Abstract
Pluripotency and cell fates can be modulated through the regulation of super-enhancers; however, the underlying mechanisms are unclear. Here, we showed a novel mechanism in which Ash2l directly binds to super-enhancers of several stemness genes to regulate pluripotency and self-renewal in pluripotent stem cells. Ash2l recruits Oct4/Sox2/Nanog (OSN) to form Ash2l/OSN complex at the super-enhancers of Jarid2, Nanog, Sox2 and Oct4, and further drives enhancer activation, upregulation of stemness genes, and maintains the pluripotent circuitry. Ash2l knockdown abrogates the OSN recruitment to all super-enhancers and further hinders the enhancer activation. In addition, CRISPRi/dCas9-mediated blocking of Ash2l-binding motifs at these super-enhancers also prevents OSN recruitment and enhancer activation, validating that Ash2l directly binds to super-enhancers and initiates the pluripotency network. Transfection of Ash2l with W118A mutation to disrupt Ash2l–Oct4 interaction fails to rescue Ash2l-driven enhancer activation and pluripotent gene upregulation in Ash2l-depleted pluripotent stem cells. Together, our data demonstrated Ash2l formed an enhancer-bound Ash2l/OSN complex that can drive enhancer activation, govern pluripotency network and stemness circuitry.
INTRODUCTION
Cellular reprogramming and maintenance of pluripotency require a complicated and interactive regulatory network of transcription factors, including Oct4, Sox2, Nanog and Esrrb (1–5). By participating in various regulatory interactions, these transcription factors increase the expression of themselves as well as other pluripotency-related genes, and suppress the expression of genes that contribute to differentiation (6). However, the interactions and regulatory circuit among these transcription factor networks are poorly understood. A super-enhancer in the mammalian genome is a region of multiple putative enhancers bound by substantial number of mediators to drive transcription and control cell identity (7). In pluripotent stem cells such as embryonic stem cells (ESCs), super-enhancers are enriched for Oct4, Sox2 and Nanog (OSN), which form the OSN complex (8). Super-enhancers, normally away and upstream from promoters, form an enhancer–promoter loop structure and recruit p300/CBP and Chd7 to catalyze H3K27 acetylation, and Mediator (Med1) to facilitate Pol II activity through the OSN complex (8–10). A previous report showed that epigenetic modifications at super-enhancers by Tex10 regulate super-enhancer activity, leading to enhanced pluripotency and reprogramming (10). The activity of Oct4 enhancer was also implicated in the regulation of pluripotent status (11). Moreover, the chromatin remodeling of Oct4 gene has been demonstrated to be a crucial step for a successful reprogramming (12). However, the transcription factor network orchestrating super-enhancer activity in cell reprogramming and pluripotency maintenance remains mostly uncertain.
Pluripotent stem cells exhibit a relatively open chromatin structure and unique epigenetic features. Methylation of histone H3K4 correlates with open chromatin structure and active transcription. A highly conserved mixed lineage leukemia (MLL) protein complex harbors methyl-transferase activity, which depends on its core components (Wdr5, Ash2l, Rbbp5 and Dpy30, termed WARD), and is responsible for catalyzing the mono-, di- and tri-methylation of H3K4 (13). Among the members of WARD, biochemical studies suggested that Wdr5 is the central component of the MLL complex and that Ash2l is required for H3K4 trimethylation (14,15). Wdr5 interacts with Oct4 and shares gene regulatory functions with Oct4 (16), and is essential for the reprogramming of somatic cells as well as ESC self-renewal (16). Genome-wide mapping showed that Wdr5 along with Oct4 and Rbbp5 localizes to the proximal region around promoters to regulate pluripotency (16). In addition, Ash2l plays important roles in regulating pluripotency and maintaining open chromatin structure (17). Without the involvement of Ash2l, Wdr5 and other MLL components still form a complex that have distinct structures and functions from the complex containing Ash2l, suggesting a unique role of Ash2l (18). However, the detailed mechanisms and signaling networks of the Ash2l-mediated regulation of pluripotency remain unclear.
The proximal and distal enhancers of Oct4 coordinately regulate Oct4 expression during embryonic and germ cell development, highlighting the significance of the Oct4 enhancer in stemness regulation (11). Although Wdr5 was reported to interact with Oct4 and binds to the Oct4 promoter to regulate pluripotency (16), the involvement of MLL members in the regulation of super-enhancer activity in stem cells is not known. In this report, we demonstrated that Ash2l has a unique binding profile with a high propensity to be enriched on the distal, non-promoter DNA elements in pluripotent stem cells. Remarkably, Ash2l tends to co-localize with OSN at the distal regions of Jarid2, Oct4, Nanog and Sox2 in ESCs. These regions are enriched with super-enhancer features such as the acetylation of H3K27 (H3K27ac), the enrichment of Med1, Chd7 and p300. Ash2l recruits OSN to super-enhancers to form the Ash2l/OSN complex that subsequently promotes the expression of pluripotency-related genes and cellular reprogramming. Blocking Ash2l binding to super enhancers using CRISPRi/dCas9 system abrogates OSN recruitment to the super-enhancers and hinders the enhancer activation of several pluripotency-related genes. In the Ash2l/OSN complex, Ash2l interacts with Oct4 through its W118 residue. The W118A point mutation of Ash2l specifically disrupts Ash2l–Oct4 interaction, and subsequently led to the reduction of OSN recruitment to super-enhancers and the activation of these super-enhancers. Moreover, based on single cell NGS analysis, W118 mutated Ash2l could not rescue the expression of OSN in Ash2l-depleted ESC cells. Taken together, our findings unravel a Wdr5-independent regulatory role of Ash2l in modulating pluripotency through recruiting OSN to super-enhancers, shedding lights on stem cell epigenetics and reprogramming circuitry.
MATERIALS AND METHODS
Cell culture
Mouse embryonic fibroblasts (MEFs) were isolated from 13.5 d.p.c. embryos and maintained in DMEM containing 10% FBS and penicillin/streptomycin. Mouse ESC lines ESD3 and reprogrammed iPSCs were routinely cultured and expanded on mitotically inactivated MEFs (5 × 104 cells/cm2) in 10-cm culture plate (BD) in the presence of 0.3% leukemia inhibitory factor in an iPSC medium, consisting of DMEM (Sigma Aldrich) supplemented with 15% FBS (Invitrogen), 100 mM MEM, nonessential amino acids (Sigma Aldrich), 0.55 mM 2-mercaptoethanol (Gibco) and antibiotics (Invitrogen). iPSCs and mouse pluripotent teratocarcinoma stem cell line SCC-PSA1 (purchased from Bioresource Collection and Research Center) were maintained on MEF feeders in serum-containing media at 5% CO2 and 37°C. A viral-packaging cell line Plat-E cell was used to produce retroviruses and maintained in DMEM containing 10% FBS and penicillin/streptomycin. All cell cultures were maintained at 37°C with 5% CO2.
Cell reprogramming
Mouse iPSCs were generated from MEFs derived from 13.5-day-old embryos of C57/B6 mice as described previously (19). The day before transduction, Plat-E cells were seeded onto 100-mm dishes. One day later, either pMXs-based retroviral vectors (pMXs-Oct4, Sox2, Klf4 and c-Myc), pBABE-based retroviral vectors (pBABE-Ash2l) or pLKO-based lentivirus vectors (pLKO-shCtrl, and shAsh2l) were introduced into Plat-E cells using Transfection Reagent TransIT-LT1® (Mirus). Twenty-four hours after the transfection, the medium was replaced with fresh culture medium. MEFs were seeded 24 h before the viral infection. After 24 h, virus-containing supernatant from the Plat-E culture was filtered through a 0.45 mm cellulose acetate filter (Pall) and then supplemented with 8 μg/ml polybrene (Sigma Aldrich). Target MEFs were incubated with the supernatant and centrifuged at 2250 rpm for 1 h. After the viral infection, the supernatant was replaced with fresh culture medium. Six days after the infection, the infected MEFs were transferred onto mitomycin C-treated MEF feeders in the presence of 0.3% leukemia inhibitory factor in ESC medium. pMXs-Oct4, pMXs-Sox2, pMXs-Klf4, pMXs-c-Myc and pBABE-control vectors were purchased from Addgene. pBABE-Ash2l were constructed in our lab. shCtrl, shAsh2l were obtained from National RNAi Core Facility Platform (Academia Sinica).
Expression and construction of shRNA and site-directed mutagenesis
shRNA vector (pLKO.1) was purchased from Academic Sinica RNAi core. shRNA that targets mouse Ash2l was designed to specifically suppress endogenous Ash2l; therefore, the shRNA-targeting sequence was designed by referring to the 5′UTR sequence. Forward and reverse shRNA oligonucleotides were synthesized and subjected to pLKO.1 empty vector. shRNA control (TRCN0000072243) and shRNA against Wdr5 (TRCN0000301680) were purchased from Academic Sinica RNAi Core. The generation and infection of lentivirus were conducted according to the instruction of Addgene (http://www.addgene.org/). Sub-confluent ESCs were infected with lentivirus in the presence of 8 μg/ml polybrene (Sigma Aldrich). Infected ESCs were subsequently selected with puromycin (2 μg/ml). Quantitative RT-PCR and western blot were used to validate the knockdown efficiency by shRNAs. For Oct4 ChIP-qPCR in Oct4 knockdown ESCs and the Ash2l rescue experiments, shRNA-resistant plasmids were used to exogenously overexpress myc-tagged Oct4 and Flag-tagged Ash2lWT or Ash2lW118A. The information of vectors is listed in Supplementary Table S1. The information of targeting sequences can be found Supplementary Table S2.
Ash2l mutation variants were created using the QuickChange site-directed mutagenesis kit (Agilent), according to manufacturer’s instructions. With the two-step overlap extension PCR method on p3 × Flag Ash2l plasmid, the 5′ and 3′ portions of wild-type Ash2l were amplified in separate reactions using external primers with internal mutagenic primers. The list of oligonucleotide primers used in this study was shown in the Supplementary Table S9.
RNA extraction and real-time quantitative PCR analysis
Total RNA was isolated from the pluripotent cells using the RNeasy kit (Qiagen, Valencia, CA). About 1 μg of total RNA was subjected to first-strand complementary DNA synthesis by using the SuperScript® III Reverse Transcriptase Kit (Invitrogen) as directed by the manufacturer. PCRs were performed using the SYBR Green method in an ABI 7900 sequence detection system (Applied Biosystems) according to the manufacturer’s guidelines. Primers (listed in Supplementary Table S2) were designed using Primer Express 2.0 software (Applied Biosystems, Foster City, CA). All primer specificity was computer-tested (BLAST, National Center for Biotechnology Information, Bethesda, MD) by homology search with the human genome and later confirmed by dissociation curve analysis. The expression level of each gene was normalized to the endogenous expression level of Gapdh and experimental control through ΔΔCt methods. The information of primers is listed in Supplementary Table S3.
Alkaline phosphatase staining
For detecting the alkaline phosphatase (AP) activity, reprogrammed MEFs, iPSCs or ESCs were fixed with 80% alcohol and then stained with the Vector Blue Alkaline Phosphatase Substrate Kit III (Vector Laboratories), according to the manufacturer’s instructions.
Immunofluorescence staining
Immunofluorescence (IF) staining was performed as previously described (19). Briefly, cells were fixed with 4% paraformaldehyde, permeabilized in 1% Triton X-100 and blocked with 5% FBS. Cells were stained with primary antibodies, and then incubated with Alexa Fluor 568 conjugated anti-rabbit (1:300, Invitrogen) or FITC conjugated anti-rabbit (1:300, Invitrogen), and cell nuclei were counterstained with DAPI (Invitrogen). Cells were then washed with PBS and photographed under a fluorescence microscope (Olympus). Details of antibodies can be found in Supplementary Table S4.
Western blot and immunoprecipitation
Western blot was performed as previously described (19). Western blot was performed using the primary antibodies described in the key resource table. Secondary antibodies used in this study were listed as following: bovine anti-rabbit IgG-HRP (catalog no. 7077S; Cell signaling) and chicken anti-mouse IgG-HRP (catalog no. 7076S; Cell signaling). Immunoblots were visualized by the chemiluminescence detection system.
For immunoprecipitation, cells were generally disrupted with lysis buffer (20 mM Tris-Cl buffer [pH 7.6], 1 mM EDTA, 120 mM NaCl, 10 mM β-glycerophosphate, 1 mM NaF, 1 mM Na3VO4 and 0.5% Triton X-100) supplemented with protease inhibitors (250 μM PMSF, 5 μg/ml pepstatin A, 10 μg/ml leupeptin and 5 μg/ml aprotinin). For samples from gel filtration, we used Centricon (Millipore) to concentrate and desalt the fraction of elute. Cleared sample solution was obtained by centrifugation at 12 000 rpm for 30 min at 4°C, and 0.5–1.5 mg of the sample was used for immunoprecipitations. For identify interacting complex of Ash2l protein, sample was added into primary antibody for 2 h at 4°C followed by 2 h of further incubation with protein A/G-Sepharose beads. After washing three times with the lysis buffer, immunoprecipitated proteins were eluted from the beads by boiling for 5 min in SDS-PAGE sample buffer and analyzed by immunoblotting. Details of antibodies can be found in Supplementary Table S4.
Teratoma
All animal procedures were performed in accordance with the Taipei Veterans General Hospital Animal Committee and the principles of Laboratory Animal Care. Teratoma assay was performed as described previously (19). Briefly, 1 × 106 cells were injected into the dorsal flanks of nude mice (The Jackson Laboratory). Six weeks after the transplantation, teratomas were excised and fixed in formalin overnight, embedded in paraffin, sectioned and further stained with hematoxylin and eosin.
