Fumarates are effective immunomodulators in multiple sclerosis but their mechanism of action remains elusive. Ntranos et al. show that the immunomodulatory effect of fumarates is due to epigenetic regulation of brain-homing T cells. Treatment with fumarates leads to hypermethylation of microRNA-21, preventing its upregulation in encephalitogenic T-helper and T-cytotoxic cells.
Keywords: multiple sclerosis, DNA methylation, fumaric acid esters, CD4 T cells, metabolic-epigenetic interplay
Abstract
Cell-permeable formulations of metabolites, such as fumaric acid esters, have been used as highly effective immunomodulators in patients with multiple sclerosis and yet their mechanism of action remains elusive. Since fumaric acid esters are metabolites, and cell metabolism is highly intertwined with the epigenetic regulation of gene expression, we investigated whether this metabolic-epigenetic interplay could be leveraged for therapeutic purposes. To this end we recruited 47 treatment-naïve and 35 fumaric acid ester-treated patients with multiple sclerosis, as well as 16 glatiramer acetate-treated patients as a non-metabolite treatment control. Here we identify a significant immunomodulatory effect of fumaric acid esters on the expression of the brain-homing chemokine receptor CCR6 in CD4 and CD8 T cells of patients with multiple sclerosis, which include T helper-17 and T cytotoxic-17 cells. We report differences in DNA methylation of CD4 T cells isolated from untreated and treated patients with multiple sclerosis, using the Illumina EPIC 850K BeadChip. We first demonstrate that Krebs cycle intermediates, such as fumaric acid esters, have a significantly higher impact on epigenome-wide DNA methylation changes in CD4 T cells compared to amino-acid polymers such as glatiramer acetate. We then define a fumaric acid ester treatment-specific hypermethylation effect on microRNA MIR-21, which is critical for the differentiation of T helper-17 cells. This hypermethylation effect was attributed to the subpopulation of T helper-17 cells using a decomposition analysis and was further validated in an independent prospective cohort of seven patients before and after treatment with fumaric acid esters. In vitro treatment of CD4 and CD8 T cells with fumaric acid esters supported a direct and dose-dependent effect on DNA methylation at the MIR-21 promoter. Finally, the upregulation of miR-21 transcripts and CCR6 expression was inhibited if CD4 or CD8 T cells stimulated under T helper-17 or T cytotoxic-17 polarizing conditions were treated with fumaric acid esters in vitro. These data collectively define a direct link between fumaric acid ester treatment and hypermethylation of the MIR-21 locus in both CD4 and CD8 T cells and suggest that the immunomodulatory effect of fumaric acid esters in multiple sclerosis is at least in part due to the epigenetic regulation of the brain-homing CCR6+ CD4 and CD8 T cells.
Introduction
Evolution has led cells to develop sophisticated mechanisms that sense and integrate environmental cues into regulatory signals of transcription, so that changes in metabolites or hormones can be detected and adequately transduced to adjust gene expression (Lu and Thompson, 2012). The epigenetic cellular machinery is central in this interaction with the environment, and several metabolites have been shown to be integral for regulating epigenetic changes in cells, such as DNA methylation and demethylation (Lu and Thompson, 2012). In its extreme, metabolic and epigenetic reprogramming has been identified as a hallmark of cancer cells (Wong et al., 2017), but metabolic and epigenetic changes are seen normally in many cells in response to changing cellular states, such as proliferation or differentiation. T cells are known to significantly change their metabolism depending on their cellular state, with naïve, memory and effector cells using metabolic substrates differently to respond to their anabolic or catabolic demands (Buck et al., 2015; Almeida et al., 2016). Interestingly, apart from different metabolism, naïve and memory T cells also exhibit different levels of DNA methylation (Schmidl et al., 2009; Sellars et al., 2015; Durek et al., 2016). It remains unknown, however, if this metabolic-epigenetic interplay could be leveraged for therapeutic purposes in immune-mediated diseases.
DNA demethylation, the process by which methyl groups are removed from methyl-cytosines, is central in this metabolic-epigenetic interplay via the interaction between Krebs cycle intermediates, such as alpha ketoglutarate, fumarate and succinate, with DNA demethylases, such as Ten-eleven Translocation (TET) enzymes (Christensen et al., 2011; Lu and Thompson, 2012; Xiao et al., 2012; Sciacovelli et al., 2016). Studies performed in fumarate-hydratase deficient cancer cells show that the intracellular accumulation of fumarate causes DNA hypermethylation by inhibiting alpha ketoglutarate-dependent dioxygenases and TET enzymes (Xiao et al., 2012; Sciacovelli et al., 2016). This has also been noted in cancer cells that are unable to catabolize succinate or produce alpha-ketoglutarate due to mutations in succinate dehydrogenase and isocitrate dehydrogenase respectively (Christensen et al., 2011; Xiao et al., 2012). Interestingly, DNA demethylation is also important for T cell lineage determination, naïve T cell activation and memory T cell formation (Schmidl et al., 2009; Sellars et al., 2015; Durek et al., 2016). Furthermore, TET-mediated active DNA demethylation is a critical epigenetic mechanism for regulating CD4 T cell function (Ichiyama et al., 2015). Since naïve CD4 T cells are hypermethylated compared to memory CD4 T cells (Schmidl et al., 2009; Sellars et al., 2015; Durek et al., 2016), it is conceivable that active DNA demethylation could be considered an immunomodulatory target. This conceptual framework led us to study whether the exogenous administration of fumaric acid esters (FAEs), a cell-permeable formulation of a Krebs cycle metabolite, may modulate DNA methylation in T cells of patients with multiple sclerosis and explain the observed immunomodulatory effects.
The effectiveness of FAEs in multiple sclerosis has been demonstrated in clinical trials (Fox et al., 2012; Gold et al., 2012), but their mechanism of action remains controversial. To this end, we recruited patients with multiple sclerosis who were treatment-naïve or treated with either FAEs or glatiramer acetate (GA) and studied their immunophenotypic profile and epigenome-wide DNA methylation levels. We used the Illumina Infinium MethylationEPIC BeadChip to interrogate the epigenome of CD4 cells sorted from untreated and treated patients. By applying stringent statistical and effect size criteria, and accounting for the cellular heterogeneity between patient groups with two different methods, we uncovered a FAE treatment-specific hypermethylating effect in CD4 T cells. These findings were then validated in an independent prospective cohort of FAE-treated patients with multiple sclerosis and the mechanistic aspect was further addressed in vitro on stimulated human CD4 and CD8 T cells. Based on our findings we suggest a novel mechanism of immunomodulation in multiple sclerosis, which uses the metabolic-epigenetic interplay in brain-homing CCR6+ CD4 and CD8 T cells and distinguishes FAEs from other available multiple sclerosis therapeutics.
Materials and methods
Study design and clinical characteristics
This research was approved by the Institutional Review Board (IRB) and informed consent was obtained for all subjects according to the Declaration of Helsinki. Diagnosis of relapsing remitting multiple sclerosis was made by McDonald 2010 criteria (Polman et al., 2011). Participants were naïve to any multiple sclerosis disease-modifying therapy or treated with either GA or FAEs and had stable disease for at least 3 months. Detailed inclusion and exclusion criteria are in Supplementary Fig. 1.
Immunophenotyping
Whole blood was collected at study enrolment and was processed within 3 h by the Mount Sinai’s Human Immune Monitoring Core. Cells were stained with a pre-optimized T-cell antibody cocktail (Supplementary Table 2) and analysed on a BD LSR Fortessa. FlowJo (Treestar Inc, San Carlos, CA) was used for post‐acquisition analysis (Supplementary Fig. 2).
DNA methylation analysis
DNA from positively isolated CD4 T cells from each sample was bisulfite-treated and methylation levels were measured at ~850 000 CpG sites by using the Infinium MethylationEPIC BeadChip array. As we have described previously (Huynh et al., 2014; Watson et al., 2016), our analysis was focused on genomic regions rather than individual CpG sites as regulatory DNA modifications generally involve multiple consecutive CpGs. To identify differentially methylated regions between treated and untreated patients, we used a linear regression model to identify the contribution of treatment status in DNA methylation changes after controlling for age, gender, race, disease duration, the total CD4 T cell percentage as well as the percentage of CCR6−CCR4+, CCR6+CCR4+, CCR6+CCR4− CD4 T cells in each sample from the discovery cohort. Since CCR4 and CCR6 are expressed mainly in memory CD4 T cells (Fagin et al., 2012a) and highly correlate with the total number of memory CD4 T cells (Pearson, n = 5, r=0.9833, P = 0.0026) (Supplementary Fig. 4), our above analysis would be able to control for potential naïve/memory imbalances of our samples (details in the Supplementary material). Data analysis was performed in R Studio by utilizing the R packages ChAMP (Tian et al., 2017) for data preprocessing and normalization, LIMMA for linear regression (Ritchie et al., 2015), bumphunter for the 1 kb sliding window function (Jaffe et al., 2012) and DMRCATE for the P-value combination with the Stouffer method (Peters et al., 2015).
