Abstract
The human genome is composed of two haplotypes, otherwise called diplotypes, which denote phased polymorphisms and structural variations (SVs) that are derived from both parents. Diplotypes place genetic variants in the context of cis-related variants from a diploid genome. As a result, they provide valuable information about hereditary transmission, context of SV, regulation of gene expression and other features which are informative for understanding human genetics. Successful diplotyping with short read whole genome sequencing generally requires either a large population or parent-child trio samples. To overcome these limitations, we developed a targeted sequencing method for generating megabase (Mb)-scale haplotypes with short reads. One selects specific 0.1–0.2 Mb high molecular weight DNA targets with custom-designed Cas9–guide RNA complexes followed by sequencing with barcoded linked reads. To test this approach, we designed three assays, targeting the BRCA1 gene, the entire 4-Mb major histocompatibility complex locus and 18 well-characterized SVs, respectively. Using an integrated alignment- and assembly-based approach, we generated comprehensive variant diplotypes spanning the entirety of the targeted loci and characterized SVs with exact breakpoints. Our results were comparable in quality to long read sequencing.
INTRODUCTION
The human genome is diploid with two copies of each chromosome. The cis- and trans- arrangement of heterozygous variants (i.e. diplotype) is an important characteristic of human genomes, especially with respect to genetic disease. For example, if there are two or more deleterious variants in a gene, the diplotype indicates whether those variants are in cis or in trans (i.e. compound heterozgosity). Importantly, combinations of multiple heterozygous alleles present in the same haplotype can lead to different phenotypes (1). Although compound heterozygosity generally refers to a combination of rare variants with high disease risk, haplotypes of common variants with lower risks can also be associated with phenotypic effects. For example, a single haplotype containing four common variants in BRCA1 has a significant association with longer survival in patients with sarcomas (2). Moreover, haplotype combinations of variants across multiple genes are associated with specific phenotypes. Notably, some major histocompatibility complex (MHC) haplotypes are causative for different immune responses to an immunodeficiency virus infection in Mauritian cynomolgus macaques, a frequently used animal model for studying response to human immunodeficiency virus vaccines (3).
Ideally, the characterization of a diplotype includes both sequence variants (e.g. single nucleotide variants (SNVs)) and structural variants (SVs) (e.g. genomic rearrangements). SVs in phase with SNVs are known to modulate expression in an allele-specific fashion (1). Somatically acquired SVs are common in cancers, and recurrent somatic SVs are often in phase with activating or inactivating mutations (4). For example, deletions of gene copies are often in trans with inactivating mutations and amplifications of gene copies are often in cis with activating mutations. Our recent report on metastatic gastric cancer genomes described an allele-specific driver amplification (5).
Although diplotype information can be obtained from the sequencing of fragmented genomic DNA samples (i.e. short read sequencing), such approaches are limited by (i) sample type and size, (ii) frequency of variants to be phased and (iii) variant type. Traditionally, haplotype information has been determined by pedigree analysis (6), and haplotyping with short read sequencing is possible when related samples are available. In the case of unrelated samples, one can analytically infer haplotypes of common variants based on linkage disequilibrium obtained from large-scale population studies (7). However, this approach is not possible in the case of rare variants, although they are likely to have larger effect sizes than common variants (8).
Detecting SV events with short read sequencing is a challenge and generally requires additional validation using orthogonal methods (9). Frequently, short-read sequencing does not provide the exact breakpoints of SVs. Moreover, short read methods perform poorly at identifying breakpoints inside of segmental duplications, whose size can be as large as 400 kilobases (kb) (10,11). These complex duplications are thought to be associated with genomic instability and have known associations with disease.
Sequencing intact DNA molecules of high molecular weight (HMW) provides extended diplotypes that resolve both simple and complex SV events. The diplotype phase accuracy is improved when linkage evidence is obtained directly from sequencing reads, therefore long read sequencing produces less switch errors than short reads (bioRxiv https://www.biorxiv.org/content/10.1101/085050v2). Long sequence reads from HMW DNA molecules can be generated using the instruments from Pacific Biosciences (Menlo Park, CA, USA) (12) and Oxford Nanopore Technologies (Oxford, UK) (13). However, these single molecule sequencers have raw read error rates orders of magnitude higher than that of short read sequencing (14). Although accuracy can be improved by generating ‘sub-reads’ using circularized libraries, such consensus sequencing has a read length limitation (e.g. no more than 20 kb) (bioRxiv https://www.biorxiv.org/content/10.1101/519025v2). To handle this problem, some research groups have used a hybrid approach to generate diplotypes, using short reads with fewer sequencing errors to complement long read assemblies (15–17).
Regardless of read length, the aforementioned approaches generally require whole genome sequencing (WGS). However, WGS methods have practical limits owing to the cost of sequencing the entire genome to sufficient coverage as well as the complexities of analyzing such large datasets. Thus, there is increasing interest in targeted sequencing of HMW DNA. For regions of interest, higher sequencing coverage of intact HMW targets improves the characterization of complex genomic features such as SV breakpoints, as well as the likelihood of successfully generating contiguous haplotypes and assemblies (13,16,18,19). This is particularly true in the case of genetic mixtures where SVs or other variants are present in a low allelic fraction. In addition, sequencing of targeted HMW DNA molecules allows evaluating regions of interest more efficiently and cost-effectively than with WGS. A targeted approach also has potential implications for diagnostic tests currently reliant on low-resolution methods such as fluorescent in-situ hybridization, because low resolution methods cannot determine exact SV breakpoints, which may vary between patients with diverse phenotypes (20).
Herein, we describe a diplotype sequencing method for targeting genomic regions of interest longer than 0.1 megabases (Mb). This method can also be used for targets as large as several Mb by tiling overlapping HMW targets. Moreover, our method can simultaneously assay multiple targets, which enables more efficient targeting when the DNA sample amount is limited. While our method uses short read sequencing, using intact HMW target molecules as input to generate barcoded linked reads allows us to generate haplotypes extended across entire targets. Importantly, linked sequencing reads can be used for the local diploid assembly of targets >0.1 Mb. Many approaches rely on whole genome amplification (WGA) but this process essentially fragments target HMW DNA. Also, no direct sequencing method (i.e. without a cloning step) has yet been described that preserves the long-range information and contiguity of specific HMW DNA targets. To solve these issues, our approach uses CRISPR-Cas9 digestion and electrophoresis size selection to isolate intact DNA targets from live cells. Then, without any intermediate amplification step, the target-enriched sample is directly used as input for the preparation of a linked read sequencing library. In this study, we describe novel experimental and computational methods which combine an end-to-end enrichment of intact HMW targets and linked read sequencing. To demonstrate the utility of our method, we test three assays designed for analyzing a germline cancer susceptibility gene, a locus involved in immune disorders, and genomic rearrangements. These targets include the entire BRCA1 gene locus, the 4-Mb MHC locus, and breakpoints of 18 SVs, respectively. By using a sample individual for which the diplotype is previously known from pedigree analysis, our results show that this new method can successfully determine the diplotype for targets from single human genome samples.