Chimera mouse production by blastocyst injection
The introduction of mouse OSKA-iPSCs (derived from C57BL/6J strain, black coat color) into mouse blastocysts derived from the C57BL/6J-Tyrc2J strain (albino) was performed as previously described with some modifications (20). The production of adult chimeras was confirmed by coat color. This study was assisted by Transgenic Mouse Model Core Facility, Academia Sinica, Taiwan.
ESC nuclear extraction and gel filtration
ESC nuclear extracts were obtained according to the manufacturer’s instructions (Thermo Fisher Scientific). Briefly, ESC nuclear extracts were lysed with lysis buffer and applied onto S400 column (HiPrep 16/60 Sephycryl). Size-exclusion chromatography in S400 column was performed using ÄKTA prime system according to the manufacturer’s instruction (GE) as described previously with minor modifications (16). The S400 column was calibrated using the protein standards purchased from GE Healthcare (cat#28-4038-41 LMW and cat#28-4038-42 HMW), and the size of the protein standards was marked above each fraction. Overall, seven fractions of ESC nuclear extracts were obtained. The fractionated samples were collected and assigned for immunoprecipitation and western blot to examine the protein content and interaction among interested proteins.
Molecular docking
For identify the potential interacting residues mediating AO-interaction, we collected the protein structure of Ash2l (3RSN), Oct4 (3L1P), Sox2 (1GT0) and Nanog (2VI6) as receptor/ligand complex. We individually subjected Oct4, Sox2 or Nanog with Ash2l protein into ZDOCK 3.0.2 (http://zdock.umassmed.edu/). The docking results were plotted by using PyMOL 2.3 software (https://pymol.org/2/).
In vitro pull-down assay
Full-length Oct4, Sox2, Nanog, p300 (Taz) and Rbbp5 were cloned into the pGex-5 × 1 plasmids (Addgene), and Ash2l variants (full-length, N-terminus and C-terminus) were cloned into the p3 × Flag plasmids (Sigma Aldrich). The plasmids were transformed into BL21 Escherichia coli. The GST-tagged Oct4, Sox2, Nanog, p300 (Taz) and Rbbp5 recombinant proteins were expressed and purified with Glutathione (GSH)-sepharose beads (Amersham Biosciences, Piscataway, NJ). Flag-tagged recombinant Ash2l variants were expressed and purified with anti-Flag beads (A2220, Sigma-Aldrich). Purified GST-tagged recombinant proteins and purified Flag-tagged recombinant Ash2l variants were then incubated together for 2 h at 4°C ambient temperature. First, we used fixed amount of purified GST-tagged recombinant proteins to pull down the interacted Ash2l variants. The pull-down protein mixture was assigned for analyzing the protein amount of Flag-tagged Ash2l variants using western blot. Second, we fixed the amount of recombinant Ash2l variants to pull down the interacted GST-tagged proteins. Five hundred μg/ml of 3 × Flag peptide (F4799, Sigma-Aldrich) was used to elute the 3 × Flag-tagged Ash2l variants and their interacting GST-tagged proteins. The eluted protein mixture was subjected to SDS-PAGE analysis, and the gel was stained by Coomassie blue.
Flag-peptide immunoprecipitation
Flag-tagged Ash2l-mutant variants encoding plasmid were transfected into the ESCs and subjected to immunoprecipitation experiments. The ESCs were transfected with 3 μg plasmid by using TransIT-LT1 reagent (Mirus) and disrupted with immunoprecipitation lysis buffer. The lysate was incubated with anti-Flag antibody-conjugated beads (A2220, Sigma-Aldrich) for 2 h at 4°C. Beads were then washed three times by IP buffer and eluted by boiling in 20 μl SDS-containing protein loading buffer. Twenty microliters of eluted samples were individually assigned for western blot with 5% input. Flag-tagged bead without peptide conjugation was used as a negative control.
ChIP-Seq analysis
ChIP-seq datasets, if not generated by us, were downloaded from either the GEO or ENCODE portal. We processed and analyzed all the datasets from scratch if the raw FASTQ files were available. For some datasets, of which only the alignment results (in SAM or BED format) were available, we used the Liftover tool to convert the coordinates to mm10 genome assembly. The raw reads were aligned to mm9 genome using Bowtie, allowing at most one mismatch and only one alignment was randomly picked for multi mappers. The peaks were called using MACS 1.4 along with corresponding input background. Different shiftsize parameters were used according to the type of antibodies (shiftsize = 100, for transcription factors; and shiftsize = 150, for histone marks; other parameters were set as default). The peaks from replicates were first called with a relaxed P-value cutoff (0.001) and then pooled together; only those peaks that overlapped with peaks called from at least one other replicate were kept. The aggregation plots and heat maps were generated similarly as described (21).
We used FIMO (22) to infer the binding sites in each peak of Oct4. These Oct4-binding sites that had other peaks of different transcription factors or histone modifications called from ChIP-seq data in the vicinity were deemed co-binding. Each binding site was assigned to the nearest protein-coding gene according to the distance from the midpoint of the binding site to the transcription start site. The binding site-gene pairs were categorized into four groups by the distance in between: near (<1 kb), Distal I (between 1 and 2 kb), Distal II (between 2 and 200 kb), and others (>200 kb). The GO term enrichment analysis was performed using Panther web service (23). Details of data source can be found in Supplementary Table S5.
RNA-Seq and microarray
Total RNA was prepared as described (19). Strand-specific libraries were generated from 500 ng total RNA using the TruSeq Stranded Total RNA Library Prep Kit (Illumina). cDNA libraries were pair-end sequenced (50 bp) on an Illumina HiSeq 2000. Reads were aligned to the mouse genome (NCBI37/mm9) with TopHat v1.3.3 and allowed one alignment with up to two mismatches per read. mRNA RPKM values were calculated using Seqmonk’s mRNA quantitation pipeline. We selected the genes with the following criteria: the minimum RPKM higher than 10 units, fold change >1.5-fold or <0.85-fold from knockdown control sample, and the expression was significantly different from knockdown control sample (P < 0.01).
For the measurement of enhancer RNA (eRNA) production, we used the nascent RNA sequencing analysis to assess eRNA production as previously described by Core and Hah, with brief modifications (24,25). Generally, the application scope of nascent RNA sequencing analysis ranged from enhancer identification to a thorough enhancer-centered analysis. Nascent RNA sequencing analysis includes a novel algorithm to prioritize enhancers by integrating RNA-seq data with the binding profiles of regulators to narrow down interesting enhancers for further experiments. The algorithm can simultaneously undergo the gene analysis of known genes and the enhancer-related analysis, including detecting, quantification and annotation of enhancers.
For microarray analysis, we selected the genes with the following criteria: the minimum log2 intensity higher than 4, fold change >1.5-fold or <0.75-fold from knockdown control sample, and the expression was significantly different from knockdown control sample (P < 0.01). Details of data source can be found in Supplementary Table S5.
Gene ontology and GSEA
The AO-affected gene was obtained from the overlap analysis among AO-binding genes from ChIP-seq analysis, downregulated gene in microarray analysis and downregulated gene in RNA-SEQ analysis. The analysis was performed using the Orange software (https://orange.biolab.si). The gene meets the following criteria: with AO-binding motif, significant fold (1.5-fold up-regulation/0.85 downregulation) in microarray and significant fold (1.5-fold up-regulation/0.85 downregulation) in RNA sequencing. These genes were subjected to gene ontology by using DAVID software (http://david.abcc.ncifcrf.gov). For GSEA analysis, we used these overlapped genes with log2 intensity as dataset and subjected to GSEA analysis (http://www.broadinstitute.org/gsea/).
Chromatin immunoprecipitation (ChIP) and Re-ChIP qPCR
Sub-confluent cells were re-suspended in PBS at the density of 1 × 107 cell/ml and crosslinked at room temperature with 1% formaldehyde for 8 min, and then quenched by incubating in 125 mM glycine. ChIP-qPCR experiments were performed according to the manufacturer’s instruction (Diagenode). Chromatin was sonicated using a Bioruptor (Diagenode) according to the manufacturer’s protocol and examined with electrophoresis assay. Sonicated chromatin was diluted to a final volume of 1000 μl in ChIP lysis buffer supplemented with protease inhibitors before pre-clearing with 30 μl protein A Dynabeads (Thermo Fisher Scientific) for 2 h at 4°C. Chromatin samples were incubated with antibody overnight at 4°C. Immunocomplexes were captured by incubating with 30 μl protein A Dynabeads for 4 h at 4°C. Sample was washed three times with ChIP lysis buffer, and eluted in elution buffer (50 mM Tris, pH 8, 10 mM EDTA, 1% SDS) at 65°C for 15 min. For Re-ChIP-qPCR assay, the samples after de-crosslink were subjected to qPCR assay and remains were diluted with ChIP lysis buffer. Diluted remains were subjected to second round immunoprecipitation against the second protein. The DNA sample was incubated overnight at 65°C to reverse crosslinks, diluted 2-fold in 50 mM Tris, pH 8, plus 10 mM EDTA, and then sequentially digested with RNase A for 2 h at 37°C and proteinase K at 55°C. DNA sample was extracted with phenol/chloroform/isoamyl alcohol and ethanol precipitated. Then, DNA enrichment was quantified by real-time PCR (ABI 7900) using SYBR Green Master Mix (Thermo Fisher Scientific). The antibodies and primers used in ChIP-qPCR assays are listed in Supplementary Tables S4 and S6. Occupancy was quantified using qPCR and normalized to input DNA. ChIP-Seq libraries were prepared using the Kapa LTP Library Preparation Kit (Kapa Biosystems). Reads were aligned to the mouse genome (NCBI37/mm9) with Bowtie2. HOMER was used for motif discovery in sequences ± 100 bp from the ChIP-seq peaks.
CRISPRi/dCas9-mediated interference of Ash2l binding in mouse ESCs
The plasmids were obtained from Academia Sinica. For transient transfection experiments, HEK293 cell line was transfected with sgRNA- and dCas9-encoding plasmid using TransIT-LT1® transfection reagent (Mirus) according to the manufacturer’s protocol. Briefly, HEK293T cells were transfected with 1 μg expression plasmid (pX334-D10A, Addgene) that can stably drive the expression of the first sgRNA and dCas9. Meanwhile, another dish of HEK293 cells was simultaneously transfected with the expression plasmid (pX334-D10A, Addgene) to drive the expression of the second sgRNA. The supernatant obtained from both transfected cultures were well mixed and subjected to the infection of ESCs. These infected ESCs were subsequently selected by the culture medium containing 1 μg/ml puromycin. Two weeks after puromycin selection, infected ESCs were cultivated onto inactivated MEFs for 2 weeks. Infected ESCs with sgRNAs guiding to the Ash2l-binding motifs at Jarid2 or Nanog super-enhancers gradually regained the stemness signature. ESCs infected with sgRNAs guiding to Oct4 super-enhancer did not regain ESC-like morphology. Quantitative RT-PCR, alkaline phosphatase staining, ChIP-qPCR and western blot were used for the validation of the expression of dCas9 and successful interference of indicated genes. The information of sgRNA design is listed in Supplementary Table S7.
Luciferase activity assay
The enhancer regions of Jarid2 (chr13: 44817842-44817933), Nanog (chr6: 122652519-122652659) and Oct4 (chr17: 35496810-35497666) were cloned by PCR amplification of genomic DNA from C57BL/6 mice, and then inserted into the BamHI/SalI of the pGL4.19 vector with minimal promoter (Promega) to generate the plasmid parental constructs for luciferase activity assay. Deletion of Jarid2 and Nanog enhancer fragments was conducted by PCR amplification. HEK293T cells were grown in 12-well tissue culture plate to 70% confluence and then co-transfected with either empty vector or 0.2 μg p3 × Flag-Ash2l or pMyc-Oct4, in the presence of 0.2 μg luciferase plasmids and 10 ng SV40 Renilla luciferase plasmids (Promega). TransIT-LT1 transfection kit was used for transfection of ESCs or HEK293T cells, and the Luciferase activity was detected using the Dual-Glo Luciferase Assay luciferase kit (Promega) according to the manufacturer’s instructions. Firefly luciferase activity was normalized to Renilla luciferase activity, and data are represented as the mean and standard deviation of three independent experiments, each performed in triplicate.
DNA affinity pull-down assay (DAPA)
The biotinylated DNA fragments were incubated with streptavidin beads (S-1638; Sigma, St Louis, MO, USA) at 4°C overnight and washed three times with DAPA buffer (137 mM NaCl, 2.7 mM KCl, 7.7 mM NaH2PO4, 1.5 mM KH2PO4, 0.1% NP-40, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol). Subsequently, the DNA-conjugated beads were incubated with 2 mg of cell lysates and 10 μg of Poly dI-dC (Sigma; P4929) at 4oC overnight. Following the incubation, 30 μl of streptavidin-agarose beads (Millipore) was added to the reaction and incubated at 4°C for 1 h. The beads were then collected by centrifugation and washed four times with DAPA buffer containing 0.5% NP-40. The pulled down complexes were then resolved by 15% SDS–PAGE and analyzed by immunoblotting. The DNA probe sequences are listed in Supplementary Table S8.