Decomposition of DNA methylation values
To determine the contribution of each cell type to the DNA methylation changes observed at the MIR-21 locus in our analysis, we decomposed the measured β-values of the CpG sites in that locus to each cell type by using a constrained least squares regression model that used the cell type proportions from our immunophenotyping analysis and the measured β-values to infer the cell type specific β-values. To obtain a P-value for each cell type, we determined the treatment-naïve distribution of these decomposed β-values by bootstrapping residuals from the treatment-naïve decomposition model and then compared them with the decomposed β-values from FAE-treated patients using the cumulative distribution function (details in the Supplementary material). Data analysis was performed in python 2.7 by using the packages scipy 0.19.0 (for constrained least squares: scipy.optimize.lsq_linear), numpy 1.13.1 and pandas 0.20.2
Isolation and in vitro culture of naïve and memory CD4 T cells
Peripheral blood mononuclear cells (PBMCs) were collected from healthy donors by the Mount Sinai’s Human Immune Monitoring Core and stored in liquid nitrogen until further use. Naïve CD45RO−CCR7+ CD4+ or CD45RO−CCR7+ CD8+ T cells and memory CD45RO+ CD4+ T cells were isolated on a BD FACSAria Fusion. CD4 T cells were then cultured for 3 days (for DNA methylation and RNA studies) or 6 days (for protein expression by flow cytometry) in X-VIVO™ 15 media (Lonza) and stimulated with antiCD3/CD28 coated beads (Dynabeads, ThermoFisher). Th17 or T cytotoxic-17 polarization was performed with 12.5 ng/ml IL-1b, 25 ng/ml IL-6, 25 ng/ml IL-23, 1 ng/ml TGFbeta (Peprotech) and 1 µg/ml anti-IL4 (Invitrogen). CD8 T cells were activated and cultured for 3 days for all analyses in X-VIVO™ 15 media also supplemented with 1 ng/ml IL7 and 10 ng/ml IL15 (Peprotech) (Montes et al., 2005; Ghassemi et al., 2016). MMF (Sigma) 20 µM or 50 µM was added twice daily for 3 days and once daily from Day 3 to Day 5. Viability was measured with eBioscience Fixable Viability Dye eFluor® 780. Genomic DNA or total RNA (including microRNA) was isolated with AllPrep™ DNA/RNA micro kit (Qiagen) and miRNeasy mini kit (Qiagen) respectively. Flow cytometry was performed after staining cells with anti-CCR6-BV421 (BD), anti-SMAD7-FITC (SantaCruz). Cells were fixed and permeabilized with the Foxp3/Transcription Factor Staining Buffer Set (ThermoFisher).
EpiTYPER MassArray® analysis
DNA methylation analysis of ex vivo and in vitro stimulated cells was performed with EpiTYPER™ MassARRAY® system (Agena Bioscience) as previously described (Moyon et al., 2016) at the Epigenetics Core facility at the CUNY Advanced Science Research Center (ASRC). Briefly, genomic DNA was bisulfite-treated with Methylamp One-Step DNA Modification Kit (EpiGentek) and HotStarTaq® DNA Polymerase kit (Qiagen) was used in a touchdown PCR reaction to amplify the MIR-21 promoter and regular PCR reaction to amplify the TNF promoter (primers in the Supplementary material). All samples were run in agarose gels to confirm the presence of a single band before running them on EpiTYPER™ MassARRAY®.
Quantitative PCR analysis
Quantitative (q)PCR for miR-21-5p, miR-21-3p, SNORD44, SNORD48 and RNU6 was performed by using the qscript miRNA cDNA synthesis kit (Quantabio). qPCR for CCR6, SMAD7, HPRT1, B2M and ACTB was performed by using the qscript cDNA synthesis kit (Quantabio) and PerfeCTa SYBR® Green Supermix, Low ROX (Quantabio). All microRNA primers were obtained from Quantabio miRNA assays. Primers for CCR6, SMAD7, HPRT1, B2M and ACTB were purchased from IDT PrimeTime qPCR Assays. qPCR was run and analysed at the Epigenetics Core facility at the CUNY Advanced Science Research Center (ASRC).
Statistical analyses
Immunophenotyping data were analysed with R Studio and Age, Gender and Race were identified as important covariates by stepwise regression and were controlled for in a linear regression model. Disease duration was also added in our linear regression model as it was significantly different between treatment naïve and treated patient groups. GraphPad Prism 7 (GraphPad Software, La Jolla, CA) was used for ANOVA and two-tailed t-test with or without Welch correction for the in vitro data analysis. Goodness of fit was determined by R2 and F-test. P-values < 0.05 were considered significant.
Data availability
The data have been deposited in NCBI’s Gene Expression Omnibus (Edgar, 2002) and are accessible through GEO Series accession number GSE112596 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112596). The R or python scripts are available upon request.
Results
Patient population and study design
We recruited 47 treatment-naïve patients with relapsing-remitting multiple sclerosis, 35 FAE-treated patients and 16 GA-treated patients as our discovery cohort (Supplementary Table 1). Whole blood was immunophenotyped and DNA from isolated CD4 T cells was used for the epigenome wide studies using the Illumina Infinium MethylationEPIC BeadChip. For our immunophenotyping analysis we measured the percentage of CD3, CD4 and CD8 T cells, as well as the chemokine receptors CCR6 and CCR4 in CD4 and CD8 T cells. CCR6 is important for T cell trafficking to the gut and brain (Reboldi et al., 2009; Wang et al., 2009), and CCR4 has been implicated in T cell trafficking to the skin (Campbell et al., 2007). Furthermore, CCR4 in combination with CCR6 could define T helper 17 (Th17) cells (CCR6+CCR4+) (Dunay et al., 2016), which have been implicated in multiple sclerosis pathogenesis (Tzartos et al., 2008), as well as Th2 cells (CCR6−CCR4+) (Rivino et al., 2004). CCR6+CCR4− and CCR6−CCR4− CD4 T cells are less well-defined but include Th1-like Th17 cells (Th1Th17) and naïve CD4 T cells, respectively (Fagin et al., 2012b; Van Langelaar et al., 2018) (Supplementary Fig. 2). Th1 cells were not measured as CXCR3 was not included in the panel. CCR6+ CD8 T cells represent early effector memory cytotoxic T cells that include T cytotoxic-17 (Tc17) cells (Kondo et al., 2007, 2009; Liang et al., 2015). The three patient groups were similar in demographic and disease characteristics except disease duration, which was shorter in treatment-naïve than treated patients [ANOVA; F(2,95) = 15.65, P < 0.0001] (Supplementary Table 1). GA-treated patients also had a longer treatment duration compared to FAE-treated patients [Welch’s t-test; t(15.337) = −2.9171, P = 0.0104] (Supplementary Table 1). We also recruited eight patients with multiple sclerosis as our validation cohort, from whom we prospectively collected CD4 T cells prior to treatment and then after an average of 10.5 months of therapy with FAEs. One baseline sample did not pass quality control and thus we used the remaining seven paired samples for our pair-wise epigenome-wide DNA methylation analysis (see ‘Materials and methods’ section) with the Illumina Infinium MethylationEPIC BeadChip (Supplementary Fig. 1).
CCR6+ CD4 and CD8 T cell reduction is the most prominent immunomodulatory effect of FAE therapy
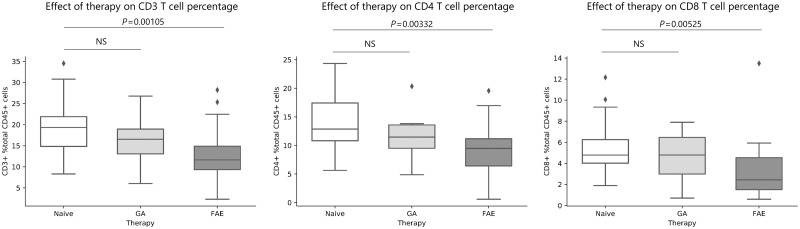
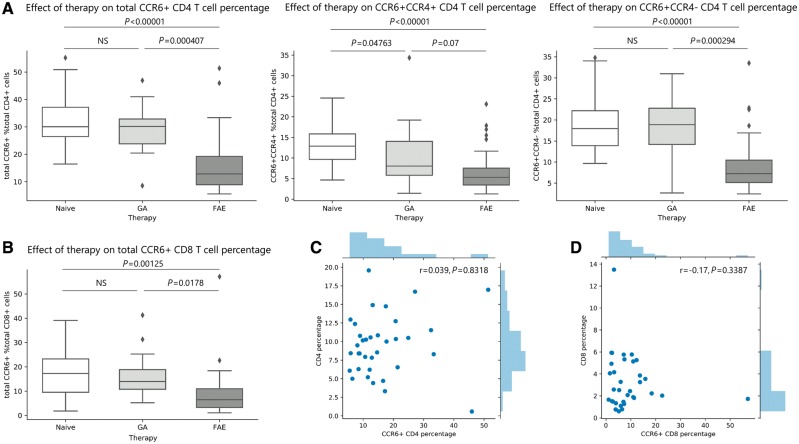
Immunophenotyping of 47 treatment-naïve, 35 FAE and 16 GA-treated patients with relapsing-remitting multiple sclerosis (Supplementary Table 1 and Supplementary Fig. 1) revealed that FAE treatment was significantly associated with a lower percentage of total CD3, CD4 and CD8 T cells in the FAE group compared to untreated patients with multiple sclerosis (Fig. 1). This finding was consistent with the previously described lymphopenic effect of FAEs (Longbrake et al., 2015; Fox et al., 2016; Gross et al., 2016; Ghadiri et al., 2017; Wu et al., 2017) [linear regression controlling for age, gender, race and disease duration; CD3 t(76) = −3.407, P = 0.00105; CD4 t(76) = −3.032, P = 0.00332; CD8 t(76) = −2.874, P = 0.00525, respectively]. We also identified a highly significant immunomodulatory effect of FAEs on CCR6+ CD4 and CD8 T cells. Both total CCR6+ CD4 and CD8 T cells were significantly lower in the FAE group compared to treatment naïve or GA-treated patients [linear regression; CCR6+ CD4: t(76) = −6.788, P < 0.00001 and t(45) = −3.819, P = 0.000407, respectively; CCR6+ CD8: t(76) = −3.354, P = 0.00125 and t(44) = −2.461, P = 0.0178, respectively] (Fig. 2A and B). CCR6+CCR4+ CD4 T cells (Th17) and CCR6+CCR4− CD4 T cells (Th1Th17), were also significantly lower in the FAE group compared to treatment naïve multiple sclerosis patients [linear regression; Th17 t(76) = −5.964, P = 7.27 × 10−8; Th1Th17 t(76) = −5.285, P = 1.16 × 10−6] (Fig. 2A). A milder reduction of CCR6−CCR4+ CD4 T cell (Th2) frequency was observed in the FAE treatment group compared to controls [linear regression; Th2 t(76) = −2.870, P = 0.00532] (Supplementary Fig. 5). FAE therapy was also associated with higher proportion of CD4 T cells that were double-negative for these chemokine receptors (CCR6−CCR4−), which comprise mostly naïve CD4 T cells [linear regression; t(76) = 6.171, P = 3.05 × 10−8] (Supplementary Fig. 5). CCR6+CCR4− and CCR6+CCR4+ CD8 T cells were both significantly lower in FAE-treated patients compared to treatment naïve patients [linear regression; CCR6+CCR4− t(76) = −3.249, P = 0.00172; CCR6+CCR4+ t(76) = −2.024, P = 0.0465]. FAE therapy did not change CCR6−CCR4+ CD8 T cells (P > 0.05) but was associated with increased proportion of CCR6−CCR4− CD8 T cells, similarly to the effect on CD4 T cells [linear regression; t(76) = 3.068, P = 0.00299] (Supplementary Fig. 6).
