Abstract
Defective placental implantation and vascularization with accompanying hypoxia contribute to preeclampsia (PE), a leading cause of maternal and neonatal morbidity and mortality. Genetic and epigenetic mechanisms underlying differentiation of proliferative cytotrophoblasts (CytTs) to multinucleated syncytiotrophoblast (SynT) are incompletely defined. The SynT performs key functions in nutrient and gas exchange, hormone production, and protection of the fetus from rejection by the maternal immune system. In this study, we used chromatin immunoprecipitation sequencing of midgestation human trophoblasts before CytT and after SynT differentiation in primary culture to analyze changes in binding of RNA polymerase II (Pol II) and of active and repressive histone marks during SynT differentiation. Our findings reveal that increased Pol II binding to promoters of a subset of genes during trophoblast differentiation was closely correlated with active histone marks. This gene set was enriched in those controlling immune response and immune modulation, including interferon-induced tetratricopeptide repeat and placenta-specific glycoprotein gene family members. By contrast, genes downregulated during SynT differentiation included proinflammatory transcription factors ERG1, cFOS, and cJUN, as well as members of the NR4A orphan nuclear receptor subfamily, NUR77, NURR1, and NOR1. Downregulation of proinflammatory transcription factors upon SynT differentiation was associated with decreased promoter enrichment of endogenous H3K27Ac and H3K9Ac and enhanced binding of H3K9me3 and histone deacetylase 1. However, promoter enrichment of H3K27me3 was low in both CytT and SynT and was not altered with changes in gene expression. These findings provide important insight into mechanisms underlying human trophoblast differentiation and may identify therapeutic targets for placental disorders, such as PE.
Proper implantation and development of the placenta are crucial for successful pregnancy. The basic functional unit of the human placenta is the chorionic villus. Cells lining the inner layer of the villus comprise dividing mononuclear cytotrophoblasts (CytT). These detach from the basement membrane and fuse to form the multinuclear syncytiotrophoblast (SynT), which comprises the outer layer of the villus and is bathed in maternal blood. The SynT mediates nutrient, gas, and waste exchange between mother and fetus and produces a variety of hormones, including estrogens and progesterone (1, 2). The SynT actively invades the maternal decidua. Defective trophoblast invasion with placental hypoxia is an underlying cause of preeclampsia (PE), a hypertensive disorder of pregnancy (2, 3). The SynT also may serve an important role in pregnancy maintenance by producing immunomodulatory factors, which protect the hemiallogeneic fetus from rejection by the maternal immune system (4, 5).
Human trophoblasts in primary culture provide a robust and biologically relevant system for defining the molecular mechanisms underlying altered gene expression during differentiation of CytT to SynT. Using this system, we previously observed that expression of hCYP19A1/aromatase P450, the key regulatory enzyme in estrogen biosynthesis (6), and the critical transcription factors estrogen-related receptor γ (7), estrogen receptor α (8), and glial cells missing 1 (9, 10) were dramatically upregulated during CytT to SynT differentiation, whereas the E-box–binding transcription factors MASH2/ASCL2 and USF1/2 declined (11, 12). Thus, changes in transcription factor expression and DNA binding and the underlying epigenetic modifications may be primary mediators of trophoblast differentiation. Importantly, the genetic and epigenetic mechanisms underlying human SynT differentiation remain incompletely defined.
Gene transcription is regulated by changes in chromatin structure mediated by a number of histone and DNA modifications (13–17). In its inactive state, DNA is tightly coiled around nucleosomes, which are the basic building blocks of chromatin (18, 19). This is facilitated, in part, by repressive histone marks, including trimethylation of lysines 9 and 27 of histone H3 (13, 20). The binding of activating transcription factors to DNA promotes the recruitment of coregulators and chromatin remodelers, which catalyze activating histone modifications and remove repressive histone marks. These activating modifications include monomethylation and trimethylation of lysine 4 of histone H3 (21) and acetylation of histone H3 on lysines 9 and 27 (13, 20). This facilitates the opening of chromatin, which allows recruitment of a preinitiation complex to the promoter that includes RNA polymerase II (Pol II), which catalyzes RNA transcription. In this study, chromatin immunoprecipitation sequencing (ChIP-seq) analysis of binding of Pol II and key modified histones was used to identify differentially regulated genes and underlying mechanisms involved in differentiation of CytT to SynT in culture.
Materials and Methods
Isolation of trophoblasts
This study was approved by the Institutional Review Board of the University of Texas Southwestern Medical Center. Informed consent was obtained from all participants. CytT were isolated from midgestation (16 to 20 weeks’ gestation) (Advanced Biosciences Resources, Alameda, CA) and term (Parkland Hospital) human placentas and placed in primary culture as described in detail previously (8, 22, 23). All ChIP-seq experiments were performed using trophoblasts isolated from 16 to 20 weeks’ gestation placentas. When trophoblasts from term placentas were used, this is indicated in the figure legends. Briefly, the placental tissues were washed with Hanks balanced salt solution (pH 7.4) (Life Technologies, Grand Island, NY), finely minced, and digested with 2.5% trypsin in Hanks balanced salt solution at 37°C for 20 minutes. The supernatant was collected, layered over 10 mL of serum, and then briefly centrifuged at 1000g. These steps were repeated twice with the pellet from the original trypsin digestion. The resulting pellet was suspended in DMEM (Life Technologies), filtered, and layered over a Percoll gradient (70% to 5%). The gradients were centrifuged at 1800g for 20 minutes at room temperature, and cells in the middle layer (density 1.045 to 1.062 g/mL) were collected, washed, and counted. The cells were then resuspended in DMEM supplemented with 10% fetal bovine serum and 1.2% antibiotic/antimycotic solution (Life Technologies). An aliquot of the cells was flash frozen as starting CytT, and the remainder was plated at a density of 2 × 106 cells per 35-mm culture dish or 15 × 106 cells per 100-mm dish and incubated for 18 hours in a standard CO2 incubator in 20% O2.
Morphological analysis of trophoblast fusion
As described previously (10), midgestation CytT were plated onto coverslips and cultured in DMEM/F12 containing 10% fetal bovine serum for 4 hours or 18 hours. The cells were rinsed in PBS, fixed in 4% paraformaldehyde, and blocked with 3% BSA in 1× PBS for 30 minutes at room temperature. Cell fusion was assessed by immunostaining using a primary antibody to plakoglobin (1:250; clone 15; BD Biosciences, San Jose, CA) (24) overnight at 4°C. The coverslips were washed three times with 1× PBS for 5 minutes each and incubated with a secondary antibody conjugated to Alexa Fluor 488 (25) (1:500) for 1 hour at room temperature. The coverslips were then washed three times with 1× PBS for 5 minutes each and mounted on glass slides in mounting medium containing 4′,6-diamidino-2-phenylindole (Fluoroshield; Sigma-Aldrich, St. Louis, MO). Images were captured using a Zeiss confocal microscope at ×400 magnification (Carl Zeiss, Oberkochen, Germany).
RNA isolation
Total RNA was purified using QIAzol lysis reagent and an miRNeasy Mini Kit (Qiagen, Valencia, CA) using a QIAcube platform (Qiagen).
Reverse transcription quantitative PCR
RNA expression was determined by reverse transcription quantitative PCR (RT-qPCR) using SYBR Green PCR Master Mix (Bio-Rad Laboratories, Hercules, CA) and normalized to 18S rRNA (8, 26) on a Bio-Rad CFX384 real-time PCR detection system with cDNA in triplicate. PCR primer sequences used for RT-qPCR are listed in Table 1.
Table 1.