Single cell RNA-seq
ScRNA-seq libraries of shCtrl, shAsh2l, shAsh2l+WT and shAsh2l+W118A were generated using the 10X Genomics Chromium Controller Instrument (10X Genomics) and Chromium™ Single Cell 30 Reagent Kits v2 according to manufacturer’s instructions. RNA of every single cell was reverse transcribed and index using the C1000 Touch Thermal cycler with 96-Deep Well Reaction Module. One thousand cells were loaded on a Chromium controller Single-Cell Instrument to first generate single-cell Gel Bead-In-Emulsions (GEMs). After the GEMS were breaking, cDNA of every single cell was barcoded, purified and amplified. After ligated with adaptors, the cDNA was converted into 3′ RNA-Seq libraries by using PCR amplification. The libraries were then sequenced using Illumina Sequencing.
Statistical analysis
Data are presented as mean ± SD and analyzed using Microsoft Excel and Prism (GraphPad). Student’s t-test was used for single variable comparison between two groups. One-way ANOVA followed by post hoc Tukey test was used for multiple comparisons versus the control group. The criterion for significance was set as P < 0.01.
RESULTS
Ash2l is crucial for reprogramming and pluripotency maintenance
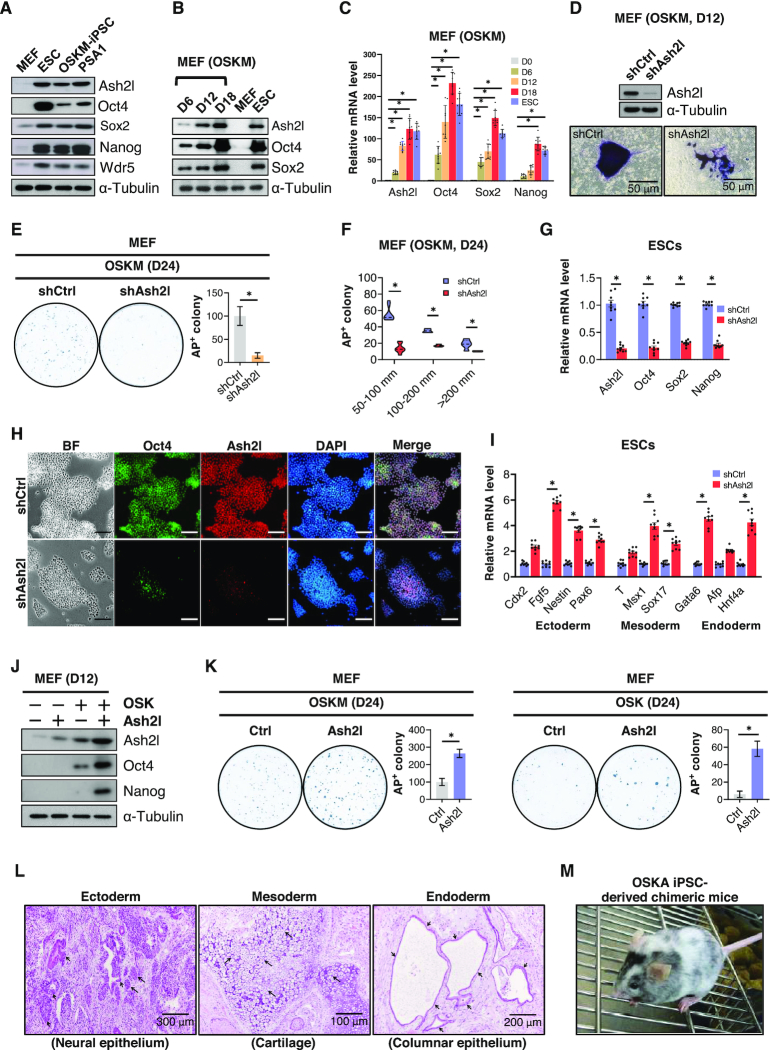
Master transcription factors such as Oct4, Sox2 and Nanog (OSN) orchestrate the epigenetic landscape to regulate pluripotency and cellular reprogramming (8). The expression of these master pluripotency factors is also tightly controlled by epigenetic regulation during differentiation and reprogramming. To elucidate the involvement of Ash2l in the regulation of the pluripotent state and reprogramming, we first analyzed the protein expression of Ash2l, Wdr5 and OSN stemness factors in pluripotent stem cells. OSN, Ash2l and Wdr5 were all highly expressed in pluripotent stem cells including ESCs, induced pluripotent stem cells (iPSCs) and teratocarcinoma stem cells (PSA-1), but not in mouse embryonic fibroblasts (MEFs) (Figure 1A). Differentiation stimuli, including retinoic acid (RA) treatment, LIF withdrawal and induction of embryoid body formation, led to a decline of Oct4 and Ash2l mRNA expression levels in iPSCs (Supplementary Figure S1A). During the Oct4/Sox2/Klf4/c-Myc (OSKM)-induced reprogramming process, Ash2l and stemness factors including Oct4, Sox2 and Nanog were all upregulated both at their protein and mRNA levels in a time-dependent manner (Figure 1B and C).
Figure 1.
Ash2l expression is essential for iPSC generation. (A) Western blot shows the protein expression of Ash2l, Oct4, Sox2, Nanog and Wdr5 in mouse fibroblasts (MEFs), mouse embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs) and embryonal carcinoma cell line PSA1. α-Tubulin was used as loading control. (B) Time-course western blot shows the changes in the protein content of Ash2l, Oct4, Sox2, Wdr5 and Klf4 during the reprogramming process of MEFs. The reprogramming was induced by infecting conventional reprogramming factors Oct4, Sox2, Klf4 and c-Myc (OSKM) in MEFs. Non-infected MEFs and ESCs were used as negative and positive controls, respectively. α-Tubulin was used as loading control. (C) A time-course qPCR shows the changes in the mRNA expression of Ash2l, Oct4, Sox2, Nanog, Nr5a2 and Wdr5 during reprogramming. (D) Alkaline phosphatase (AP) staining shows the reprogramming colony size of control or Ash2l knockdown MEFs at day 12 post-reprogramming. Control (shCtrl) and Ash2l knockdown (shAsh2l) MEFs were subjected to the reprogramming process using OSKM factors. Twelve days after reprogramming, the efficiency of Ash2l knockdown was confirmed by western blot (upper), and MEFs were then subjected to AP staining (lower). (E) Reprogramming efficiency of MEFs with or without Ash2l knockdown. MEFs were reprogrammed by OSKM with or without Ash2l knockdown. Reprogrammed MEFs were stained with AP 24 days after reprogramming (left), and the numbers of AP-positive colonies (right) in each plate were calculated. (F) Measurements of the colony numbers of OSKM-reprogrammed MEFs with or without Ash2l knockdown at all given colony size at day 24 post-reprogramming. Data are presented as mean ± SD; *P< 0.01. (G) qPCR analysis of pluripotency genes (Oct4, Sox2 and Nanog) in ESCs with or without Ash2l knockdown. (H) Immunofluorescence staining showing the Oct4 protein expression in ESCs with or without Ash2l-knockdown. DAPI was used for staining the nucleus; scale bar = 100 μm. (I) qPCR analysis of development-related genes (ectoderm: Cdx2, Fgf5, Nestin, Pax6; mesoderm endoderm: T, Msx1, Sox17; endoderm: Gata6, Afp, HNF4α) in ESCs with or without Ash2l knockdown. (J) Western blot shows the expression of endogenous Ash2l, Oct4 and Nanog protein in OSK-reprogrammed MEFs with or without Ash2l overexpression. α-Tubulin was used as loading control. (K) Reprogramming efficiency of MEFs infected with OSKM (left) or OSK (right), with or without Ash2l overexpression. Reprogrammed MEFs were subjected to AP staining 24 days after infection, and the numbers of AP-positive colonies in each plate were calculated. Data are presented as mean ± SD; *P< 0.01. (L) Ex vivo biopsies and histological analysis reveal the teratoma formation in the grafts of OSK- and Ash2l-reprogrammed iPSCs in the dorsal flanks of nude mice. Sections of the teratoma tissues were subjected to hematoxylin and eosin staining for the examination of the formation of three germ layers. Ectoderm: neuronal epithelium; Mesoderm: cartilage and keratinocytes; Endoderm: columnar epithelium. (M) Competence of OSK/Ash2l-reprogrammed iPSCs to generate chimeric mice is confirmed by coat color. Western blot in panels (A), (B), (D) and (J) are representatives of three separate experiments using independent cell preparations. The quantification of alkaline phosphatase staining shown in panels (E) and (K) are the mean ± SD of six independent experiments. Data are normalized to Gapdh and are presented as mean ± SD; *P< 0.01; see also Supplementary Figure S1.
To examine the role of Ash2l in reprogramming, we first knocked down Ash2l expression in MEFs using a shRNA against its transcript and then subjected these MEFs to OSKM-mediated reprogramming (Figure 1D). At day 12 after the reprogramming, the Ash2l-knockdown MEFs formed smaller colonies and exhibited lower alkaline phosphatase (AP) activity, compared with scrambled shRNA-infected control cells (Figure 1D, lower panel). At day 24 post-reprogramming, depletion of Ash2l significantly reduced the efficiency of iPSC generation in OSKM-infected MEFs (Figure 1E). Meanwhile, Ash2l knockdown tended to reduce the colony numbers of OSKM-reprogrammed iPSCs at all given size (Figure 1F). These effects induced by Ash2l knockdown on these reprogrammed MEFs suggested that Ash2l serves a role in the regulation of pluripotency. We further tested the effect of Ash2l knockdown in ESCs, another cell type that carries all pluripotent cell signatures. In ESCs, inhibition of Ash2l suppressed the mRNA expression levels of Oct4, Sox2 and Nanog (Figure 1G). Immunofluorescence also revealed that ESCs infected with shAsh2l became flat, lost their ESC morphology and largely reduced the numbers of Oct4-positive cells (Figure 1H). The numbers of Rex1-positive cells were also decreased (Supplementary Figure S1B). Meanwhile, Ash2l knockdown simultaneously increased the levels of several differentiation-related genes that are closely associated with tridermal differentiation (Figure 1I; ectoderm: Cdx2, Fgf5, Nestin, Pax6; mesoderm endoderm: T, Msx1, Sox17; endoderm: Gata6, Afp, HNF4α). Furthermore, in MEFs, Ash2l overexpression showed no obvious effect on MEF growth, while Ash2l knockdown led to an ∼20% decrease in MEF growth (Supplementary Figure S1C), accompanied with a mild G1-phase cell cycle arrest (Supplementary Figure S1D). These data indicated that Ash2l serves a regulatory role in pluripotency.
To validate the role of Ash2l in regulating pluripotency, we further manipulated Ash2l levels by overexpression in MEFs, and examined the effect of Ash2l overexpression on the expression of Oct4 and Nanog, and the efficiency of iPSC generation. Western blot analysis at day 12 post-reprogramming showed that, overexpression of Ash2l alone was not sufficient to increase the Oct4 and Nanog protein amount (Figure 1J). However, in MEFs Ash2l co-infected with Oct4/Sox2/Klf4 was able to enhance the protein expression of Oct4 and Nanog (Figure 1J). At day 24 post-reprogramming, Ash2l overexpression significantly increased the efficiency of iPSC generation in OSKM-infected MEFs (Figure 1K, left). Moreover, even without c-Myc, Ash2l overexpression still enhanced the efficiency of iPSC generation from Oct4/Sox2/Klf4-infected MEFs, compared with MEFs infected with only Oct4/Sox2/Klf4 (Figure 1K, right). To test pluripotency in vivo, iPSCs reprogrammed by transfection of Oct4/Sox2/Klf4 plus Ash2l (OSKA-iPSCs) were implanted into the subcutaneous space of immunocompromised mice. Six weeks after transplantation, ex vivo biopsies and hematoxylin and eosin staining revealed the teratoma formation in the neuronal epithelium (ectoderm), cartilage and keratinocytes (mesoderm) and smooth muscle (endoderm; Figure 1L). Moreover, we injected the iPSCs induced by Oct4/Sox2/Klf4 plus Ash2l into blastocytes and then transferred these blastocytes to the uteri of pseudopregnant recipient mice. The coat color of the resultant mice indicated that OSKP-iPSCs were competent to produce chimeric mice (Figure 1M). Collectively, these findings highlighted the crucial role of Ash2l in regulating the pluripotency and the establishment of stem cell identity in pluripotent stem cells.
Ash2l forms a complex with OSN in the absence of Wdr5
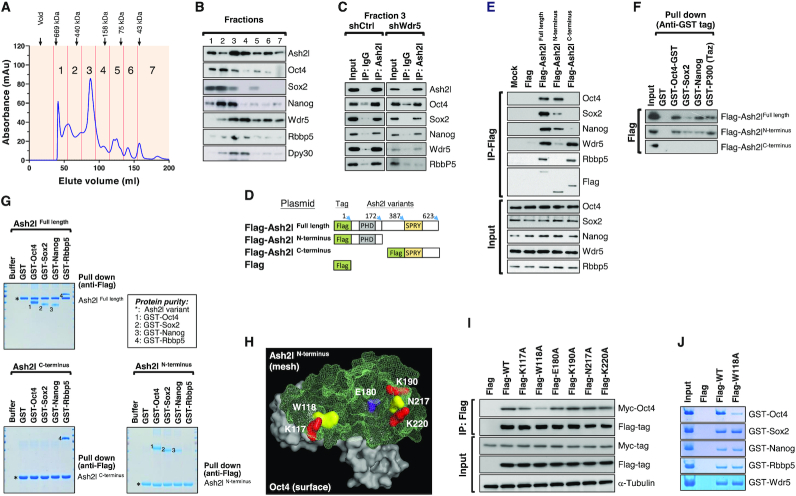
To further understand the role of Ash2l in stemness regulation, we used gel filtration analysis (Figure 2A) followed by western blot to investigate the profiles of molecular complexes in the nuclei of ESCs (Figure 2B). We separated the ESC nuclear extracts into seven fractions based on the filtration order. OSN was abundant in fractions 1–3 (OSN-enriched fractions), whereas Wdr5, Rbbp5 and Dpy30 (WRD) were enriched in fractions 3–4 (WRD-enriched fractions) (Figure 2B). Subsequently, we assessed these OSN-enriched (fraction 1), OSN/WRD-enriched (fraction 3) and WRD-enriched fractions (fraction 4) in an immunoprecipitation assay. In fraction 1 (an OSN-enriched fraction), we found an interaction between Ash2l and OSN (Figure 2A and Supplementary Figure S2A). Note that the expression of Wdr5 was extremely low or barely undetectable in fraction 1 (Supplementary Figure S2A). In line with previous report (14,15), an interaction between Ash2l and WRD was observed in fraction 4 (Supplementary Figure S2A). To elucidate whether Wdr5 affects Ash2l–OSN interaction, we repeated the gel filtration and fractionation experiments in the nuclear extracts of ESCs with or without Wdr5 knockdown to examine the molecular complexes profiles (Figure 2C). We compared the interaction between Ash2l and other interacted proteins in the OSN/WRD-enriched fraction (fraction 3) from ESCs infected with shCtrl or shWdr5 (Figure 2C). Wdr5 knockdown efficiency was confirmed by western blot (Supplementary Figure S2B). And in the absence of Wdr3, Ash2l still interacted with OSN (Figure 2C), indicating the OSN/Ash2l complex occurred independently of Wdr5.
