Abstract
Current research indicates that beta cell loss in type 2 diabetes may be attributed to beta cell dedifferentiation rather than apoptosis; however, the mechanisms by which this occurs remain poorly understood. Our previous study demonstrated that elevation of microRNA-24 (miR-24) in a diabetic setting caused beta cell dysfunction and replicative deficiency. In this study, we focused on the role of miR-24 in beta cell apoptosis and dedifferentiation under endoplasmic reticulum (ER) stress conditions. We found that miR-24 overabundance protected beta cells from thapsigargin-induced apoptosis at the cost of accelerating the impairment of glucose-stimulated insulin secretion (GSIS) and enhancing the presence of dedifferentiation markers. Ingenuity® Pathway Analysis (IPA) revealed that elevation of miR-24 had an inhibitory effect on XBP1 and ATF4, which are downstream effectors of two key branches of ER stress, by inhibiting its direct target, Ire1α. Notably, elevated miR-24 initiated another pathway that targeted Mafa and decreased GSIS function in surviving beta cells, thus guiding their dedifferentiation under ER stress conditions. Our results demonstrated that the elevated miR-24, to the utmost extent, preserves beta cell mass by inhibiting apoptosis and inducing dedifferentiation. This study not only provides a novel mechanism by which miR-24 dominates beta cell turnover under persistent metabolic stress but also offers a therapeutic consideration for treating diabetes by inducing dedifferentiated beta cells to re-differentiation.
Keywords: microRNA-24, ER stress, apoptosis, dedifferentiation, type 2 diabetes, pancreatic beta cells
Introduction
Pancreatic beta cells are specialized for the synthesis and secretion of sufficient insulin in response to blood glucose challenges. A well-developed endoplasmic reticulum (ER) ensures the effective production of insulin under physiological stimuli. However, when confronted with persistent metabolic stress, such as obesity, the rapidly increased demand of insulin secretion activates the unfolded protein response (UPR) to relieve ER stress (Eizirik et al., 2008; Hasnain et al., 2016). However, high ER stress induces beta cell dysfunction and apoptosis through sensors, such as the inositol-requiring enzyme 1 (IRE1), protein kinase-like ER kinase (PERK), and the activation of transcription factor 6 (ATF6) (Oslowski and Urano, 2010). The unique roles of IRE1α in beta cells include the promotion of differentiation, enhancement of postprandial proinsulin biosynthesis, adaptive proliferation, and anti-apoptosis upon ER stress (Lipson et al., 2006; Lerner et al., 2012; Sharma et al., 2015). Upon activation, IRE1α initiates the non-conventional splicing of the mRNA encoding X-box-binding protein-1 (XBP1), which upregulates IRE1α target genes to mitigate ER stress (Sha et al., 2011). However, prolonged activation of IRE1α causes phosphorylation of the c-Jun N-terminal kinase (JNK) and increased expression of DNA-damage inducible transcript 3 (Ddit3, also known as Chop), which can lead to beta cell dysfunction and apoptosis (Brozzi et al., 2014; Chan et al., 2015).
New evidence increasingly indicates that the main contributor of beta cell loss in type 2 diabetic patients is dedifferentiation, rather than apoptosis, since beta cell failure can be partly and temporarily reversed by dietary or pharmacological interventions (Talchai et al., 2012; Fiori et al., 2013; Blum et al., 2014; Wang et al., 2014; Cummings et al., 2016). Notably, remission of type 2 diabetes can be achieved by bariatric surgeries operated on diabetic patients with morbid obesity (Mingrone et al., 2015), which suggests that cellular loss of beta cells is not permanent. Chronic ER stress in diabetic settings causes gradual loss of beta cell-specific transcriptional factors, including PDX1, MAFA, and FOXO1 (Sachdeva et al., 2009; Kluth et al., 2011; Guo et al., 2013), and reoccurrence of endocrine progenitor markers, such as NGN3 and OCT4 (Talchai et al., 2012). These changes stimulate beta cells to exit their mature state into a dedifferentiated state manifested by defective GSIS function, decreased insulin positive cells, and consequently lessened beta cell mass. Talchai et al. (2012) first confirmed that loss of beta cell mass is mostly due to beta cell dedifferentiation in diabetes and in the discussion part, they pointed that those dedifferentiated beta cells were ‘selfish beta cells’, since dedifferentiation could facilitate their survival. Direct evidence was found in type 1 diabetes, which showed that under inflammatory stress, a group of beta cells decreased their characteristics of mature beta cells but showed increased stem-like features to escape from T-cell-mediated death with an unknown mechanism (Rui et al., 2017).
Even though beta cell dedifferentiation has been proposed as an important mechanism of beta cell failure in type 2 diabetes (Talchai et al., 2012; Kitamura, 2013), the underlying mechanism has not been clarified. Prior studies focused on finding evidence to support the theory that beta cell dedifferentiation exists in diabetic animals and patients (Talchai et al., 2012; White et al., 2013). In later animal studies, lineage tracing experiments were widely utilized to track the fate of beta cells, which might revert to progenitor-like cells or transdifferentiate to glucagon-producing ‘alpha-like’ cells to confront chronic glucolipotoxicity (Talchai et al., 2012; Blum et al., 2014). Recently, evidence drawn from type 2 diabetic patients has also supported the view that beta cells become dedifferentiated and convert to ‘alpha-like’ and ‘delta-like’ cells (Cinti et al., 2016). However, the leading factors of beta cell dedifferentiation were still undetected. Further studies to uncover whether ER stress is contributed to beta cell dedifferentiation are needed. More specifically, the mechanisms that cause ER stress, such as how IRE1α induces beta cell survival, apoptosis, and dedifferentiation under ER stress conditions, are still unknown.
MicroRNAs (miRNAs) are endogenous noncoding RNAs (~22 nucleotides) that regulate gene expression by binding to the 3′UTR of their target mRNAs to degrade and/or inhibit the mRNAs translation. These miRNAs target potentially hundreds of mRNAs and posttranscriptionally control complex cellular processes, such as proliferation, differentiation, and apoptosis (Brown and Sanseau, 2005; Kim, 2005; Filios and Shalev, 2015). Our previous study demonstrated that microRNA-24 (miR-24) was elevated in islets isolated from diabetic mice and in human islets, as well as MIN6 cells treated with palmitate—a factor known to induce beta cell ER stress. Ectopic expression of miR-24 caused beta cell dysfunction and replicative deficiency by modulating two MODY gene expressions (Zhu et al., 2013). miR-24 elevation also resulted in an inhibitory effect on the translation of XBP1 and ATF4, which are two downstream effectors of two key branches of ER stress. In the present study, we tested the effect of miR-24 on treatment with the ER stress activator thapsigargin (TG) and explored the potential mechanism by which beta cells avoid ER stress-induced apoptosis and undergo dedifferentiation.