Primer Sequences Used for RT-qPCR
| Gene Name | Forward Primer Sequence (5′ to 3′) | Reverse Primer Sequence (5′ to 3′) |
|---|---|---|
| CYP19AI.1 | ACGGAAGGTCCTGTGCTCG | GTATCGGGTTCAGCATTTCCA |
| C10orf10 | AGAGAGGAGAGGAGGAAGATTG | GAATAACCTAGACTGACAGCTCAC |
| EGFR | TTTGCCAAGGCACGAGTAA | CAAGGACCACCTCACAGTTATT |
| EGR1 | TAAAGGACAGGAGGAGGAGATG | GGAAGTGGGCAGAAAGGATT |
| FN1 | CCGAGTGGGTGACACTTATG | GTTTGCGATGGTACAGCTTATTC |
| FOS | GGTGCATTACAGAGAGGAGAAA | GTGTGTTTCACGCACAGATAAG |
| FOSB | CCTGACGGCTTCTCTCTTTAC | CAGGTGAGGACAAACGAAGA |
| GDF15 | ATGCACGCGCAGATCAA | ATGAGCACCATGGGATTGTAG |
| GULP1 | CAGGCAGTATGACACCTAAGTC | CAGGTCCCGTTTAATCTCAGTAG |
| HSPA5 | CCTTCGATGTGTCTCTTCTCAC | ACGCTGGTCAAAGTCTTCTC |
| JUN | CCTGATGTACCTGATGCTATGG | CCTCCTGAAACATCGCACTAT |
| KIAA1683 | ACGGTCACCATCAGATTTCC | TCACTGAGTCACCCTCTATGT |
| LRRC32 | CCTGTAAGATGGTGGACAAGAA | GCTGGTTCCCAGATAGATCAAG |
| PSG2 | CAGGGACCTCTACCATTACATTAC | TGCATTGGAATATGCTGTTTCTC |
| PSG4 | CAGAGGAGAACACACAAGCA | GGGCGGATTCCAGAAGTTTA |
| TEAD1 | CTTCTCTTTGTCTCTCCCAGATTAG | GGAGACTGCCATACAGAATGAA |
| h36B4 | TGCATCAGTACCCCATTCTATCA | AAGGTGTAATCCGTCTCCACAGA |
| 18S | GTAACCCGTTGAACCCCATT | CCATCCAATCGGTAGTAGCG |
Chromatin immunoprecipitation assay
Freshly isolated CytT before (0 hours) and after (18 hours) SynT differentiation in culture were cross-linked with freshly prepared 1% formaldehyde in PBS at room temperature for 10 minutes; the reaction was stopped by the addition of glycine (125 mM final). Cells (2 × 107) were washed twice with PBS, resuspended in 1 mL of nuclear extraction buffer [5 mM PIPES pH 8.0, 85 mM KCl, 0.5% Nonidet P-40, 1 mM PMSF, and protease inhibitors (Sigma-Aldrich)], and incubated for 10 minutes at 4°C. Nuclei were collected by centrifugation at 3000 rpm for 5 minutes at 4°C and resuspended (2 × 107 nuclei/mL) in nuclear lysis buffer [50 mM Tris-HCl, pH 8.1, 10 mM EDTA, 1% SDS, 1 mM PMSF, and protease inhibitors (Sigma-Aldrich)]. Nuclear lysates were either sonicated (for RNA Pol II ChIP) or digested with micrococcal nuclease (for modified histone ChIP). Nuclear lysates were sonicated using a Qsonica Q800R2 (Qsonica, Newtown, CT) at 70% intensity for 20 seconds on, 40 seconds off, for 20 minutes to generate 100- to 300-bp DNA fragments (27). Chromatin DNA was quantified using a Qubit® 3.0 Fluorometer (Life Technologies). In parallel, formaldehyde cross-linked nuclei (4 × 107 nuclei/mL) were resuspended in micrococcal nuclease digestion buffer [10 mM Tris-HCl, pH 8.0, 100 mM NaCl, and protease inhibitors (Sigma-Aldrich)] and briefly sonicated using the Qsonica Q800R2 at 30% intensity for 20 seconds on, 40 seconds off, for 2.5 minutes. The sonicated nuclei were incubated with 2000 gel units of micrococcal nuclease (New England Biolabs, Ipswich, MA) in 2 mM CaCl2 and 100 μg/mL BSA for 20 minutes at 37°C to produce mononucleosomes to trinucleosomes. Sonicated chromatin (3 μg) was incubated with 1 to 3 μg of antibody against RNA Pol II (N-20) (sc-899; Santa Cruz Biotechnology, Santa Cruz, CA) (28) for 16 hours at 4°C. Micrococcal nuclease-treated chromatin (1 μg) was incubated with 1 μg of antibody against histones H3K4me3 (39159; Active Motif, Carlsbad, CA) (29), H3K9Ac (ab4441; Abcam, Cambridge, MA) (30), H3K27Ac (39133; Active Motif) (31), or H3K27me3 (CS9733; Cell Signaling Technology, Danvers, MA) (32). Protein A or G dynabeads (20 μL) were then added to the chromatin-antibody mixtures and then incubated for 2 hours at 4°C. The beads were sequentially washed twice with the following buffers: low-salt buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, and 2 mM EDTA), high-salt buffer (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 1% Triton X-100, 0.1% SDS, and 2 mM EDTA), LiCl wash buffer (0.25 M LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mM EDTA, and 20 mM Tris-HCl, pH 8.0), and TE buffer (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA). Chromatin immunocomplexes were eluted by incubation for 10 minutes at 65°C with 1% SDS and 100 mM NaHCO3 and incubated with RNase at 1 mg/mL concentration. Cross-linking was reversed by incubation in the solution adjusted to 200 mM NaCl and proteinase K (20 μg) for 16 hours at 65°C. RT-qPCR was performed on CFX384 real-time PCR detection system (Bio-Rad Laboratories) to validate antibodies for ChIP assay.
ChIP-seq library preparation and sequencing
ChIP DNA was quantified on a Qubit® 3.0 Fluorometer (Life Technologies). Purified ChIP DNAs from five independent experiments were combined for each time point and used for ChIP-seq library preparation in duplicate. DNA (5 ng) was used to prepare ChIP-seq libraries using methods previously described in detail (27). Briefly, genomic DNA fragments were end-repaired, 3′-end adenylated, and ligated to Y-adaptors containing bar-coded sequences. After agarose-based size selection and purification with Ampure XP beads (Beckman Coulter, Brea, CA), the DNA was PCR-amplified (8 to 12 cycles) using KAPA HiFi HotStart Ready Mix (Kapa Biosystems, Wilmington, MA), size-selected with Ampure XP beads, and quantified on the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The final ChIP-seq libraries were subjected to quality control assessment using a Bioanalyer (Agilent Technologies) and submitted to the McDermott Center Next Generation Sequencing Core (University of Texas Southwestern Medical Center) for high-throughput sequencing using a HiSeq 2500 v3 (Illumina, San Diego, CA). Twenty to 50 million 50-nucleotide sequencing reads were generated for all ChIP experiments.
ChIP-seq data quality control, alignment, and peak calling
Quality of the ChIP-seq data sets was assessed using the FastQC tool. The ChIP-seq reads were aligned to the human reference genome (hg19) using BOWTIE (33). We uniquely mapped between 70% and 90% of the reads to the human genome. Uniquely mappable reads were converted into BigWig files using BEDTools for visualization in Integrative Genomics Viewer (34, 35). Peak calling was performed using MACS software (36) using a default P value of 0.01 and input as the control for each replicated sample. The resulting peaks were filtered by peak height to reduce false positives.
Differential binding analysis of RNA Pol II, H3K4me3, H3K9Ac, H3K27me3, and H3K27Ac in CytT vs SynT
Differential binding analyses were performed using the Bioconductor package edgeR (35). To capture prominent changes in Pol II binding to the promoters and gene body regions in CytT (0 hours) vs SynT (18 hours), only the significantly regulated genes based on P value and false discovery rate (FDR) were considered for further analyses. Genes manifesting increased Pol II binding in SynT vs CytT comprise those with a P value <2.99E-05 and FDR <0.001; genes manifesting decreased Pol II binding in SynT vs CytT comprise those with a P value <2.94E-05 and FDR <0.001. Lists of significantly up- and downregulated genes were submitted to the Gene Expression Omnibus and are shown as supplemental tables in an online repository [accession number GSE127288 (37)]. Lists of genes that manifested differential binding (up and down) of H3K4me3, H3K9Ac, and K3K27Ac in SynT vs CytT are also shown in this repository (37).
Gene ontology analysis
Gene ontology analyses were performed using Database for Annotation, Visualization, and Integrated Discovery tool (38).
Average profiles
Bigwig files were used to generate average ChIP-seq profiles of the CytT and SynT conditions of Pol II and other modifications under Pol II upregulated and downregulated genes using the next-generation sequencing data analysis suite of Python called Deep Tools.
Box plots
Box plot representations were used to quantitatively assess the read distribution in a fixed window around the transcription start site (TSS) for recruitment of Pol II and modified histones to each gene expressed in CytT and SynT. The read distribution in a 6-kb window (±3 kb) surrounding the TSS of genes was calculated and plotted using the box plot function in R. Wilcoxon rank-sum tests were performed to determine the statistical significance for all comparisons.
Venn diagrams
Venn diagrams were plotted using custom R scripts to assess the overlap of genes that showed increased Pol II binding and increased binding of the active histone H3 modifications H3K4me3, H3K9Ac, and H3K27Ac during trophoblast differentiation.