Figure 2.
Ash2l forms complexes with Oct4, Sox2 and Nanog in the absence of Wdr5. (A) The ESC nuclear extracts were separated into seven fractions by gel filtration assay based on the molecular weight of filtrated nuclear extracts. Based on the filtration order of the fraction, these fractions were defined as fractions 1–7. Migration of molecular markers is indicated above the panels. (B) Western blot shows the protein expression pattern among the seven fractions of ESC nuclear extracts. Note that Oct4, Sox2 and Nanog are enriched in fractions 1–3 and Wdr5, Rbbp5 and Dpy30 are enriched in fractions 2–4. Oct4, Sox2, Nanog, Wdr5, Rbbp5 and Dpy30 are all enriched in fraction 3. (C) Immunoprecipitation of fraction 1 shows the interaction of Ash2l with Oct4, Sox2 and Nanog (left). In the fraction 3 filtrated from the ESC nuclear extracts, immunoprecipitation shows that Ash2l still interacts with Oct4, Sox2 and Nanog in the ESC extracts with or without Wdr5 knockdown (right). To examine whether Wdr5 is involved in the interaction of Ash2l and other assayed proteins, the nuclear extracts from ESCs with or without Wdr5 knockdown were subjected to gel filtration and fractionation experiments. The fraction 3 from ESCs with or without Wdr5 knockdown was subjected to immunoprecipitation assay. Ash2l were immunoprecipitated, and the amount of co-precipitated proteins were analyzed by western blot. (D) Scheme depicting the design of different constructs of Flag-tagged Ash2l with truncation (Ash2l-N terminus: 1–172, the PHD domain; Ash2l-C terminus: 387–623, the SPRY domain) or full-length Ash2l (Ash2l-Full length: 1–623). (E) Immunoprecipitation assay indicates that N terminus of Ash2l interacts with Oct4, Sox2 and Nanog. ESCs were transfected with full-length or truncated Flag-tagged Ash2l and subjected to the immunoprecipitation assay. Truncated or full-length Flag-tagged Ash2l were immunoprecipitated, and the protein amount of co-precipitated Oct4, Sox2, and Nanog were analyzed by Western blot. Rbbp5 was used as the control protein that recognizes the C terminus of Ash2l. (F) GST Pull-down assay indicates the direct interaction among Ash2l and various recombinant GST-fusion proteins. Recombinant GST-fusion proteins of Oct4, Sox2, Nanog and p300 (Taz) were subjected to a GST pull-down assay and individually incubated with Flag-tagged Ash2l full-length and truncated forms of Ash2l. Western blot of the immunoprecipitants confirmed the direct binding of Oct4, Sox2 and Nanog to the full-length and N terminus of Ash2l. (G) In vitro pull-down assay with full-length or truncated Ash2l for evaluating the functional domain(s) of Ash2l for the interaction between Ash2l and other proteins. Recombinant GST-fusion proteins of Oct4, Sox2, Nanog and Rbbp5 were subjected to a Flag pull-down assay and individually incubated with Flag-tagged Ash2l-Full length, Ash2l-N terminus and Ash2l-C terminus. The co-precipitated complexes were separated on SDS-PAGE and stained with Coomassie blue. (H) Simulation of the molecular docking of Oct4 and Ash2l using the ZDOCK software. Several residues at the N terminus of Ash2l, including K117, W118, E180A, K190A, N217A and K220A, were predicted as the crucial position for the Ash2l-Oct4 interaction. (I) Validating the crucial residue of Ash2l for Ash2l–Oct4 interaction in ESCs using immunoprecipitation. ESCs were transfected with full-length or truncated Flag-tagged Ash2l and the cell lysates were purified. Flag-tagged Ash2l with distinct mutated residues (K117A, W118A, E180A, K190A, N217A and K220A) were immunoprecipitated with Myc-tagged Oct4. W118A point mutation of Ash2l significantly reduced 79.2% of Ash2l interaction with Oct4. (J) In vitro pull-down assay with wild-type or W118A-mutated Ash2l for evaluating the interaction of Ash2l variants with indicated GST-tagged proteins. Recombinant GST-fusion proteins of Oct4, Sox2, Nanog, Rbbp5 and Wdr5 were subjected to a Flag pull-down assay and individually incubated with Flag-tagged wild-type Ash2l or Ash2l W118A mutant. The co-precipitated complexes were separated on SDS-PAGE and stained with Coomassie blue.
To identify the domain(s) of Ash2l responsible for this protein interaction, we constructed expression plasmids for Flag-tagged truncated Ash2l (Ash2l-N-terminus: 1–172, the PHD domain; and Ash2l-C-terminus: 387–623, the SPRY domain) and full-length Ash2l (Ash2l-full length: 1–623; Figure 2D) for various experiments. First, we overexpressed truncated and full-length Ash2l proteins in HEK293T cells, and subjected the nuclear extracts to immunoprecipitation (Figure 2E). Rbbp5 associated with full-length Ash2l and Ash2l-C, which is consistent with previous findings (26). Remarkably, Oct4, Sox2 and Nanog associated with full-length Ash2l and Ash2l-N terminus, but not Ash2l-C terminus (Figure 2E). To further assess the direct protein–protein interaction, Ash2l-N, Ash2l-C and full-length Ash2l recombinant proteins were purified from bacteria (Figure 2G) and independently incubated with various recombinant proteins, including Oct4, Sox2 and Nanog, and the p300 (Taz domain) protein. As shown in the in vitro pull-down assays with further analysis using western blot or Coomassie staining (Figure 2F and G), Ash2l directly interacted with the master stemness factors, Oct4, Sox2 and Nanog through its N terminus (1–172) (Figure 2F and G). We then used the ZDOCK software to simulate the molecular docking between Ash2l and Oct4 and further predicted that several residues at the N terminus of Ash2l, including K117, W118, E180, K190, N217 and K220, may be crucial for the recognition and interaction with Oct4 (Figure 2H). To determine which residue(s) of Ash2l was important for its interaction with Oct4, we used immunoprecipitation to assess the binding abilities of different Flag-tagged Ash2l mutants (K117A, W118A, E180A, K190A, N217A and K220A) to Myc-tagged Oct4. We found that among all Ash2l mutations, W118A point mutation significantly reduced by 79.2% of Ash2l interaction with Oct4 (Figure 2I), indicating that the W118 residue of Ash2l was crucial for the Ash2l/Oct4 interaction. We further examined whether W118 residue of Ash2l is also crucial for Ash2l/Sox2 and Ash2l/Nanog interaction. Molecular docking simulation indicated that W118 residue was not predicted as candidate binding sites of Sox2 and Nanog (Supplementary Figure S2C). In vitro pull-down assay further confirmed that W118 mutation only interfered with Ash2l binding with Oct4 but not with Sox2, Nanog, Rbbp5 and Wdr5 (updated Figure 2J). The purity of all recombinant proteins used in these in vitro pull-down assays was shown in Supplementary Figure S2D. Together, these findings demonstrated that besides the well-known Wdr5-Ash2l-Rbbp5-Dpy30 (WARD) complex, Ash2l physically interacts with Oct4, Sox2 and Nanog to form a novel Wdr5-independent Ash2l-Oct4-Sox2-Nanog (Ash2l/OSN) complex, in which the W118 residue of Ash2l is critical for the Ash2l–Oct4 interaction.
Co-localization of Ash2l, Oct4, Sox2 and Nanog to distal cis-elements in a Wdr5-independent manner
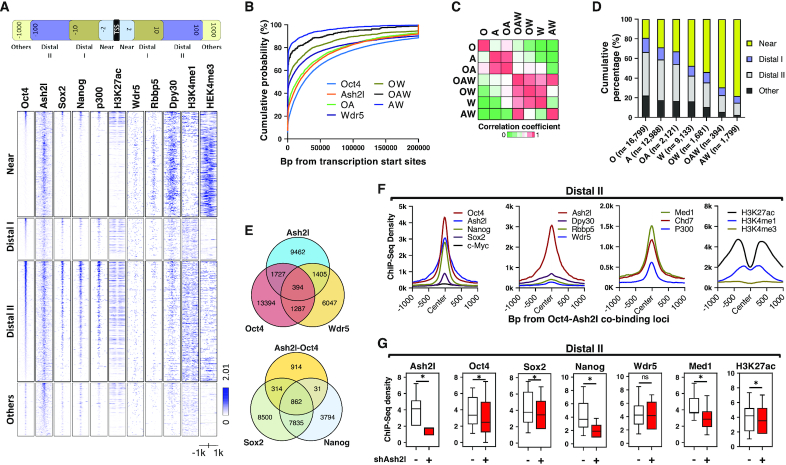
Oct4 is the most pivotal transcription factor for the initiation of reprogramming and is crucial for the maintenance of pluripotency/self-renewal (27). Enhancers function as critical regulatory elements that integrate genomic information for cell fate transition (28). Oct4 can bind to enhancer elements and regulate distal genes (8–10). Following the identification of Ash2l/OSN complex, we investigated whether Ash2l could cooperate with Oct4 to regulate the distal elements of core pluripotent factors. We analyzed bioinformatics data on public domains to seek a potential interactive relationship between Ash2l and Oct4. We collected genome-wide chromatin immunoprecipitation (ChIP) binding profiles of OSN, Ash2l and other members of the WARD complex (Wdr5, Rbbp5 and Dpy30) from previous reports (16,17) and from the ENCODE consortium to generate the sorted heat map binding profile of OSN, Ash2l and other members of the WARD complex (Wdr5, Rbbp5 and Dpy30), H3K27ac, H3K4me1 and H3K4me3 at different genomic regulatory elements (Figure 3A). Using Oct4-Ash2l co-binding sites as the reference, we found that Oct4, Ash2l, Sox2 and Nanog were highly enriched at distal elements while Wdr5, Rbbp5, and Dpy30 were less enriched on Oct4-Ash2l co-binding gene loci (Figure 3A). We further organized elements bound by Oct4-Ash2l (OA), Oct4-Wdr5 (OW), Ash2l-Wdr5 (AW) and Oct4-Ash2l-Wdr5 (OAW; see Supplementary Excel file) and generated the cumulative probability of the distance to the closest TSS (Figure 3B), the similarity matrix (Figure 3C), the stacked bar plot (Figure 3D) and the Venn diagram (Figure 3E). The cumulative probability of the distance to the closest TSS revealed that elements bound by OA predominantly localized far from the TSS, while the W, OW, AW and OAW bound elements were relatively close to the TSS (Figure 3B). Similarity matrix showed that the binding patterns of A and OA categories were similar while the patterns of W-bound elements (W, OAW and OW) were also relatively comparable but distinct from those of A and OA categories (Figure 3C). The stacked bar plot showed that Oct4, Ash2l and OA binding predominantly occurred at distal elements (i.e. Distal I and Distal II) rather than the TSS; while Wdr5-bound elements (i.e. W, OW, OAW and AW) were generally localized at Near elements near the TSSs (Figure 3D and Supplementary Figure S3). The corresponding numbers of binding/co-binding loci for the given genes are shown in a Venn diagram (Figure 3E). Among the total 2121 of OA co-binding loci, 862 gene loci were Ash2l/OSN co-binding loci (Figure 3E). A Venn pie chart further showed that the binding of Oct4 or Ash2l predominantly localized at Distal II elements, and that their most comment binding pattern at Distal II elements was OA co-binding (Supplementary Figure S3, left and middle); by contrast, Wdr5 mainly localized at Near elements (Supplementary Figure S3, right). Taken together, these results showed that loci bound by Ash2l without the involvement of Wdr5 (i.e. OA and A) had high tendency to be located at distal regions, while Wdr5-bound loci (OW, AW and OAW) were TSS-adjacent.
Figure 3.