Figure 1.
The FAE-induced lymphopenic effect. Whole blood from 47 treatment-naïve, 35 FAE and 16 GA treated multiple sclerosis (MS) patients was analysed by FACS. FAE treatment was associated with significantly reduced percentage of total CD3, CD4 and CD8 T cells [linear regression controlling for age, gender, race and disease duration; t(76) = −3.407, P = 0.00105; t(76) = −3.032, P = 0.00332; t(76) = −2.874, P = 0.00525, respectively]. GA was not associated with changes in these T cell populations. NS = not-significant (P > 0.05). The lines in the box plot represent the quartiles of the dataset and the whiskers show the rest of the distribution, except for ‘outliers’ that were determined by a function of the inter-quartile range based on the package seaborn in python.
Figure 2.
CCR6+ CD4 and CD8 T cell reduction is a novel immunomodulatory effect of FAE therapy. Whole blood from 47 treatment-naïve, 35 FAE and 16 GA treated multiple sclerosis (MS) patients was analysed by FACS. (A) FAE treatment was associated with significantly reduced percentage of total CCR6+ CD4 T cells compared to both treatment naïve and GA-treated patients, whereas GA had no effect. FAE therapy also reduced both CCR6+CCR4+ (Th17) and CCR6+CCR4− (Th1Th17) CD4 T cells. GA was only associated with reduced percentage of CCR6+CCR4+ (Th17) CD4 T cells, but with a trend towards a smaller magnitude compared to FAEs. (B) FAE therapy was also associated with significantly reduced CCR6+ CD8 T cells compared to both treatment naïve and GA-treated patients, whereas GA had no effect. (C) The total CD4 T cell percentage and CCR6+ CD4 percentage as well as (D) the total CD8 T cell percentage and CCR6+ CD8 percentage of each FAE-treated patient (n = 35) was plotted together with their corresponding histogram of distribution. The percentage of these cell types did not significantly correlate with each other. NS = not significant (P > 0.05). The lines in the box plot represent the quartiles of the dataset and the whiskers show the rest of the distribution, except for ‘outliers’ that were determined by a function of the interquartile range based on the package seaborn in python.
GA did not significantly change CD3, CD4 or CD8 T cell percentages compared to controls (P > 0.05 for all) (Fig. 1), but it was associated with a mildly reduced percentage of Th17 cells compared to treatment naïve multiple sclerosis patients [linear regression; t(57) = −2.024, P = 0.04763] (Fig. 2A). Total CCR6+ or Th1Th17 cell percentages did not significantly change in GA-treated compared to untreated multiple sclerosis patients (P > 0.05) (Fig. 2A). GA also had a minimal effect on the CCR6 and CCR4 expression of CD8 T cells and was associated with only a mild reduction of CCR6–CCR4+ CD8 T cells [linear regression; t(56) = −2.070, P = 0.0431] (Supplementary Fig. 6). Therefore, while both immunomodulatory therapies reduced Th17 cells, FAE treatment showed a trend towards a greater effect compared to GA [linear regression; t(45) = −1.856, P = 0.07] and was also uniquely associated with a stronger reduction of Th1Th17 cells [linear regression; t(45) = −3.926, P = 0.000294], thereby affecting CCR6+ CD4 T cells more broadly (Fig. 2A). FAE treatment was also uniquely associated with a significant reduction in CCR6+ CD8 T cells compared to GA-treated patients (Fig. 2B), including CCR6+CCR4− CD8 T cells (Supplementary Fig. 6).
This immunomodulatory effect of FAEs on CCR6 cells did not correlate with their overall lymphopenic effect, as measured by total CD4 and CD8 T cell percentage. Low CCR6+ CD4 or CD8 percentage did not explain the presence of low total CD4 or CD8 percentage in the FAE-treatment group, and these cell counts did not correlate with each other (Pearson partial correlation controlling for age, gender and race; CD4: r = 0.039, P = 0.8318, CD8: r = −0.17, P = 0.3387) (Fig. 2C and D). It has been proposed that the lymphopenic effect of FAEs is consequent to its preferential cytotoxicity of memory T cells, which does not correlate with clinical response and relapse reduction (Fox et al., 2016; Ghadiri et al., 2017). Our results provide a potential alternative explanation by suggesting the existence of an additional immunomodulatory effect of FAE treatment, which is independent from a generalized reduction of memory T cells, and preferentially targets CCR6+ CD4 and CD8 T cells.
FAE treatment hypermethylates the MIR-21 locus in CD4 T cells of patients with multiple sclerosis
To investigate the mechanism of this immunomodulatory effect of FAE treatment, we conducted an epigenome-wide DNA methylation analysis of CD4 T cells obtained from the same discovery cohort of patients with multiple sclerosis by using the Illumina Infinium MethylationEPIC BeadChip. Methylation values for ~850 000 individual CpG sites in each sample were measured as β-values, which represent the ratio of signal intensities between methylated and unmethylated alleles in each sample. As regulatory modifications of DNA methylation generally involve multiple consecutive CpGs, we focused our differential DNA methylation analysis on genomic regions, rather than focusing on isolated changes in individual CpGs, as we have previously described (Huynh et al., 2014; Watson et al., 2016). To this end, we first obtained site-specific P-values by using a linear regression model to identify the contribution of treatment status in DNA methylation changes after controlling for age, gender, race, disease duration and the measured cell proportions from our immunophenotyping analysis. This allowed us to account for cellular heterogeneity of samples, including potential naïve/memory CD4 T cell imbalances. We then used a 1 kb sliding window to define genomic regions with closely located CpG sites, and combined the previously obtained site-specific P-values from each region with the Stouffer’s method (Whitlock, 2005). This was followed by the Bonferroni correction for multiple hypothesis testing. Based on this analysis we identified that FAE treatment was associated with 228 hypermethylated and only 13 hypomethylated differentially methylated regions (DMRs) compared to untreated patients with multiple sclerosis. On the other hand, GA treatment was associated with only 26 DMRs, 24 of which were hypermethylated compared to untreated patients with multiple sclerosis. This suggests that Krebs cycle intermediates (e.g. FAEs) have a significantly higher impact on epigenome-wide DNA methylation changes in CD4 T cells compared to amino-acid polymers such as GA.
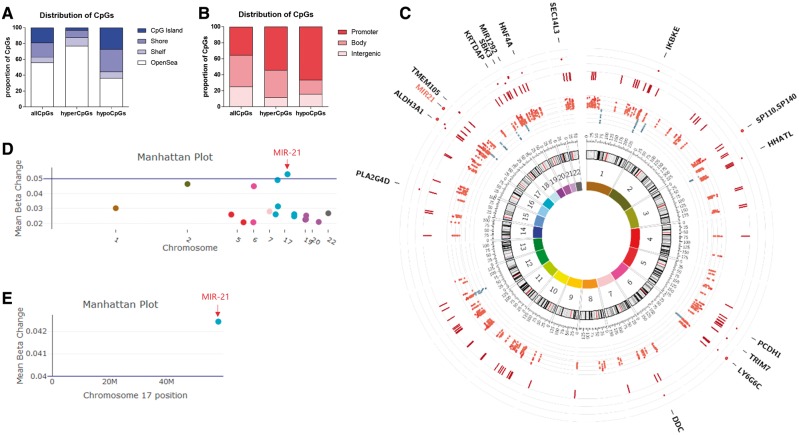
To increase the specificity of our previously identified DMRs for FAE therapy, we only kept DMRs that are differentially methylated in the analysis between FAE-treated versus untreated multiple sclerosis patients (adjusted P-value < 0.01) but are not differentially methylated in the comparison between GA-treated and untreated patients with multiple sclerosis (adjusted P-value > 0.05). Based on this analysis, we uncovered 202 hypermethylated DMRs (containing 1545 CpGs) and only 13 hypomethylated DMRs (containing 158 CpGs) that were uniquely changed in FAE-treated patients with multiple sclerosis. We then compared the CpG distribution of these hypermethylated and hypomethylated DMRs to that of the CpG distribution in the Illumina Infinium MethylationEPIC BeadChip (Fig. 3A and B) based on their relation to CpG islands (shores = regions 0–2 kb from CpG islands, shelves = regions 2–4 kb from CpG islands, open sea = regions of CpGs that do not have a specific designation) and transcriptional start sites. We noted that hypermethylated DMRs from FAE-treated patients were preferentially located in open sea CpGs rather than CpG islands and a significant proportion of them were spanning promoter regions, as defined by 2 kb before and after transcriptional start sites. The DNA methylation differences detected in the genome of CD4 cells from FAE-treated and untreated multiple sclerosis patients were widely distributed in the genome as shown in the graphical circular representation of the genome (Fig. 3C). Among the largest genomic loci identified (CpG number >10), the region with the greatest differential methylation was a hypermethylated region that contained 12 CpG sites and spanned the promoter of microRNA 21 (MIR-21), a microRNA that has been reported to be critical for Th17 differentiation and CCR6 expression in mouse models of multiple sclerosis (Murugaiyan et al., 2015) (Fig. 3D).