Results
Midgestation human CytT begin to syncytialize and express genes involved in cell fusion and SynT differentiation within 18 hours of culture
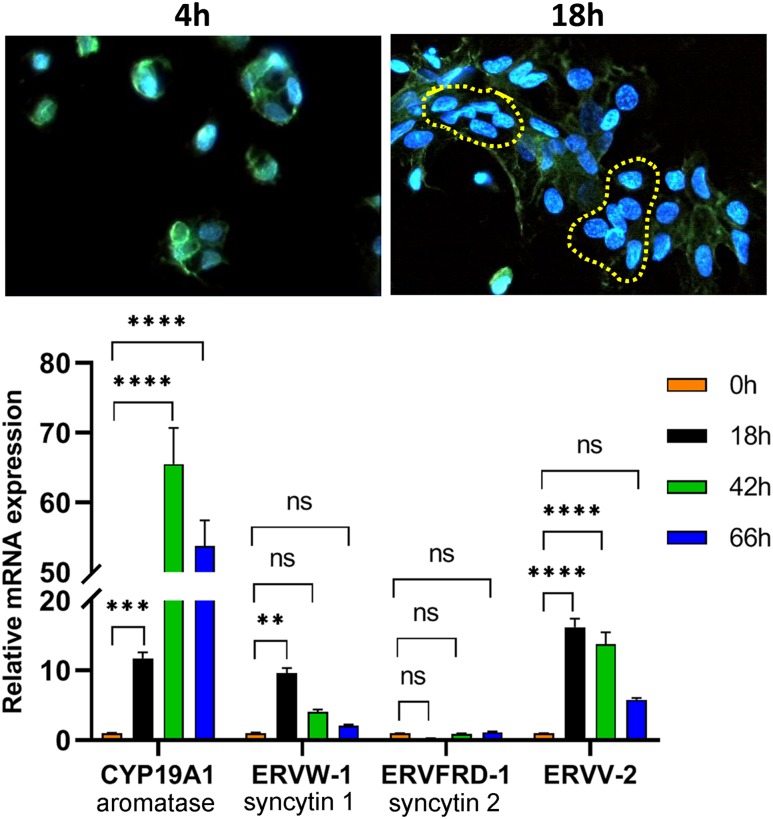
Freshly isolated midgestation human CytT were plated on coverslips and cultured for 4 hours or 18 hours. The cells were immunostained using antibodies to plakoglobin (γ-catenin, green stain), a component of both adherens junctions and desmosomes that plays a vital role in the regulation of cell-cell adhesion and trophoblast fusion (39). Nuclei were stained with 4′,6-diamidino-2-phenylindole (blue). After 4 hours of culture, we mainly observed single cells without evidence of fusion (Fig. 1, top). After 18 hours of culture, numerous cell clusters were evident, and a number of clustered nuclei without surrounding cell membranes (encircled by yellow dashes), indicative of syncytialization, were apparent. This was accompanied by a considerable induction of CYP19A1/aromatase mRNA, which is exclusively expressed in SynT (40), as well as the human endogenous retroviral genes ERVW-1/syncytin 1 and endogenous retrovirus group V member 2, envelope (ERVV-2/ENVV2) (Fig. 1, bottom). Syncytin 1 is believed to serve a role in human trophoblast fusion (41), whereas ERVV-2 is specifically and highly expressed in SynT of human placenta, but serves as a syncytin in placentas of Old World monkeys. Notably, syncytin 2 (ERVFRD-1) was expressed at very low levels and did not change during SynT differentiation (Fig. 1, bottom). Thus, midgestation human trophoblasts cultured for 18 hours manifest a number of indices of SynT differentiation.
Figure 1.
Trophoblast fusion and induction of SynT-specific CYP19A1 and the fusogenic proteins ERVW-1 (syncytin 1) and ERVV-2 are significantly increased within 18 h of human trophoblast culture. (Top) Freshly isolated midgestation human CytT were plated on coverslips and cultured for 4 h or 18 h. The cells were subjected to immunostaining using a primary antibody to plakoglobin and a secondary antibody conjugated to Alexa Fluor 488. The yellow dashed lines encircle cell clusters in which the cell membranes have fused to form syncytia. Images were captured using a Zeiss confocal microscope at ×400 magnification. (Bottom) Midgestation CytT were analyzed before (0 h) or after 18, 42, and 66 h of culture for expression of CYP19A1, ERVW-1, ERVFRD-1, and ERVV-2 by RT-qPCR using specific primers. Data are the mean ± SEM of triplicate samples. Significantly (**P < 0.01; ***P < 0.001; ****P < 0.0001) different from 0 h. ns, not significant.
ChIP-seq of Pol II binding identifies novel target genes regulated during human trophoblast differentiation
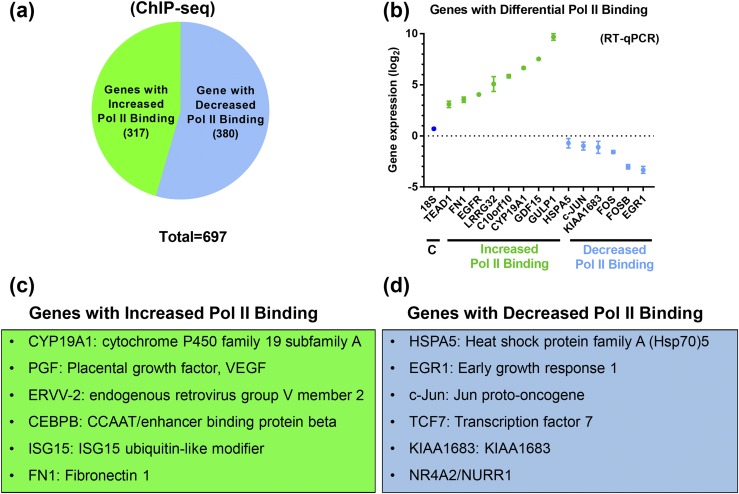
Pol II is a major component of the RNA transcription machinery involved in initiation and elongation of transcription; thus, its binding to the gene promoter and gene body serves as a marker of transcriptional activity (42–45). We performed Pol II ChIP-seq to identify genes that are differentially regulated during human trophoblast differentiation genome-wide. To conduct these studies, CytT were isolated from five independent midgestation human placentas of 16 to 20 weeks’ gestation and harvested before culture (0 hours) or after 18 hours of culture, when SynT differentiation is occurring. CYP19A1 expression was analyzed by RT-qPCR to verify SynT differentiation. ChIP-seq libraries prepared in duplicate from pooled ChIP DNA from these five samples were sequenced and analyzed for binding of Pol II to gene promoter and gene body as an index of transcriptional activity and global changes in gene expression. Computational analysis of Pol II ChIP-seq data revealed that 697 genes manifested significant differential binding of Pol II at their promoter and gene body regions during SynT differentiation, based on P value and FDR. Of these, Pol II binding was upregulated at 317 genes and downregulated at 380 genes [Fig. 2(a)] (37). Selected genes that showed upregulation and downregulation of Pol II binding were validated by RT-qPCR [Fig. 2(b)], confirming that Pol II binding to the promoters of these genes was correlated with their expression. Genes that showed increased Pol II binding in SynT compared with CytT included CYP19A1/aromatase (in which Pol II binding was evident upstream of placenta-specific exon I.1), human placental growth factor (PGF), and the cell fusion gene ERVV-2, as well as transcription factor C/EBPB, which was previously shown to play an essential role in mouse labyrinthine trophoblast development (46) and to be upregulated during differentiation of human SynT in culture (23) [Fig. 2(c)]. Top ranked genes on the increased Pol II–binding gene list include fibronectin 1 (FN1), leucine-rich repeat containing protein 32 (LRRC32), and interferon-inducible gene 15 (ISG15). Genes that showed decreased Pol II binding in SynT compared with CytT comprised negative regulators of trophoblast differentiation, including early growth response gene 1 (EGR1) (47) and proinflammatory transcription factor NR4A2/NURR1 (48) [Fig. 2(d)]. In addition, Pol II binding to the proto-oncogenes cFOS and cJUN was significantly decreased, which is consistent with previous reports that showed rapid downregulation of expression of these genes in primary trophoblast cultures (49).
Figure 2.
Pol II is differentially bound to promoters of a subset of genes during SynT differentiation. In these studies, ChIP-seq libraries were prepared from CytT isolated from midgestation human placentas before and after SynT differentiation in culture. (a) Numbers of genes that manifested a more than twofold increase (green) or more than twofold decrease (blue) in Pol II binding in SynT vs CytT. (b) RT-qPCR was used to assess expression of selected genes manifesting increased (green) and decreased (blue) binding of endogenous Pol II by ChIP-seq. (c) Representative genes that showed increased Pol II binding in SynT vs CytT. (d) Representative genes that showed decreased Pol II binding in SynT vs CytT. VEGF, vascular endothelial growth factor.
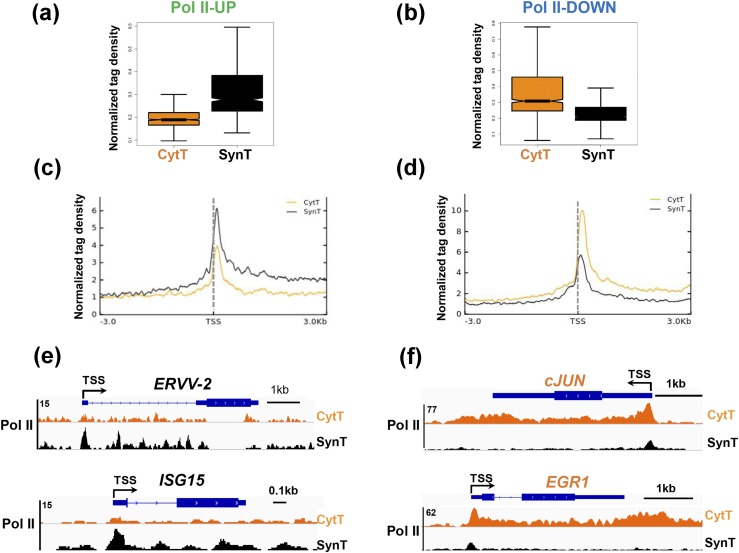
Box plot [Fig. 3(a) and 3(b)] and metagene analysis [Fig. 3(c) and 3(d)] revealed altered Pol II binding in CytT and SynT of up- and downregulated genes. Pol II-UP genes are those that showed increased Pol II binding in SynT compared with CytT [Fig. 3(a) and 3(c)], and Pol II-DOWN genes are those that showed decreased Pol II binding in SynT compared with CytT [Fig. 3(b) and 3(d)]. In the Pol II-UP genes, Pol II density at the promoter and gene body was higher in SynT compared with CytT [Fig. 3(c)]. By contrast, in the Pol II-DOWN genes, Pol II binding within the same areas of the gene was decreased in SynT compared with CytT [Fig. 3(d)]. Genome browser views of selected genes showed that the Pol II-UP genes included the endogenous retroviral gene ERVV-2/ENVV2, expressed specifically in human and nonhuman primate placentas that is involved in SynT fusion (50, 51), and interferon-induced gene ISG15 [Fig. 3(e)]. By contrast, Pol II-DOWN genes included the proinflammatory transcription factor, cJUN, and early growth response 1, EGR1 [Fig. 3(f)]. These findings suggest that genome-wide changes in Pol II binding at the TSS and gene body result in differential gene expression during human SynT differentiation.