Ash2l, Oct4, Sox2 and Nanog colocalized to distal cis-elements in a Wdr5-independent manner. (A) Heat map analysis shows the genome-wide chromatin immunoprecipitation-sequencing (ChIP-seq) binding profiles of Oct4, Sox2, Nanog, Ash2l, Wdr5, Rbbp5, Dpy30, p300, H3K27ac, H3K4me1 and H3K4me3 from existing ChIP-seq data of the ENCODE consortium and previously reported studies. Near, 0 to 2 k bp; Distal I, 2 to 10k bp; Distal II, 10 to 100 k bp; other, >100 k bp. (B) Cumulative probability of the distance to the closest TSS reveals that the binding loci for Oct4, Ash2l and OA predominantly localize farther from the TSS, while the binding loci for Wdr5-related peaks (i.e. W, OW, AW and OAW) were relatively closer to the TSS. OA: Oct4-Ash2l; OW: Oct4-Wdr5; AW: Ash2l-Wdr5; OAW: Oct4-Ash2l-Wdr5. (C) A similarity matrix calculated based on the ChIP-seq data shows that the binding patterns for Ash2l and OA are similar, while the patterns of Wdr5, OAW and OW were similar. (D) A stacked bar plot shows that the binding loci for Oct4, Ash2l and OA are predominantly localized to distal elements (i.e. Distal I and Distal II), while Wdr5-related binding loci (i.e. Wdr5, OW, OAW and AW) are generally localized to Near elements. (E) Venn diagrams show the corresponding numbers of binding loci for Ash2l, Oct4 and Wdr5 (upper) and the numbers of binding loci for the Ash2l-Oct4 overlap, Sox2 and Nanog (lower). (F) Aggregation profiles around all Oct4-Ash2l co-binding loci show the comparison of binding patterns among stemness-related factors (first panel from left); WARD components (second panel from left); enhancer-binding proteins (second panel from right); and histone marks (first panel from right). (G) Box plots of Ash2l, Oct4, Sox2, Nanog, Wdr5, Med1 and H3K27ac ChIP-seq density at Distal II elements in ESCs with or without Ash2l knockdown.
We further analyzed the binding proteins on the Oct4-Ash2l (OA) co-binding gene loci with a focus on master pluripotent genes, WARD components, enhancer-binding proteins and histone marks (Figure 3F). The aggregation profiles showed that, at Distal II elements, the OA co-binding gene loci were enriched for Oct4, Sox2 and Nanog (Figure 3F, first panel from left), as well as several well-known enhancer-binding proteins, including Med1, p300 and Chd7 (Figure 3F, second panel from right). For the components of WARD complex, only Ash2l but not Wdr5, Rbbp5 and Dpy30 was enriched in Distal II elements (Figure 3F, second panel from left). Notably, a prominently strong H3K27ac mark and moderate H3K4me1 were detected at the Distal II elements, whereas H3K4me3 marks were not detected (Figure 3F, first panel from right), indicating an active and poised chromatin environment around the OA-co-binding gene loci at distal elements. Next, we performed ChIP-seq analysis to evaluate the binding of Oct4, Sox2, Nanog, Wdr5, Med1 and H3K27ac in both control and Ash2l knockdown ESCs. At Distal II elements, Ash2l knockdown reduced the enrichment of Oct4, Sox2, Nanog, Med1 and H3K27ac, whereas the enrichment of Wdr5 was not affected (Figure 3G). The colocalization of Ash2l, Oct4, Sox2 and Nanog at Distal II elements was Wdr5-independent, distinct from the WARD complex prone to be near the TSS.
Ash2l/OSN complex locates on the super-enhancers to regulate enhancer activation
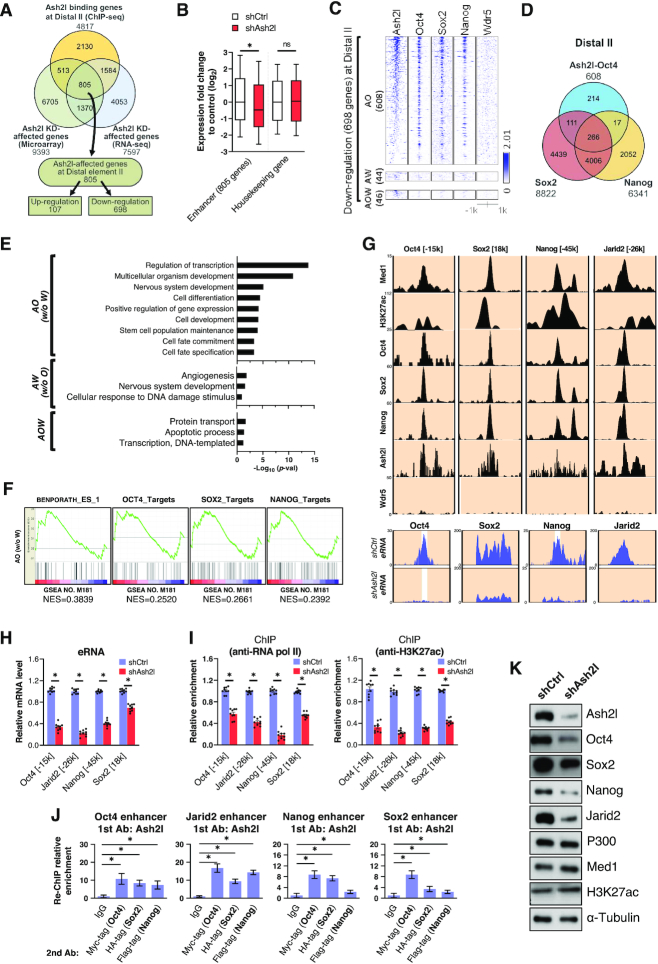
Super-enhancer activity was reported to control pluripotency and reprogramming, as well as to regulate genes critical for cell identity (10,29). Given that our genome-wide analysis showed the colocalization of Ash2l and OSN at Distal II elements, we hypothesized that Ash2l might cooperate with the master transcription factors at Distal II elements to regulate genes involving in pluripotency and cell fate determination. To verify this hypothesis, we first overlapped ChIP-seq data of Ash2l-binding genes with microarray data and RNA-seq data from Ash2l knockdown cells to identify the downstream genes that were bound by Ash2l at Distal II elements with expression affected by Ash2l (Figure 4A). Overall, the expression of a total of 805 super-enhancer-driven genes was affected by Ash2l depletion (Figure 4A and B). Among these 805 genes, we found 698 Ash2l-targeting gene loci down-regulated upon Ash2l knockdown at Distal II elements (Figure 4A). We then categorized these 698 gene loci into binding targets of Ash2l-Oct4 (AO), Ash2l-Wdr5 (AW) and Ash2l-Oct4-Wdr5 (AOW) and found 608 AO-targeted gene loci at Distal II elements (Figure 4C; See also Supplementary Table S5 and Supplemental Excel file). Among the total 608 AO-targeted gene loci, 266 gene loci were co-bound by Ash2l/OSN at Distal II elements (Figure 4D). We also performed the same analysis and categorized the ChIP-seq data of Near elements into Ash2l-Oct4 (AO), Ash2l-Wdr5 (AW) and Ash2l-Oct4-Wdr5 (AOW) (Supplementary Figure S4A). The number of overlapping peaks is shown in Supplementary Figure S4A (left). Among the total 117 AO-targeted gene loci at Near elements, 14 gene loci were co-bound by Ash2l/OSN (Supplementary Figure S4B).
Figure 4.
Ash2l/Oct4/Sox2/Nanog complex locates on the super-enhancers to regulate enhancer activation. (A) A Venn diagram shows the corresponding numbers of Ash2l-bound genes at Distal II elements (4817 genes, obtained from ChIP-seq data), genes that were affected by Ash2l knockdown (9393 genes, obtained from microarray data; 7597 genes, obtained from RNA-seq data). Ash2l-bound genes at Distal II elements were detected by ChIP-seq, and the genes that were affected by Ash2l knockdown were analyzed by microarray and RNA-seq. A total of 805 genes in the intersection were identified as Ash2l-affected genes at Distal II element, including 107 upregulated genes and 698 downregulated genes. (B) A box plot showing the 805 super-enhancer-driven genes that are affected by Ash2l knockdown at Distal II elements. (C) Categorization of the 698 genes that bound by Ash2l at Distal II elements and down-regulated upon Ash2l knockdown into the binding targets of Ash2l-Oct4 (AO; 608 genes), Ash2l-Wdr5 (AW; 64 genes) and Ash2l-Oct4-Wdr5 (AOW; 46 genes). (D) A Venn diagram shows the numbers of binding loci for the Ash2l-Oct4 overlap, Sox2, Nanog at Distal II elements. (E) Gene Ontology analysis shows the downregulated genes in Ash2l knockdown ESCs and co-bound by Ash2l and Oct4 (AO), Ash2l and Wdr5 (AW), or Ash2l, Oct4 and Wdr5 (AOW) at Distal II elements. (F) Gene set enrichment analysis of the 608 AO co-bound genes at Distal II elements and down-regulated upon Ash2l knockdown. These genes include upregulated genes in ESCs (the first on the left) and downstream genes targeted by Oct4, Sox2 and Nanog, respectively (the second/third/fourth on the left). (G) Genome browser tracks showing the ChIP-seq results of Med1, H3K27ac, Oct4, Sox2, Nanog, Ash2l and Wdr5 at Jarid2, Oct4, Sox2 and Nanog enhancers from the existing ChIP-seq database (Upper) RNA-seq results showing the production of enhancer RNA (eRNA) at Ash2l-bound super-enhancers (lower). (H) qPCR of the eRNA production levels at super-enhancers of Oct4, Jarid2 and Nanog in ESCs with or without Ash2l-knockdown. The eRNA expression at each super-enhancer in Ash2l-knockdown ESCs was shown as relative levels to that in ESCs with shCtrl. (I) ChIP-qPCR shows the enrichment of RNA polymerase II and H3K27ac at the super-enhancers of Oct4, Jarid2 and Nanog in ESCs with or without Ash2l knockdown. Data were normalized with IgG control and shown as relative enrichment to that in ESCs with shCtrl. (J) Re-ChIP-qPCR analysis revealed the binding relationship of Ash2l, Oct4, Sox2 and Nanog at Jarid2, Oct4, Sox2 and Nanog super-enhancer in ESCs with or without Ash2l knockdown. The first ChIP-qPCR with anti-Ash2l antibody revealed the direct binding of Ash2l at the super-enhancer of Oct4. The second ChIP-qPCR with indicated antibodies showed that Ash2l, Oct4, Sox2 and Nanog virtually co-bound to the gene loci at the super-enhancer of Jarid2, Oct4, Sox2 and Nanog. Data were shown as relative fold change to the IgG control. (K) Western blot reveals that Ash2l knockdown suppressed the protein levels of Oct4, Sox2 and Nanog, but not p300, Med1 and H3K27ac in ESCs with shAsh2l. Data are presented as mean ± SD; *P< 0.01.
The Gene Ontology analysis revealed that these 608 AO-binding gene loci were responsible for genes important for regulation of transcription, multicellular organism development and cell differentiation etc. while AW- and AOW-binding loci carried relatively poor biological relevance (Figure 4E). By gene set enrichment analysis (GSEA), we also observed that the genes enriched for AO binding at Distal II elements and downregulated by shAsh2l are primarily those genes that are upregulated in ESCs and the downstream genes targeted by Oct4, Sox2 and Nanog (Figure 4F). We performed ChIP followed by quantitative PCR (ChIP-qPCR) to evaluate the binding ability of Ash2l and the enrichment of H3K27ac, the histone mark that identifies active enhancer, at these Ash2l-regulated genes. We observed that the enhancers of Jarid2 and Nanog genes had strong enrichment for Ash2l and H3K27ac, while Oct4 and Sox2 genes were moderately bound by Ash2l and enriched for H3K27ac (Supplementary Figure S4C). Using ChIP-qPCR, we next showed that Ash2l, Oct4, Sox2 and Nanog were individually bound to the enhancers of Oct4, Jarid2, Nanog and Sox2 (Supplementary Figure S4D).
Subsequently, we used ChIP-seq and RNA-seq to examine whether these four Ash2l-bound enhancers (i.e. Jarid2, Oct4, Sox2 and Nanog enhancers) carry enhancer-specific characteristics. First, we added the genome browser tracks to show the ChIP-seq results of Med1, H3K27ac, Oct4, Sox2, Nanog, Ash2l and Wdr5 from the existing ChIP-seq database (Figure 4G, upper). In the ChIP-seq results, Ash2l, Oct4, Sox2 and Nanog, but not Wdr5, did bind to aforementioned enhancer regions that are enriched for enhancer marks H3K27ac and co-activator Med1 (Figure 4G, upper). RNA-seq also detected that the regions producing enhancer RNA (eRNA) colocalized with these Ash2l-bound enhancers (Figure 4G, lower). Notably, as examined by qPCR and ChIP-qPCR, Ash2l knockdown reduced eRNA production, RNA polymerase II and H3K27ac histone mark enrichment, indicating that Ash2l drove the activation of these enhancers (Figure 4H and I). Super-enhancers are transcriptional regulatory regions distant from TSS, known to be enriched for OSN and to recruit various enhancer-binding proteins to catalyze H3K27 acetylation (8–10). Based on the enhancer features of these Ash2l-bound regions and the binding of Oct4, Sox2 and Nanog at distal elements distant from the TSS (>10k), our findings indicated that these Ash2l-bound regions on Oct4, Jarid2, Nanog and Sox2 genes are super-enhancers. To validate the existence of Ash2l/OSN complex at the super-enhancers of Jarid2, Oct4, Sox2 and Nanog, we further used Re-ChIP assay to examine the co-binding of Ash2l and Oct4, Sox2 and Nanog at these sites. Ash2l did form the aforementioned Ash2l/OSN complex at these super-enhancers (Figure 4J). Furthermore, we analyzed the effects of Ash2l knockdown on the expression of stemness-associated genes in ESCs and showed that Ash2l depletion led to the decrease in mRNA and protein levels of Jarid2, Oct4, Sox2 and Nanog (Supplementary Figure S4E and K). Ash2l knockdown did not affect p300 mRNA expression (Supplementary Figure S4E) and the protein amount of p300, Med1 and H3K27ac (Figure 4K). These data revealed an Ash2l-mediated regulation on the protein expression levels of Jarid2, Oct4, Sox2 and Nanog. Ash2l bound to the super-enhancers at the distal elements of stemness-associated genes to regulate genes expression through enhancer activation.