MATERIALS AND METHODS
Guide RNA design
To design our 20-bp target sequences of guide RNAs (gRNAs), we considered all 20-bp sequences (20-mers) in the regions surrounding our target cut sites. The 20-mers had to occur directly adjacent to a Cas9 binding motif to be considered as a possible gRNA target sequence. We compared our candidate gRNA 20-mers to all 20-mers that exist in the reference human genome, and retained only those candidate targets that: (i) appeared only once in the human genome, and (ii) had few 20-mer matches with one or two mismatched bases. Sequence uniqueness was examined with respect to both strands of the human genome. To select optimal 20-mer gRNA targets, we evaluated their positions relative to known genetic variants in the NA12878 genome, including SNVs, insertion-deletions and SVs. We also evaluated the location of the 20-mer gRNA targets relative to genomic features such as repetitive sequences (e.g. microsatellite regions), pseudogenes and highly variable sequences (e.g. T-cell receptor hypervariability sites). To avoid variability in cutting efficiency and off-target cleavage, we selected the gRNA 20-mers which were free from polymorphic and non-unique sequences.
For our assay targeting the 4-Mb MHC locus (Assay 2), we designed two sets of gRNAs, referred to as Sets 1 and 2 (Supplementary Figure S1a). Both sets generated tandem tiled cuts in 0.2 Mb intervals across the 4 Mb MHC locus. The two gRNA sets were designed with an offset, such that the Set 1 cut sites were separated by 0.1 Mb from the Set 2 cut sites. This strategy generated overlapping segments, which was critical for phasing and assembly. Two separate processes were performed, i.e. one for each pooled gRNA set. The CRISPR-DNA fragments were pooled to prepare a linked read sequencing library.
Guide RNA preparation
For the BRCA1 assay (Assay 1), synthetic CRISPR RNAs (crRNAs) and a trans-activating CRISPR RNA (tracrRNA) were purchased from Integrated DNA Technologies (Coralville, IA, USA). Five crRNAs were used to excise the 200 kb BRCA1 fragments, three crRNAs targeting the 5′ flanking region and two targeting the 3′ flanking region (Supplementary Table S1). The crRNAs were annealed to the tracrRNA in duplex buffer (each crRNA at 20uM, tracrRNA at 13 uM) at 95°C for 10 min, followed by cooling at room temperature for 5 min.
Array-synthesized oligonucleotide pools (CustomArray, Bothell, WA, USA) were used as templates to prepare gRNA pools for the MHC and multiplex SV assays (Supplementary Table S1). Each oligonucleotide/gRNA consisted of four components: an adapter, a T7 promoter, a target-specific region and a tracrRNA region. For the MHC assay, a total of 126 gRNAs were prepared for the two 100 kb-offset sets of gRNAs (Set 1 and Set 2); gRNAs in Set 1 and Set 2 had distinct (i.e. set-specific) adapters. For the initial amplification, we added forward primers (5′– GAGCTTCGGTTCACGCAATG– 3′ and 5′–CAAGCAGAAGACGGCATACGAGAT– 3′) that matched to the set-specific adapter sequences and a reverse primer (5′–AAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC– 3′) complementary to the tracrRNA sequence. Primers were chemically synthesized by Integrated DNA Technologies (Coralville, IA, USA,). For the multiplex SV assays, 108 gRNAs were prepared as a single pool. We used a reverse primer complementary to the tracrRNA component (same as above), and a forward primer complementary to the T7 promoter.
As previously described, our preparation of gRNA pools used array-synthesized oligonucleotides (21). For this study, two ng of the input oligonucleotide pool was amplified in a 25-μl reaction mixture including 1× Kapa HiFi Hot Start Mastermix (Kapa Biosystems, Wilmington, MA, USA) and 1-μM of each primer. The reaction was initially denatured at 95°C for 2 min, followed by 20 cycles of 20 s of 98°C, 15 s of 65°C and 15 s of 72°C. The final steps for amplification involved an incubation at 72°C for 1 min and cooling to 4°C. The amplified product was purified with AMPure XP beads (Beckman Coulter, Brea, CA, USA) in a bead solution to sample ratio of 1.8. A total of 200 ng of the purified product was used as a template for in vitro transcription using the MEGAscript T7 transcription kit (Thermo Fisher Scientific, Waltham, MA, USA). After the transcription reaction, the RNA products were purified using RNAClean XP beads (Beckman Coulter) in a bead solution to sample ratio of 3.0. The final gRNAs were quantified with the Qubit RNA High Sensitivity kit (Thermo Fisher Scientific). The RNA reagent kit on a LabChip GX (Perkin-Elmer, Waltham, MA, USA) was used to confirm the product size per the manufacturer's protocol.
Isolation of target HMW DNA molecules
The 0.1 Mb GM12878 targets and the 0.2 Mb BRCA1 target were isolated using the ‘CATCH 100–300kb extr1h sep3h.shflow’ workflow on the SageHLS instrument (Sage Science, Beverly, MA, USA). Isolation of the 4-Mb MHC locus was performed with the ‘CATCH 100–300kb extr1h sep4h.shflow’ workflow on the SageHLS instrument. Intact GM12878 cells (∼1.5 million) were loaded into the sample well, and a lysis buffer containing 3% sodium dodecyl sulphate (SDS) was loaded into a reagent well just upstream of the sample well. Electrophoresis was carried out for 1 h, thereby driving the SDS through the sample well, where the cells were rapidly lysed. Along with the SDS, proteins and membrane components were carried away from the sample well to the bottom electrode chamber. The genomic DNA was very large, generally more than 2 Mb, and was embedded in the agarose wall of the sample well during the extraction electrophoresis. At the end of the extraction stage, the electrophoresis was halted, the reagent well was emptied and refilled with the Cas9–gRNA reaction mixture.
For the treatment stage, a 40-μl Cas9-gRNA mixture had the following components: 1× SAGE enzyme buffer, 10-μM of the gRNA pool and 4-μM of Cas9 enzyme (New England Biolabs, Ipswich, MA, USA). The reaction was pre-incubated at 37°C for 10 min, and then mixed with 40-μl 1× SAGE enzyme buffer. Electrophoresis was carried out for 1 min to drive the Cas9 enzyme into contact with the genomic DNA inside the sample well wall. Then, the electrophoresis was stopped, followed by Cas9 digestion of the genomic DNA at room temperature for 30 min. After Cas9 digestion, the reagent well was emptied and refilled with the SDS lysis reagent, and size selection electrophoresis was carried out for 3 h. The electrophoresis process used a pulsed field waveform designed for optimal resolution of DNA fragments 100- to 300-kb in size. After size separation, a second orthogonal set of electrodes was used to elute the size-separated DNA into a series of elution modules located along one side of the gel column. Eluted DNA was removed from the cassette within 1 h of run termination, and the Qubit HS assay (Thermo Fisher Scientific) was used to measure the total DNA.
Quantitation of isolated DNA targets
We used a TaqMan qPCR Copy Number assay (Thermo Fisher Scientific) to measure the DNA concentration after extraction. The 10-μl reaction included 2-μl of eluted target DNA sample, 1× TaqMan Genotyping Mix, 1× TaqMan RNaseP reference and 1× TaqMan assay for a specific target. The samples were denatured at 95°C for 10 min, followed by 50 cycles of 15 s at 95°C and 60 s at 60°C. For a relative quantification (i.e. target versus RNaseP reference), we used a modified ΔΔCt method (22). A total of 1 ng of NA12878 genomic DNA was used as a control. For an estimation of absolute copy number in Supplementary Figure S2, we assumed that there were 290 genome copies in each ng of the control sample. Supplementary Table S2 shows the list of TaqMan assays used for all CRISPR-linked read assays in this study.