Figure 3.
Epigenome-wide DNA methylation analysis reveals MIR-21 as the top hypermethylated region in CD4 T cells from FAE-treated patients. CD4 T cell DNA from 47 treatment-naïve, 35 FAE-treated and 16 GA-treated multiple sclerosis patients was analysed by using the Infinium MethylationEPIC BeadChip array. To identify differentially methylated regions between treated and untreated patients, we utilized a linear regression model at each individual CpG site to identify the contribution of treatment status in DNA methylation changes after controlling for age, gender, race, disease duration, the total CD4 T cell percentage as well as the percentage of CCR6−CCR4+, CCR6+CCR4+ and CCR6+CCR4− CD4 T cells in each sample. We then used a 1-kb sliding window to define genomic regions with closely located CpG sites, and then combined each CpG specific P-value from our previous linear regression model within a single region with the Stouffer’s method. This was followed by the Bonferroni correction for multiple hypothesis testing. DMRs were defined as regions with more than four CpG sites that have an absolute median β-value change > 0.02 and an adjusted combined P-value of < 0.01. Based on this analysis we uncovered 202 hypermethylated DMRs (containing 1545 CpGs) and only 13 hypomethylated DMRs (containing 158 CpGs) in FAE-treated patients compared to controls. (A and B) The CpGs distribution of these hypermethylated (hyperCpGs) and hypomethylated (hypoCpGs) DMRs was compared to that of the CpG distribution in the Illumina Infinium MethylationEPIC BeadChip (allCpGs). (C) Circos plot showing (from inside out); innermost first circle: chromosomal colors, numbers and size; second circle: a scatter plot with each point representing the genomic location and mean change in β-value of hypomethylated (in blue) and hypermethylated (in red) CpG sites; third circle: dark red perpendicular lines represent the genomic location of significantly hypermethylated DMRs; fourth circle: dark red scatter plot with each point representing the genomic location and mean beta change of significantly hypermethylated DMRs spanning more than 10 CpGs; outermost fifth circle: contains the names of genes whose promoters are located in these large DMRs (MIR-21 is shown in red). (D) Manhattan plot of the discovery cohort analysis showing mean beta change of significantly hypermethylated DMRs spanning more than 10 CpGs, where MIR-21 was the top differentially methylated locus. (E) Manhattan plot from the pair-wise longitudinal analysis showing that MIR-21 was the hypermethylated DMR with the most consistent and greatest methylation change in the validation cohort.
To validate our findings in an independent cohort, we prospectively followed seven patients before and after FAE treatment (Supplementary Table 1). The prospective design of this cohort allowed us to do a pair-wise analysis of DNA methylation changes before and after FAE therapy at the whole epigenome level. We obtained site specific P-values by using a pair-wise linear regression model to identify the contribution of FAE treatment in DNA methylation changes and used our 1 kb sliding window approach to define DMRs as described above. Given that DNA methylation can be influenced by many different factors across patients, this design and sample size allowed us to identify the genomic region with the most consistent and greatest methylation change in each patient after treatment with FAEs and provided sufficient power to detect a significant difference at the MIR-21 locus (Supplementary material). This analysis validated the significant hypermethylation of the MIR-21 locus in CD4 T cells after FAE therapy (adjusted combined P < 0.01). The other hypermethylated regions from our discovery cohort analysis did not survive the Bonferroni correction in this pair-wise analysis (Fig. 3E). This does not exclude a potential effect of FAEs on the other hypermethylated regions from our discovery cohort but rather reinforces the consistency and effect size of MIR-21 hypermethylation by FAEs.
The MIR-21 locus is significantly hypermethylated in Th17 cells of FAE-treated patients
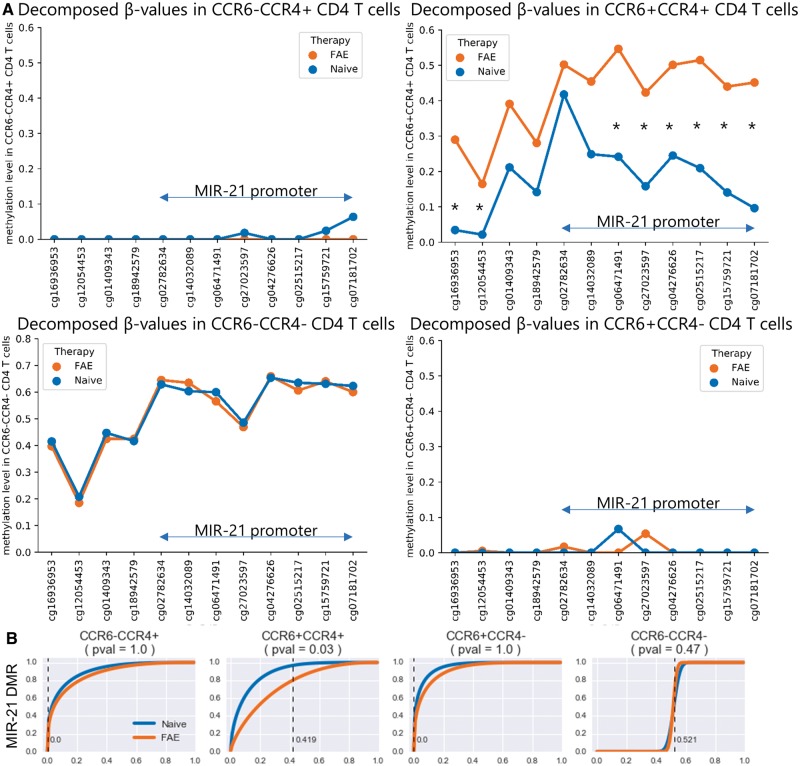
To determine the contribution of each cell type to the DNA methylation changes observed at the MIR-21 locus, we decomposed the β-values in that locus to the cell types that we identified from our immunophenotyping analysis of our discovery cohort (Supplementary Fig. 7). Given that the measured β-value of each sample is a linear combination of the β-values of each cell type and their proportions in the sample, we used a constrained least squares regression model to determine the methylation value of each cell type at each CpG site. Using this β-value decomposition analysis, we attributed the MIR-21 methylation change only to the Th17 cells of FAE-treated patients. Interestingly, FAE therapy hypermethylated most CpG sites spanning the MIR-21 promoter of Th17 cells, but it did not affect the other CD4 T cell subtypes (Fig. 4A and B). As Th17 cells do not include naïve T cells, this change could not have been due to potential naïve/memory imbalances in our samples, but rather suggests that FAEs directly affect MIR-21 methylation in Th17 cells.
Figure 4.
MIR-21 is hypermethylated specifically in Th17 cells from FAE-treated patients. Given that the measured β-value of each sample is a linear combination of the β-values of each cell type and their respective proportions in the sample, decomposition of the β-values of the CpG sites in the MIR-21 locus was done by using a constrained least squares regression model that used the measured β-values as the dependent variable and the FACS-obtained proportions of the cell types as the independent variables. The coefficients of this model then represent the average methylation value for each cell type at a given CpG site. (A) Based on this analysis, CCR6+CCR4+ CD4 T cells (Th17) exhibited a hypermethylated MIR-21 locus in FAE-treated patients compared to treatment naïves. Both CCR6-CCR4+ (Th2) and CCR6+CCR4- (Th1Th17) CD4 T cells had MIR-21 locus with almost zero methylation values, whereas CCR6-CCR4- (Naïve) CD4 T cells exhibited average methylation values without any significant difference between treated and untreated patients. (B) The distribution of these cell-type specific methylation values was obtained by bootstrapping the residuals of our constrained least squares regression model. The cumulative distribution function of the average methylation value of the MIR-21 DMR from treatment naïve patients is shown in blue. The cumulative distribution function of the MIR-21 DMR from FAE-treated patients is shown in orange. Each point on the cumulative distribution function represents the probability (y-axis) that a given cell type has a methylation value below a certain β-value (x-axis). The dotted line represents the average methylation value of the MIR-21 locus in each cell type that was obtained from fitting the model in all FAE-treated patients. The P-value, as the probability of obtaining a methylation value from treatment naïve patients that is higher than FAE-treated patients, was obtained from the cumulative distribution function of the treatment naïve distribution from each cell type. *P < 0.05.
FAEs directly modulate DNA methylation at the MIR-21 promoter in a specific and dose-dependent manner
To investigate whether FAEs directly modulate DNA methylation levels at the MIR-21 locus in CD4 or CD8 T cells, we isolated naïve (CCR7+ CD45RO−) and memory (CD45RO+) CD4 T cells or naïve (CCR7+ CD45RO−) CD8 T cells from human peripheral blood mononuclear cells by fluorescence-activated cell sorting (FACS). We then analysed their DNA methylation at the MIR-21 promoter directly after isolation ex vivo, as well as after 3 days of activation with antiCD3/CD28 beads and culture with or without FAEs in vitro. From the two available FAEs, monomethyl-fumarate (MMF) and dimethyl-fumarate (DMF), we chose to use MMF in our in vitro experiments because it is the FAE with the greatest bioavailability and in vivo relevance (Mrowietz et al., 2017). Finally, to assess the specificity of FAE’s effect, we analysed DNA methylation at the promoter of TNF, a cytokine that is essential for immune function against several pathogens.