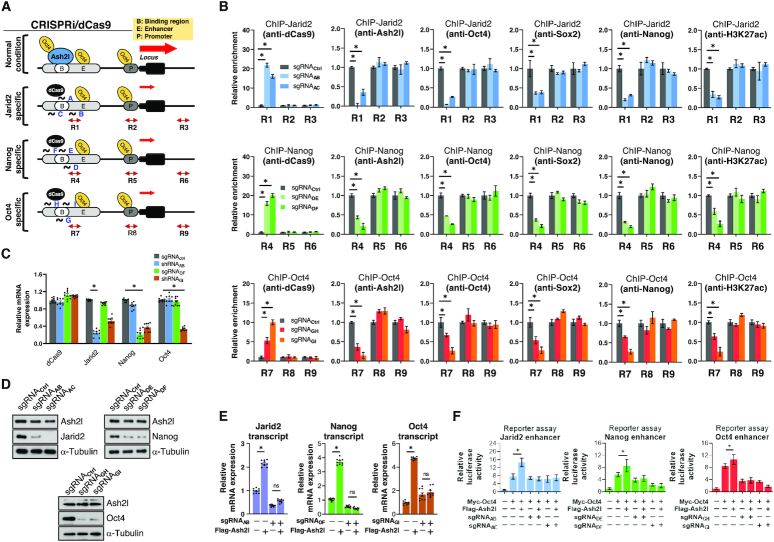
Disruption of Ash2l-binding motifs at super-enhancers abrogated OSN enrichment and enhancer activation
Considering the crucial role of Ash2l in pluripotency maintenance and the enrichment of Ash2l at the super-enhancers of Jarid2, Nanog and Oct4, we sought to further examine the effects of Ash2l binding on OSN recruitment to super-enhancers and on the regulation of pluripotency genes. Ash2l-binding motifs were deleted to interrupt Ash2l binding to Jarid2 and Nanog enhancers. First, we used HEK293T cells and introduced reporter plasmids containing either wild-type Jarid2 or Nanog enhancer, or the same enhancers with deleted Ash2l-binding motifs (Supplementary Figure S5A and S5B). HEK293T cells per se did not express endogenous Oct4 and provided an excellent platform for the investigation of the interaction between Ash2l and Oct4. These cells were subsequently subjected to ChIP-qPCR experiments and luciferase reporter assays (Supplementary Figure S5C–F). For the ChIP-qPCR experiments, we found that overexpression of Flag-tagged Ash2l-WT enhanced the binding of Myc-tagged Oct4 to both Jarid2 and Nanog enhancers (Supplementary Figure S5C and E, upper). Overexpression of Flag-tagged Ash2l with W118 point mutation (Flag-tagged Ash2l-W118A) abrogated the Ash2l-mediated enhancement effect on Myc-tagged Oct4 binding (Supplementary Figure S5C and S5E, upper). In addition, deletion of the Ash2l-binding sites on Jarid2 and Nanog super-enhancers also abolished this Ash2l-mediated enhancement effect on Myc-tagged Oct4 binding (Supplementary Figure S5C and S5E, lower). For the reporter assay conducted in the same cells, we observed that either Flag-tagged Ash2l or Myc-tagged Oct4 alone were capable of direct activation of Jarid2 and Nanog expression (Supplementary Figure S5D and S5F, upper). Co-expression of Flag-tagged Ash2l and Myc-tagged Oct4 led to a synergistic effect on the enhancer activity of Jarid2 (Supplementary Figure S5D, upper) and Nanog (Supplementary Figure S5F, upper), while deletion of Ash2l-binding site and overexpression of Flag-tagged Ash2l-W118A abrogated this synergistic effect (Supplementary Figure S5D, upper and lower; Supplementary Figure S6F, upper and lower). These findings indicated that the Ash2l-binding motifs are required for the enhanced Oct4 recruitment at Jarid2 and Nanog super-enhancers, and for the activation of Jarid2 and Nanog enhancers. The Ash2l–Oct4 interaction via Ash2l W118 residue is critical for mediating this Ash2l effect.
To further validate the effects of Ash2l binding to Jarid2 and Nanog super-enhancers in pluripotent stem cells, we applied CRISPRi/dCas9 genomic editing technology to block the accessibility of Ash2l-binding sites on Jarid2, Nanog and Oct4 super-enhancers in ESCs. We designed three sets of sequence-specific short guide RNAs (sgRNAs) for each gene to recognize the Ash2l-binding motifs at Jarid2, Nanog or Oct4 enhancer (Supplementary Figure S5G) through either monoallelic silencing (labeled as sgRNAAB) or biallelic silencing (labeled as sgRNAAC, sgRNADE, sgRNADF, sgRNAGH and sgRNAGI) (Figure 5A). Stably infected clones were selected by antibiotics. After the transfection and selection, infected ESCs were cultivated onto inactivated MEFs for 2 weeks. Infected ESCs with sgRNAs guiding to the Ash2l-binding motifs at Jarid2 or Nanog super-enhancers gradually regained the self-renewal/stemness signature (Supplementary Figure S5H) and ESC-specific morphology (Supplementary Figure S5I). However, ESCs infected with sgRNAs guiding to Oct4 super-enhancer did not regain the stemness/self-renewal properties (Supplementary Figure S5H–I). CRISPRi/dCas9 system can drive the expression of dCas9 protein and directs it to the designed regions. Therefore, the appearance of dCas9 protein and decreased Ash2l binding at all given regions in the genome was examined to confirm a successful CRISPRi/dCas9-mediated blocking at indicated sites in the genome. ChIP-qPCR results showed that the three distinct sets of sgRNAs specifically increased dCas9 protein and interfered with Ash2l binding at the R1, R4 and R7 regions of Jarid2, Nanog and Oct4 super-enhancers, respectively (Figure 5B). These sgRNAs also simultaneously blocked the binding of Ash2l to these motifs, and hindered the recruitment of Oct4, Sox2 and Nanog to the enhancer regions (R1, R4 and R7), but not the promoter regions (R2, R5 and R8) and Ash2l non-binding regions (R3, R6 and R9) (Figure 5B). A decrease of H3K27ac histone mark at enhancer regions (R1, R4 and R7), but not promoter regions (R2, R5 and R8) and Ash2l non-binding regions (R3, R6 and R9), was also observed upon CRISPR/dCas9-mediated blocking on Jarid2, Nanog and Oct4 super-enhancers (Figure 5B).
Figure 5.
Ash2l binding motif is crucial for the recruitment of OSN to Jarid2, Oct4 and Nanog super-enhancers. (A) Schematic illustration of the design of sequence-specific sgRNA for blocking of Jarid2, Nanog or Oct4 super-enhancers using monoallelic silencing (labeled as sgRNAAB) or biallelic silencing (labeled as sgRNAAC, sgRNADE, sgRNADF, sgRNAGH and sgRNAGI). The dCas9 nuclease is targeted to super-enhancer of Jarid2, Nanog or Oct4 by either set of sgRNA. Three amplicons for each enhancer (Jarid2: R1: enhancer; R2: promoter, R3: desert; Nanog: R4: enhancer; R5: promoter, R6: desert; Oct4: R7: enhancer; R8: promoter, R9: desert) were designed to evaluate protein enrichment in ChIP-qPCR assay. (B) ChIP-qPCR analysis showing the enrichment of indicated protein at Jarid2, Nanog and Oct4 super-enhancers in ESCs with CRISPRi/dCas9-mediated interference of Ash2l binding. ChIP-qPCR analysis with indicated antibodies was conducted in CRISPRi/dCas9-modified ESCs, and the enrichment of each protein at R1–3 regions, R4–6 regions or R7-R9 regions was evaluated with specific primers. Data showed that CRISPRi/dCas9-mediated interference of Ash2l binding at Jarid2 enhancer (upper), Nanog enhancer (middle) and Oct4 enhancer (lower), specifically increased dCas9 binding and decreased the binding of Ash2l, Oct4, Sox2, Nanog and H3K27ac, to R1, R4 and R7, but not R2, R3, R5, R6, R8 and R9. (C) qPCR results showing the effect of CRISPRi/dCas9-mediated interference of Ash2l binding on mRNA expression of Jarid2, Nanog and Oct4. (D) Western blot showing the effect of CRISPRi/dCas9-mediated interference of Ash2l binding on the protein content of Jarid2, Nanog and Oct4. (E) qPCR shows CRISPRi/dCas9-mediated interference of Ash2l binding at indicated enhancers abrogate the upregulation of Jarid2, Nanog, Oct4 transcripts induced by Ash2l overexpression. (F) Reporter assay shows that Flag-Ash2l and Myc-Oct4 induce a synergistic effect on enhancer activity in Jarid2, Nanog and Oct4 super-enhancers. The Ash2l-mediated enhancement of Jarid2, Nanog and Oct4 enhancer activation can be abrogated by specific sgRNA for indicated enhancer. Data are presented as mean ± SD; *, P< 0.01.
Among the distinct sets of designed sgRNAs, CRISPR/dCas9-mediated blocking by sgRNAAB, sgRNADF and sgRNAGI led to the maximal inhibition on Ash2l binding to Jarid2, Nanog and Oct4 super-enhancers (Figure 5B). Therefore, sgRNAAB, sgRNADF and sgRNAGI were used in the residual experiments for CRISPR/dCas9-mediated blocking of the Ash2l binding. qPCR indicated that CRISPRi/dCas9-mediated blocking with sgRNAAB, sgRNADF and sgRNAGI led to the expected inhibition of Jarid2, Nanog and Oct4 expression (Figure 5C). This blocking of Ash2l-binding motifs at Jarid2, Nanog and Oct4 super-enhancers also abrogated Jarid2, Nanog and Oct4 protein without affecting Ash2l levels (Figure 5D). Similar results of western blot were also obtained by blocking the binding motifs with sgRNAAC, sgRNADE and sgRNAGH (Figure 5D).
As shown by qPCR, Jarid2, Nanog and Oct4 transcripts were upregulated by overexpression of Flag-Ash2l in ESCs (Figure 5E). The CRISPRi/dCas9-mediated blocking of the Ash2l-binding motif at each enhancer abrogated this upregulation of Jarid2, Nanog and Oct4 transcripts (Figure 5E). Furthermore, the effects of CRISPRi/dCas9-mediated blocking on Ash2l-binding motifs were also tested in a luciferase reporter assay of the Jarid2, Nanog or Oct4 super-enhancers. Overexpression of Flag-tagged Ash2l synergistically enhanced the Myc-tagged Oct4-induced enhancer activity in Jarid2, Nanog or Oct4 enhancer (Figure 5F), and this enhancement can be abolished by the CRISPRi/dCas9-mediated blocking on corresponding Ash2l-binding motifs at Jarid2, Nanog or Oct4 super-enhancer, respectively (Figure 5F). Taken together, these data indicated that Ash2l-binding motifs are essential for Ash2l binding to super-enhancers, for the subsequent recruitment of OSN, and for the activation of corresponding enhancer.
Ash2l enhances OSN recruitment to the Oct4 super-enhancer in ESCs
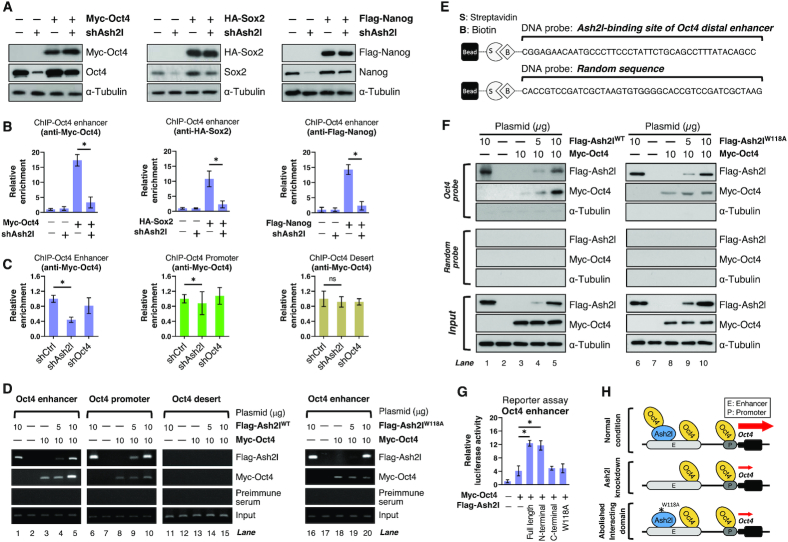
In addition to Jarid2 and Nanog enhancer, we also observed a moderate enrichment of Ash2l and OSN at Oct4 super-enhancer. Oct4 has been extensively accepted to be the master pluripotent factor whereas the regulation of Oct4 remained not fully understood. In addition to the CRISPRi/dCas9-mediated interference experiments at Oct4 super-enhancer (Figure 5), we conducted further experiments to validate whether Ash2l could also recruit OSN and form Ash2l/OSN complex at Oct4 super-enhancer to increase Oct4 enhancer activity. We evaluated and compared the OSN recruitment to Oct4 super-enhancer in ESCs with or without Ash2l knockdown. As Ash2l is also an upstream regulator of Oct4, Sox2 and Nanog (Figure 4K), exogenously expressed Myc-tagged Oct4, HA-tagged Sox2 and Flag-tagged Nanog were employed to carry out the experiments without the confounding by down-regulation of endogenous OSN. Ash2l depletion had no effect on the protein levels of exogenous Myc-tagged Oct4, HA-tagged Sox2 and Flag-tagged Nanog (Figure 6A) but reduced their binding ability to Oct4 super-enhancer (Figure 6B). The similar effect of Ash2l depletion on OSN recruitment was observed at Jarid2 and Nanog super-enhancers (Supplementary Figure S6A and B). Moreover, Ash2l depletion reduced the recruitment of p300 and Med1 to these super-enhancers (Supplementary Figure S6C) without affecting the p300 and Med1 protein amounts (Figure 4K). Remarkably, transfection of recombinant wild-type Ash2l (Flag-Ash2lWT), but not recombinant Ash2l with W118A mutation (Flag-Ash2lW118), rescued the decreased OSN recruitment in Ash2l depleted cells (Supplementary Figure S6D and E). These findings supported that Ash2l recruited OSN to Oct4, Jarid2 and Nanog super-enhancers, and that Ash2l–Oct4 interaction is crucial for OSN recruitment at super-enhancers.