Library preparation, sequencing and alignment-based phasing
Using a 1.25-μl aliquot of target-enriched DNA sample from the automated SageHLS (Sage Science) process (typically 0.2 ng), we prepared linked read libraries using the Chromium Gel Bead and Library Kit (10× Genomics, Pleasanton, CA, USA). We sequenced the libraries on a NextSeq 500 sequencer (Illumina, San Diego, CA, USA) with 2 × 151 base-pair paired-end reads using a Mid Output v2 kit. With the Chromium library preparation, all resulting read pairs contain a 16-base barcode that indicates their HMW DNA molecule of origin. We used Long Ranger v2.1.6 (10× Genomics) to: (i) demultiplex and convert the resulting BCL files to FASTQ files, (ii) align the barcoded reads in the FASTQ files to the human genome reference and (iii) phase variants using the barcode information attached to each read. We used a Long Ranger-compatible version (v2.1.0) of the human GRCh38 reference (downloaded from: ‘http://cf.10xgenomics.com/supp/genome/refdata-GRCh38-2.1.0.tar.gz’). For Assay 2, which targets a larger region than the other assays, we tested different filtering parameters to get a phase block covering the entire target locus. Specifically, we applied a filtering process to exclude the 1% of barcodes with higher read counts than the other 99%, and then conducted random read downsampling using the ‘downsample’ argument of Long Ranger (Supplementary Methods and Table S3).
Local assembly
We assembled the linked reads derived from the target DNA with the Supernova assembler v2.0.0 (10× Genomics) (23). Barcoded reads were selected as input for the local assembly based on their alignment to the GRCh38 human reference genome, and thus, use of a genome reference more closely related to the sample may improve the assembly. We first collected all the barcodes that had at least one read mapping inside the target. Then, for each barcode, the number of mapped reads were counted in windows of a defined size (e.g. 1 kb) across the entire target region. To facilitate assembly, we removed barcodes with 20 or less mapped reads (Supplementary Figure S3). A downsampling method was then used in order to further improve assembly quality and size (Supplementary Methods and Table S3). To achieve 70× on-target coverage, we applied one of two methods: (i) random downsampling of reads or (ii) elimination of barcodes with a mapped read density greater than an iteratively determined threshold. For the first method, we used the ‘maxreads’ argument of Supernova. For the second method, we pre-processed the linked read data as follows: (i) For each barcode, the size of the genomic region spanned was determined by the mapping locations of reads labeled with that barcode, and calculated as the difference between the largest read mapping coordinate and the smallest read mapping coordinate. The size of the genomic region spanned provided a proxy for the size of the original HMW DNA molecule for any given droplet partition. (ii) We then assigned the sequence-imputed HMW molecules into 10-kb bins, i.e. 0–10 kb, 10–20 kb etc. (iii) Within each bin, we eliminated barcodes with a mapped read density of on-target mapped reads above the n-th percentile, where n was iteratively determined to achieve an on-target coverage below 70×. The mapped read density was calculated by dividing the number of unique reads with the barcode by the size of the genomic region spanned by the barcode. (iv) If a target was tiled by multiple HMW molecules (e.g. Assay 2 for the MHC locus), we repeated this downsampling for the windows separated by all of the CRISPR cuts. The barcodes filtered in individual windows were subsequently merged and used for the assembly.
After the barcode filtering step, we used the final on-target linked read data to perform assembly. We extracted reads with these barcodes from the original FASTQ files generated by Long Ranger (10× Genomics), then used these subset FASTQ files as input to the Supernova assembler (10× Genomics) (23) to generate assembled scaffolds. The subset FASTQ files included not only the on-target reads but also off-target reads sharing barcodes with on-target reads. The parameters used to run the assembler are described in Supplementary Table S3. To assess the scaffold structures of the Supernova output, we compared where each raw FASTQ read aligned to the reference genome GRCh38 with where it aligned to the assembled scaffold; plotting this comparison in R provided a visual representation of scaffold structure (Supplementary Figure S4). For any downstream analysis (e.g. haplotype validation, HLA genotyping), we used only scaffolds which were longer than 50 kb in size and which had fewer than 10% N bases.
Haplotype validation and assignment
To assess the quality of the Supernova assemblies for each haplotype, we compared the allelic variants of the assembled scaffolds with the ground truth haplotypes as determined across multiple studies of NA12878. For allelic variants according to haplotyped assembly, we aligned each scaffold to the human reference genome GRCh38 using mappy v2.15, a python interface for minimap2 (24). The ground truth data used for comparison were the Platinum Genome phased variants (downloaded from: ‘ftp://ussd-ftp.illumina.com/2017-1.0/hg38/hybrid/hg38.hybrid.vcf.gz’). Although other well-known truth variant sets are also available for NA12878, the Platinum Genome has the highest coverage over our target regions. For example, the most recent version of the Genome in a Bottle Consortium set (v3.3.2) covered only 70% of the region targeted by the BRCA1 assay (Assay 1). The ground truth variant dataset was filtered to obtain only phased heterozygous SNVs. Afterward, we compared the shared positions between the CRISPR-linked read scaffold variant calls and the filtered ground truth datasets, and calculated the number of shared alleles between the datasets/number of shared variant positions between the datasets. We expected that each assembled scaffold should share all of its alleles with one of the two ground truth haplotypes in the target genomic region. This process was also used to assign haplotypes to assembled scaffolds when the assembly process failed to generate a single contiguous scaffold for a target locus (Supplementary Figure S5). In this case, instead of the ground truth haplotypes, the haplotypes obtained by the alignment-based process (i.e. the output of Long Ranger) were used for the comparison.
Comparison with other assemblies
We compared our assembled scaffolds with assemblies generated from long read sequencing and linked read WGS. We downloaded assemblies from the following URLs:
Pacific Biosciences assembly (16):
Oxford Nanopore assembly (13):
(‘http://s3.amazonaws.com/nanopore-human-wgs/canu.30x.contigs.fasta’).
To locate where the sequences from our targeted assays occurred in these other assemblies, we aligned GRCh38 target sequences to the assemblies using minimap2 v2.7 (24). Alignment of our target sequences provided a unique location for each target (Supplementary Table S4 for Assays 1 and 2, Supplementary Table S5 for Assay 3). To obtain the structure and allelic content of the assemblies, we used the same method described above in ‘Local assembly’ and ‘Haplotype validation and assignment’.
Identification of HLA genotypes
All the reported HLA gene alleles (downloaded from: ‘ftp://ftp.ebi.ac.uk/pub/databases/ipd/imgt/hla/hla_gen.fasta.txt’) were aligned to our assembled scaffolds using minimap2 v2.7 (24). Among the alignments having a percent match greater than 90%, we selected the closest allele sequence, considering both the edit distance and overall length of insertions and deletions, which were provided in the alignment output.
We also repeated the same genotyping on binned Oxford MHC scaffolds (downloaded from: ‘http://s3.amazonaws.com/nanopore-human-wgs/mhc_haplotypeA.pilon.fasta’, and ‘http://s3.amazonaws.com/nanopore-human-wgs/mhc_haplotypeB.pilon.fasta’).