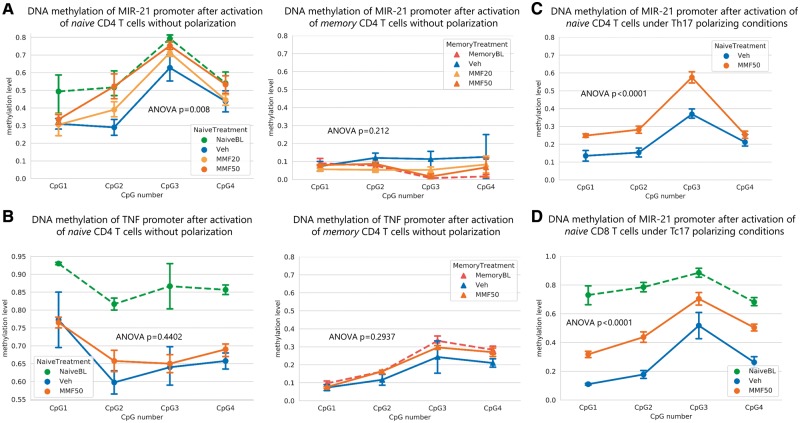
EpiTYPER™ MassARRAY® DNA methylation analysis of the MIR-21 promoter in DNA isolated from ex vivo human naïve and memory CD4 T cells revealed that it was hypermethylated in naïve compared to memory CD4 T cells, whose methylation values were lower than 0.1 [two-way ANOVA; n = 3, F(1,16) = 225.1; P < 0.0001] (Supplementary Fig. 8A). After activation and culture of naïve CD4 T cells with FAEs (20 µM MMF or 50 µM MMF) or vehicle for 3 days, the promoter of MIR-21 was hypermethylated in the FAE group compared to vehicle in a dose dependent manner, with the highest difference observed in the 50 µM group [two-way ANOVA; naïve n = 4, F(2,36) = 5.53, P = 0.008]. Of note, the methylation level in the high FAE dose group (50 µM MMF) was unchanged and remained at the same pre-culture baseline level, contrary to vehicle-treated cells that showed a demethylated promoter compared to their baseline (Fig. 5A). Memory CD4 T cells, on the other hand, did not exhibit any change in their methylation level at the MIR-21 promoter and remained with DNA methylation levels that were similar to their pre-culture baseline [two-way ANOVA memory n = 3 F(2,23) = 1.661, P = 0.212] (Fig. 5A). Furthermore, the hypermethylating effect of FAE treatment at the MIR-21 promoter was even greater when naïve CD4 T cells were activated and cultured under Th17 conditions [two-way ANOVA n = 3, F(1,16) = 39.58, P < 0.0001] (Fig. 5C). Finally, FAEs were also able to inhibit the demethylation of the MIR-21 promoter in naïve CD8 T cells that were activated with antiCD3/CD8 beads and cultured with or without FAE (50 µM MMF) for 3 days in vitro (Fig. 5D) [two-way ANOVA; n = 4, F(1,24) = 50.52, P < 0.0001]. These results demonstrate that FAE treatment has a direct effect on MIR-21 demethylation during activation of naïve CD4 and CD8 T cells.
Figure 5.
FAEs inhibit DNA demethylation at the MIR-21 promoter in a specific and dose-dependent manner. Naïve (CD45RO−CCR7+) and memory (CD45RO+) CD4 T cells were isolated from human PBMCs by FACS. The baseline DNA methylation levels of naïve (NaïveBL) and memory CD4 T cells (MemoryBL) at the (A) MIR-21 and (B) TNF promoters were measured ex vivo by EpiTYPER™ MassARRAY®. Naïve and memory CD4 T cells were also cultured with or without MMF at the specified concentration and stimulated with anti-CD3 and anti-CD28 coated beads for 3 days either without polarization or under Th17 polarizing conditions, after which DNA methylation levels at the (A) MIR-21 and (B) TNF promoters were measured by EpiTYPER™ MassARRAY®. (A) MMF was able to inhibit DNA demethylation of the MIR-21 promoter in naïve CD4 T cells, but not in memory CD4 T cells, in a dose dependent manner. (B) DNA methylation at the TNF promoter was not significantly changed by MMF in either naïve or memory CD4 T cells. (C) MMF had an even greater hypermethylating effect on the MIR-21 locus of naïve CD4 T cells that were stimulated under Th17 polarizing conditions. (D) MMF was also able to inhibit the demethylation of the MIR-21 promoter of naïve (CD45RO−CCR7+) CD8 T cells after 3 days of activation with anti-CD3 and anti-CD28 coated beads in vitro under Tc17 polarizing conditions. MMF20 = 20 µM of MMF; MMF50 = 50 µM of MMF; Veh = vehicle.
We then asked whether the effect of FAE treatment on DNA methylation was selective for the MIR-21 locus and analysed methylation of the TNF promoter in DNA isolated from naïve and memory CD4 T cells treated either with vehicle or with FAE (50 µM MMF). Consistent with the previously described role of DNA methylation during immune cell differentiation, EpiTYPER™ MassARRAY® analysis of the TNF promoter revealed significant differences in methylation levels of ex vivo naïve compared to memory CD4 T cells [two-way ANOVA; n = 3, F(1,16) = 812.5, P < 0.0001] (Supplementary Fig. 8B). However, no differences were detected between the FAE-treated and untreated group of either naïve or memory CD4 T cells [two-way ANOVA naïve n = 4, F(1,24) = 0.616, P = 0.4402; memory n = 3, F(1,16) = 1.179, P = 0.2937] (Fig. 5B). These results suggest that FAE treatment in vitro does not impact DNA demethylation broadly and non-specifically, but on the contrary FAE’s hypermethylation effects are targeted to the MIR-21 locus whereas other genomic regions, such as the TNF promoter, are being demethylated normally.
FAEs prevent the upregulation of miR-21 under Th17 polarizing conditions and reduce CCR6 production
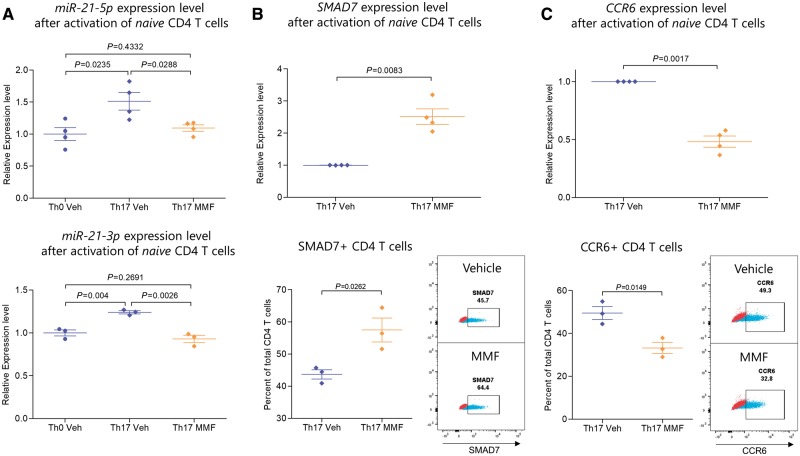
Since MIR-21 was highly methylated in Th17 cells of FAE-treated patients and this microRNA had been previously found to be important for Th17 differentiation in mice (Murugaiyan et al., 2015), we hypothesized that its expression levels could be impacted in FAE-treated human Th17 cells. To this end, we treated naïve human CD4 T cells with either FAEs (50 µM MMF) or vehicle, under Th17 polarizing conditions and measured the expression of both mature miR-21 transcripts, miR-21-5p and miR-21-3p. We then compared these levels to miR-21 expression under no polarizing conditions (Th0). As expected, Th17 polarization resulted in higher miR-21 expression compared to no polarization [two-tailed t-test; miR-21-5p n = 4, t(6) = 3.015 P = 0.0235; miR-21-3p n = 3, t(4) = 5.966, P = 0.004] (Fig. 6A). FAE treatment resulted in reduced expression of both mature transcripts of miR-21 in CD4 T cells under Th17 polarizing conditions in vitro (two-tailed t-test, miR-21-5p, n = 4, t(6) = 2.860, P = 0.0288; miR-21-3p, n = 3, t(4) = 6.671, P = 0.0026] (Fig. 6A). miR-21 expression in FAE-treated CD4 T cells under Th17 polarizing conditions was in fact similar to the miR-21 level in Th0 CD4 T cells [two-tailed t-test; miR-21-5p t(6) = 0.8399 P = 0.4332; miR-21-3p t(4) = 1.282, P = 0.2691] (Fig. 6A), thereby suggesting that FAEs were able to block the effect of Th17 polarization on miR-21 expression. Of note, CD4 T cell viability was not affected by this in vitro treatment schedule [n = 3; two-tailed t-test, t(4) = 0.5954, P = 0.5836] (Supplementary Fig. 9). Furthermore, we measured the expression of SMAD7, a previously-identified miR-21 target in CD4 T cells that serves as an inhibitor of the TGF-beta signalling pathway and Th17 differentiation (Yan et al., 2009; Murugaiyan et al., 2015). We found that FAE treatment of naïve CD4 T cells under Th17 polarizing conditions increased SMAD7 transcript levels [Welch’s t-test, n = 4, t(3) = 6.231, P = 0.0083] as well as the percentage of SMAD7+ CD4 T cells [two-tailed t-test, n = 3, t(4) = 3.445, P = 0.0262] compared to vehicle, which is in accordance with the down regulation of miR-21 in these cells (Fig. 6B). Finally, FAEs were able to reduce the expression of CCR6 measured by qPCR [Welch’s t-test, n = 4, t(3) = 10.89, P = 0.0017] as well as the percentage of CCR6+ CD4 T cells measured by flow cytometry [two-tailed t-test, n = 3, t(4) = 4.094, P = 0.0149] (Fig. 6C).
