Epilepsia 2008; 49: 50–7. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff M, Johannesen KM, Hedrich UBS, Masnada S, Rubboli G, Gardella E, et al. Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain 2017; 140: 1316–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the Epi25 Consortium, upon reasonable request. The biobank data are available from the UKB, BioVU, and BBJ upon successful project application.