Figure 6.
Ash2l recruits OSN to Oct4 super-enhancer to modulate Oct4 expression. (A) Western blot shows the restoration of endogenous Oct4, Sox2 and Nanog protein by exogenous introduction of Myc-tagged Oct4, HA-tagged Sox2 and Flag-tagged Nanog in ESCs with Ash2l knockdown. To examine the effect of Ash2l knockdown on OSN recruitment without affecting cellular protein content of Oct4, Sox2 and Nanog, Myc-tagged Oct4, HA-tagged Sox2 and Flag-tagged Nanog were exogenously introduced into ESCs with or without Ash2l knockdown. (B) ChIP-qPCR shows that the binding affinity of exogenously introduced Oct4 (left), Sox2 (middle) and Nanog (right) to the Oct4 enhancer in ESCs with or without Ash2l knockdown. Data were normalized with IgG control and shown as relative levels to that in shCtrl-transfected ESCs. (C) ChIP-qPCR reveals that Ash2l knockdown decreases Oct4 enrichment to Oct4 enhancer. Gene knockdown in ESCs was conducted with indicated lentivirus-based shRNA, followed by ChIP-qPCR analysis with specific antibodies. ChIP-qPCR shows that knockdown of Ash2l hampered Oct4 binding to the enhancer. Data were normalized with IgG control and presented as relative fold changes to the enrichment at enhancer in control ESCs. Data are presented as mean ± SD; *P< 0.01. (D) ChIP-RT-PCR analysis validates the regulatory effect of Ash2l on Oct4 binding ability to Oct4 enhancer. Flag-tagged Ash2l variants (Flag-Ash2lWT and Flag-Ash2lW118A) and Myc-tagged Oct4 were expressed in HEK293T cells. The amount of each transfected plasmid was indicated at the top. The nuclear extracts from the transfected HEK293T cells were purified and subjected to a ChIP-qPCR assay with indicated antibodies. The co-precipitated sequence of Oct4 enhancer and promoter were analyzed. Flag-Ash2lWT alone directly bound to Oct4 enhancer and promoter (lanes 1 and 6). Increasing amount of Flag-Ash2lWT enhanced Myc-Oct4 binding to Oct4 enhancer (lanes 4 and 5) but not to Oct4 promoter (lanes 9 and 10). Mutation of W118 position of Ash2l (Flag-Ash2lW118A) abrogated the Ash2l effect on Oct4 binding to enhancer (lanes 18–20). The pre-immune serum served as a negative control. (E) The design of DNA probes for Ash2l-binding site of Oct4 super-enhancer [-15k] and random sequence. (F) Validating the binding relationship between Ash2l and Oct4 at Oct4 super-enhancer by a DNA Affinity pull-down (DAPA) assay. The oligonucleotide probe containing Oct4 super-enhancer sequence was incubated with recombinant Ash2l and Oct4 proteins in vitro. The probe was precipitated and the co-precipitated Flag-Ash2l and Myc-Oct4 proteins were analyzed by western blot. Flag-Ash2lWT directly bound to Oct4 probe and enhanced the binding of recombinant Oct4 to Oct4 probe, whereas Flag-Ash2lW118A failed to exert the same effect. (G) A reporter plasmid containing the Oct4 enhancer sequence was transfected into HEK293T cells along with Oct4 and full-length (Ash2l-Full length), truncated (Ash2l-N terminus or Ash2l-C terminus) or mutated (Ash2lW118A) Ash2l expression plasmids. The luciferase activity representing Oct4 enhancer activity was assayed and shown as relative fold change to that of the empty vector-transfected cells. The Ash2l-Full length and Ash2l-N terminus enhanced Oct4 enhancer activity while Ash2l-C terminus and Ash2lW118A failed to increase Oct4 enhancer activity. (H) Illustrative scheme for the Ash2l-mediated Oct4 recruitment to Oct4 super-enhancer. The Ash2l-mediated Oct4 recruitment could be abrogated by Ash2l knockdown or the point mutation of W118A at Ash2l-N terminus. Data are presented as mean ± SD; *P< 0.01.
We next attempted to elucidate in detail how Ash2l regulated Oct4 expression in ESCs through the cis-regulatory elements. We found that Ash2l knockdown only reduced Oct4 recruitment to its own enhancer but not promoter and Desert (Figure 6C). To confirm the importance of Ash2l on Oct4 binding to its own super-enhancer, we transfected a plasmid containing the Oct4 enhancer sequence into HEK293T cells and assessed the binding capacity of Flag-tagged Ash2l and Myc-tagged Oct4 to this sequence. ChIP-reverse transcription-PCR (ChIP-RT-PCR) experiments showed that Ash2l enhanced the binding of Myc-tagged Oct4 to its own enhancer (Figure 6D, comparing lane 5 with lane 3), but not at the Oct4 promoter (Figure 6D, comparing lane 10 with lane 8). We further validated that recombinant Ash2l with W118A mutation (Flag-Ash2lW118) failed to recruit Oct4 to enhancer (Figure 6D, comparing lane 20 with lane 18). Similar results were obtained in ESCs (Supplementary Figure S6F). We further performed an in vitro DNA-affinity pull-down assay (DAPA) by incubating an oligonucleotide probe containing the Oct4 enhancer sequence with Ash2l and Oct4 recombinant proteins (Figure 6E). Without recombinant Oct4 protein, recombinant Ash2l per se directly bound the Oct4 probe (Figure 6F, lane 1). Increasing the amount of recombinant Ash2l protein enhanced the binding of recombinant Oct4 protein to Oct4 probe in a dose-dependent manner (Figure 6F, lanes 4 and 5). Flag-Ash2lW118 also bound the Oct4 probe (Figure 6F, lane 6), but failed to recruit recombinant Oct4 protein to Oct4 probe (Figure 6F, lane 10 compared with lane 8). Moreover, we sought to demonstrate whether Ash2l-recruited Oct4 indeed has the function to stimulate Oct4 enhancer activity. A reporter plasmid containing the Oct4 enhancer was transfected into HEK293T cells with Oct4 and full-length Ash2l, truncated Ash2l (Ash2l-N terminus or Ash2l-C terminus) or Flag-Ash2lW118 expression vectors; the luciferase activity representing the activity of Oct4 enhancer was then quantified. We observed that full-length Ash2l or Ash2l-N terminus indeed enhanced Oct4-stimulated luciferase activity, while Ash2l-C terminus or Ash2l with W118A mutation had no effect on Oct4-stimulated luciferase activity (Figure 6G). These observations were consistent with the capability of each Ash2l variant to recruit Oct4 as we previously showed (Figure 2E, F and I). Furthermore, Re-ChIP assay revealed that W118 mutation of Ash2l specifically interfered with Ash2l binding to Oct4 at Oct4 enhancer but did not affect its binding to Wdr5 at Oct4 promoter (Supplementary Figure S6G). Collectively, our findings demonstrated that Ash2l acts as an upstream regulator of Oct4 expression in a pluripotent state, predominantly by enhancing OSN recruitment to the Oct4 super-enhancer via its N-terminus. Upon Ash2l knockdown or disruption of Ash2l–Oct4 interaction by W118 point mutation, Oct4 is neither recruited nor bound to its own super-enhancer, and therefore the Oct4 super-enhancer was not activated (Figure 6H).
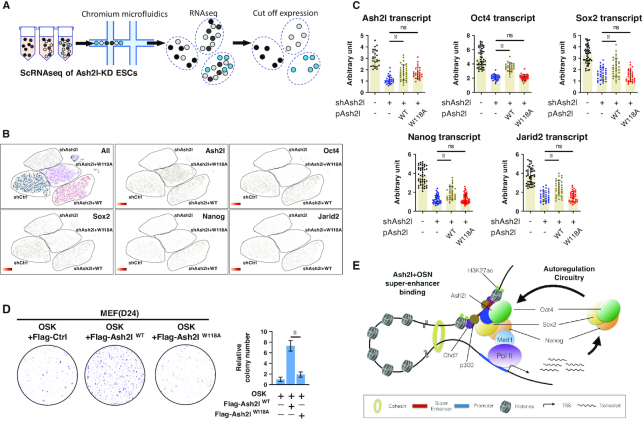
Ash2l–Oct4 interaction via W118 residue is crucial for the Ash2l-mediated pluripotent network
Based on the data of molecular docking simulation and point mutation studies, we identified that W118 is the crucial residue responsible for the Ash2l–Oct4 interaction and the Ash2l-mediated enhancement of Oct4 binding at the super-enhancers for stimulating the activity of enhancers of Oct4, Jarid2 and Nanog genes (Figure 6D–G; Supplementary Figure S5C–F and S6D–G). These results suggested that Ash2l-recruited enhancer-bound Oct4 is crucial for Ash2l-mediated stemness circuitry and pluripotency maintenance. To illustrate the role of Ash2l–Oct4 interaction in pluripotency, we next investigated the functional consequences of disrupting Ash2l–Oct4 interaction in the whole picture of Ash2l-mediated pluripotency network. Single cell RNA-seq is a powerful tool for the measurement of gene expression at individual cell level and the study of cellular heterogeneity. To elucidate the transcriptional networks among Jarid2, Nanog and stemness factors, ESCs with Ash2l knockdown and concomitant overexpression of Flag-Ash2lWT or Flag-Ash2lW118A were subjected to single cell-seq analysis (scRNA-seq; Figure 7A). We analyzed transcriptomes from 3000 ESCs with various treatments, and t-distributed stochastic neighbor embedding (t-SNE) was used to reduce dimension for clustering and visualization. Ash2l knockdown decreased the transcripts of Oct4, Sox2, Nanog and Jarid2 (Figure 7B and C). Comparing with scRNA-seq profiles in ESCs with Ash2l knockdown, we observed that only Flag-Ash2lWT but not Flag-Ash2lW118A could rescue the expression of Ash2l, Oct4, Sox2, Jarid2 and Nanog transcripts (Figure 7B and C). These data indicated that Ash2l–Oct4 interaction is crucial for the upregulation of Oct4, Sox2, Jarid2 and Nanog in pluripotent state. To further validate the functions of Ash2l–Oct4 interaction in pluripotency, Flag-Ash2lWT or Flag-Ash2lW118A was overexpressed in MEFs and these cells were then subjected to OSK-mediated reprogramming (Figure 7D). At day 24 post-reprogramming, overexpression of Flag-Ash2lWT, but not Flag-Ash2lW118A, enhanced the reprogramming efficiency, compared with MEFs infected with Oct4/Sox2/Klf4 only (Figure 7D). Collectively, these data demonstrated that Ash2l–Oct4 interaction through Ash2l W118 residue governs the recruitment of Oct4 to the super-enhancers of downstream target genes, the subsequent activation of downstream genes and pluripotent network.
Figure 7.
Ash2l–Oct4 interaction via W118 residue governs the overall Ash2l-mediated pluripotent network. (A) Illustrative scheme shows the procedure of scRNA-seq. (B) After analyzing transcriptomes from ESCs with Ash2l knockdown and simultaneous overexpression of Flag-Ash2lWT or Flag-Ash2lW118A. t-distributed stochastic neighbor embedding (t-SNE) was used to reduce dimension for clustering and visualization. The transcriptome of indicated genes was shown. (C) Quantification of the gene transcripts (Ash2l, Oct4, Sox2, Nanog and Jarid2) from ESCs with various Ash2l knockdown and simultaneous overexpression of Flag-Ash2lWT or Flag-Ash2lW118A. (D) Examining the effect of point-mutation of Ash2l W118 residue on the reprogramming efficiency in MEFs. MEFs were reprogrammed with Oct4, Sox2, and Klf4 plus Flag-Ash2lWT or Flag-Ash2lW118A. The reprogrammed colonies were stained with alkaline phosphatase, and the numbers of alkaline phosphatase-positive colonies were counted. Data are presented as mean ± SD; *P< 0.01. (E) Schematic presentation of the working model of Ash2l-dependent regulation of super-enhancer activity to modulate the expression of stemness-related genes and pluripotent state. Ash2l binds to its binding motifs at the super-enhancers of downstream genes, such as Oct4, Jarid2 and Nanog, and recruits a serial of stemness factors and enhancer-binding proteins to these super-enhancers. This complex creates an open chromatin structure by modifying histone marks, leading to enhanced transcription of the target genes. This enhanced gene expression further contributes to facilitate cell pluripotency and reprogramming.