Figure 6.
FAE treatment reduced miR-21 and CCR6 expression and increased SMAD7 expression of CD4 T cells under Th17 polarizing conditions. Naïve (CD45RO−CCR7+) CD4 T cells were isolated from human PBMCs by FACS and were stimulated with anti-CD3 and anti-CD28 coated beads and cultured either without polarization (Th0) or under Th17 polarizing conditions with or without 50 µM of MMF. The expression of mature miR-21 transcripts (miR-21-5p and miR-21-3p) was measured by qPCR, which was first normalized to the geometric mean of SNORD44, SNORD48 and RNU6 expression level and then to the average relative expression level of the Th0 condition from all human donors. CCR6 and SMAD7 transcript levels were first normalized to the geometric mean of ACTB, B2M and HPRT1 expression level and then to the relative expression level of the vehicle treated Th17 condition from each human donor. Protein expression was measured by flow cytometry. (A) MMF resulted in the inhibition of upregulation of both miR-21-5p and miR-21-3p in Th17 polarizing conditions and kept their expression at Th0 levels. (B) SMAD7, a previously-identified miR-21 target that is known to inhibit the TGF-beta signalling pathway, was upregulated by MMF treatment as measured by qPCR and flow cytometry. (C) MMF reduced the expression of CCR6 measured by qPCR as well as the percentage of CCR6+ CD4 T cells measured by flow cytometry. MMF = 50 µM of MMF; Veh = vehicle. Flow cytometry plots show a representative sample of either vehicle or MMF treated CD4 T cells in blue and the fluorescence minus one (FMO) control in red, which was used for gating the positive population.
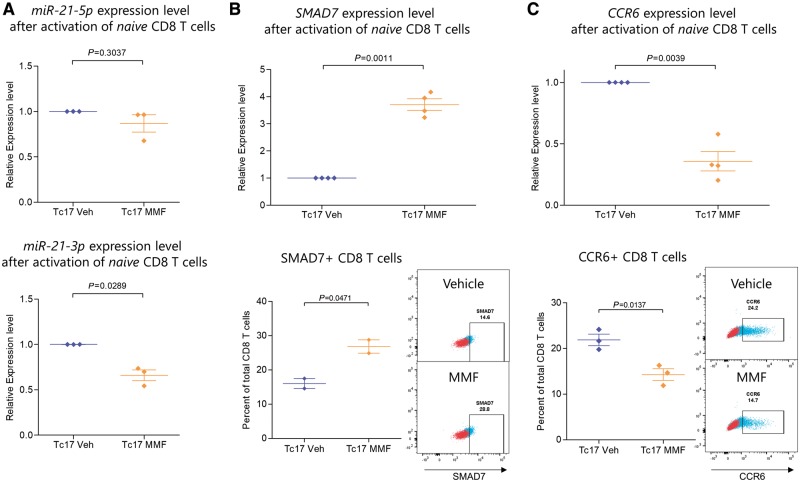
FAEs exerted a similar effect on CD8 T cells that were activated under Tc17 polarizing conditions, which exhibited reduced miR-21-3p transcript levels compared to vehicle, however miR-21-5p was unchanged [Welch’s t-test, miR-21-5p t(2) = 1.372, P = 0.3037; miR-21-3p t(2) = 5.754, P = 0.0289] (Fig. 7A). Furthermore, FAE treatment was able to upregulate SMAD7, which can also be targeted by miR-21-3p, suggesting that the change in mature miR-21 transcripts by FAEs has a functional effect in CD8 T cells as well [Welch’s t-test, n = 4, t(3) = 12.61, P = 0.0011] (Fig. 7B). FAEs also increased the percentage of SMAD7+ CD8 T cells [two-tailed t-test, n = 2, t(2) = 4.444, P = 0.0471], as well as reduced the expression of CCR6 transcripts [Welch’s t-test, n = 4, t(3) = 8.128, P = 0.0039] and lowered the percentage of CCR6+ CD8 T cells [two-tailed t-test, n = 3, t(4) = 4.199, P = 0.0137] (Fig. 7B and C). Again, the viability of naïve CD8 T cells was not affected by MMF (Supplementary Fig. 9). Collectively these results provide a potential mechanistic explanation for our immunophenotyping results from multiple sclerosis patients.
Figure 7.
CD8 T cells treated with FAEs under Tc17 polarizing conditions also exhibited reduced miR-21 and CCR6 expression and higher SMAD7 levels. Naïve (CD45RO−CCR7+) CD8 T cells were isolated from human peripheral blood mononuclear cells by FACS and were stimulated with anti-CD3 and anti-CD28 coated beads under Tc17 polarizing conditions with or without 50 µM of MMF for 3 days. The expression of mature miR-21 transcripts (miR-21-5p and miR-21-3p) was measured by qPCR, which were first normalized to the geometric mean of SNORD44, SNORD48 and RNU6 expression level and then to the relative expression level of the vehicle treated Tc17 condition from each human donor. CCR6 and SMAD7 transcript levels were first normalized to the geometric mean of ACTB, B2M and HPRT1 expression level and then to the relative expression level of the vehicle treated Tc17 condition from each human donor. Protein expression was measured by flow cytometry. (A) MMF resulted in the downregulation primarily of miR-21-3p in CD8 T cells under Tc17 polarizing conditions. (B) MMF treatment resulted in the upregulation of SMAD7 transcripts and increased the percentage of SMAD7+ CD8 T cells. (C) MMF treatment reduced the expression of CCR6 transcripts in CD8 T cells as well as the percentage of CCR6+ CD8 T cells. MMF = 50 µM of MMF; Veh = vehicle. Flow cytometry plots show a representative sample of either vehicle or MMF treated CD8 T cells in blue and the fluorescence minus one (FMO) control in red, which was used for gating the positive population.
Discussion
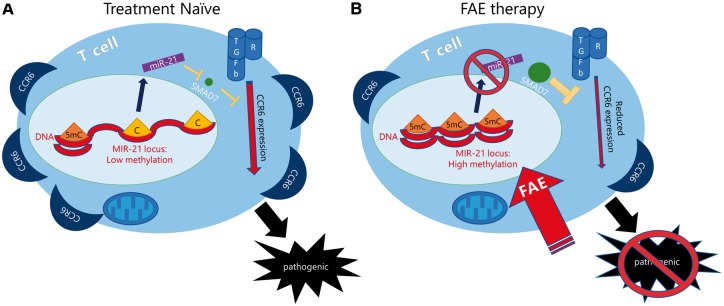
FAEs are an effective oral multiple sclerosis therapeutic, with a mechanism of action that is not completely understood. Here, we propose a novel mechanism of immunomodulation in multiple sclerosis by FAEs, which leverage the epigenetic effect of a metabolite (i.e. fumaric acid) on DNA methylation in CD4 and CD8 T cells to produce an immunomodulatory effect in multiple sclerosis patients. Based on our stringent epigenome-wide DNA methylation analysis, FAE therapy specifically hypermethylates a genomic locus in CD4 T cells of multiple sclerosis patients that encodes for miR-21, a microRNA critical for Th17 differentiation, CCR6 production and Th17-mediated autoimmunity (Murugaiyan et al., 2015). MIR-21 was found to be specifically hypermethylated in Th17 cells of FAE-treated patients after our decomposition analysis, thereby linking its hypermethylation with the Th17 cell reduction that we observed in FAE-treated patients with multiple sclerosis. FAEs induced DNA hypermethylation at the MIR-21 promoter of both CD4 and CD8 T cells in vitro and were associated with decreased levels of miR-21 transcripts and consequent upregulation of its target SMAD7 in developing Th17 and Tc17 cells in vitro. Finally, FAEs were able to reduce CCR6+ CD4 and CD8 T cells in patients with multiple sclerosis and also inhibit CCR6 expression in developing Th17 and Tc17 cells in vitro, thereby reducing the production of potentially pathogenic brain-homing CD4 and CD8 T cells (Cao et al., 2015; Liang et al., 2015) (Fig. 8).
Figure 8.
FAE therapy modifies the metabolic-epigenetic interplay in multiple sclerosis to reduce pathogenic CCR6+ CD4 and CD8 T cells. (A) CD4 T cells from treatment naïve patients with multiple sclerosis exhibit low DNA methylation levels at the MIR-21 locus and high expression of CCR6. CD8 T cells from treatment naïve multiple sclerosis patients also exhibit high levels of CCR6. (B) FAE therapy epigenetically modulates the MIR-21 locus in CD4 and CD8 T cells, which results in hypermethylation and downregulation of miR-21 expression in FAE-treated Th17 and Tc17 cells. This in turn allows for the upregulation of SMAD7 in both CD4 and CD8 T cells, a miR-21-5p and miR-21-3p target with inhibitory feedback on the TGF-b signalling pathway. Finally, FAEs reduce CCR6 expression and lower this potentially pathogenic brain-homing CCR6+ CD4 and CD8 T cell population in FAE-treated multiple sclerosis patients.
CCR6+ CD4 T cells have come to the forefront of multiple sclerosis research after Reboldi et al. (2009) demonstrated that CCR6 is essential for Th17 cell entry in the uninflamed CNS and initiation of EAE. CCR6 has also been found to be required by Th17 cells to cooperate with CD8+ T cells for infiltration into the CNS (Huber et al., 2013). Moreover, myelin-reactive CD4 T cells from the blood of patients with multiple sclerosis have been found to be CCR6+ CD4 T cells (Cao et al., 2015). Finally, CCR6+ CD4 T cells have been implicated in the pathogenesis of psoriasis (Lee and Hwang, 2012), another immune-mediated disease for which FAEs are an effective treatment.