Results
miR-24 alleviates ER stress-induced beta cell apoptosis while causing dysfunction via dedifferentiation
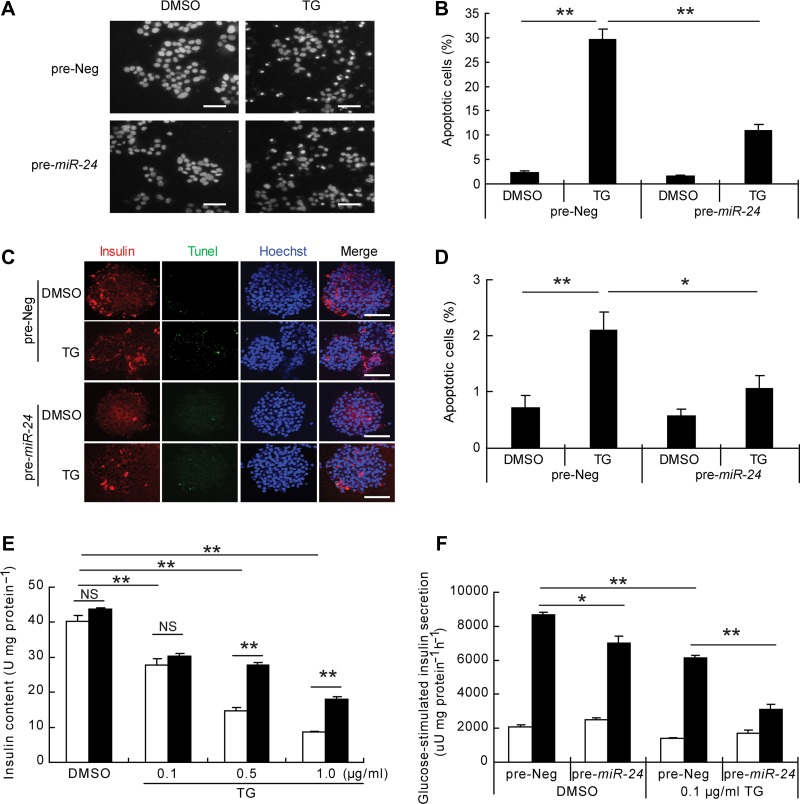
TG is a widely used ER stress activator that induces beta cell dysfunction and apoptosis by increasing cellular free Ca2+ levels. We assessed apoptosis of MIN6 cells after treatment with 1 μg/ml of TG at different time intervals. A Hoechst 33342 staining assay showed that the number of pycnotic nuclei significantly increased in a time-dependent manner (Supplementary Figure S1A). PARP1, an endogenous caspase 3 cleavage substrate, was also time-dependently cleaved in the presence of TG treatment (Supplementary Figure S1B). We then transfected MIN6 cell with pre-miR-24 and evaluated the effects of TG. About 90% cells were successfully transfected verified by the fluorescently labeled pre-miR-24 (Supplementary Figure S1C). Surprisingly, miR-24-overexpressed MIN6 cells were resistant to TG-induced apoptosis (Figure 1A and B) as well as palmitate-induced apoptosis (Supplementary Figure S2). The anti-apoptotic effect of miR-24 was also reproduced in primary mouse islets (Supplementary Figure S1D; Figure 1C and D). Moreover, insulin content was preserved in these cells after exposure to high doses of TG, while no between-group alterations of insulin content were observed with low dosages (0.1 μg/ml) of TG (Figure 1E). We took advantage of this relatively low-dose TG and assessed GSIS function on miR-24-transfected MIN6 cells. Intriguingly, miR-24 elevation and 0.1 μg/ml TG treatment synergistically impaired GSIS function in MIN6 cells (Figure 1F).
Figure 1.
miR-24 inhibits TG-induced apoptosis and GSIS dysfunction. (A) MIN6 cells were transfected with pre-Neg or pre-miR-24 for 48 h, treated with DMSO or TG (1 μg/ml) for 24 h, and then stained with Hoechst 33342. Scale bar, 40 μm. (B) Columns demonstrate the percentage of apoptotic cells in A. (C) Islets from 8-week-old C57BL6/J mice were transfected with pre-Neg or pre-miR-24 for 72 h and then treated with DMSO or TG (1 μg/ml) for further 24 h. Tunel combined with insulin and Hoechst 33342 staining was performed. Scale bar, 50 μm. (D) Columns show the percentage of apoptotic cells in C. (E) MIN6 cells were transfected with pre-Neg (clear bar) or pre-miR-24 (black bar) for 48 h, treated with DMSO or the indicated concentrations of TG (0.1, 0.5, or 1.0 μg/ml) for 12 h, and then the insulin content was detected. (F) The GSIS assay was performed after 48 h post-transfection and a further 12-h treatment with DMSO or 0.1 μg/ml TG. Insulin secretion in low glucose (clear bar) and high glucose (black bar) was measured. *P < 0.05 and **P < 0.01.
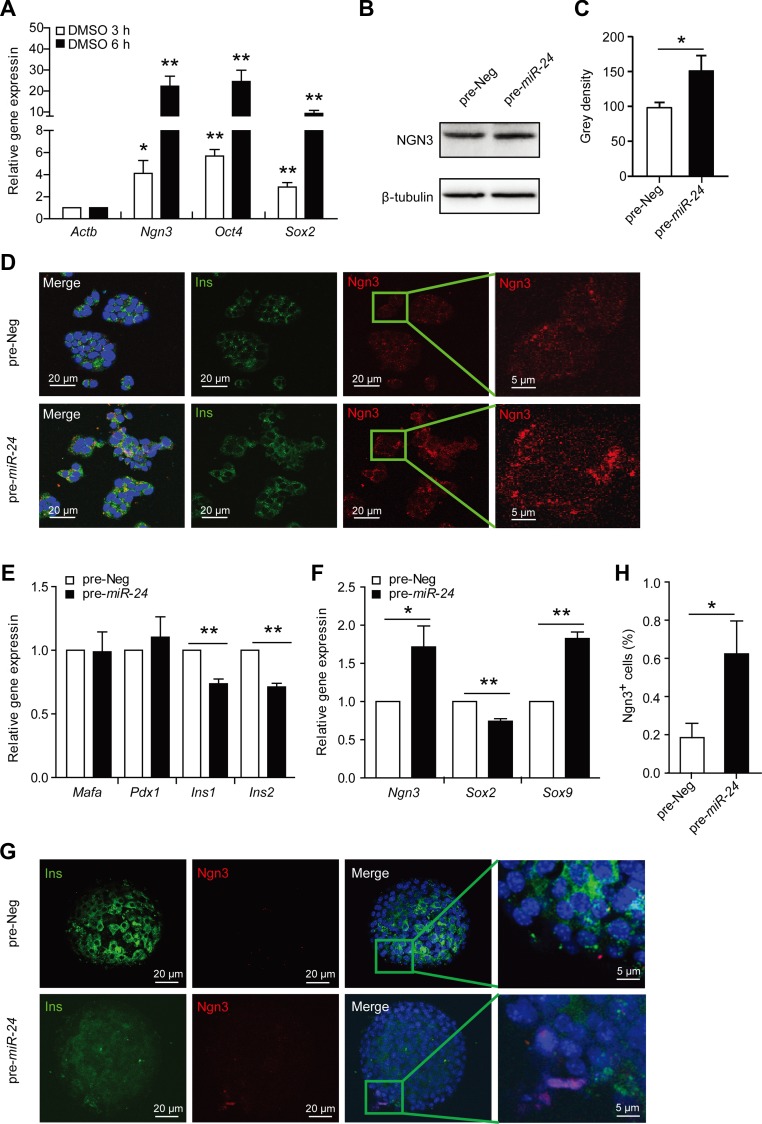
Also, elevation of miR-24 increased gene expression levels of Ngn3, Oct4, and Sox2, which are known dedifferentiation markers of beta cells (Figure 2A). The level of NGN3 protein was also examined by western blot and immunofluorescence assays (Figure 2B–D). Ectopic elevation of miR-24 inhibited expression of two insulin genes (Figure 2E) and changed dedifferentiation markers in primary isolated islets (Figure 2F). Although the expression level of Sox2 was slightly decreased, those of Ngn3 and Sox9 were significantly increased (Figure 2F). Using islet immunofluorescence, we also found a higher expression of Ngn3 after 96 h post-transfection with miR-24 (Figure 2G and H). The elevated expression of Ngn3 suggests that beta cells dedifferentiate into proendocrine cells, while Sox9 elevation means that they convert into pancreatic progenitor cells (Lynn et al., 2007; Seymour et al., 2007). Sox2 overexpression indicates the pancreatic beta cells might return to pluripotent stem cells (Wilson et al., 2005). Taken together, elevated miR-24 prevented ER stress-induced beta cell apoptosis while accelerated beta cell dysfunction, thus leading to beta cell dedifferentiation under diabetic conditions.