Validation with Pacific Biosciences circular consensus sequence (CCS) reads
The CCS sequencing data from the Genome in a Bottle Consortium (downloaded from: https://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/NA12878/PacBio_SequelII_CCS_11kb/HG001.SequelII.pbmm2.hs37d5.whatshap.haplotag.RTG.trio.bam) was used to validate our assemblies. The CCS WGS reads were aligned to both maternal and paternal haploid assemblies using minimap2 v2.15 (24). To remove false alignments, we used only primary alignments with (i) more than 1 kb of matched alignment, (ii) a modified edit distance less than 100 and (iii) a fraction of soft-clipped bases <0.5. We subtracted the number of deleted and inserted bases from the edit distance determined by minimap2 to evaluate only the contribution of substitutions. At last, using the filtered alignments, variants were called with freebayes v1.0.2 (arXiv https://arxiv.org/abs/1207.3907). We selected only variants with an allelic fraction >0.8 because switch errors would only generate homozygous variants.
RESULTS
CRISPR-linked read sequencing and assembly
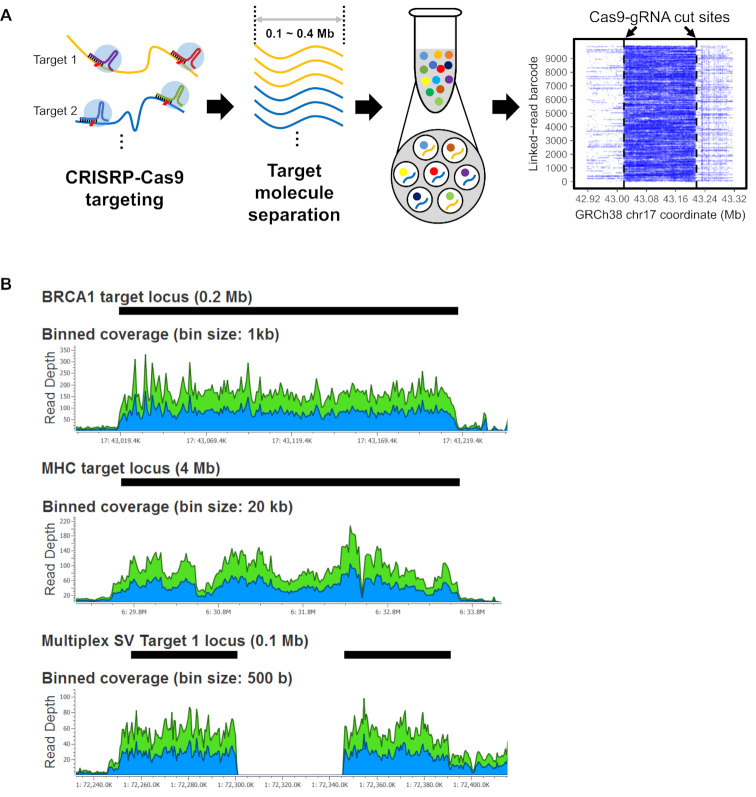
Our approach to targeted sequencing of HMW DNA molecules involves several discrete steps (Figure 1A). First, we isolate chromosome-length HMW DNA from intact cells using a rapid gel electrophoresis method. This method (Cas9-Assisted Targeting of CHromosome segments (CATCH)) (25,26) enriches the targets via size selection. This step eliminates a significant fraction of off-target molecules (Supplementary Figure S6), and provides adequate yields of target DNA without any intermediate processing steps such as WGA. Second, the HMW DNA targets are processed into a linked read library with the automated Chromium microfluidics platform (10× Genomics). The HMW DNA is distributed across approximately one million droplets, each with an oligonucleotide barcode reagent. This step produces a synthetic DNA molecule that incorporates the droplet-specific barcodes. Third, we use an Illumina sequencer to generate high-coverage sequencing data from the linked read targeted libraries. The barcodes link individual reads back to their originating HMW DNA molecule. The bioinformatic elements of the process involve an alignment-based approach to delineate variant haplotypes, and assembly methods for producing diploid scaffolds from target regions (Supplementary Figure S5).
Figure 1.
CATCH targeting and linked read sequencing of HMW DNA. (A) Overview of the process is illustrated. First, guide RNAs target and cut multiple genomic regions of interest. Second, target HMW DNA within the specific size range is isolated by an electrophoresis-based process. At last, the target DNA is used for linked read library preparation and sequencing. The alignment of barcode linked reads shows how sequence coverage is increased across the target segment. In the alignment plot, the X-axis indicates the reference coordinates and the Y-axis shows different barcodes representing individual HMW molecules. Dashed vertical lines indicate Cas9-gRNA cut sites. (B) Sequencing coverage for the target regions is shown for the three assays. For Assays 1 and 2, BRCA1-R2 and MHC-30 libraries are shown. For Assay 3, an example of a homozygous deletion (SV1) is shown. Black bars indicate the target regions. Blue and green areas in plots indicate coverage for forward and reverse reads, respectively.
We designed three assays and evaluated their performance on NA12878 DNA, a well-characterized genome. Guide RNAs were designed to cut 0.1 to 0.2 Mb targets, either individually or in a tiled fashion. The assays targeted the following: (i) a 0.2 Mb region encompassing the entire BRCA1 gene locus; (ii) a 4 Mb segment containing the entire MHC locus (27); (iii) 18 SVs of different classes present within 0.1 Mb segments (Supplementary Table S4). After DNA preparation, we conducted linked read sequencing and data analysis. All three assays showed an enrichment of the given target region after aligning all reads to the human reference genome (Figure 1B).
Low sequence diversity in targeted linked read sequencing
The 10× Chromium linked read system used in this study is designed for whole genome sequence data, and thus for high sequence diversity. However, our CRISPR-linked read assays targeted <0.1% of the genome, and thus encountered problems related to low sequence diversity. For example, barcode collision, when a droplet is occupied by both allelic copies of a target DNA molecule, occurred more frequently in our target- enriched DNA samples than in WGS samples. We, therefore, modified the 10× Chromium experimental protocol and developed a bioinformatic process to minimize such side effects.
First, we increased sequence diversity by retaining some non-target genomic DNA fragments released from non-viable cells. In the extraction stage before the CRISPR-based enrichment, we optimized the electrophoresis duration such that randomly fragmented DNA was not completely removed from the electrophoresis channel. The large non-target fragments remaining in the channel were later eluted with the target fragments, and resulted in 3–5× whole genome coverage (Supplementary Table S6). A longer post-lysis electrophoresis removed these large non-target fragments and increased the fold target enrichment up to several hundred times. However, the modification resulted in more frequent barcode collisions.
Second, we loaded the droplets with less than the recommended amount of input DNA in order to reduce the DNA molecule to droplet ratio. While 1 ng of input DNA is recommended, we loaded 0.2–0.4 ng. However, this under-loading increased the number of barcodes with relatively few mapped reads (e.g. 20 or less), even when the size of the genomic region spanned by the reads was as large as the size of the target (Supplementary Figure S3). The majority of these barcodes had one or two mapped reads on-target and were likely to be the result of empty droplet partitions. Thus, we excluded these barcodes from our bioinformatic analysis to improve the downstream assembly process.