CCR6+ CD8 T cells include Tc17 cells, which have been found to be CCR6+ and CCR4− (Kondo et al., 2009), the subtype of CD8 T cells most affected by FAE in our study. Tc17 cells are present in increased numbers in the CSF of multiple sclerosis patients (Huber et al., 2013) and it has been shown that they can render Th17 cells more encephalitogenic during induction of EAE in a cell contact-dependent manner (Huber et al., 2013; Liang et al., 2015). Finally, Tc17 cells have been found in inflamed tissues and peripheral blood of many immune-mediated diseases, such as multiple sclerosis, psoriasis, systemic lupus erythematosus, rheumatoid arthritis, and inflammatory bowel disease (Liang et al., 2015).
miR-21 has been shown to be upregulated in PBMCs from patients with multiple sclerosis, especially during the acute relapse phase (Fenoglio et al., 2011), and in CNS-infiltrating T cells in mouse models of multiple sclerosis (Mycko et al., 2012), as well as in T cells from patients with psoriasis (Meisgen et al., 2012). Since miR-21 expression is required for CCR6 production in T cells (Murugaiyan et al., 2015), these observations further support our proposed mechanism of action of FAEs in patients with multiple sclerosis. Hypermethylation of the MIR-21 locus by FAEs could be a mechanism to prevent its upregulation in encephalitogenic CD4 and CD8 T cells and thereby contribute to remission and prevention of relapses in patients with multiple sclerosis taking the drug. Of note, some studies have observed downregulation of miR-21 in relapsing-remitting multiple sclerosis patients compared to healthy controls (Muñoz-Culla et al., 2016; Ruhrmann et al., 2017); however, this contradiction could be the result of cell-type proportion differences between multiple sclerosis and healthy control samples, the use of steroids for treatment of relapses or the initiation of other disease modifying therapies in these patients.
Interestingly, our DNA methylation analysis in multiple sclerosis patients revealed a specific effect of FAE treatment on the MIR-21 locus. Other loci important for Th17 function, such as IL17 or RORC, as well as promoters of master cytokines, such as TNF, were not affected by FAEs, a property that could potentially minimize the disruption of T cell function and reduce its overall immunosuppressant effect. Moreover, the specificity of the effect suggests that our analysis effectively controlled for potential naïve/memory CD4 T cell imbalances, as cellular heterogeneity in samples of DNA methylation analyses is expected to lead to broad changes in the average DNA methylation of the samples. Furthermore, despite the fact that GA was able to reduce Th17 cells in multiple sclerosis patients, it did not affect DNA methylation at the MIR-21 locus. Therefore, our results demonstrate a novel immunomodulatory effect of FAEs that is mediated via a unique hypermethylating effect at the MIR-21 locus of CD4 and CD8 T cells. Our results also suggest that FAEs cannot modulate DNA methylation levels in already polarized cells with unmethylated DNA, given the lack of effect in memory CD4 T cells in vitro. However, FAEs could specifically modulate DNA methylation levels during activation or polarization of naïve CD4 and CD8 T cells with hypermethylated DNA. Although this pattern of methylation changes suggests an effect of FAEs on DNA demethylases during T cell activation, more studies need to be done to define the effect of FAEs on DNA methylases or demethylases.
Our findings collectively suggest FAEs as epigenetic immunomodulators of brain-homing CCR6+ CD4 and CD8 T cells in multiple sclerosis, which separates them from other multiple sclerosis therapeutics, such as GA. Our results also suggest that the metabolic-epigenetic interplay in T cells could be harnessed for therapeutic purposes, as FAEs affect both T cell metabolism by inhibiting aerobic glycolysis (Kornberg et al., 2018) and also influence their epigenetic regulation. This metabolic-epigenetic interplay could also be targeted in other immune-mediated diseases that are dependent on miR-21 overexpression. Apart from multiple sclerosis and psoriasis, miR-21 has been found to be overexpressed in systemic lupus erythematosus, where its levels correlate with disease activity (Stagakis et al., 2011). miR-21 has also been implicated in the pathogenesis of delayed-type-hypersensitivity (Lu et al., 2011) and in mouse models of colitis (Shi et al., 2013). Indeed, FAEs have already been successfully used in mouse models of colitis (Casili et al., 2016), and case reports of successful treatment of SLE exist in the literature (Balak and Thio, 2011).
Since DNA methylation changes could be regarded a somewhat stable epigenetic modification, our findings raise the question of how permanent this epigenetic immunomodulation is. Do these epigenetically modified T-cells revert to an autoimmune phenotype after discontinuation of therapy or is the epigenetic manipulation of microRNA expression with therapeutic metabolites a potential long-lasting treatment of autoimmune disease? Interestingly, a significantly delayed lymphocyte repopulation has already been observed in multiple sclerosis patients following discontinuation of FAEs (Khatri et al., 2017). More research is needed to be done, however, to answer these questions and further delineate the link between DNA methylation changes in T cells and cellular metabolism.
Supplementary Material
Acknowledgements
We would like to thank the Epigenetics Core at the CUNY Advanced Science Research Center for the provision of resources and expertise for the EpiTYPER MassArray and qPCR analyses. We would also like to thank Eran Halperin PhD and Sam Horng MD PhD for their insightful comments. Finally, the authors would like to thank all of those who helped in patient recruitment, especially the multiple sclerosis clinicians (Fred Lublin, Aaron Miller, Stephen Krieger, Michelle Fabian, Sylvia Klineova, Aliza Ben-Zacharia, and Gretchen Mathewson) and research coordinators at Mount Sinai.
Glossary
Abbreviations
- DMR
differentially methylated region;
- FAE
fumaric acid ester
- GA
glatiramer acetate
Funding
Funds were provided by the Friedman Brain Institute of Mount Sinai and NIH-R37-NS42925.
Competing interests
The authors report no competing interests.
References
- Almeida L, Lochner M, Berod L, Sparwasser T. Metabolic pathways in T cell activation and lineage differentiation. Semin Immunol 2016; 28: 514–24. [DOI] [PubMed] [Google Scholar]
- Balak DM, Thio H. Treatment of lupus erythematosus with fumaric acid ester derivatives: two case-reports. J Transl Med 2011; 9: P15. [Google Scholar]
- Buck MD, O’Sullivan D, Pearce EL. T cell metabolism drives immunity. J Exp Med 2015; 212: 1345–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JJ, O’Connell DJ, Wurbel M-A. Cutting edge: chemokine receptor CCR4 is necessary for antigen-driven cutaneous accumulation of CD4 T cells under physiological conditions. J Immunol 2007; 178: 3358–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Goods BA, Raddassi K, Nepom GT, Kwok WW, Love JC, et al. Functional inflammatory profiles distinguish myelin-reactive T cells from patients with multiple sclerosis. Sci Transl Med 2015; 7: 287ra74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casili G, Cordaro M, Impellizzeri D, Bruschetta G, Paterniti I, Cuzzocrea S, et al. Dimethyl fumarate reduces inflammatory responses in experimental colitis. J Crohn’s Colitis 2016; 10: 472–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen BC, Smith AA, Zheng S, Koestler DC, Houseman EA, Marsit CJ, et al. DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. J Natl Cancer Inst 2011; 103: 143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunay GA, Tóth I, Eberhard JM, Degen O, Tolosa E, van Lunzen J, et al. Parallel assessment of Th17 cell frequencies by surface marker co-expression versus ex vivo IL-17 production in HIV-1 infection. Cytom Part B - Clin Cytom 2016; 90: 486–92. [DOI] [PubMed] [Google Scholar]
- Durek P, Nordström K, Gasparoni G, Salhab A, Kressler C, de Almeida M, et al. Epigenomic profiling of human CD4+T cells supports a linear differentiation model and highlights molecular regulators of memory development. Immunity 2016; 45: 1148–61. [DOI] [PubMed] [Google Scholar]
- Fagin U, Pitann S, Gross WL, Lamprecht P. Increased frequency of CCR4+ and CCR6+ memory T-cells including CCR7+CD45RA med very early memory cells in granulomatosis with polyangiitis (Wegener’s). Arthritis Res Ther 2012a; 14: R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagin U, Pitann S, Gross WL, Lamprecht P. Increased frequency of CCR4+ and CCR6+ memory T-cells including CCR7+CD45RA med very early memory cells in granulomatosis with polyangiitis (Wegener’s). Arthritis Res Ther 2012b; 14: R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenoglio C, Cantoni C, De Riz M, Ridolfi E, Cortini F, Serpente M, et al. Expression and genetic analysis of miRNAs involved in CD4+ cell activation in patients with multiple sclerosis. Neurosci Lett 2011; 504: 9–12. [DOI] [PubMed] [Google Scholar]
- Fox RJ, Chan A, Gold R, Phillips JT, Selmaj K, Chang I, et al. Characterizing absolute lymphocyte count profiles in dimethyl fumarate’treated patients with MS Patient management considerations. Neurol Clin Pract 2016; 6: 220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox RJ, Miller DH, Phillips JT, Hutchinson M, Havrdova E, Kita M, et al. Placebo-controlled phase 3 study of oral BG-12 or glatiramer in multiple sclerosis. N Engl J Med 2012; 367: 1087–97. [DOI] [PubMed] [Google Scholar]
- Ghadiri M, Rezk A, Li R, Evans A, Luessi F, Zipp F, et al. Dimethyl fumarate-induced lymphopenia in MS due to differential T-cell subset apoptosis. Neurol Neuroimmunol NeuroInflammation 2017; 4: e340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemi S, Bedoya F, Nunez-Cruz S, June C, Melenhorst J, Milone M. Shortened T cell culture with IL-7 and IL-15 provides the most potent chimeric antigen receptor (CAR)-modified T cells for adoptive immunotherapy. J Immunol 2016; 196: 214.23. [Google Scholar]