Figure 2.
miR-24 increases the expressions of dedifferentiation markers. MIN6 cells were transfected with pre-Neg (control) or pre-miR-24 for 48 h, treated with DMSO for further 3 h or 6 h, and then expression levels of Ngn3, Oct4, and Sox2 were analyzed. (A) The mRNA levels of Ngn3, Oct4, and Sox2 were analyzed by qRT-PCR assay. β-actin was used as internal control. (B) The protein level of NGN3 was examined by western blot. β-tubulin was used as internal standard. (C) Grey density of B. (D) An immunofluorescence assay was used to detect protein levels and distribution of INS and NGN3. (E and F) Primary isolated islets were transfected with pre-Neg or pre-miR-24 for 72 h, and then mRNA levels of Mafa, Pdx1, Ins1, Ins2, Ngn3, Sox2, and Sox9 were detected by qRT-PCR. *P < 0.05 and **P < 0.01 vs. control. (G) Primary isolated islets were transfected with pre-Neg or pre-miR-24 for 96 h. An immunofluorescence assay was used to detect protein levels and distribution of INS and NGN3. (H) The percentage of NGN3+ cells in islets of G. *P < 0.05 vs. control.
miR-24 inhibits XBP1 and ATF4 pathways
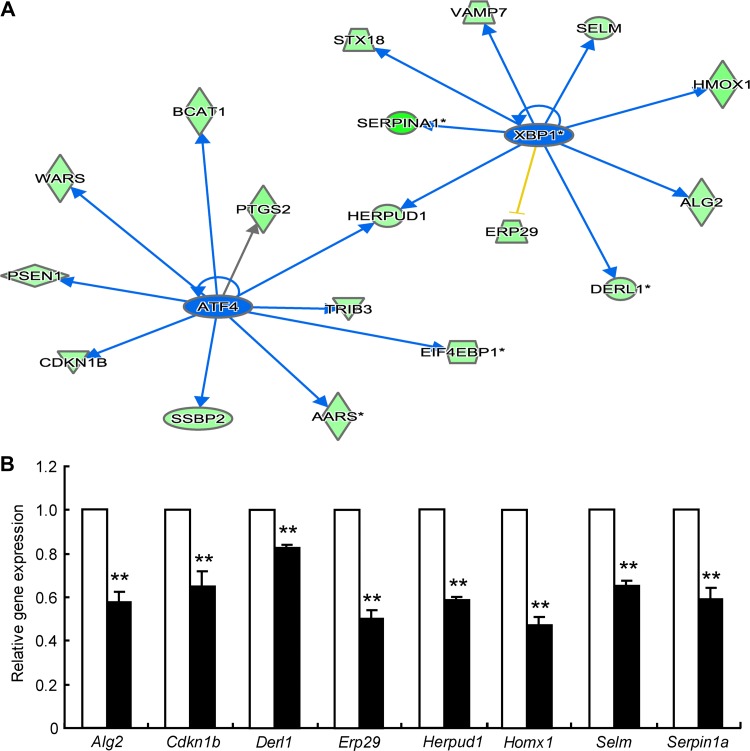
Considering the effects of miR-24 on ER stress, we used Ingenuity Pathway Analysis (IPA), an integration and search tool for pathways analyses, to re-evaluate the data from the Affimatrix mRNA chip that were obtained previously. We found that both the XBP1 and ATF4 pathways were inhibited by miR-24 overexpression in MIN6 cells as shown by clustering of expression levels of downstream genes (Figure 3A). Three ATF4 transcriptional target genes and seven XBP1-associated genes were validated by qRT-PCR (Figure 3B), suggesting their involvement in miR-24-modulated, ER stress-associated effects.
Figure 3.
miR-24 inhibits XBP1 and ATF4 signaling. (A) Expression levels of ER stress-associated genes were reanalyzed based on the previous mRNA microarray data by IPA. (B) MIN6 cells were transfected with pre-Neg (control, clean bar) or pre-miR-24 (black bar) for 48 h, and mRNA levels were analyzed by qRT-PCR assay. Data shown are mean ± SEMs and represent three separate experiments. β-actin was used as internal control. **P < 0.01 vs. control.
Ire1α is a target of miR-24
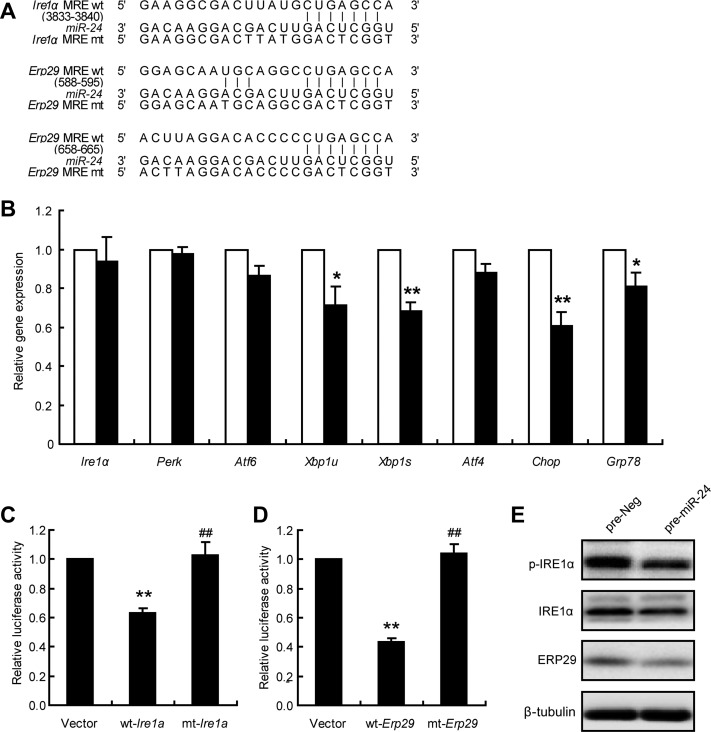
To identify the direct targets of miR-24, Targetscan was used for target gene prediction. Since no conserved seed sequences were found on the 3′UTRs of either Xbp1 and Atf4, the prediction work was then extended to their upstream regulators. Ire1α, Perk, Atf6, and Erp29, are related genes involved in XBP1 signaling. As shown in Figure 4A, the miRNA response elements (MRE) of miR-24 were located on 3′UTR of Ire1α and Erp29. The gene expression levels of these three key branches and their main effectors were examined, and only Xbp1, Chop, and Grp78 were modestly decreased. Ire1α, Perk, Atf6, and Atf4 did not change with the expression of miR-24 (Figure 4B). Next, the sequences containing wild-type (wt) and mutant (mt) miR-24 MRE were subcloned into pmiR-REPORTOR. The luciferase activity assay showed that miR-24 inhibited the activity of the wt-Ire1α construct when compared with a vector control, whereas no alteration was observed with the mt-Ire1α construct (Figure 4C). miR-24 was also found to inhibit MRE from Erp29 (Figure 4D) and Pdi (Supplementary Figure S3A and B).
Figure 4.
miR-24 regulates Ire1α and Erp29. (A) The 3′UTR sequences of Ire1α and Erp29 predicted to include miR-24 MREs were aligned with miR-24, and both wt and mt sequences are listed. (B) MIN6 cells were transfected with pre-Neg (control, clean bar) or pre-miR-24 (black bar) for 48 h, and mRNA levels were analyzed by qRT-PCR assay. β-actin was used as internal control. *P < 0.05 and **P < 0.01 vs. control. (C) MIN6 cells were co-transfected with pre-miR-24 and Vector, wt-Ire1α, or mt-Ire1α; after 48 h, dual luciferase reporter activities were measured. **P < 0.01 vs. Vector and ##P < 0.01 vs. wt-Ire1α. (D) MIN6 cells were co-transfected with pre-miR-24 and Vector, wt-Erp29, or mt-Erp29; after 48 h, dual luciferase reporter activities were measured. **P < 0.01 vs. Vector and ##P < 0.01 vs. wt-Erp29. (E) MIN6 cells were transfected with pre-Neg or pre-miR-24 for 48 h and protein levels were analyzed by western blot. β-tubulin was used as internal standard.