At last, we performed read downsampling when the targeted sequencing coverage was higher than 70× (e.g. BRCA1-R2 library). Barcodes with an over-abundance of mapped reads had a higher probability of barcode collisions. To reduce the negative effect of this subset of barcodes and their associated reads on analysis, we evaluated two methods of downsampling to improve the assembly quality and size: (i) random downsampling of on-target linked reads and (ii) elimination of barcodes with a higher likelihood of having a barcode collision. Using the total number of assembled bases excluding gaps (i.e. N bases) as a criterion, both methods improved the net assembly size (Supplementary Figure S7A). For both downsampling approaches, the optimal on-target coverage range was 60–80×, which produced both a high assembly contiguity and high phasing quality (Supplementary Figure S7B). Therefore, we aimed for an average coverage below 70× when downsampling.
Diplotype and assembly of a 0.2-Mb target including BRCA1
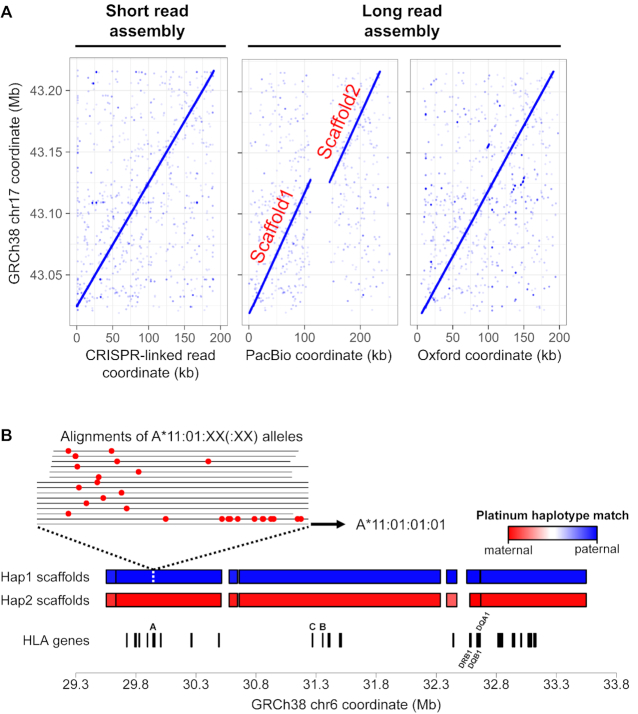
For the first assay, targeting the entire BRCA1 locus on the long arm of chromosome 17 (17q21.31), we ran two replicate experiments (BRCA1-R1 and BRCA1-R2). We observed a 16- and 36-fold increase for sequences from the intact BRCA1 DNA segment compared to off-target sequences across the two assays (Figure 1B and Supplementary Table S6). Moreover, the parental haplotypes obtained by the alignment-based approach were 100% concordant with the previously described NA12878 phased variants (Supplementary Table S7). For both replicates, the diploid assembly generated scaffold was 0.2 Mb, nearly the same size as the original target. BRCA1-R2 required a downsampling of sequence data due to high coverage (155×). The scaffold haplotypes were concordant with reported haplotypes (28) (Supplementary Table S8). When comparing with published long read assemblies, our assembly had comparable or better contiguity (Figure 2A). There are two long read NA12878 assemblies based on WGS (13,16), in which we could identify only one of the two diploid copies for the BRCA1 target (Supplementary Table S4). The Oxford Nanopore assembly had comparable contiguity with our targeted assembly. However, in the Pacific Biosciences assembly, there were two scaffolds each of which had a match to the first or second half of the target.
Figure 2.
Assembly results from assays targeting a single continuous region. (A) The 0.2 Mb BRCA1 assembly (BRCA1-R1) was compared with other long read assemblies. The X-axis indicates the coordinates of each NA12878 assembly across different platforms. The Y-axis indicates the corresponding segment from the GRCh38 reference. Each point indicates where a CRISPR-linked read aligned to the reference versus where it aligned to the NA12878 assemblies. (B) The assembled MHC scaffolds with assigned haplotype blocks where red and blue indicate the parental haplotype. The X-axis represents the GRCh38 reference genome on which the assembly scaffold is aligned. The HLA genes are indicated below the scaffolds, with the labels only for the major class I (HLA-A, HLA-B and HLA-C) and II (HLA-DRB1, HLA-DQA1 and HLA-DQB1) genes. All the NA12878 genotypes of these genes from our assembly are available in Table 1. For HLA-A, alignment of all the alleles coding the same protein sequence [A*11:01:XX(:XX)] are shown. The red dots indicate mismatch bases to the allele in Haplotype 1 (A*11:01:01:01).
Diplotype and assembly of a 4-Mb MHC region
For the second assay, our goal was to characterize haplotypes larger than the 0.2 Mb DNA molecule size by using a set of overlapping targets (Supplementary Figure S1A). We targeted the entire MHC locus, which spans 4 Mb on the short arm of chromosome 6 (6p21.3) (Supplementary Table S4). To optimize phasing and assembly, we tested different read-per-barcode ratios for linked read sequencing. From the prepared linked read sequencing droplets, we made a 90-μl aliquot (MHC-90) with more droplets and a 30-μl volume portion with fewer droplets (MHC-30). Loaded with the same input amount, MHC-90 had twice as many barcodes compared to MHC-30. Both libraries provided 80× on-target average coverage (Figure 1B and Supplementary Table S6). Therefore, MHC-30 had a higher read-per-barcode ratio than MHC-90.
We assessed the quality of our diplotypes from both alignment- and assembly based approaches. With the alignment-based approach, both MHC-90 and MHC-30 provided high quality diplotypes throughout the entire 4-Mb region with a high correlation to the Platinum Genome haplotypes. Overall, we observed a 96% sensitivity and >98% concordance (Supplementary Table S7). With the assembly based approach, we generated 16 and 14 large scaffolds from MHC-90 and MHC-30, respectively (Supplementary Table S9 and Figure S8). For both libraries, the N50 scaffold sizes were consistently >0.88 Mb, and more than 90% of the 4-Mb MHC region was assembled. The scaffolds were binned according to the haplotypes obtained by the alignment-based approach, and compared with Platinum genome haplotypes for haplotype accuracy. Although the concordance was consistently >96%, the sensitivity was better in MHC-30 scaffolds (90 versus 83% in MHC-90 scaffolds). Therefore, we concluded that the library with fewer barcodes (MHC-30) provided a higher quality diploid assembly in terms of haplotype content.
Comparison with an Oxford Nanopore MHC diplotype
For NA12878, we identified genotypes for over 27 HLA genes (Supplementary Table S10). Because our assembly provided both coding and intronic variants, the genotyping was based on the alignment of all reported HLA allele sequences to our assemblies. In this analysis, we excluded any alignment with a percent match <90%. The HLA genotypes were phased into parental haplotypes encompassing the entire 4-Mb region (Figure 2B). For the six major MHC class I and II genes, MHC-30 generated haplotypes that matched the allele haplotype predictions based on the Oxford Nanopore assembly (13) (Table 1). The Nanopore-based predictions were based on one or two highly variable exons, and did not include intronic variants. When using the entire gene sequence, alleles of only 18 HLA genes aligned to Oxford Nanopore scaffolds (Supplementary Table S11) versus our assembly which had a diplotype across 27 HLA genes. Moreover, for the Oxford-based analysis, only nine of the 18 HLA genes had alignments to both haplotypes. Among the major MHC genes, none of the HLA-A and HLA-DRB1 alleles aligned to Oxford Nanopore scaffolds (Table 1). In the cases where an alignment was available, the edit distance between HLA gene alleles and Oxford Nanopore scaffolds was larger than between the HLA gene alleles and our assemblies.