- Gold R, Kappos L, Arnold DL, Bar-Or A, Giovannoni G, Selmaj K, et al. Placebo-controlled phase 3 study of oral BG-12 for relapsing multiple sclerosis. N Engl J Med 2012; 367: 1098–107. [DOI] [PubMed] [Google Scholar]
- Gross CC, Schulte-Mecklenbeck A, Klinsing S, Posevitz-Fejfár A, Wiendl H, Klotz L. Dimethyl fumarate treatment alters circulating T helper cell subsets in multiple sclerosis. Neurol - Neuroimmunol Neuroinflammation 2016; 3: e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M, Heink S, Pagenstecher A, Reinhard K, Ritter J, Visekruna A, et al. IL-17A secretion by CD8+T cells supports Th17-mediated autoimmune encephalomyelitis. J Clin Invest 2013; 123: 247–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh JL, Garg P, Thin TH, Yoo S, Dutta R, Trapp BD, et al. Epigenome-wide differences in pathology-free regions of multiple sclerosis-affected brains. Nat Neurosci 2014; 17: 121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyama K, Chen T, Wang X, Yan X, Kim BS, Tanaka S, et al. The Methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity 2015; 42: 613–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Murakami P, Lee H, Leek JT, Fallin MD, Feinberg AP, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol 2012; 41: 200–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri BO, Tarima SS, Essig B, Sesing J, Olapo T. Delayed lymphocyte re-population following discontinuation of dimethyl fumarate and after switching to other disease modifying drug therapies. Mult Scler Relat Disord 2017; 18: 60–4. [DOI] [PubMed] [Google Scholar]
- Kondo T, Takata H, Matsuki F, Takiguchi M. Cutting edge: phenotypic characterization and differentiation of human CD8+ T cells producing IL-17. J Immunol 2009; 182: 1794–8. [DOI] [PubMed] [Google Scholar]
- Kondo T, Takata H, Takiguchi M. Functional expression of chemokine receptor CCR6 on human effector memory CD8+ T cells. Eur J Immunol 2007; 37: 54–65. [DOI] [PubMed] [Google Scholar]
- Kornberg MD, Bhargava P, Kim PM, Putluri V, Snowman AM, Putluri N, et al. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 2018; 360: 449–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Langelaar J, Van Der Vuurst De Vries RM, Janssen M, Wierenga-Wolf AF, Spilt IM, Siepman TA, et al. T helper 17.1 cells associate with multiple sclerosis disease activity: Perspectives for early intervention. Brain 2018; 141: 1334–49. [DOI] [PubMed] [Google Scholar]
- Lee CH, Hwang STY. Pathophysiology of chemokines and chemokine receptors in dermatological science: a focus on psoriasis and cutaneous T-cell lymphoma. Dermatologica Sin 2012; 30: 128–35. [Google Scholar]
- Liang Y, Pan HF, Ye DQ. Tc17 cells in immunity and systemic autoimmunity. Int Rev Immunol. 2015; 34: 318–31. [DOI] [PubMed] [Google Scholar]
- Longbrake EE, Naismith RT, Parks BJ, Wu GF, Cross AH. Dimethyl fumarate-associated lymphopenia: Risk factors and clinical significance. Mult Scler J – Exp Transl Clin 2015; 1: 205521731559699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab 2012; 16: 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TX, Hartner J, Lim E-J, Fabry V, Mingler MK, Cole ET, et al. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN- pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J Immunol 2011; 187: 3362–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltby VE, Lea RA, Sanders KA, White N, Benton MC, Scott RJ, et al. Differential methylation at MHC in CD4+ T cells is associated with multiple sclerosis independently of HLA-DRB1. Clin Epigenet 2017; 9: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisgen F, Xu N, Wei T, Janson PC, Obad S, Broom O, et al. miR-21 is up-regulated in psoriasis and suppresses T cell apoptosis. Exp Dermatol 2012; 21: 312–4. [DOI] [PubMed] [Google Scholar]
- Michels KB, Binder AM, Dedeurwaerder S, Epstein CB, Greally JM, Gut I, et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat Methods 2013; 10: 949–55. [DOI] [PubMed] [Google Scholar]
- Montes M, Rufer N, Appay V, Reynard S, Pittet MJ, Speiser DE, et al. Optimum in vitro expansion of human antigen-specific CD8+ T cells for adoptive transfer therapy. Clin Exp Immunol 2005; 142: 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyon S, Huynh JL, Dutta D, Zhang F, Ma D, Yoo S, et al. Functional characterization of DNA methylation in the oligodendrocyte lineage. Cell Rep 2016; 15: 748–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrowietz U, Morrison PJ, Suhrkamp I, Kumanova M, Clement B. The pharmacokinetics of fumaric acid esters reveal their in vivo effects. Trends Pharmacol Sci 2017; 39: 1–12. [DOI] [PubMed] [Google Scholar]
- Muñoz-Culla M, Irizar H, Sáenz-Cuesta M, Castillo-Triviño T, Osorio-Querejeta I, Sepúlveda L, et al. SncRNA (microRNA &snoRNA) opposite expression pattern found in multiple sclerosis relapse and remission is sex dependent. Sci Rep 2016; 6: 20126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugaiyan G, Da Cunha AP, Ajay AK, Joller N, Garo LP, Kumaradevan S, et al. MicroRNA-21 promotes Th17 differentiation and mediates experimental autoimmune encephalomyelitis. J Clin Invest 2015; 125: 1069–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mycko MP, Cichalewska M, Machlanska A, Cwiklinska H, Mariasiewicz M, Selmaj KW. microRNA-301a regulation of a T-helper 17 immune response controls autoimmune demyelination. Proc Natl Acad Sci 2012; 109: E1248–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, V Lord R, et al. De novo identification of differentially methylated regions in the human genome. Epigenet Chromatin 2015; 8: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polman CH, Reingold SC, Banwell B, Clanet M, Cohen JA, Filippi M, et al. Diagnostic criteria for multiple sclerosis: 2010 Revisions to the McDonald criteria. Ann Neurol 2011; 69: 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboldi A, Coisne C, Baumjohann D, Benvenuto F, Bottinelli D, Lira S, et al. C-C chemokine receptor 6-regulated entry of TH-17 cells into the CNS through the choroid plexus is required for the initiation of EAE. Nat Immunol 2009; 10: 514–23. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivino L, Messi M, Jarrossay D, Lanzavecchia A, Sallusto F, Geginat J. Chemokine receptor expression identifies Pre–T Helper (Th)1, Pre–Th2, and nonpolarized cells among human CD4 + central memory T cells. J Exp Med 2004; 200: 725–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhrmann S, Ewing E, Piket E, Kular L, Cetrulo Lorenzi JC, Fernandes SJ, et al. Hypermethylation of MIR21 in CD4+ T cells from patients with relapsing-remitting multiple sclerosis associates with lower miRNA-21 levels and concomitant up-regulation of its target genes. Mult Scler J 2017; 24: 1288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl C, Klug M, Boeld TJ, Andreesen R, Hoffmann P, Edinger M, et al. Lineage-specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome Res 2009; 19: 1165–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciacovelli M, Gonçalves E, Johnson TI, Zecchini VR, Da Costa ASH, Gaude E, et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature 2016; 537: 544–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellars M, Huh JR, Day K, Issuree PD, Galan C, Gobeil S, et al. Regulation of DNA methylation dictates Cd4 expression during the development of helper and cytotoxic T cell lineages. Nat Immunol 2015; 16: 746–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Liang Y, Yang J, Xia Y, Chen H, Han H, et al. MicroRNA-21 knockout improve the survival rate in DSS induced fatal colitis through protecting against inflammation and tissue injury. PLoS One 2013; 8: e66814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagakis E, Bertsias G, Verginis P, Nakou M, Hatziapostolou M, Kritikos H, et al. Identification of novel microRNA signatures linked to human lupus disease activity and pathogenesis: MiR-21 regulates aberrant T cell responses through regulation of PDCD4 expression. Ann Rheum Dis 2011; 70: 1496–506. [DOI] [PubMed] [Google Scholar]
- Tian Y, Morris TJ, Webster AP, Yang Z, Beck S, Feber A, et al. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics 2017; 33: 3982–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzartos JS, Friese MA, Craner MJ, Palace J, Newcombe J, Esiri MM, et al. Interleukin-17 production in central nervous system-infiltrating T cells and glial cells is associated with active disease in multiple sclerosis. Am J Pathol 2008; 172: 146–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Kang SG, Lee J, Sun Z, Kim CH. The roles of CCR6 in migration of Th17 cells and regulation of effector T-cell balance in the gut. Mucosal Immunol 2009; 2: 173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CT, Roussos P, Garg P, Ho DJ, Azam N, Katsel PL, et al. Genome-wide12 DNA methylation profiling in the superior temporal gyrus reveals epigenetic signatures associated with Alzheimer’s disease. Genome Med 2016; 8: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC. Combining probability from independent tests: the weighted Z-method is superior to Fisher’s approach. J Evol Biol 2005; 18: 1368–73. [DOI] [PubMed] [Google Scholar]
- Wong CC, Qian Y, Yu J. Interplay between epigenetics and metabolism in oncogenesis: mechanisms and therapeutic approaches. Oncogene 2017; 36: 3359–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Wang Q, Mao G, Dowling CA, Lundy SK, Mao-Draayer Y. Dimethyl fumarate selectively reduces memory T cells and shifts the balance between Th1/Th17 and Th2 in multiple sclerosis patients. J Immunol 2017; 198: 3069–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev 2012; 26: 1326–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Liu Z, Chen Y. Regulation of TGF-β signaling by Smad7. Acta Biochim Biophys Sin (Shanghai) 2009; 41: 263–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data have been deposited in NCBI’s Gene Expression Omnibus (Edgar, 2002) and are accessible through GEO Series accession number GSE112596 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112596). The R or python scripts are available upon request.