Furthermore, miR-24 overexpression significantly reduced the protein levels of pIRE1α, IRE1α, and ERP29 (Figure 4E). The gene expression and protein levels of PDI—a disulfide isomerase that catalyzes the formation, breakage, and rearrangement of disulfide bonds—were also significantly decreased by miR-24 overexpression (Supplementary Figure S3C and D). Thus, we identified that the three ER-resident proteins, Ire1α, Erp29, and Pdi, were targets of miR-24.
Ire1α mediates the anti-apoptotic effect of miR-24
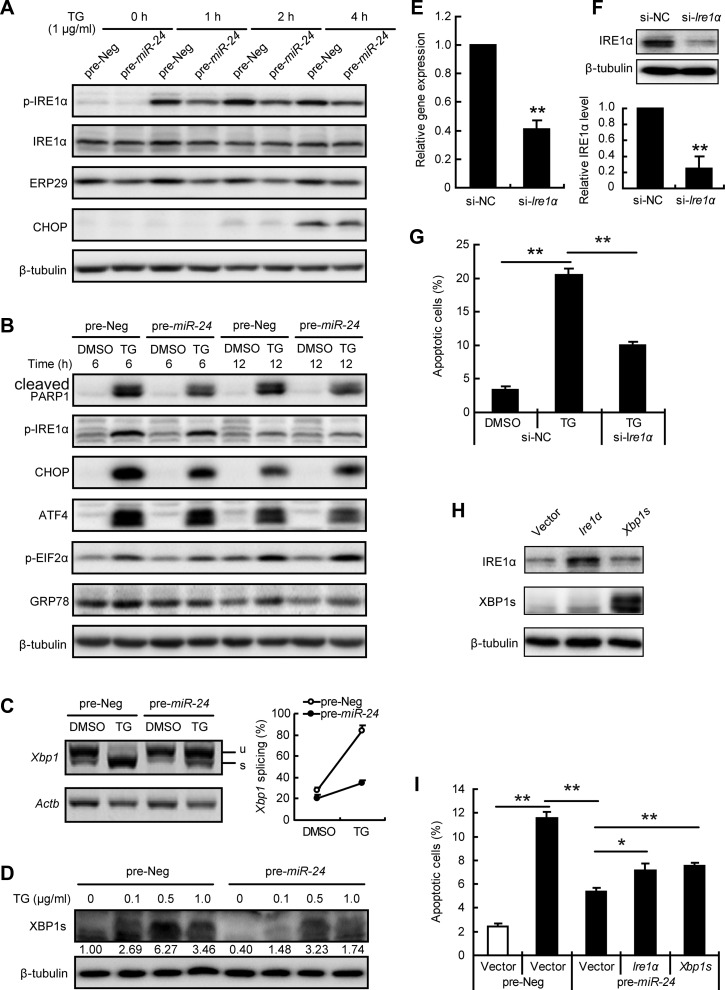
To further understand the role of Ire1α on miR-24 mediated effects, MIN6 cells transfected with miR-24 were treated short-term and long-term with TG. As shown in Figure 5A, short-term TG treatment significantly increased the phosphorylation of IRE1α. p-IRE1α was at the highest level after a 2-h treatment when the level of CHOP started to increase. The levels of both p-IRE1α and CHOP can be inhibited by miR-24 overexpression.
Figure 5.
miR-24 inhibits IRE1α/XBP1s signaling. (A–D) MIN6 cells were transfected with pre-Neg or pre-miR-24 for 48 h. (A and B) Transfected cells were further treated with or without 1 μg/ml TG for indicated time and protein levels were analyzed by western blot. β-tubulin was used as internal standard. (C) A semi-quantitative RT-PCR assay was performed after a 3-h treatment with or without 1 μg/ml TG, and the PCR products were separated with a 3% agarose gel. Xbp1 splicing efficiency was calculated as the ratio of the spliced Xbp1 level divided by the total Xbp1 level. (D) Transfected cells were treated with the indicated concentrations of TG for 3 h and protein level of XBP1s was determined. Relative XBP1s level was shown under the band. (E–G) MIN6 cells were transfected with si-NC or si-Ire1α for 48 h. (E) mRNA level of Ire1α was analyzed by qRT-PCR assay. β-actin was used as internal control. **P < 0.01 vs. si-NC. (F) Protein level of IRE1α was analyzed by western blot. β-tubulin was used as internal standard. Grey density data are shown below. **P < 0.01 vs. si-NC. (G) Transfected cells were further treated with or without 1 μg/ml TG for 24 h, and apoptotic cells were calculated by Hoechst33342 staining. (H) MIN6 cells were transfected with Ire1α or Xbp1s for 48 h and protein levels were measured. (I) MIN6 cells were co-transfected with the indicated plasmid and miRNA for 48 h, treated with (black bar) or without (clean bar) 1 μg/ml TG for an additional 16 h, and then apoptotic cells were counted. *P < 0.05 and **P < 0.01.
Meanwhile, the protein levels of ERP29 and PDI were unchanged by TG treatment, even though miR-24 can decrease protein levels. Prolonged treatment with TG for 8 h can induce MIN6 cell apoptosis (Supplementary Figure S1); thus, miR-24-transfected cells were treated with TG for 6 and 12 h to assess the molecular alterations of the three key ER branches (Figure 5B). The decreased level of cleaved PARP1 at both timepoints showed the anti-apoptotic role of miR-24 at the molecular level. Again, p-IRE1α, CHOP, and GRP78 levels were reduced by miR-24 overexpression at 6 h. However, p-IRE1α was decreased to normal levels after a 12-h treatment with TG, by which time, the protein levels of CHOP and GRP78 remained high. No between-group alterations of ATF4 and p-eIF2α were discovered (Figure 5B), nor were there any alterations of other ER stress-associated molecules of interest (Supplementary Figure S4).
IRE1α modulates the gene expression level and splicing of Xbp1 mRNA, and miR-24 overexpression inhibited both unspliced-Xbp1 (Xbp1u) and spliced-Xbp1 (Xbp1s) mRNA levels (Figure 3B). In particular, TG provoked a high expression level of Xbp1s that was significantly inhibited in miR-24-transfected MIN6 cells (Figure 5C). The inhibitory effect of miR-24 was confirmed by the XBP1s protein level (Figure 5D). Collectively, miR-24 elevation inhibited IRE1α/XBP1 signaling at both basal and TG-stimulated levels.
To assess the role of IRE1α on miR-24-associated effects, siRNA interference of Ire1α was introduced. The interference efficiency was examined in both mRNA and protein levels (Figure 5E and F). Knockdown of Ire1α significantly inhibited the number of apoptotic cells induced by TG, mimicking the anti-apoptotic effect of miR-24 (Figure 5G). In contrast, overexpression of Ire1α had no additional effect on TG-induced MIN6 cell apoptosis (Supplementary Figure S5). Notably, overexpression of either Ire1α or Xbp1s counteracted the anti-apoptotic effect of miR-24 against TG exposure (Figure 5H and I). Taken together, these results demonstrate that elevated miR-24 under metabolic stress could confer resistance to ER stress-induced apoptosis via inhibiting the IRE1α/XBP1 pathway.
miR-24 induces insulin secretion defect by targeting Mafa
The role of IRE1α on specialized beta cell functions was also examined. Knockdown of Ire1α inhibited insulin content and basal insulin secretion without altering glucose-stimulated insulin secretion (Supplementary Figure S6A and B). TG-induced GSIS defects could not be potentiated by Ire1α-knockdown (Supplementary Figure S6C). This suggested that Ire1α did not contribute to miR-24-excess-induced insulin secretion defects.