Figure 3.
Genome-wide changes in Pol II binding occur at the TSS and gene body during SynT differentiation. Box plots of Pol II density at genes that manifested (a) increased Pol II binding in SynT compared with CytT (Pol II-UP) and (b) decreased Pol II binding in SynT compared with CytT (Pol II-DOWN). Metagene analysis of Pol II distribution in (c) Pol II-UP and (d) Pol II-DOWN genes in CytT vs SynT. Genome browser views of representative (e) Pol II-UP and (f) Pol II-DOWN genes in CytT vs SynT.
Genes regulated during human trophoblast differentiation are enriched in functions involved in human placental development and pregnancy
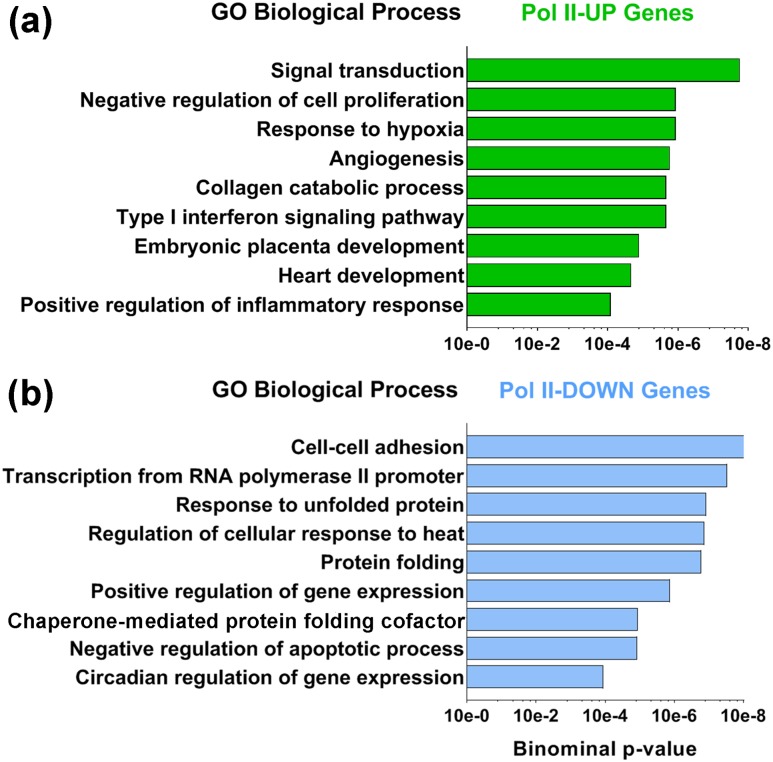
Functional annotation of biological process of Pol II-UP and Pol II-DOWN genes showed that Pol II-UP genes are enriched in functions involved in response to hypoxia [adrenomedullin (ADM), EPAS1, PGF, heme oxygenase 1 (HMOX1), and CITED2], angiogenesis (CYP1B1, PGF, HMOX1, VAV2, MMP14, RNF213, EPHB2, SERPINE1, TYMP, and FN1), and placental development (EGFR, GATA2, C/EBPB, EPAS1, and CITED2) [Fig. 4(a)]. By contrast, Pol II-DOWN gene functions include cellular adhesion (HSP90AB1, STX5, YWHAZ, HSPA5, and PTPN1) and negative regulation of apoptosis (CLDN7, PPARD, MCL1, ANXA1, BIRC6, CD44, and DUSP1), which would be more typical of CytT [Fig. 4(b)]. Therefore, genes that show a differential increase in Pol II binding during trophoblast differentiation are enriched in functions involved in angiogenesis and placental development.
Figure 4.
Gene ontology (GO) analyses were performed using the Database for Annotation, Visualization, and Integrated Discovery tool to assess biological processes that are represented in the Pol II-UP and Pol II-DOWN genes in SynT vs CytT. (a) Functional annotation of biological processes showed that Pol II-UP genes are enriched in functions involved in hypoxia, angiogenesis, and placental development. (b) By contrast, Pol II-DOWN gene functions included cellular adhesion and negative regulation of apoptosis.
Upregulated Pol II binding during SynT differentiation is correlated with increased active histone marks H3K4me3, H3K9Ac, and H3K27Ac
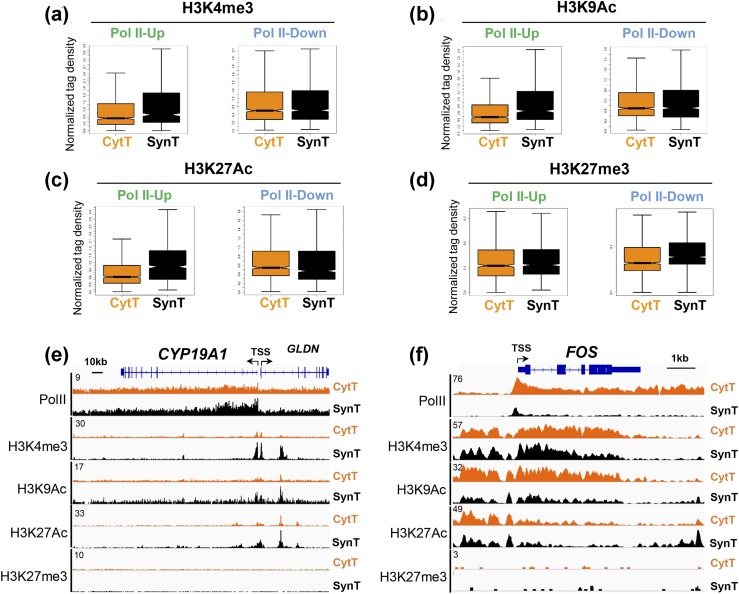
Various histone modifications regulate transcriptional activation by Pol II (14, 15). These include the active histone marks H3K4me3, H3K9Ac, and H3K27Ac, at the promoter and enhancer regions of genes, which promote transcriptional activation, as well as the repressive histone mark H3K27me3, which is increased in inactive chromatin. To determine whether up- and downregulation of Pol II binding was correlated with active and repressive histone modifications genome-wide, we performed ChIP-seq analysis of H3K4me3, H3K9Ac, H3K27Ac, and H3K27me3 binding in CytT and SynT. Box plot analysis indicates that Pol II-UP genes (increased Pol II binding in SynT compared with CytT) showed increased binding of active histone marks H3K4me3, H3K9Ac, and H3K27Ac; however, no change in the repressive histone mark H3K27me3 was observed [Fig. 5(a)–5(d), Pol II-UP]. By contrast, Pol II-DOWN genes, with decreased Pol II binding in SynT compared with CytT, showed no change in the binding of these active or repressive histone marks [Fig. 5(a)–5(d), Pol II-DOWN] overall. Genome browser tracts of selected genes showed that for the CYP19A1 gene, which is markedly induced during SynT differentiation, Pol II binding was increased at the promoter and within the gene body in SynT compared with CytT. This was closely correlated with increased binding of the histone marks H3K4me3, H3K9Ac, and H3K27Ac, but not with repressive histone mark H3K27me3 [Fig. 5(e)]. In contrast, the FOS gene, which is downregulated during SynT differentiation, manifested decreased Pol II binding in SynT compared with CytT. This was correlated with decreased binding of the active histone marks H3K9Ac and H3K27Ac, but not with changes in the repressive histone mark H3K27me3 [Fig. 5(f)]. These results indicate that upregulated Pol II binding is tightly correlated with increased binding of positive histone marks, but not with the repressive histone mark H3K27me3.
Figure 5.
Genes with increased Pol II promoter binding in SynT vs CytT show increased binding of active histone marks. (a–d) Box plot analyses of density of active and repressive histone marks in the promoter regions of Pol II-UP and Pol II-DOWN genes in SynT vs CytT. Genome browser views of binding of Pol II and postranslationally modified histones in representative (e) Pol II-UP (CYP19A1) and (f) Pol II-DOWN (cFOS) genes.