Table 1.
Alignment of reported HLA gene alleles to NA12878 assemblies
| CRISPR-linked read assembly | Oxford assembly | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype 1 | Haplotype 2 | Haplotype 1 | Haplotype 2 | |||||||
| HLA gene | Aligned allele | Edit distanceb | Aligned allele | Edit distanceb | Aligned allele | Edit distanceb | Allele predictiona by Jain et al. (13) | Aligned allele | Edit distanceb | Allele predictiona by Jain et al. (13) |
| A | 11:01:01:01 | 0 | 01:01:01:01 | 0 | n.a. | 11:01:01G | n.a. | 01:01:01G | ||
| B | 56:01:01:01 + 2 alleles | 1 | 08:01:01:01 | 0 | n.a. | 56:01:01G | 08:177, 08:182 | 9 | 08:01:01G | |
| C | 01:02:01:02 + 28 alleles | 1 | 07:01:01:01 | 0 | 01:148 | 16 | 01:02:01G | 07:01:01:14Q | 10 | 07:01:01G |
| DQA1 | 01:01:01:03 | 2 | 05:01:01:02 | 0 | 01:16N | 82 | 01:01:01G | 05:03:01:01 | 129 | 05:01:01G |
| DQB1 | 05:01:01:03 | 0 | 02:01:01 | 0 | 05:01:01:03 | 4 | 05:01:01G | 02:02:01:01 | 45 | 02:01:01G |
| DRB1 | 01:01:01 | 2 | 03:01:01:01 | 0 | n.a. | 01:01:01G | n.a. | 03:01:01G | ||
Only the six major HLA genes are shown. All alignments for both assemblies are available in Supplementary Tables S10 and 11.
aThe prediction is based on exons 2 and 3 for MHC class I genes and exon 2 for MHC class II genes, and information regarding the ‘G’ group is available at ‘http://hla.alleles.org/alleles/g_groups.html’.
bThe edit distance is between assembly versus allele.
n.a.: no alignment having a percent match >90%.
Multiplex characterization of SVs
The third assay was used for a multiplexed characterization of SVs that were previously reported in NA12878 (Supplementary Table S5). We selected a ground truth set of 18 SVs previously reported using two or more WGS approaches—either with linked reads (18), long reads with Pacific Biosciences (16) or long reads with Oxford Nanopore (13). These SVs had one or more reported assemblies that covered their locations. We designed a multiplexed set of gRNAs with cut sites flanking each SV, encompassed within a 0.1 Mb segment. The SVs included 13 deletions varying in size from 30 to 150 kb (Figure 1B), and five inversions (Supplementary Figure S1B).
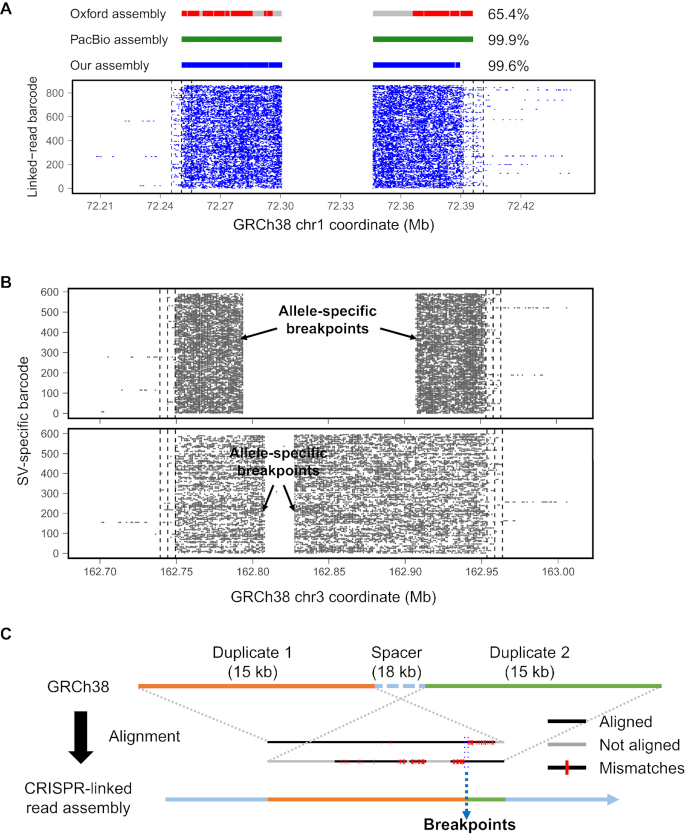
We compared our targeted local assemblies to the reported WGS assemblies based on Oxford Nanopore and Pacific Biosciences sequencing (13,16). We demonstrated concordant assemblies for 14 of 18 SVs (77.8%) while Pacific Biosciences had 13 (72.2%) and Oxford Nanopore had 15 (83.3%); our CRISPR-linked read method and Pacific Biosciences generated assemblies that aligned to the reference genome with more coverage than the Oxford assembly (Figure 3A). Four SV events did not generate an assembly scaffold due to inefficient Cas9-gRNA cleavage activity. Nevertheless, for one inversion event among the four (SV15 on Chromosome 12), alignment of the barcode-specific reads of the locus showed both breakpoints of the inversion (Supplementary Figure S9), suggesting better gRNA design might improve the results. The other SV events in our assay generated one or two assembled scaffolds. Because we size-selected SV alleles, both alleles were recovered only when the target and non-target allele were similar in length. SNV haplotypes in all of the assembled scaffolds matched well to either the maternal or paternal Platinum genome haplotype (Supplementary Table S12), which demonstrated that our CRISPR-linked read assembly could describe haplo- or diplotypes of target SV events.
Figure 3.
Assembly results from multiplex SV assay. (A) CRISPR-linked read assembly for SV1 in Assay 3 was aligned to the reference genome and compared with other long read assemblies. Red, green and blue bars indicate the portion of the assembly that aligns to the reference while a gray gap indicates no alignment. Although the gray regions have some similarity to the reference, they generally have too many homopolymer errors to successfully align. Fraction of aligned bases in the assembly is indicated at the end of the bars. (B) Two different sets of deletion breakpoints were determined for SV5. CRISPR-linked read SV assay captured the two SV alleles with different deletion sizes. For (A and B), the X- and Y-axes indicate the reference coordinates and the alignment of barcoded linked reads, respectively. Dashed vertical lines indicate Cas9-gRNA cut sites. (C) Illustration of how the breakpoints are determined in segmental duplications. Duplicated copies from GRCh38 reference genome were aligned to CRISPR-linked read assemblies. Breakpoint ranges in the reference duplicates were determined by alignment and mismatches. The example shown here is from our SV17 assembly. The two 15-kb segments have 93% similarity.