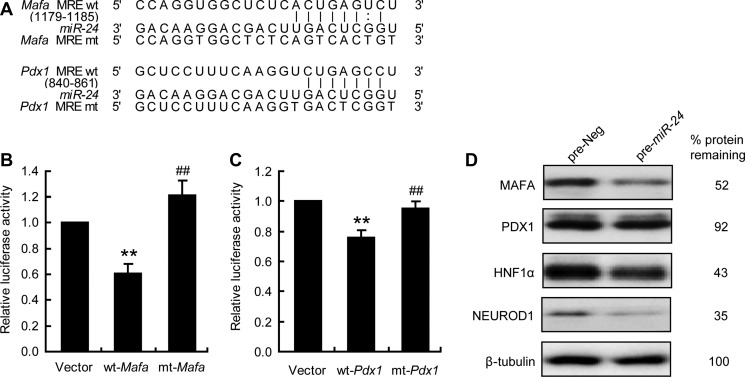
Beta cell-specific transcriptional factors, including MAFA, PDX1, NEUROD1, and HNF1α, are well-recognized factors contributing to ER stress-induced beta cell dysfunction. We previously reported that miR-24 MREs were located on 3′UTR of Neurod1, Hnf1α, and Pdx1. Mafa was also identified as a target by the Targetscan software (Figure 6A). The regulatory role between miR-24 and Mafa was confirmed by a luciferase activity assay (Figure 6B), as was the connection between miR-24 and Pdx1 (Figure 6C). The protein levels of MAFA, PDX1, NEUROD1, and HNF1α were significantly reduced by miR-24 overexpression (Figure 6D). The above results showed that, in addition to the three previously reported transcriptional factors, Mafa was also a target of miR-24.
Figure 6.
miR-24 targets Mafa and Pdx1. (A) 3′UTR sequences of Mafa and Pdx1 predicted to include miR-24 MREs were aligned with miR-24, and both wt and mt sequences were listed. (B) MIN6 cells were co-transfected with pre-miR-24 and Vector, wt-Mafa, or mt-Mafa. After 48 h, dual luciferase reporter activities were measured. **P < 0.01 vs. Vector and ##P < 0.01 vs. wt-Mafa. (C) MIN6 cells were co-transfected with pre-miR-24 and Vector, wt-Pdx1, or mt-Pdx1. After 48 h, dual luciferase reporter activities were measured. **P < 0.01 vs. Vector and ##P < 0.01 vs. wt-Pdx1. (D) MIN6 cells were transfected with pre-Neg or pre-miR-24 for 48 h and protein levels were analyzed by western blot. β-tubulin was used as internal standard.
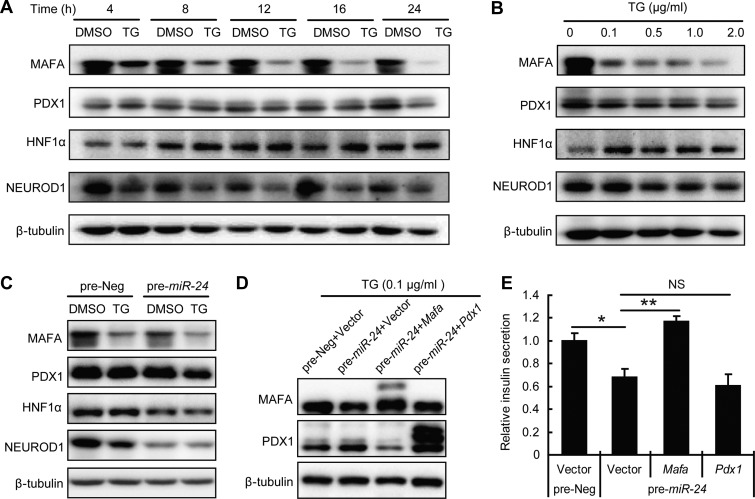
Next, mRNA and protein levels of these four transcriptional factors were sequentially examined after TG treatment. No significant alterations of their mRNA levels were detected (data not shown), but the protein level of MAFA rapidly declined after 4 h of TG treatment. In fact, the protein level was even affected by a low TG dose, while the other three transcriptional factors decreased later and were only affected by a high-dose TG treatment (Figure 7A and B).
Figure 7.
MAFA contributes to the miR-24-induced GSIS defect under ER stress condition. (A) MIN6 cells were treated with or without 0.1 μg/ml TG for the indicated times and protein levels were determined by western blot. (B) MIN6 cells were treated with the indicated concentrations of TG for 12 h, and protein levels were determined by western blot. (C) MIN6 cells were transfected with pre-Neg or pre-miR-24 for 48 h and further treated with or without 0.1 μg/ml TG for 12 h when protein levels were analyzed by western blot. (D and E) MIN6 cells were co-transfected with the indicated miRNA and plasmid for 48 h and further treated with 0.1 μg/ml TG for 12 h. (D) Protein levels were analyzed by western blot. β-tubulin was used as internal standard. (E) GSIS assay was performed and relative insulin secretion levels were normalized by the Vector and pre-Neg group. *P < 0.05 and **P < 0.01.
Moreover, the protein level of MAFA exhibited the most obvious synergistic decrease in miR-24-transfected cells treated with TG (Figure 7C). PDX1 level also displayed a small degree of synergistic decline (Figure 7C). Afterwards, recoveries of MAFA and PDX1 by plasmid-based overexpression were performed (Figure 7D). Surprisingly, only Mafa overexpression could rescue the effects of elevated miR-24 when combined with TG-treatment-induced GSIS defects (Figure 7E). Taken together, miR-24 inhibits MAFA to guide the survived beta cells towards dysfunction under ER stress conditions.
Discussion
Our study explored the role of miR-24 in regulating beta cell turnover under diabetic settings. Phenotypically, miR-24 elevation was resistant to ER stress-induced apoptosis but at the price of inhibiting GSIS function, thus guiding beta cell survival from ER stress towards dedifferentiation. Mechanistically, miR-24 targeted Ire1α to protect beta cells from ER stress-induced apoptosis. In contrast, miR-24 also targeted Mafa to induce beta cell dysfunction and dedifferentiation. These factors represent a possible mechanism through which beta cell loss in type 2 diabetes could be attributed to dedifferentiation rather than apoptosis.
It is widely appreciated that pancreatic beta cells experience dramatic ER stress in type 2 diabetes because of the increased insulin secretion demand that is primarily caused by hyperglycemia and obesity (Fonseca et al., 2010; Biden et al., 2014). We have previously reported that miR-24 abundance in pancreatic islets could be significantly elevated when confronting an ER stress condition. Ectopic overexpression of miR-24 resulted in inhibition of both insulin secretion and beta cell growth by modulating Neurod1-MODY and Hnf1α-MODY gene expression (Zhu et al., 2013). It seems that the elevated miR-24 initiates protective processes to avoid excess insulin secretion and relieve the severe metabolic burden imposed on the beta cells. The efforts made by miR-24 are likely to antagonize ER stress conditions by reducing insulin biosynthesis and secretion, despite it possibly not having any benefit on maintaining blood glucose homeostasis in diabetic settings. This provides indirect evidence to support the theory that miR-24 can mitigate ER stress conditions.
Therefore, we reanalyzed the data from the mRNA gene chip analysis that previously reported on to search for any direct connection between miR-24 and ER stress. Surprisingly, downstream effectors of both XBP1 and ATF4 were significantly decreased by miR-24 elevation, suggesting a potential inhibitory role of miR-24 on ER stress. By using software prediction, luciferase activity assay, and western blotting, we found that Ire1α is a target gene of miR-24. This provides a direct link between miR-24 and ER stress through IRE1α.
IRE1α plays an important role in secretory cells, such as pancreatic beta cells and plasma cells (Jurkin et al., 2014; Zhang et al., 2014). Physical activation of IRE1α by transient exposure to high glucose positively regulates proinsulin biosynthesis (Lipson et al., 2006). Alternatively, severe activation of IRE1α caused by a chronic high-glucose and ER stress inducer leads to ER-resident mRNA decay, such as insulin, thus causing defective insulin biosynthesis, beta cell dysfunction, and apoptosis (Hou et al., 2008; Yang et al., 2013; Zhang et al., 2014).
The divergent role of IRE1α on insulin biosynthesis relies on the ability of autophosphorylation-coupled RNase activity. Acute exposure to high glucose only initiates an increase in autophosphorylation of IRE1α (Lipson et al., 2006). In contrast, chronic exposure of beta cells to high glucose levels provokes both phosphorylation and RNase activities in IRE1α, thereby causing adaptive splicing of the Xbp1 mRNA to promote survival (Lipson et al., 2006; Chan et al., 2015), but otherwise enforcing endonucleolytic decay of many ER-resident mRNAs, including insulin and ER chaperones to promote dysfunction and apoptosis (Hollien and Weissman, 2006; Yang et al., 2013; Zhang et al., 2014).