Analysis of genes upregulated in SynT by active histone marks and increased Pol II binding reveals a gene set involved in immunity
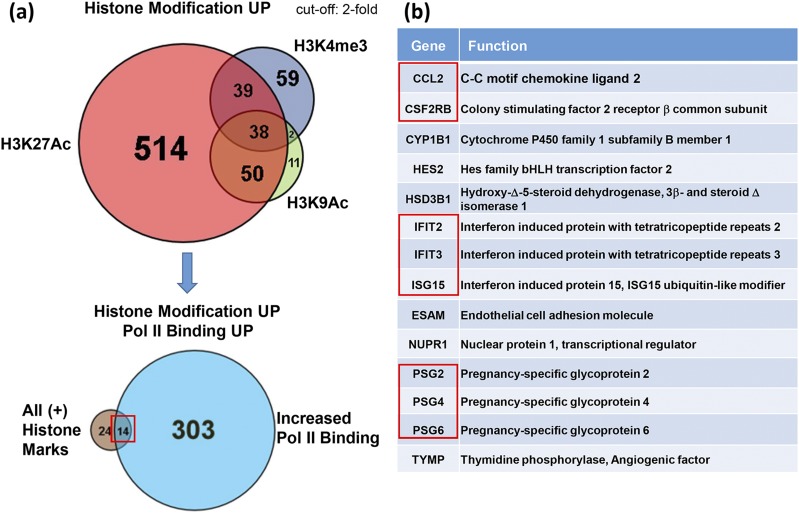
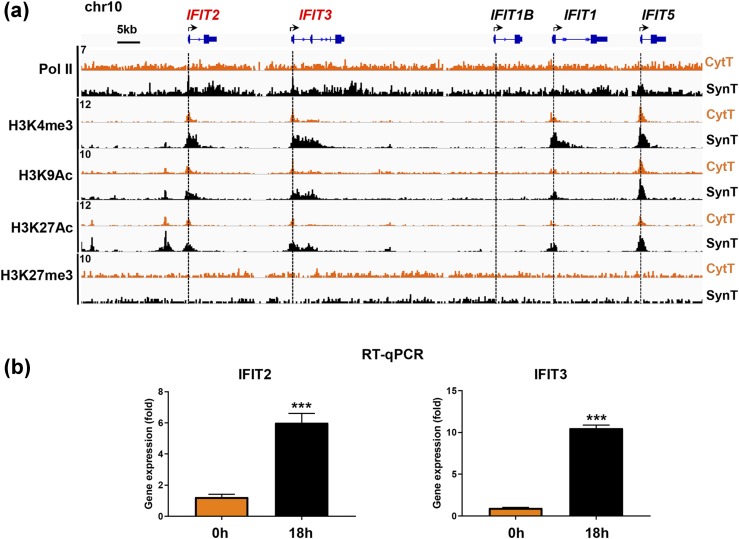
Because the above findings indicate that genes with increased Pol II binding during SynT differentiation manifested increased binding of active histone marks, we compared genes in SynT that had increased binding of one or more the active histone marks. This analysis identified 38 genes that had increased binding of all three active marks upon SynT differentiation, as shown in the top Venn diagram [Fig. 6(a)]. As shown in the bottom Venn diagram [Fig. 6(a)], we then analyzed these 38 genes for those that also showed increased binding of Pol II and found 14 genes that had all 3 active histone marks and increased Pol II binding [Fig. 6(a)]. One of these genes (HSD3B1) [Fig. 6(b)] encodes 3β-hydroxysteroid dehydrogenase, a key placental enzyme that is critical for the synthesis of progesterone, required for the maintenance of pregnancy (52). HSD3B1 expression rapidly increases in human trophoblasts during differentiation in culture (49). Interestingly, this gene list also includes those involved in the immune response (CCL2 and CSF2RB), the interferon signaling pathway (interferon-induced protein with tetratricopeptide repeat, IFIT2 and IFIT3), and immune modulation (placenta-specific glycoprotein genes PSG2, PSG4, and PSG6) [Fig. 6(b)]. These findings indicate that genes that are upregulated by active histone marks and increased Pol II binding are largely involved in immune regulation. Genome browser views show binding of Pol II and active and repressive histone marks to the promoters and gene bodies of the cluster of interferon-inducible IFIT family members on chromosome 10 during SynT differentiation [Fig. 7(a)]. The binding of Pol II and of active histone marks was increased at the promoters of these genes in SynT compared with CytT; however, binding of the repressive histone mark H3K27me3 was low and remained unchanged. This was associated with a statistically significant increase in mRNA expression of IFIT2 and IFIT3 [Fig. 7(b)].
Figure 6.
Genes that manifest enrichment of all three active histone marks (H3K4me3, H3K9Ac, and H3K927Ac) and increased Pol II binding in SynT vs CytT comprise a unique subset of genes involved in immune modulation. (a) Venn diagram of genes that manifest upregulated binding of one or more active histone marks in SynT compared with CytT (top). Venn diagram of genes that manifest enrichment of all three active histone marks and increased Pol II binding (bottom) comprise a unique subset of (b) genes primarily involved in immune response and immune modulation.
Figure 7.
(a) Genome browser view of ChIP-seq analysis of interferon-inducible gene IFIT family members on human chromosome 10 indicates that increased binding of Pol II to the gene promoters in SynT vs CytT is correlated with increased active histone marks. (b) These changes in binding of endogenous Pol II and histone H3 modifications indicative of active chromatin are correlated with upregulated expression of the IFIT2 and IFIT3 genes in SynT (18 h of culture) vs CytT (0 h) analyzed by RT-qPCR. Data are the mean ± SEM of triplicate samples. Significantly different from 0 h (***P < 0.001).
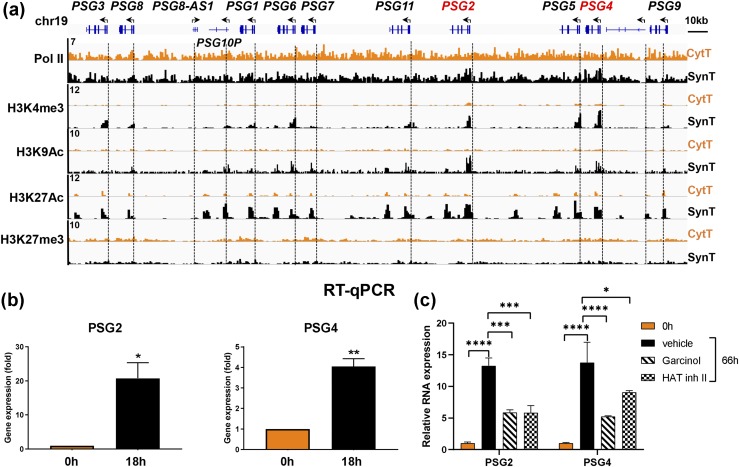
Genome browser tracks for binding of Pol II, the active and the repressive histone marks to the cluster of immunomodulatory pregnancy-specific glycoprotein genes (PSGs) on chromosome 19 in CytT vs SynT also indicated increased binding of Pol II and active histone marks within the upstream regions of several of these PSG genes [Fig. 8(a)]. Using RT-qPCR, we confirmed temporal induction in PSG2 and PSG4 expression during SynT differentiation [Fig. 8(b)]. Thus, binding of Pol II and active histone marks is tightly correlated with expression of these genes during human trophoblast differentiation. Again, no changes in the repressive histone mark H3K27me3 were observed. To assess effects of histone-modulating chemicals on PSG2 and PSG4 expression during SynT differentiation, we analyzed effects of garcinol, an inhibitor of the histone acetyltransferases (HATs) p300 and PCAF (53), and HAT inhibitor II, a selective inhibitor of p300 (54), which catalyzes H3K27 acetylation. Culture of freshly isolated CytT with either of these inhibitors (5 μM) for 66 hours caused a significant reduction in the levels of induced expression of PSG2 and PSG4 during SynT differentiation [Fig. 8(c)]. Thus, increased H3K27 acetylation serves a role in the induction of PSG expression during SynT differentiation.
Figure 8.
Binding of Pol II and active histone marks is closely correlated with PSG gene expression during trophoblast differentiation. (a) Genome browser view of binding of Pol II and of active and repressive histone marks at the promoters of the cluster of PSG genes on the long arm of chromosome 19. (b) RT-qPCR was performed to assess expression of PSG2 and PSG4 genes during differentiation of freshly isolated CytT from midgestation placenta. (c) Freshly isolated CytT from term human placenta were analyzed for PSG2 and PSG4 mRNA expression before (0 h) or after 66 h in culture in the absence (vehicle) or presence of 5 μM garcinol or HAT inhibitor (inh) II. Data are the mean ± SEM of triplicate samples. Significantly different from 0 h or from vehicle (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
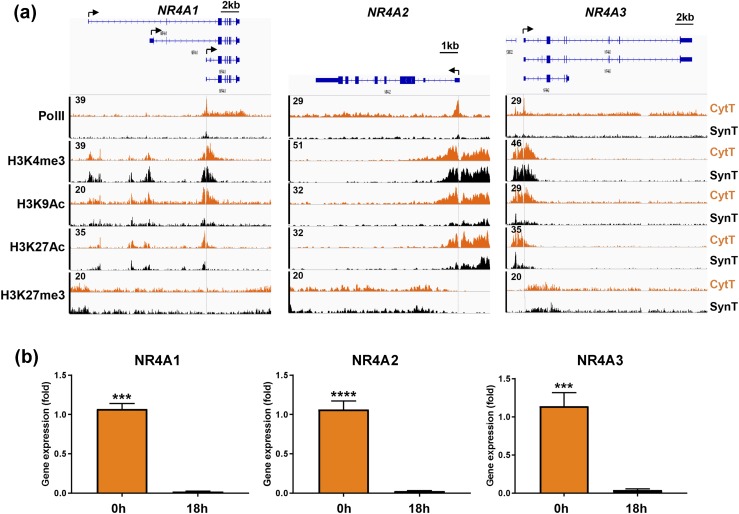
SynT differentiation is associated with a marked downregulation of members of the NR4A family, NR4A1 (NUR77), NR4A2 (NURR1), and NR4A3 (NOR1)
We were interested to find that Pol II binding to the TSS of genes encoding all three members of the NR4A orphan nuclear receptor subfamily was markedly downregulated during human SynT differentiation [Fig. 9(a)]. Of the putative NUR77 transcripts, we discovered using specific primers that one or both of the shortest transcripts was transcribed. The TSS for the shortest NURR77 transcript corresponded to the peak for Pol II binding. The NURR1 gene has only one transcript, whereas the NOR1 gene has three potential transcripts that share the same TSS. NR4A family members NUR77, NURR1, and NOR1 are immediate early genes involved in inflammation, immune modulation, and cancer (55). The decline in Pol II binding to their promoters during SynT differentiation was associated with a profound decrease in mRNA expression, assessed by RT-qPCR [Fig. 9(b)]. As observed for other downregulated genes, this was associated with a marked decrease in binding of H3K9Ac and H3K27Ac, with little change in the promoter activation mark H3K4me3 or the repressive mark H3K27me3 [Fig. 9(a)].