Base pair-resolution breakpoint determination by assembly
From our assemblies, we were able to precisely determine SV breakpoints and additional events associated with them. Most of the breakpoints identified by our CRISPR-linked read assemblies matched breakpoints from reported long read assemblies, but there were some discrepancies (Supplementary Table S13 and Figure S4). For example, breakpoints for SV8 and SV9 matched the Oxford Nanopore assembly but not the Pacific Biosciences assembly. Some minor breakpoint-associated events occurred, such as insertions and deletions (indels) of several bases. These small indels were not identified by alignment-based analysis and are likely to be the result of microhomologies or non-homologous end-joining (29,30). For SV5, we assembled two alleles with two different sets of breakpoints (Figure 3B), only one of which was confirmed by the Oxford Nanopore assembly (Supplementary Figure S4). On the deletion allele concordant with the Nanopore results, we also detected a 2-kb inversion starting at the second deletion breakpoint (Supplementary Table S13). In addition, the haplotypes of heterozygous SNVs in the two SV5 scaffolds with different deletions perfectly matched to either the maternal or paternal haplotype of the NA12878 Platinum genome (Supplementary Table S12).
According to the GRCh38 human reference genome, breakpoints in seven SVs were located within segmental duplications, of which our CRISPR-linked read analysis generated assemblies for five (Supplementary Table S14). In all five assemblies, as a result of deletions, no duplication was found, i.e. only a single homologous copy remained. To examine these sequences in more detail, we aligned the duplicated copies in the GRCh38 reference (31) to our targeted assemblies. We identified the breakpoints for three of the cases: SV7, SV16 and SV17 (Figure 3C and Supplementary Figure S10). We did not identify breakpoints for two of the five cases (SV12 and SV14) because their duplicated copies had a sequence similarity >99%; we therefore concluded that we had assembled only one of the two identical copies.
Validation with Pacific Biosciences CCS reads
We additionally used CCS read data to further validate the contiguity and parental haplotype assignment of our diploid assemblies (Supplementary Table S15). Among existing long read technologies, CCS reads are known to have the highest sequencing accuracy (bioRxiv https://www.biorxiv.org/content/10.1101/519025v2). In this validation, read alignment generated similar coverage for both haplotype assemblies, with few exceptions. In addition, only 1.4% of total assembled bases were not covered which, together with the balanced coverage, suggested reasonably good sequence contiguity for both haplotype assemblies. To validate the parental haplotypes of our assembly, we inspected variants and soft-clip events. We found that the counts of homozygous variants and soft-clipping events were linearly correlated with the length of the assembly (Supplementary Figure S11A). These events are likely to represent switch errors in our assemblies, but generally occurred locally and at a small fraction and therefore did not affect overall haplotype assignment. Interestingly, the variants found by the CCS reads were also strongly correlated with the number of haplotype switch errors found in the Platinum Genome validation (Supplementary Figure S11B), meaning both validation methods were in agreement.
DISCUSSION
Our study successfully demonstrated a targeted sequencing method that enables assembly of genomic regions as large as 0.2 Mb. Using assembly, both SNVs and SVs were successfully phased to diplotypes. When multiple overlapping HMW targets were tiled, the diplotype of multiple genes clustered in several Mb could be determined. This represents a novel method for sequencing targeted HMW DNA, and has a number of advantages. Our targeted CRISPR-linked read method: (i) selects intact target HMW DNA without polymerase chain reaction (PCR), (ii) overcomes limitations of WGS-based linked read sequencing, (iii) provides accurate diplotype across Mb regions without a hybrid approach and (iv) enables assembly-based base pair resolution analysis of SVs.
Our use of CRISPR as an in vitro technology makes possible amplification-free targeting of intact HMW DNA segments. We have already published for the first time the application of CRISPR-Cas9 for highly parallel in vitro targeted DNA fragmentation (21). In the current study, we applied this principle to the characterization of multi-Mb regions of the human genome with potentially complex genetic events across multiple genes. With an automated instrument, our approach provided sufficient target molecule yield for downstream sequencing without any further enrichment. Methods that require amplification, however, such as WGA, preclude preserving the phased genetic information of the original HMW DNA target. Alternatively, long-range PCR can generate amplicons of up to 10 kb (32), which can then be sequenced via a long read technology (15). Since the mean size of a human gene is 67 kb (33), this method is generally insufficient for phasing even a single gene. Compared to long-range PCR, our approach increases the potential length of targets by more than an order of magnitude. Moreover, for highly homologous duplications where duplicate copies are longer than 10 kb, primer design may be impossible, but because of the larger target size, our method may still allow successful analysis.
Combined with linked short read sequencing, our targeted approach provides highly accurate variant haplotypes of up to 4-Mb regions in the human genome. One problem with WGS approaches generally relying on random fragmentation is that linkage among variants in a target of interest are frequently broken. Our targeted approach uses barcoded linked reads to guarantees high sequencing coverage, generating single phased blocks covering entire HMW targets. Linked read sequencing does rely on barcode sharing between heterozygous variants to obtain long range haplotype information, so long stretches of homozygosity can cause interruptions in phasing. However, in our targeted approach, we generate a sufficient number of barcodes to support the long-distance linkage of heterozygous variants even across long stretches of homozygosity. Targeted CRISPR-linked read sequencing overcomes the major problem of WGS-based linked read sequencing because we use highly specific CRISPR fragmentation instead of random fragmentation.
The high on-target sequence coverage enables the short read sequencing-based assembly of HMW DNA targets, and thus can make use of the high accuracy of short read methods. Our study shows assembly contiguity comparable to, and better SNV haplotype quality than, long read methods. For the highly polymorphic MHC genes occurring across 4 Mb, we genotyped a CRISPR-linked read assembly at a resolution that distinguished intronic variations. Because of the high-sequencing accuracy, even a simple alignment method was able to generate the diplotype of MHC genes. Unlike conventional approaches relying on pedigree information (6), our method generated the MHC diplotype from a single individual sample. Against long read methods which generally must be complemented by short reads (15), our method had adequate accuracy by itself and thus no hybrid approach was required.
In addition, our approach also served to delineate multiple types of SVs with base-pair breakpoint resolution, whereas conventional short read sequencing would likely not delineate these SV structures. Moreover, even breakpoints inside highly homologous segmental duplications were precisely determined by our high-quality assemblies. By comparing our assembly against reference duplications, we were able to identify the hybrid copy in the assembly, of which a part matched to each of the duplicated reference copies. It has been suggested that a hybrid copy can be generated when a deletion or inversion occurs between two segmentally duplicated copies (10,11), and our assembly results suggest this also. There are many well-known hereditary syndromes associated with SV events in segmental duplications (10,11), with potentially different SV event-derived hybrid copies at hotspots. The locations of the hybrid junctions vary within these hotspots, which may be a cause of phenotypic variation among patients. Our CRISPR-linked read approach would be an efficient method to study this question.
In summary, CRISPR-linked read sequencing is an efficient and cost-effective substitute for current long read technologies in targeted sequencing. With a single assay on an individual sample, a high accuracy diplotype of SNV and SV alleles can be obtained. In future studies, we will continue to evaluate if this approach has utility for detecting clinically actionable gene diplotypes and genomic rearrangements causative for a variety of genetic disorders.
DATA AVAILABILITY
Sequence data is available at the NIH’s Sequence Read Archive with accession code SRP148930.
The scripts used in this study are available in an online repository (‘https://github.com/sgtc-stanford/CRISPR-LinkedReads’).
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Dr Anuja Sathe for generous assistance with tissue culture.