Reports have shown that IRE1α kinase activation modes control alternate RNase outputs to determine divergent cell fates (Han et al., 2009; Tohmonda et al., 2015). Nevertheless, allosteric inhibition of the IRE1α RNase by KIRA6 could protect beta cells from severe ER stress-induced dysfunction, growth inhibition, and apoptosis in vitro and in vivo (Ghosh et al., 2014). Moreover, deletion of Ire1α or reduction of IRE1α through RNAi also repressed ER-localized mRNA decay during ER stress. This suggests that the protein level of IRE1α directly controls its RNase activity (Han et al., 2009; So et al., 2012).
As a verified target, our research showed that elevation of miR-24 reduced the protein and phosphorylation levels of IRE1α under both basic and TG-imposed ER stress conditions. The inactivation of IRE1α by miR-24 caused a decrease in insulin secretion but preserved insulin biosynthesis in beta cells during ER stress situations, perfectly representing the physical and pathological characteristics of IRE1α. Thus, the ER stress-induced miR-24 has a negative feedback on ER stress conditions in pancreatic beta cells.
Overactivation of IRE1α promotes cell apoptosis through the JNK and CHOP pathways. Reports have shown that IRE1α recruits TRAF2 to cause ASK1 activation under ER stress conditions (Urano et al., 2000; Nishitoh et al., 2002). Activated ASK1 mediates JNK activation and results in neuronal cell apoptosis (Nishitoh et al., 2002). However, their roles in ER stress-mediated beta cell apoptosis remain unclear. In our observations, the phosphorylation of JNK only increased after treatment with TG for 8 h when the apoptosis of beta cells had already occurred at an elevated level. This helped us rule out JNK for further investigation.
CHOP is an important component to ER stress-mediated apoptosis (Oyadomari and Mori, 2004). Deletion of CHOP promotes beta cell survival and improves beta cell function in the Akita diabetes mouse model, which expresses a mutant (C96Y) proinsulin that is unable to complete oxidative folding and thus causes chronic ER stress (Oyadomari et al., 2002). According to our results, the protein level of CHOP started to increase at 1 h and continued to climb to its peak at 6 h of treatment with TG. Moreover, elevated miR-24 inhibited the TG-induced CHOP increase, suggesting the anti-apoptotic role of miR-24 is commanded by an IRE1α/CHOP pathway. It is well accepted that CHOP is primarily regulated by the PERK/ATF4 pathway (Rozpedek et al., 2016). However, the ATF4 level was not be inhibited by miR-24. We speculate that CHOP might also be modulated by IRE1α, since the downstream effectors of three ER branches are indistinguishable under severe ER stress (Szegezdi et al., 2006).
The identification of CHOP as a downstream effector of IRE1α does not rule out the contribution of XBP1s in ER stress-induced apoptosis. Considerable evidence indicates that the transcription factor, XBP1s, drives the expression of ER chaperones and components of the ER-associated degradation (ERAD) to restore ER homeostasis (Gupta et al., 2010; Wu et al., 2015; Sun et al., 2016). However, overexpression of XBP1s counteracted the anti-apoptotic effect of miR-24, which suggests that XBP1s is a proapoptotic molecule under irremediable ER stress conditions in pancreatic beta cells.
Consistent with our data, Allagnat et al. (2010) reported that sustained production of XBP1s, but not XBP1u, promotes pancreatic beta cell dysfunction and apoptosis. These impairments engage with beta cell-specific molecules, such as insulin, PDX1, and MAFA, thus declaring a unique role for XBP1s in beta cell apoptosis. Nevertheless, mild ER stress could augment the proinflammatory effect of IL-1β through the IRE1α–XBP1s pathway in pancreatic beta cells (Miani et al., 2012).
Although miR-24 promoted beta cell survival from ER stress-induced apoptosis and inhibited insulin mRNA degradation, the GSIS function was somehow synergistically restrained by miR-24 elevation and TG exposure, thereby causing beta cell dedifferentiation. To solve this problem, we identified the maturation factors, MAFA and PDX1, as two additional targets of miR-24. Protein levels of both MAFA and PDX1 were reduced by TG in dose- and time-dependent manners. The emergence of MAFA reduction started at 4 h and could be synergistically decreased by TG treatment, which indicates its special role. The in vitro phenotype of beta cell dedifferentiation induced by increased miR-24 could only be detected by DMSO pre-treatment, since DMSO itself can promote differentiation (Pal et al., 2012). This could also explain why those dedifferentiation markers were not elevated much in isolated primary mouse islets compared to MIN6 cells, which have not completed development and contain other pancreatic endocrine hormones (Nakashima et al., 2009). Using islets immunofluorescence of Ngn3, we found those dedifferentiated cells were located in the margin of islets, a pancreatic neogenic niche with plasticity in which alpha cells could transdifferentiate into beta cells (van der Meulen et al., 2017). Cells in this niche have dedifferentiation and transdifferentiation potentials; so, we speculate that early dedifferentiation occurs from this location.
Indeed, overexpression of MAFA, but not PDX1, effectively rescued TG-induced GSIS defects and recovered insulin-positive cells. Mafa is critical for the homeostasis of mature beta cells and regulates cell plasticity (Nishimura et al., 2015). Mafa KO mice promote beta cell dedifferentiation, while overexpression of Mafa preserves beta cells and improves hyperglycemia in db/db diabetic mice (Matsuoka et al., 2015; Nishimura et al., 2015). Our results support the same conclusion: that the transcription factor, MAFA, is important for maintaining beta cell identity in diabetic settings.
In conclusion, our investigation revealed that the ER stress effector miR-24 could counteract ER stress-induced apoptosis at the cost of defective GSIS function. The anti-apoptotic capacity of miR-24 provided a favorable explanation for the low detection of apoptosis rates in islets from type 2 diabetic patients and rodent animals, despite the presence of severe insulin defects. Our study strongly supports the theory that beta cell dedifferentiation is a dominant contributor to beta cell loss in type 2 diabetes. Once the hyperglycemia controls and the ER stress conditions remit, inhibition of miR-24 might be an attractive therapeutic approach to treating diabetes.
Materials and methods
Materials
Cell culture
Cells of the MIN6, a mouse pancreatic beta cell line, were cultured in DMEM with 15% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 10 mM HEPES, and 50 μM β-mercaptoethanol (Sigma-Aldrich). The cells were incubated at 37°C in a suitable atmosphere containing 95% O2 and 5% CO2. TG or DMSO (Sigma-Aldrich) was added to the appropriate experiments for the indicated time and at the indicated dose.
Pancreatic islet isolation
Male 8-week-old C57BL6/J mice were purchased from Nanjing Medical University Laboratory Animal Center. Islet isolation and culturing methods were performed as previously described (Zhu et al., 2013). Isolated islets were equilibrated in RPMI 1640 containing 11.1 mM glucose with 10% FBS, 10 mM HEPES, 100 U/ml penicillin, and 100 μg/ml streptomycin overnight. The cells were transfected with pre-miR-24 or the pre-Negative control (pre-Neg) (Ambion) using Lipofectamine 2000 (Invitrogen) for 72 h, incubated with 1 μg/ml TG or DMSO for 24 h, and then fixed with 4% paraformaldehyde before use in further experiments. All animal studies were performed according to guidelines established by the Research Animal Care Committee of Nanjing Medical University.
Plasmid construction
The mouse expression plasmids, pcDNA3.1-Xbp1s and pCMV5-flag-Ire1α, were gifts from Prof. Yong Liu (Wuhan University) (Xu et al., 2014). The wild-type (wt) and mutant (mt) 3′UTR-luciferase constructs of mouse Ire1α, Erp29, Mafa, and Pdx1 were generated as previously described and inserted into the pMIR-REPORT luciferase vector (Ambion) between the HindIII and SpeI cutting sites (Zhu et al., 2013). The full-length coding region sequences of mouse Pdx1 and Mafa were inserted into pCMV5-flag vector. All constructs were confirmed by sequencing. The primer sequences are shown in Supplementary Table S1.