Figure 9.
Decreased binding of Pol II and of the active histone marks H3K9Ac and H3K27Ac to the promoters and enhancers of the NR4A1 (NUR77), NR4A2 (NURR1), and NR4A3 (NOR1) genes is correlated with downregulation of their expression during SynT differentiation, whereas binding of H3K4me3 and H4K27me3 remain unchanged. (a) Genome browser views of binding of Pol II and of active and repressive histone marks to the promoters/enhancers of the NR4A1, NR4A2, and NR4A3 genes. (b) RT-qPCR of RNA isolated from midgestation placenta before and after 18 h of culture revealed marked downregulation of all three genes during SynT differentiation. Data are the mean ± SEM of triplicate samples. Significantly different from 0 h (***P < 0.001; ****P < 0.0001).
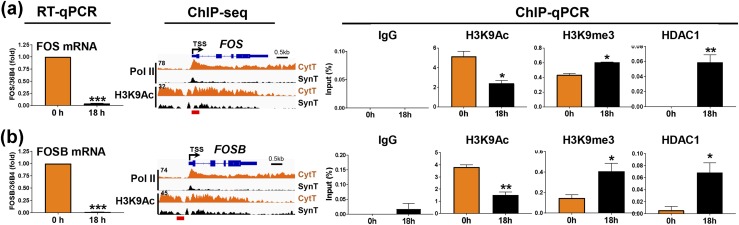
Differentiation of CytT to SynT occurs with decreased enrichment of H3K9Ac, increased binding of H3K9me3, and histone deacetylase 1 binding to the promoters of downregulated genes
Because H3K27me3 binding was low and remained unchanged at the promoters of a number of downregulated genes during SynT differentiation, we used ChIP-qPCR to compare binding of the repressive mark H3K9me3 with the activating mark H3K9Ac to promoters of several Pol II–downregulated genes during trophoblast differentiation. Results are shown for the proinflammatory transcription factor genes cFOS and FOSB, which were markedly downregulated during SynT differentiation. Decreased mRNA expression of cFOS and FOSB in SynT compared with CytT was associated with significantly decreased H3K9Ac recruitment to their promoter regions, whereas association of H3K9me3 was increased. This was correlated with a pronounced increase in promoter enrichment of histone deacetylase (HDAC) 1 (Fig. 10a and 10b). Taken together, these findings indicate that binding of the active histone mark H3K9Ac and the repressive mark H3K9me3 are reciprocally regulated. Moreover, increased HDAC1 enrichment was associated with decreased binding of H3K9Ac and increased binding of H3K9me3 to repressed genes during trophoblast differentiation.
Figure 10.
Decreased expression of the cFOS and FOSB genes during SynT differentiation is associated with decreased binding of Pol II and the active histone mark H3K9Ac and with increased recruitment of the repressive histone mark H3K9me3 and of HDAC1 to their promoters. RT-qPCR of (a) cFOS and (b) FOSB mRNA expression before (0 h, CytT) and after (18 h, SynT) differentiation of midgestation human trophoblasts in culture. Genome browser views of binding of Pol II and of the active histone mark H3K9Ac to the (a) cFOS and (b) FOSB gene promoters/enhancers before (0 h, CytT) and after (18 h, SynT) differentiation in culture. ChIP-qPCR of binding of H3K9Ac, H3K9me3, and HDAC1 to the (a) cFOS and (b) FOSB gene promoters/enhancers before (0 h, CytT) and after (18 h, SynT) differentiation of trophoblasts isolated from term human placenta. Antibody to nonimmune IgG was used as a control for ChIP. Data are the mean ± SEM of triplicate samples. Significantly different from 0 h (*P < 0.05; **P < 0.01; ***P < 0.001).
Discussion
Genome-wide analyses using microarray and high-throughput sequencing methods have previously been used to identify differentially expressed genes in maternal and fetal villous parenchyma of the placenta (56, 57) using primary cultures of human trophoblasts during differentiation of CytT along the invasive pathway (58) and during differentiation to SynT (59, 60) and in trophoblast cell lines (61, 62). Single-cell RNA sequencing also was used to analyze transcriptomes of various placental cell types (63, 64). In the current study, we defined the epigenetic mechanisms that underlie early changes in gene expression during human SynT differentiation. To this end, we used high-throughput next-generation ChIP-seq to investigate genome-wide changes in binding of Pol II and associated modified histones indicative of active and repressed chromatin during differentiation of midgestation human CytT (0 hours) to SynT (18 hours) in primary culture. Differential Pol II binding was analyzed as an index of transcriptional activation or repression.
As a validation of our approach, ChIP-seq revealed expected changes in Pol II binding to several critical genes previously shown to be up- and downregulated during SynT differentiation. For example, increased Pol II binding to the TSS and gene bodies of CYP19A1 and C/EBPB, which are important markers of SynT differentiation (6, 65), was observed in SynT compared with CytT. During SynT differentiation, we also observed strong upregulation of Pol II binding to the promoter of the gene encoding fibronectin (FN1), which is involved in cell adhesion and motility and promotes formation of a syncytium (66). The upregulation of FN1 expression was somewhat surprising, because this gene is known to be highly expressed in extravillous trophoblast (67, 68). However, because the CytT used in our study were isolated from midgestation human placenta, it is speculated that they may have the capacity to differentiate into extravillous as well as villous trophoblasts, as reported for first-trimester CytT (66). Accordingly, we found a number of similarities when we compared our differential Pol II–binding gene lists to published microarray data from midtrimester CytT before and after culture on Matrigel to mimic differentiation along the invasive pathway (58). Genes that were upregulated during differentiation in the study by Robinson et al. (58) and that showed increased RNA Pol II binding in our ChIP-seq analysis included those involved in cell migration and vascular development. Genes that were downregulated during differentiation in the Robinson et al. (58) study and that showed decreased RNA Pol II binding in our ChIP-seq analysis included those involved in inflammation and cell proliferation. Although trophoblasts cultured on Matrigel do not syncytialize, they do manifest induction of genes that are involved in migration and formation of multicellular aggregates. Clearly, these genes are also important in the formation of syncytia.
Genes that manifested increased Pol II binding in SynT compared with CytT were enriched in functions involved in placental development and response to hypoxia. These included placental development genes EGFR, GATA2, C/EBPB, EPAS1, and CITED2. The genes enriched in hypoxia function included ADM and HMOX1, which serve a protective role against exposure to marked alterations in O2 tension. ADM protects mesenchymal stem cells against hypoxia (69). HMOX1, which protects the liver against ischemia-induced hypoxia (70), has been found to serve roles in placental vascular development and spiral artery remodeling, is anti-inflammatory and promotes immune tolerance of the mother to the hemiallogeneic fetus during pregnancy (71). We speculate that increased expression of these genes in SynT may protect against inflammation and hypoxic insult and serve important roles in trophoblast differentiation and function.
Notably, genes that manifested increased binding of both Pol II and active histone marks during SynT differentiation included those involved in immune response and immune modulation. In addition to HMOX1, the IFIT and PSG gene clusters showed increased binding of Pol II and the active histone modifications H3K4me3, H3K9Ac, and H3K27Ac. In humans, there are four IFIT family members, which are interferon-induced, regulated by the JAK-STAT signaling pathway and involved in host antiviral immunity (17, 72). The IFIT proteins inhibit viral replication by binding to and regulating the function of cellular and viral proteins and RNAs. RNA-seq analysis has shown that IFIT genes are induced in extravillous trophoblasts (68). The PSG genes, previously shown to be rapidly induced during differentiation of CytT to SynT in primary culture (49, 66), were prominent on our list of genes that manifested increased binding of all three active histone marks and of Pol II during SynT differentiation. The PSG genes are involved in immunoregulatory, angiogenic, and antiplatelet functions (73, 74). Ten of the PSG genes exist as a cluster on chromosome 19 (75). PSGs are the most abundant trophoblastic proteins in maternal blood during human pregnancy; decreased serum concentrations of PSGs are associated with fetal growth restriction and PE (73). Thus, the IFIT and PSG proteins may serve a vital function in pregnancy maintenance by protecting the fetus from viral infection and from rejection by the maternal immune system. Because we observed that expression of the PSG gene family was strongly associated with recruitment of the activating histone acetylation marks H3K9Ac and H3K27Ac, we examined the effects of histone-modifying chemicals on PSG gene expression. We observed that incubation of human trophoblasts with the p300 HAT inhibitors garcinol or HAT inhibitor II significantly repressed the induction of PSG2 and PSG4 mRNA expression during SynT differentiation. In light of these findings, it would be of interest to determine whether HDAC inhibitors could provide a potential therapeutic strategy for pregnancy maintenance (76, 77). In this regard, we previously observed that treatment of timed-pregnant mice with the HDAC inhibitor trichostatin A, late in gestation, increased histone H3 acetylation in the myometrium and delayed parturition by 24 to 48 hours (78).