Authors’ contributions: G.S., H.L., C.B. and H.P.J. designed the experiments. G.S. conducted the experiments with technical assistance from J.Z. and C.B. in the BRCA1 study. G.S and C.S. optimized the isolation process. G.S. developed the analysis algorithms. G.S., L.X. and S.U.G. analyzed the data. G.S., S.U.G. and H.P.J. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [2R01HG006137-04 to H.P.J., P01HG00205ESH to G.S., H.P.J.]; American Cancer Society [RSG-13-297-01-TBG to H.P.J.]; Clayville Foundation (to H.P.J.); Howard Hughes Medical Institute Early Career Grant [57006499 to H.P.J.]. Funding for open access charge: Clayville Foundation (to H.P.J.).
Conflict of interest statement. J.Z. and C.B. are employees of Sage Science, Inc.
REFERENCES
- 1. Tewhey R., Bansal V., Torkamani A., Topol E.J., Schork N.J.. The importance of phase information for human genomics. Nat. Rev. Genet. 2011; 12:215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Laroche-Clary A., Chaire V., Le Morvan V., Neuville A., Bertucci F., Salas S., Sanfilippo R., Pourquier P., Italiano A.. BRCA1 haplotype and clinical benefit of trabectedin in soft-tissue sarcoma patients. Br. J. Cancer. 2015; 112:688–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borsetti A., Ferrantelli F., Maggiorella M.T., Sernicola L., Bellino S., Gallinaro A., Farcomeni S., Mee E.T., Rose N.J., Cafaro A. et al.. Effect of MHC haplotype on immune response upon experimental SHIVSF162P4cy infection of Mauritian cynomolgus macaques. PLoS One. 2014; 9:e93235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leary R.J., Lin J.C., Cummins J., Boca S., Wood L.D., Parsons D.W., Jones S., Sjoblom T., Park B.H., Parsons R. et al.. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. PNAS. 2008; 105:16224–16229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Greer S.U., Nadauld L.D., Lau B.T., Chen J., Wood-Bouwens C., Ford J.M., Kuo C.J., Ji H.P.. Linked read sequencing resolves complex genomic rearrangements in gastric cancer metastases. Genome Med. 2017; 9:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jensen J.M., Villesen P., Friborg R.M., Danish Pan-Genome C., Mailund T., Besenbacher S., Schierup M.H.. Assembly and analysis of 100 full MHC haplotypes from the Danish population. Genome Res. 2017; 27:1597–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barrett J.C., Fry B., Maller J., Daly M.J.. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005; 21:263–265. [DOI] [PubMed] [Google Scholar]
- 8. Bomba L., Walter K., Soranzo N.. The impact of rare and low-frequency genetic variants in common disease. Genome Biol. 2017; 18:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xia L.C., Sakshuwong S., Hopmans E.S., Bell J.M., Grimes S.M., Siegmund D.O., Ji H.P., Zhang N.R.. A genome-wide approach for detecting novel insertion-deletion variants of mid-range size. Nucleic Acids Res. 2016; 44:e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Emanuel B.S., Shaikh T.H.. Segmental duplications: an ‘expanding’ role in genomic instability and disease. Nat. Rev. Genet. 2001; 2:791–800. [DOI] [PubMed] [Google Scholar]
- 11. Dennis M.Y., Eichler E.E.. Human adaptation and evolution by segmental duplication. Curr. Opin. Genet. Dev. 2016; 41:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H. et al.. An integrated map of structural variation in 2,504 human genomes. Nature. 2015; 526:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A., Tyson J.R., Beggs A.D., Dilthey A.T., Fiddes I.T. et al.. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018; 36:338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Dijk E.L., Jaszczyszyn Y., Naquin D., Thermes C.. The third revolution in sequencing technology. Trends Genet. 2018; 34:666–681. [DOI] [PubMed] [Google Scholar]
- 15. Fuselli S., Baptista R.P., Panziera A., Magi A., Guglielmi S., Tonin R., Benazzo A., Bauzer L.G., Mazzoni C.J., Bertorelle G.. A new hybrid approach for MHC genotyping: high-throughput NGS and long read MinION nanopore sequencing, with application to the non-model vertebrate Alpine chamois (Rupicapra rupicapra). Heredity. 2018; 121:293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pendleton M., Sebra R., Pang A.W., Ummat A., Franzen O., Rausch T., Stutz A.M., Stedman W., Anantharaman T., Hastie A. et al.. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat. Methods. 2015; 12:780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edge P., Bafna V., Bansal V.. HapCUT2: robust and accurate haplotype assembly for diverse sequencing technologies. Genome Res. 2017; 27:801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zheng G.X., Lau B.T., Schnall-Levin M., Jarosz M., Bell J.M., Hindson C.M., Kyriazopoulou-Panagiotopoulou S., Masquelier D.A., Merrill L., Terry J.M. et al.. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat. Biotechnol. 2016; 34:303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xia L.C., Bell J.M., Wood-Bouwens C., Chen J.J., Zhang N.R., Ji H.P.. Identification of large rearrangements in cancer genomes with barcode linked reads. Nucleic Acids Res. 2018; 46:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bittel D.C., Yu S., Newkirk H., Kibiryeva N., Holt A. 3rd, Butler M.G., Cooley L.D.. Refining the 22q11.2 deletion breakpoints in DiGeorge syndrome by aCGH. Cytogenet. Genome Res. 2009; 124:113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shin G., Grimes S.M., Lee H., Lau B.T., Xia L.C., Ji H.P.. CRISPR-Cas9-targeted fragmentation and selective sequencing enable massively parallel microsatellite analysis. Nat. Commun. 2017; 8:14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 23. Weisenfeld N.I., Kumar V., Shah P., Church D.M., Jaffe D.B.. Direct determination of diploid genome sequences. Genome Res. 2017; 27:757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018; 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bennett-Baker P.E., Mueller J.L.. CRISPR-mediated isolation of specific megabase segments of genomic DNA. Nucleic Acids Res. 2017; 45:e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gabrieli T., Sharim H., Fridman D., Arbib N., Michaeli Y., Ebenstein Y.. Selective nanopore sequencing of human BRCA1 by Cas9-assisted targeting of chromosome segments (CATCH). Nucleic Acids Res. 2018; 46:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium. Nature. 1999; 401:921–923. [DOI] [PubMed] [Google Scholar]
- 28. Eberle M.A., Fritzilas E., Krusche P., Kallberg M., Moore B.L., Bekritsky M.A., Iqbal Z., Chuang H.Y., Humphray S.J., Halpern A.L. et al.. A reference data set of 5.4 million phased human variants validated by genetic inheritance from sequencing a three-generation 17-member pedigree. Genome Res. 2017; 27:157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moore J.K., Haber J.E.. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell Biol. 1996; 16:2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McVey M., Lee S.E.. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008; 24:529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schneider V.A., Graves-Lindsay T., Howe K., Bouk N., Chen H.C., Kitts P.A., Murphy T.D., Pruitt K.D., Thibaud-Nissen F., Albracht D. et al.. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017; 27:849–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jia H., Guo Y., Zhao W., Wang K.. Long-range PCR in next-generation sequencing: comparison of six enzymes and evaluation on the MiSeq sequencer. Sci. Rep. 2014; 4:5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Piovesan A., Caracausi M., Antonaros F., Pelleri M.C., Vitale L.. GeneBase 1.1: a tool to summarize data from NCBI gene datasets and its application to an update of human gene statistics. Database. 2016; 2016:baw153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data is available at the NIH’s Sequence Read Archive with accession code SRP148930.
The scripts used in this study are available in an online repository (‘https://github.com/sgtc-stanford/CRISPR-LinkedReads’).