Transient transfection and luciferase reporter assay
MIN6 cells were cultured and transfected with luciferase reporter plasmids or overexpression plasmids as well as pre-miR-24 or pre-Neg using Lipofectamine 2000 according to the manufacturer’s instructions. Luciferase activities were measured 48 h after transfection with a dual-luciferase reporter assay system (Promega) and normalized to Renilla activity expressed with the PRL-SV40 plasmid (Promega).
Cell apoptosis assay
After different treatments, MIN6 cells were stained with Hoechst 33342 (Sigma-Aldrich) for 10 min at room temperature, washed with PBS, and then observed using a fluorescence microscope (Imager A1, Zeiss). All the cell nuclei were stained with a blue color. Cells with high blue fluorescence and fragmented nuclei were considered to be apoptotic, while weak blue signal indicated live cells. Images were obtained in random fields, and quantification of apoptotic cells was determined for at least 2000 cells in eight fields for each well. Apoptotic cells in primary islets were measured using a Tunel kit (Boster) and performed according to the manufacturer’s instructions.
Glucose-stimulated insulin secretion assay
MIN6 cells were transfected with pre-Neg or pre-miR-24 and incubated with DMSO or TG. The cell culture medium was refreshed 12 h before initiating this assay to diminish the influence of the insulin accumulated in the cell supernatant. MIN6 cells were then pre-incubated for 1 h in KRBH buffer supplemented with 2 mM glucose and 1 g/L bovine serum albumin (BSA). Subsequently, the cells were incubated for 1 h in KRBH containing basal glucose (2 mM) or stimulatory glucose (20 mM). The supernatants were collected and frozen at −80°C for subsequent measurement of insulin concentration, and whole-cell insulin contents were extracted using an acid-ethanol solution (0.15 M HCL in 75% ethanol in H2O) overnight at 4°C. The collected supernatants were measured by radioimmunoassay (RIA) as previously described (Zhu et al., 2013). Insulin content and glucose-stimulated insulin secretion were normalized to the total protein concentration.
RNA extraction and quantitative real-time PCR
Total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. The reverse transcriptions of miRNA and mRNA were performed as previously described (Zhu et al., 2013). Quantitative real-time PCR was performed using the SYBR Green PCR Master Mix and Roche Lightcycle480 II Sequence Detection System (Roche Diagnostics). U6 and β-actin were used to normalize miRNAs and mRNA, respectively. Sequences of the primers used here are available in Supplementary Table S2.
For semi-quantitative detective of spliced and un-spliced Xbp1, the following primers were used: forward, AAACAGAGTAGCAGCGCAGACTGC; reverse, TCCTTCTGGGTAGACCTCTGGGAG. PCR products were resolved on 3% agarose gels. β-actin was used as an internal control. The size difference between spliced and un-spliced Xbp1 is 26 nucleotides.
Western blot analysis
After culturing and transfecting as described above, MIN6 cells were lysed with ice-cold lysis buffer (50 mM Tris-HCL, pH 7.4, 1% NP-40, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride). After protein content determination, western blotting was performed as previously described (Zhu et al., 2013). See Supplementary Methods for more details.
Immunofluorescence
MIN6 cells were grown on coverslips in 24-well plates, cultured, and transfected. Islets were cultured and transfected after isolation. Cells or islets were then fixed with 4% paraformaldehyde and blocked with 1 g/L BSA for 30 min. Cells or islets were incubated overnight in 4°C with a rabbit-anti Ngn3 antibody (ThermoFisher) that had been diluted 1:20 and further stained with an Alexa Fluor 594 AffiniPure Donkey anti-Rabbit IgG (YEASEN) in 37°C for 1 h. After that, cells or islets were co-stained with an goat-anti Insulin antibody (Santa Cruz) diluted 1:100 in 37°C for 1.5 h and then an Alexa Fluor 488 AffiniPure Donkey anti-Goat IgG (YEASEN) in 37°C for 1 h. All the antibodies were diluted with 1% BSA. Nuclei were stained with Hoechst 33342 and then observed by confocal microscopy (FV1200, Olympus).
Statistical analysis
Data are expressed as the mean ± SEM. P-values were calculated with a Student’s two-tail t-test, and P < 0.05 was considered to be significant.
Supplementary Material
Acknowledgements
We thank Prof. Yong Liu from Wuhan University for providing plasmids of pcDNA3.1-Xbp1s and pCMV5-Ire1α. The authors gratefully acknowledge the generous support of the Collaborative Innovation Center for Cardiovascular Disease Translational Medicine of Jiangsu Province. We would like to express our gratitude to the illustrator Yuyang Sun for painting a graphic abstract.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2016YFC1304804) and the National Natural Science Foundation of China (81420108007) to X.H. and the National Natural Science Foundation of China (81670703) and China Postdoctoral Science Foundation (2016M590479) to Y.Z.
Conflict of interest
none declared.
References
- Allagnat F., Christulia F., Ortis F., et al. (2010). Sustained production of spliced X-box binding protein 1 (XBP1) induces pancreatic β cell dysfunction and apoptosis. Diabetologia 53, 1120–1130. [DOI] [PubMed] [Google Scholar]
- Biden T.J., Boslem E., Chu K.Y., et al. (2014). Lipotoxic endoplasmic reticulum stress, β cell failure, and type 2 diabetes mellitus. Trends Endocrinol. Metab. 25, 389–398. [DOI] [PubMed] [Google Scholar]
- Blum B., Roose A.N., Barrandon O., et al. (2014). Reversal of β cell de-differentiation by a small molecule inhibitor of the TGFβ pathway. eLife 3, e02809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.R., and Sanseau P. (2005). A computational view of microRNAs and their targets. Drug Discov. Today 10, 595–601. [DOI] [PubMed] [Google Scholar]
- Brozzi F., Gerlo S., Grieco F.A., et al. (2014). A combined ‘omics’ approach identifies N-Myc interactor as a novel cytokine-induced regulator of IRE1 protein and c-Jun N-terminal kinase in pancreatic β cells. J. Biol. Chem. 289, 20677–20693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.Y., Luzuriaga J., Maxwell E.L., et al. (2015). The balance between adaptive and apoptotic unfolded protein responses regulates β-cell death under ER stress conditions through XBP1, CHOP and JNK. Mol. Cell. Endocrinol. 413, 189–201. [DOI] [PubMed] [Google Scholar]
- Cinti F., Bouchi R., Kim-Muller J.Y., et al. (2016). Evidence of β-cell dedifferentiation in human type 2 diabetes. J. Clin. Endocrinol. Metab. 101, 1044–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings D.E., Arterburn D.E., Westbrook E.O., et al. (2016). Gastric bypass surgery vs intensive lifestyle and medical intervention for type 2 diabetes: the CROSSROADS randomised controlled trial. Diabetologia 59, 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizirik D.L., Cardozo A.K., and Cnop M. (2008). The role for endoplasmic reticulum stress in diabetes mellitus. Endocr. Rev. 29, 42–61. [DOI] [PubMed] [Google Scholar]
- Filios S.R., and Shalev A. (2015). β-cell microRNAs: small but powerful. Diabetes 64, 3631–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiori J.L., Shin Y.K., Kim W., et al. (2013). Resveratrol prevents β-cell dedifferentiation in nonhuman primates given a high-fat/high-sugar diet. Diabetes 62, 3500–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S.G., Urano F., Burcin M., et al. (2010). Stress hypERactivation in the β-cell. Islets 2, 1–9. [DOI] [PubMed] [Google Scholar]
- Ghosh R., Wang L., Wang E.S., et al. (2014). Allosteric inhibition of the IRE1α RNase preserves cell viability and function during endoplasmic reticulum stress. Cell 158, 534–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Dai C., Guo M., et al. (2013). Inactivation of specific β cell transcription factors in type 2 diabetes. J. Clin. Invest. 123, 3305–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Deepti A., Deegan S., et al. (2010). HSP72 protects cells from ER stress-induced apoptosis via enhancement of IRE1α-XBP1 signaling through a physical interaction. PLoS Biol. 8, e1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Lerner A.G., Vande Walle L., et al. (2009). IRE1α kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell 138, 562–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasnain S.Z., Prins J.B., and McGuckin M.A. (2016). Oxidative and endoplasmic reticulum stress in β-cell dysfunction in diabetes. J. Mol. Endocrinol. 56, R33–R54. [DOI] [PubMed] [Google Scholar]