We observed that ∼50% of the genes differentially regulated during trophoblast differentiation manifested decreased Pol II binding in SynT compared with CytT. These included the proinflammatory transcription factors EGR1, cFOS, and cJUN as well as members of the NR4A orphan nuclear receptor family, which were previously reported to be upregulated in placentas of women with gestational diabetes (48). In the case of genes in which Pol II binding was markedly downregulated during SynT differentiation (e.g., cFOS and NR4A family), there was an associated decrease in promoter recruitment of the active histone marks H3K9Ac and H3K27Ac. By contrast, binding of the repressive histone mark H3K27me3 remained very low and unchanged, despite a marked decrease in Pol II binding and gene expression. On the other hand, ChIP-qPCR revealed that binding of the repressive mark H3K9me3 and of the histone deacetylase HDAC1 were dynamically upregulated at the promoters of genes that manifested decreased binding of Pol II and active histone marks (cFOS and FOSB) during SynT differentiation. Binding of H3K4me3 is enriched at the promoters of essentially all expressed genes and is a marker of an open chromatin state (79). We observed that H3K4me3 recruitment was induced at the promoters of genes upregulated during SynT differentiation (e.g., CYP19A1, IFIT, and PSG clusters). H3K4me3 binding was also elevated at the promoters of proinflammatory genes that were upregulated in CytT compared with SynT (e.g., NR4A family and cFOS). Notably, H3K4me3 remained elevated at the promoters of these genes, despite their suppression upon SynT differentiation. This interesting finding suggests that H3K4me3 binding to the promoters of proinflammatory genes upregulated in CytT maintains an open chromatin state, despite other epigenetic changes (e.g., H3K9me3) that promote gene repression during SynT differentiation. We speculate that this may permit dynamic changes in expression of these genes in response to inflammatory mediators and environmental factors. Collectively, these findings provide important insight into the epigenetic mechanisms underlying human trophoblast differentiation and may reveal therapeutic strategies for human placental disorders, such as PE and intrauterine growth restriction.
Acknowledgments
The authors thank Ms. Jo Smith for assistance in isolating human trophoblast cells and Mr. Rohit Setlem for help with some of the computational aspects of this study.
Financial Support: This work was supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development Grant 5P01-HD087150.
Glossary
Abbreviations:
- ADM
adrenomedullin
- ChIP-seq
chromatin immunoprecipitation sequencing
- CytT
cytotrophoblast
- EGR1
early growth response gene 1
- FDR
false discovery rate
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HMOX1
heme oxygenase 1
- PE
preeclampsia
- PGF
placental growth factor
- Pol II
polymerase II
- PSG
placenta-specific glycoprotein gene
- RT-qPCR
reverse transcription quantitative PCR
- SynT
syncytiotrophoblast
- TSS
transcription start site
Additional Information
Current Affiliation: S. Muralimanoharan’s current affiliation is the Department of Molecular Medicine, Institute of Biotechnology, University of Texas Health Science Center, San Antonio, San Antonio, Texas 78229.
Disclosure Summary: The authors have nothing to disclose.
Data Availability:
All data generated or analyzed during this study are included in this published article or in the data repositories listed in References.
References and Notes
- 1. Everett RB, MacDonald PC. Endocrinology of the placenta. Annu Rev Med. 1979;30(1):473–488. [DOI] [PubMed] [Google Scholar]
- 2. Red-Horse K, Zhou Y, Genbacev O, Prakobphol A, Foulk R, McMaster M, Fisher SJ. Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J Clin Invest. 2004;114(6):744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McMaster MT, Zhou Y, Fisher SJ. Abnormal placentation and the syndrome of preeclampsia. Semin Nephrol. 2004;24(6):540–547. [DOI] [PubMed] [Google Scholar]
- 4. Arck PC, Hecher K. Fetomaternal immune cross-talk and its consequences for maternal and offspring’s health. Nat Med. 2013;19(5):548–556. [DOI] [PubMed] [Google Scholar]
- 5. PrabhuDas M, Bonney E, Caron K, Dey S, Erlebacher A, Fazleabas A, Fisher S, Golos T, Matzuk M, McCune JM, Mor G, Schulz L, Soares M, Spencer T, Strominger J, Way SS, Yoshinaga K. Immune mechanisms at the maternal-fetal interface: perspectives and challenges. Nat Immunol. 2015;16(4):328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kamat A, Alcorn JL, Kunczt C, Mendelson CR. Characterization of the regulatory regions of the human aromatase (P450arom) gene involved in placenta-specific expression. Mol Endocrinol. 1998;12(11):1764–1777. [DOI] [PubMed] [Google Scholar]
- 7. Kumar P, Mendelson CR. Estrogen-related receptor γ (ERRgamma) mediates oxygen-dependent induction of aromatase (CYP19) gene expression during human trophoblast differentiation. Mol Endocrinol. 2011;25(9):1513–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumar P, Kamat A, Mendelson CR. Estrogen receptor α (ERalpha) mediates stimulatory effects of estrogen on aromatase (CYP19) gene expression in human placenta. Mol Endocrinol. 2009;23(6):784–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumar P, Luo Y, Tudela C, Alexander JM, Mendelson CR. The c-Myc-regulated microRNA-17∼92 (miR-17∼92) and miR-106a∼363 clusters target hCYP19A1 and hGCM1 to inhibit human trophoblast differentiation. Mol Cell Biol. 2013;33(9):1782–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang M, Muralimanoharan S, Wortman AC, Mendelson CR. Primate-specific miR-515 family members inhibit key genes in human trophoblast differentiation and are upregulated in preeclampsia. Proc Natl Acad Sci USA. 2016;113(45):E7069–E7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiang B, Kamat A, Mendelson CR. Hypoxia prevents induction of aromatase expression in human trophoblast cells in culture: potential inhibitory role of the hypoxia-inducible transcription factor Mash-2 (mammalian achaete-scute homologous protein-2). Mol Endocrinol. 2000;14(10):1661–1673. [DOI] [PubMed] [Google Scholar]
- 12. Jiang B, Mendelson CR. USF1 and USF2 mediate inhibition of human trophoblast differentiation and CYP19 gene expression by Mash-2 and hypoxia. Mol Cell Biol. 2003;23(17):6117–6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. [DOI] [PubMed] [Google Scholar]
- 14. Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–719. [DOI] [PubMed] [Google Scholar]
- 15. Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130(1):77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014;15(11):703–708. [DOI] [PubMed] [Google Scholar]
- 17. Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12(1):7–18. [DOI] [PubMed] [Google Scholar]
- 18. Olins DE, Olins AL. Chromatin history: our view from the bridge. Nat Rev Mol Cell Biol. 2003;4(10):809–814. [DOI] [PubMed] [Google Scholar]
- 19. Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98(3):285–294. [DOI] [PubMed] [Google Scholar]
- 20. Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. [DOI] [PubMed] [Google Scholar]
- 21. Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA. 2002;99(13):8695–8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kliman HJ, Nestler JE, Sermasi E, Sanger JM, Strauss JF III. Purification, characterization, and in vitro differentiation of cytotrophoblasts from human term placentae. Endocrinology. 1986;118(4):1567–1582. [DOI] [PubMed] [Google Scholar]
- 23. Muralimanoharan S, Kwak YT, Mendelson CR. Redox-sensitive transcription factor NRF2 enhances trophoblast differentiation via induction of miR-1246 and aromatase. Endocrinology. 2018;159(5):2022–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. RRID:AB_823448, http://scicrunch.org/resolver/AB_823448.
- 25. RRID:AB_141362, http://scicrunch.org/resolver/AB_141362.
- 26. Hardy DB, Janowski BA, Corey DR, Mendelson CR. Progesterone receptor plays a major anti-inflammatory role in human myometrial cells by antagonism of nuclear factor-κB activation of cyclooxygenase 2 expression. Mol Endocrinol. 2006;20(11):2724–2733. [DOI] [PubMed] [Google Scholar]
- 27. Franco HL, Nagari A, Kraus WL. TNFα signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol Cell. 2015;58(1):21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. RRID:AB_632359, http://scicrunch.org/resolver/AB_632359.
- 29. RRID:AB_2615077, http://scicrunch.org/resolver/AB_2615077.
- 30. RRID:AB_2118292, http://scicrunch.org/resolver/AB_2118292.
- 31. RRID:AB_2561016, http://scicrunch.org/resolver/AB_2561016.
- 32. RRID:AB_2616029, http://scicrunch.org/resolver/AB_2616029.