DISCUSSION
Super-enhancers are the genomic region that contains multiple putative enhancers to drive gene expression and control cell identity, as well as regulate stem cell-specific phenotypes in pluripotent cells. Pluripotent transcription factors are reported to functionally pre-mark and activate the cell-type-restricted enhancers (28). A dynamic interplay of enhancer elements plays crucial roles in the maintenance of pluripotent state via regulating the expression of stemness factors such as Klf4 (30). It has been shown that super-enhancers are enriched for Oct4, Sox2 and Nanog (OSN), which can subsequently form an OSN complex. However, the mechanisms for the enrichment of OSN on the super-enhancers and the role of this complex were not fully understood. In the present study, we demonstrated that Ash2l recruited OSN to the super-enhancers and formed an Ash2l/OSN complex to regulate pluripotency. This Ash2l/OSN complex at super-enhancers is distinct from the well-known Wdr5/Ash2l/Rbbp5/Dpy30 (WARD) complex, which localizes near the TSS. The physical interaction between Ash2l and Oct4 in this complex predominantly depends on the W118 residue at N-terminus of Ash2l. After forming the Ash2l/OSN complex, several downstream stemness-associated genes were activated, including Jarid2, Nanog and Oct4. Ash2l knockdown, disruption of Ash2l-binding motifs and the CRISPRi/dCas9-mediated blocking of the binding motifs showed the crucial role of Ash2l in forming the complex and in regulating downstream pluripotency gene expression. Together, our data demonstrate that Ash2l forms a novel complex with master pluripotent transcription factors OSN to regulate Oct4-associated stemness circuitry and control the pluripotency network (Figure 7E). The protein members of WARD complex have been reported to physically and functionally interact with key stemness-related transcription factors to promote cellular pluripotency (14). Apart from the well-defined interaction between Wdr5 and Oct4 (16), Ash2l has been shown to interact with Sox2 and Klf4, evidenced by protein co-immunoprecipitation in 293T cells (31). In the human osteosarcoma U2OS cell line, Ash2l could form a complex with Myc to mediate gene transcription efficiency through their promoters (32). These reports regarding the interaction between WARD complex and stemness factors mostly focused on their regulatory role at gene promoters. Ash2l as a member of WARD complex has been previously demonstrated to maintain chromatin opening and pluripotency via its binding to promoter and the induction of H3K4 tri-methylation (15,17). In the WARD complex, Ash2l mediates and enhances the H3K4 trimethylation activity, while Wdr5 maintains the integrity of WARD (15). In this study, we found that Ash2l recruited OSN and formed a stable complex at super-enhancers without the involvement of Wdr5. This discrepancy revealed that the functional roles of Ash2l in the WARD complex and Ash2l/OSN complex were diverse. Further experiments are required to identify the differences in the binding specificity of Ash2l in the two Ash2l-associated complexes in different gene loci.
Among all stemness factors, Oct4 is known to be the most pivotal factor. However, the mechanisms by which regulates Oct4 expression is still not fully understood. We observed that Ash2l directly binds to and recruits OSN to Oct4 super-enhancer in order to stimulate Oct4 expression (Figure 7), indicating that Ash2l is the direct upstream activator of Oct4. Oct4 protein amount was largely diminished in Ash2l-knockdown ESCs (Figures 1H and 4K) also supporting this point. In addition, we found that the interaction between Ash2l and Oct4 plays a crucial role in Ash2l-mediated Oct4 activation. The Ash2l/Oct4 interaction was first demonstrated in ESCs (6). In the present study, molecular docking simulation and point mutation study revealed that the W118 residue of Ash2l is essential for the Ash2l/Oct4 interaction (Figure 2H–J; Supplementary Figure S2C and D), as it is responsible for ∼79.2% Ash2l/Oct4 binding (Figure 2I). Through the protein interaction of Ash2l with Oct4, the binding of Ash2l to super-enhancers recruit and enhance Oct4 binding to the same region, leading to augmented enhancer activation (Figures 5F and 6G). The ChIP-seq data and the aggregation profiles revealed this region co-bound by OA at the Distal II elements were enriched for OSN, H3K27 acetylation, and several enhancer-binding proteins (Med1, p300 and Chd7 etc.; Figure 3). This Ash2l binding allows these enhancer-binding proteins to localize to the same region and catalyze histone H3K27 acetylation to create an open chromatin structure and activate transcription. Mutation of Ash2l W118 residue largely hampered the Ash2l–Oct4 interaction, and hindered the Oct4 recruitment to super-enhancers for activating the stemness genes and maintaining pluripotency. These findings indicated that Ash2l drives stemness circuitry and pluripotency maintenance predominantly via its physical interaction with Oct4. Interestingly, Ash2l protein was detected in all given fractions of ESC nuclear extracts (Figure 2B), implying the existence of other unknown Ash2l-associated complexes that remain to be explored. In conclusion, we unraveled an Ash2l/OSN-driven circuitry that enhances the expression of core pluripotent genes and promotes cellular reprogramming through epigenetically activating the super-enhancer activity of downstream stemness genes. In addition to Oct4, Sox2 and Nanog are also partner proteins in the Ash2l/OSN complex. Whether the Ash2l–Sox2 and/or Ash2l–Nanog interaction are equally crucial for the pluripotency regulation remains not clear. Future works will be needed to elucidate the roles of Ash2l–Sox2 and Ash2l–Nanog interaction in the Ash2l-mediated pluripotent network.
DATA AVAILABILITY
ChIP-seq data generated for this publication was deposited at the Gene Expression Omnibus as study GSE136870 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136870).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor Wen-chang Lin, Mr. Chao-Yu Pan and Mr. Joye Li for the excellent assistance in next-generation sequencing analysis and data processing.
Author contributions: Conception and design: Tsai, P.-H., Chien, Y., Wang, M.-L., Hsu, C.-H., Laurent, B., Chiou, S.-H.
Acquisition, analysis or interpretation of data for the work: Tsai, P.-H., Chien, Y., Wang, M.-L., Hsu, C.-H., Lee, H.-C., Li, H.-Y., Huang, P.-I., Huang, Y.-H., Hung, J.-H., Huo, T.-I.
Drafting the work or revising it critically for important intellectual content: Tsai, P.-H., Chien, Y., Wang, M.-L., Hsu, C.-H., Laurent, B.
Final approval of the version to be published: Chiou, S.-H.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology (MOST) [MOST 106-2633-B-009-001, MOST 106-2321-B-010-006, MOST 106-2119-M-010-001, MOST 106-2320-B-075-002, MOST 106-3114-B-010-002]; Academia Sinica and MOST [MOST 104-0210-01-09-02, 105-0210-01-13-01, 106-0210-01-15-02]; Taipei Veterans General Hospital [V106E-004-2]; Department of Health Cancer Center Research of Excellence [MOHW106-TDU-B-211-113001, MOHW106-TDU-B-211-144003]; NRPB Human iPSC Alliance-Core Service [MOST 106-2319-B-001-003]; VGH, TSGH, NDMC, AS Joint Research Program [VTA105-V1-5-1]; National Health Research Institutes [NHRI-EX106-10621BI]; Center for Intelligent Drug Systems and Smart Bio-devices [IDS2B]; The Featured Areas Research Center Program, Higher Education Sprout Project, and Ministry of Education (MOE) in Taiwan. Funding for open access charge: Ministry of Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R.. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006; 442:533–538. [DOI] [PubMed] [Google Scholar]
- 2. Tam W.L., Lim B.. StemBook. 2008; Cambridge. [Google Scholar]
- 3. Kim J.B., Zaehres H., Wu G., Gentile L., Ko K., Sebastiano V., Arauzo-Bravo M.J., Ruau D., Han D.W., Zenke M. et al.. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008; 454:646–650. [DOI] [PubMed] [Google Scholar]
- 4. Huangfu D., Osafune K., Maehr R., Guo W., Eijkelenboom A., Chen S., Muhlestein W., Melton D.A.. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 2008; 26:1269–1275. [DOI] [PubMed] [Google Scholar]
- 5. Adachi K., Kopp W., Wu G., Heising S., Greber B., Stehling M., Arauzo-Bravo M.J., Boerno S.T., Timmermann B., Vingron M. et al.. Esrrb unlocks silenced enhancers for reprogramming to naive pluripotency. Cell Stem Cell. 2018; 23:266–275. [DOI] [PubMed] [Google Scholar]
- 6. Ding J., Xu H., Faiola F., Ma’ayan A., Wang J.. Oct4 links multiple epigenetic pathways to the pluripotency network. Cell Res. 2012; 22:155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A.. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsankov A.M., Gu H., Akopian V., Ziller M.J., Donaghey J., Amit I., Gnirke A., Meissner A.. Transcription factor binding dynamics during human ES cell differentiation. Nature. 2015; 518:344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ding J., Huang X., Shao N., Zhou H., Lee D.F., Faiola F., Fidalgo M., Guallar D., Saunders A., Shliaha P.V. et al.. Tex10 Coordinates epigenetic control of Super-Enhancer activity in pluripotency and reprogramming. Cell Stem Cell. 2015; 16:653–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choi H.W., Joo J.Y., Hong Y.J., Kim J.S., Song H., Lee J.W., Wu G., Scholer H.R., Do J.T.. Distinct enhancer activity of Oct4 in naive and primed mouse pluripotency. Stem Cell Rep. 2016; 7:911–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu P., Chen M., Liu Y., Qi L.S., Ding S.. CRISPR-Based chromatin remodeling of the endogenous Oct4 or Sox2 locus enables reprogramming to pluripotency. Cell Stem Cell. 2018; 22:252–261. [DOI] [PubMed] [Google Scholar]
- 13. Takahashi Y.H., Westfield G.H., Oleskie A.N., Trievel R.C., Shilatifard A., Skiniotis G.. Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:20526–20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ernst P., Vakoc C.R.. WRAD: enabler of the SET1-family of H3K4 methyltransferases. Brief. Funct. Genomics. 2012; 11:217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Steward M.M., Lee J.S., O’Donovan A., Wyatt M., Bernstein B.E., Shilatifard A.. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 2006; 13:852–854. [DOI] [PubMed] [Google Scholar]
- 16. Ang Y.S., Tsai S.Y., Lee D.F., Monk J., Su J., Ratnakumar K., Ding J., Ge Y., Darr H., Chang B. et al.. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011; 145:183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wan M., Liang J., Xiong Y., Shi F., Zhang Y., Lu W., He Q., Yang D., Chen R., Liu D. et al.. The trithorax group protein Ash2l is essential for pluripotency and maintaining open chromatin in embryonic stem cells. J. Biol. Chem. 2013; 288:5039–5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dou Y., Milne T.A., Ruthenburg A.J., Lee S., Lee J.W., Verdine G.L., Allis C.D., Roeder R.G.. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006; 13:713–719. [DOI] [PubMed] [Google Scholar]
- 19. Chiou S.H., Jiang B.H., Yu Y.L., Chou S.J., Tsai P.H., Chang W.C., Chen L.K., Chen L.H., Chien Y., Chiou G.Y.. Poly(ADP-ribose) polymerase 1 regulates nuclear reprogramming and promotes iPSC generation without c-Myc. J. Exp. Med. 2013; 210:85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sung L.Y., Gao S., Shen H., Yu H., Song Y., Smith S.L., Chang C.C., Inoue K., Kuo L., Lian J. et al.. Differentiated cells are more efficient than adult stem cells for cloning by somatic cell nuclear transfer. Nat. Genet. 2006; 38:1323–1328. [DOI] [PubMed] [Google Scholar]
- 21. Chen P.B., Hung J.H., Hickman T.L., Coles A.H., Carey J.F., Weng Z., Chu F., Fazzio T.G.. Hdac6 regulates Tip60-p400 function in stem cells. Elife. 2013; 2:e01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grant C.E., Bailey T.L., Noble W.S.. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mi H., Muruganujan A., Casagrande J.T., Thomas P.D.. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013; 8:1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Core L.J., Waterfall J.J., Lis J.T.. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008; 322:1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hah N., Danko C.G., Core L., Waterfall J.J., Siepel A., Lis J.T., Kraus W.L.. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011; 145:622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen Y., Cao F., Wan B., Dou Y., Lei M.. Structure of the SPRY domain of human Ash2L and its interactions with RbBP5 and DPY30. Cell Res. 2012; 22:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G. et al.. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005; 122:947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim H.S., Tan Y., Ma W., Merkurjev D., Destici E., Ma Q., Suter T., Ohgi K., Friedman M., Skowronska-Krawczyk D. et al.. Pluripotency factors functionally premark cell-type-restricted enhancers in ES cells. Nature. 2018; 556:510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blinka S., Reimer M.H. Jr, Pulakanti K., Rao S.. Super-Enhancers at the nanog locus differentially regulate neighboring Pluripotency-Associated genes. Cell Rep. 2016; 17:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xie L., Torigoe S.E., Xiao J., Mai D.H., Li L., Davis F.P., Dong P., Marie-Nelly H., Grimm J., Lavis L. et al.. A dynamic interplay of enhancer elements regulates Klf4 expression in naive pluripotency. Genes Dev. 2017; 31:1795–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang Z., Augustin J., Hu J., Jiang H.. Physical interactions and functional coordination between the core subunits of Set1/Mll complexes and the reprogramming factors. PLoS One. 2015; 10:e0145336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ullius A., Luscher-Firzlaff J., Costa I.G., Walsemann G., Forst A.H., Gusmao E.G., Kapelle K., Kleine H., Kremmer E., Vervoorts J. et al.. The interaction of MYC with the trithorax protein ASH2L promotes gene transcription by regulating H3K27 modification. Nucleic Acids Res. 2014; 42:6901–6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq data generated for this publication was deposited at the Gene Expression Omnibus as study GSE136870 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136870).