- Hollien J., and Weissman J.S. (2006). Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313, 104–107. [DOI] [PubMed] [Google Scholar]
- Hou Z.Q., Li H.L., Gao L., et al. (2008). Involvement of chronic stresses in rat islet and INS-1 cell glucotoxicity induced by intermittent high glucose. Mol. Cell. Endocrinol. 291, 71–78. [DOI] [PubMed] [Google Scholar]
- Jurkin J., Henkel T., Nielsen A.F., et al. (2014). The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J. 33, 2922–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N. (2005). MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 6, 376–385. [DOI] [PubMed] [Google Scholar]
- Kitamura T. (2013). The role of FOXO1 in β-cell failure and type 2 diabetes mellitus. Nature reviews. Endocrinology 9, 615–623. [DOI] [PubMed] [Google Scholar]
- Kluth O., Mirhashemi F., Scherneck S., et al. (2011). Dissociation of lipotoxicity and glucotoxicity in a mouse model of obesity associated diabetes: role of forkhead box O1 (FOXO1) in glucose-induced β cell failure. Diabetologia 54, 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner A.G., Upton J.P., Praveen P.V., et al. (2012). IRE1α induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab. 16, 250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson K.L., Fonseca S.G., Ishigaki S., et al. (2006). Regulation of insulin biosynthesis in pancreatic β cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 4, 245–254. [DOI] [PubMed] [Google Scholar]
- Lynn FC., Smith SB., Wilson ME., et al. (2007). Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proc. Natl Acad. Sci. USA 104, 10500–10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka T.A., Kaneto H., Kawashima S., et al. (2015). Preserving Mafa expression in diabetic islet β-cells improves glycemic control in vivo. J. Biol. Chem. 290, 7647–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miani M., Colli M.L., Ladriere L., et al. (2012). Mild endoplasmic reticulum stress augments the proinflammatory effect of IL-1β in pancreatic rat β-cells via the IRE1α/XBP1s pathway. Endocrinology 153, 3017–3028. [DOI] [PubMed] [Google Scholar]
- Mingrone G., Panunzi S., De Gaetano A., et al. (2015). Bariatric-metabolic surgery versus conventional medical treatment in obese patients with type 2 diabetes: 5 year follow-up of an open-label, single-centre, randomised controlled trial. Lancet 386, 964–973. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Kanda Y., Hirokawa Y., et al. (2009). MIN6 is not a pure β cell line but a mixed cell line with other pancreatic endocrine hormones. Endocr. J. 56, 45–53. [DOI] [PubMed] [Google Scholar]
- Nishimura W., Takahashi S., and Yasuda K. (2015). MafA is critical for maintenance of the mature β cell phenotype in mice. Diabetologia 58, 566–574. [DOI] [PubMed] [Google Scholar]
- Nishitoh H., Matsuzawa A., Tobiume K., et al. (2002). ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 16, 1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oslowski C.M., and Urano F. (2010). A switch from life to death in endoplasmic reticulum stressed β-cells. Diabetes Obes. Metab. 12, 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S., Koizumi A., Takeda K., et al. (2002). Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J. Clin. Invest. 109, 525–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S., and Mori M. (2004). Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 11, 381–389. [DOI] [PubMed] [Google Scholar]
- Pal R., Mamidi M.K., Das A.K., et al. (2012). Diverse effects of dimethyl sulfoxide (DMSO) on the differentiation potential of human embryonic stem cells. Arch. Toxicol. 86, 651–661. [DOI] [PubMed] [Google Scholar]
- Rozpedek W., Pytel D., Mucha B., et al. (2016). The role of the PERK/eIF2α/ATF4/CHOP signaling pathway in tumor progression during endoplasmic reticulum stress. Curr. Mol. Med. 16, 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui J., Deng S., Arazi A., et al. (2017). β cells that resist immunological attack develop during progression of autoimmune diabetes in NOD mice. Cell Metab. 25, 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M.M., Claiborn K.C., Khoo C., et al. (2009). Pdx1 (MODY4) regulates pancreatic β cell susceptibility to ER stress. Proc. Natl Acad. Sci. USA 106, 19090–19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour P.A., Freude K.K., Tran M.N., et al. (2007). SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc. Natl Acad. Sci. USA 104, 1865–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha H., He Y., Yang L., et al. (2011). Stressed out about obesity: IRE1α-XBP1 in metabolic disorders. Trends Endocrinol. Metab. 22, 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R.B., O’Donnell A.C., Stamateris R.E., et al. (2015). Insulin demand regulates β cell number via the unfolded protein response. J. Clin. Invest. 125, 3831–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So J.S., Hur K.Y., Tarrio M., et al. (2012). Silencing of lipid metabolism genes through IRE1α-mediated mRNA decay lowers plasma lipids in mice. Cell Metab. 16, 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Lin D.C., Guo X., et al. (2016). Inhibition of IRE1α-driven pro-survival pathways is a promising therapeutic application in acute myeloid leukemia. Oncotarget 7, 18736–18749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegezdi E., Logue S.E., Gorman A.M., et al. (2006). Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 7, 880–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talchai C., Xuan S., Lin H.V., et al. (2012). Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell 150, 1223–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohmonda T., Yoda M., Iwawaki T., et al. (2015). IRE1α/XBP1-mediated branch of the unfolded protein response regulates osteoclastogenesis. J. Clin. Invest. 125, 3269–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F., Wang X., Bertolotti A., et al. (2000). Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287, 664–666. [DOI] [PubMed] [Google Scholar]
- van der Meulen T., Mawla AM., DiGruccio MR., et al. (2017). Virgin β cells persist throughout life at a neogenic niche within pancreatic islets. Cell Metab. 25, 911–926.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., York N.W., Nichols C.G., et al. (2014). Pancreatic β cell dedifferentiation in diabetes and redifferentiation following insulin therapy. Cell Metab. 19, 872–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M.G., Marshall H.L., Rigby R., et al. (2013). Expression of mesenchymal and α-cell phenotypic markers in islet β-cells in recently diagnosed diabetes. Diabetes Care 36, 3818–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ME., Yang KY., Kalousova A., et al. (2005). The HMG box transcription factor Sox4 contributes to the development of the endocrine pancreas. Diabetes 54, 3402–3409. [DOI] [PubMed] [Google Scholar]
- Wu R., Zhang Q.H., Lu Y.J., et al. (2015). Involvement of the IRE1α-XBP1 pathway and XBP1s-dependent transcriptional reprogramming in metabolic diseases. DNA Cell Biol. 34, 6–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Yang L., Yan C., et al. (2014). The IRE1α-XBP1 pathway regulates metabolic stress-induced compensatory proliferation of pancreatic β-cells. Cell Res. 24, 1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Diiorio P., Jurczyk A., et al. (2013). Pathological endoplasmic reticulum stress mediated by the IRE1 pathway contributes to pre-insulitic β cell apoptosis in a virus-induced rat model of type 1 diabetes. Diabetologia 56, 2638–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Nosak C., Sollazzo P., et al. (2014). IRE1 inhibition perturbs the unfolded protein response in a pancreatic β-cell line expressing mutant proinsulin, but does not sensitize the cells to apoptosis. BMC Cell Biol. 15, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., You W., Wang H., et al. (2013). MicroRNA-24/MODY gene regulatory pathway mediates pancreatic β-cell dysfunction. Diabetes 62, 3194–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.