- 33. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9(9):R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kwak Y-T, Muralimanoharan S, Gogate AA, Mendelson CR. Data from: Human trophoblast differentiation is associated with profound gene regulatory and epigenetic changes. Gene Expression Omnibus 2019. Deposited 2 July 2019. www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE127288. [DOI] [PMC free article] [PubMed]
- 38. Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):3. [PubMed] [Google Scholar]
- 39. Lu J, Zhang S, Nakano H, Simmons DG, Wang S, Kong S, Wang Q, Shen L, Tu Z, Wang W, Wang B, Wang H, Wang Y, van Es JH, Clevers H, Leone G, Cross JC, Wang H. A positive feedback loop involving Gcm1 and Fzd5 directs chorionic branching morphogenesis in the placenta. PLoS Biol. 2013;11(4):e1001536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fournet-Dulguerov N, MacLusky NJ, Leranth CZ, Todd R, Mendelson CR, Simpson ER, Naftolin F. Immunohistochemical localization of aromatase cytochrome P-450 and estradiol dehydrogenase in the syncytiotrophoblast of the human placenta. J Clin Endocrinol Metab. 1987;65(4):757–764. [DOI] [PubMed] [Google Scholar]
- 41. Pötgens AJ, Drewlo S, Kokozidou M, Kaufmann P. Syncytin: the major regulator of trophoblast fusion? Recent developments and hypotheses on its action. Hum Reprod Update. 2004;10(6):487–496. [DOI] [PubMed] [Google Scholar]
- 42. Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461(7261):186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13(10):720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81(1):119–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grünberg S, Hahn S. Structural insights into transcription initiation by RNA polymerase II. Trends Biochem Sci. 2013;38(12):603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bégay V, Smink J, Leutz A. Essential requirement of CCAAT/enhancer binding proteins in embryogenesis. Mol Cell Biol. 2004;24(22):9744–9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Saben J, Zhong Y, Gomez-Acevedo H, Thakali KM, Borengasser SJ, Andres A, Shankar K. Early growth response protein-1 mediates lipotoxicity-associated placental inflammation: role in maternal obesity. Am J Physiol Endocrinol Metab. 2013;305(1):E1–E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lappas M. The NR4A receptors Nurr1 and Nur77 are increased in human placenta from women with gestational diabetes. Placenta. 2014;35(11):866–875. [DOI] [PubMed] [Google Scholar]
- 49. Morrish DW, Dakour J, Li H, Xiao J, Miller R, Sherburne R, Berdan RC, Guilbert LJ. In vitro cultured human term cytotrophoblast: a model for normal primary epithelial cells demonstrating a spontaneous differentiation programme that requires EGF for extensive development of syncytium. Placenta. 1997;18(7):577–585. [DOI] [PubMed] [Google Scholar]
- 50. Kumar P, Thirkill TL, Ji J, Monte LH, Douglas GC. Differential effects of sodium butyrate and lithium chloride on Rhesus monkey trophoblast differentiation. PLoS One. 2015;10(8):e0135089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Esnault C, Cornelis G, Heidmann O, Heidmann T. Differential evolutionary fate of an ancestral primate endogenous retrovirus envelope gene, the EnvV syncytin, captured for a function in placentation. PLoS Genet. 2013;9(3):e1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Simard J, Ricketts ML, Gingras S, Soucy P, Feltus FA, Melner MH. Molecular biology of the 3β-hydroxysteroid dehydrogenase/delta5-delta4 isomerase gene family. Endocr Rev. 2005;26(4):525–582. [DOI] [PubMed] [Google Scholar]
- 53. Balasubramanyam K, Altaf M, Varier RA, Swaminathan V, Ravindran A, Sadhale PP, Kundu TK. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J Biol Chem. 2004;279(32):33716–33726. [DOI] [PubMed] [Google Scholar]
- 54. Costi R, Di Santo R, Artico M, Miele G, Valentini P, Novellino E, Cereseto A. Cinnamoyl compounds as simple molecules that inhibit p300 histone acetyltransferase. J Med Chem. 2007;50(8):1973–1977. [DOI] [PubMed] [Google Scholar]
- 55. Safe S, Jin UH, Morpurgo B, Abudayyeh A, Singh M, Tjalkens RB. Nuclear receptor 4A (NR4A) family - orphans no more. J Steroid Biochem Mol Biol. 2016;157:48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sood R, Zehnder JL, Druzin ML, Brown PO. Gene expression patterns in human placenta. Proc Natl Acad Sci USA. 2006;103(14):5478–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mikheev AM, Nabekura T, Kaddoumi A, Bammler TK, Govindarajan R, Hebert MF, Unadkat JD. Profiling gene expression in human placentae of different gestational ages: an OPRU Network and UW SCOR Study. Reprod Sci. 2008;15(9):866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Robinson JF, Kapidzic M, Gormley M, Ona K, Dent T, Seifikar H, Hamilton EG, Fisher SJ. Transcriptional dynamics of cultured human villous cytotrophoblasts. Endocrinology. 2017;158(6):1581–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aronow BJ, Richardson BD, Handwerger S. Microarray analysis of trophoblast differentiation: gene expression reprogramming in key gene function categories. Physiol Genomics. 2001;6(2):105–116. [DOI] [PubMed] [Google Scholar]
- 60. Rouault C, Clément K, Guesnon M, Henegar C, Charles MA, Heude B, Evain-Brion D, Degrelle SA, Fournier T. Transcriptomic signatures of villous cytotrophoblast and syncytiotrophoblast in term human placenta. Placenta. 2016;44:83–90. [DOI] [PubMed] [Google Scholar]
- 61. Kudo Y, Boyd CA, Sargent IL, Redman CW, Lee JM, Freeman TC. An analysis using DNA microarray of the time course of gene expression during syncytialization of a human placental cell line (BeWo). Placenta. 2004;25(6):479–488. [DOI] [PubMed] [Google Scholar]
- 62. Shankar K, Kang P, Zhong Y, Borengasser SJ, Wingfield C, Saben J, Gomez-Acevedo H, Thakali KM. Transcriptomic and epigenomic landscapes during cell fusion in BeWo trophoblast cells. Placenta. 2015;36(12):1342–1351. [DOI] [PubMed] [Google Scholar]
- 63. Pavličev M, Wagner GP, Chavan AR, Owens K, Maziarz J, Dunn-Fletcher C, Kallapur SG, Muglia L, Jones H. Single-cell transcriptomics of the human placenta: inferring the cell communication network of the maternal-fetal interface. Genome Res. 2017;27(3):349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tsang JCH, Vong JSL, Ji L, Poon LCY, Jiang P, Lui KO, Ni YB, To KF, Cheng YKY, Chiu RWK, Lo YMD. Integrative single-cell and cell-free plasma RNA transcriptomics elucidates placental cellular dynamics. Proc Natl Acad Sci USA. 2017;114(37):E7786–E7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Maltepe E, Bakardjiev AI, Fisher SJ. The placenta: transcriptional, epigenetic, and physiological integration during development. J Clin Invest. 2010;120(4):1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Morrish DW, Dakour J, Li H. Functional regulation of human trophoblast differentiation. J Reprod Immunol. 1998;39(1-2):179–195. [DOI] [PubMed] [Google Scholar]
- 67. Aplin JD, Haigh T, Jones CJ, Church HJ, Vićovac L. Development of cytotrophoblast columns from explanted first-trimester human placental villi: role of fibronectin and integrin α5β1. Biol Reprod. 1999;60(4):828–838. [DOI] [PubMed] [Google Scholar]
- 68. Okae H, Toh H, Sato T, Hiura H, Takahashi S, Shirane K, Kabayama Y, Suyama M, Sasaki H, Arima T. Derivation of human trophoblast stem cells. Cell Stem Cell. 2018;22(1):50–63.e6. [DOI] [PubMed] [Google Scholar]
- 69. Si H, Zhang Y, Song Y, Li L. Overexpression of adrenomedullin protects mesenchymal stem cells against hypoxia and serum deprivation-induced apoptosis via the Akt/GSK3β and Bcl-2 signaling pathways. Int J Mol Med. 2018;41(6):3342–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hong JM, Lee SM. Heme oxygenase-1 protects liver against ischemia/reperfusion injury via phosphoglycerate mutase family member 5-mediated mitochondrial quality control. Life Sci. 2018;200:94–104. [DOI] [PubMed] [Google Scholar]
- 71. Ozen M, Zhao H, Lewis DB, Wong RJ, Stevenson DK. Heme oxygenase and the immune system in normal and pathological pregnancies. Front Pharmacol. 2015;6:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fensterl V, Sen GC. Interferon-induced Ifit proteins: their role in viral pathogenesis. J Virol. 2015;89(5):2462–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Moore T, Dveksler GS. Pregnancy-specific glycoproteins: complex gene families regulating maternal-fetal interactions. Int J Dev Biol. 2014;58(2-4):273–280. [DOI] [PubMed] [Google Scholar]
- 74. Jones K, Ballesteros A, Mentink-Kane M, Warren J, Rattila S, Malech H, Kang E, Dveksler G. PSG9 stimulates increase in FoxP3+ regulatory T-cells through the TGF-β1 pathway [published correction appears in PLoS One. 2017;12(4):e0175636]. PLoS One. 2016;11(7):e0158050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Olsen A, Teglund S, Nelson D, Gordon L, Copeland A, Georgescu A, Carrano A, Hammarström S. Gene organization of the pregnancy-specific glycoprotein region on human chromosome 19: assembly and analysis of a 700-kb cosmid contig spanning the region. Genomics. 1994;23(3):659–668. [DOI] [PubMed] [Google Scholar]
- 76. Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014;13(9):673–691. [DOI] [PubMed] [Google Scholar]
- 77. Behera J, Jayaprakash V, Sinha BN. Histone deacetylase inhibitors: a review on class-I specific inhibition. Mini Rev Med Chem. 2015;15(9):731–750. [DOI] [PubMed] [Google Scholar]
- 78. Condon JC, Jeyasuria P, Faust JM, Wilson JW, Mendelson CR. A decline in the levels of progesterone receptor coactivators in the pregnant uterus at term may antagonize progesterone receptor function and contribute to the initiation of parturition. Proc Natl Acad Sci USA. 2003;100(16):9518–9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442(7098):86–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article or in the data repositories listed in References.