Abstract
Mammalian cells express a variety of nucleic acid sensors as one of the first lines of defense against infection. Despite extensive progress in the study of sensor signaling pathways during the last decade, the detailed mechanisms remain unclear. In our previous studies, we reported increased type I interferon expression and the upregulation of several proposed cytosolic DNA sensors after transfection of several tumor cell types with plasmid DNA (pDNA). In the present study, we sought to reveal the early events in the cytosolic sensing of this nucleic acid in a myoblast cell line. We demonstrated that DNA-dependent activator of interferon regulatory factors/Z-DNA binding protein 1 (DAI/ZBP1) bound plasmid DNA in the cytosol within 15 minutes of transfection and at consistent levels for 4 h. Interferon activated gene 204 protein (p204) and DEAH box helicase 9 (DHX9) also bound pDNA, peaking 15 and 30 min respectively. Plasmid DNA was not detectably bound by DEAD box helicase 60 (DDX60) protein, despite a similar level of mRNA upregulation to DAI/ZBP1, or by cyclic GMP-AMP synthase (cGAS), despite its presence in the cell cytosol. Taken together, these results indicate several DNA sensors may participate and cooperate in the complex process of cytosolic DNA sensing.
INTRODUCTION
The infection of mammalian cells induces the production of type I interferon (IFN) and pro-inflammatory cytokines (1,2). The central event in this process is the recognition of pathogen-associated molecular patterns by a group of cytosolic proteins termed pattern recognition receptors (PRRs). PRRs are expressed in a wide variety of mammalian cells and can be divided into several classes based on their subcellular localization and specificity (3–6). Since the first cytosolic DNA-specific PRR or DNA sensor, DNA-dependent activator of IFN-regulatory factors/Z-DNA-binding protein 1 (DAI/ZBP1), was reported to elicit a type I IFN-mediated immune response (7), many additional cytosolic DNA sensors have been proposed (6,8), suggesting a redundant or cell type specific role in DNA sensing (9–11). This response is initiated via the cyclic guanosine monophosphate (GMP)–adenosine monophosphate (AMP) synthase-cyclic GMP-AMP synthase-stimulator of IFN gene (cGAS-cGAMP-STING) pathway (12), although sensors such as polyglutamine binding protein 1 (PQBP1) (13) or interferon gamma inducible factor 16 (IFI16) (14) complex with DNA and cGAS to promote its function. The sophisticated interplay of apparently parallel sensing pathways complicates the ability to estimate the individual contributions of specific DNA sensors in the response to cytosolic DNA. Thus, detailed mechanisms of DNA detection on the cellular level still remain unclear.
Viral and non-viral methods are currently used to deliver DNA into cells for in vitro and in vivo purposes. Viral methods are effective; however, viruses by their nature bind and activate DNA sensors (9,15). Non-viral approaches to DNA delivery are less efficient and often require chemical or physical assistance (16). One physical method, electroporation, is widely used in laboratory applications for plasmid DNA (pDNA) delivery (17).
Electroporation is an almost instantaneous process (18). Plasmid DNA crosses the plasma membrane as soon as 10 min after pulse application, which makes it a valuable technique to precisely document the early events of cytosolic DNA sensing (19–21). Several groups of proteins, including nucleocytoplasmic shuttling proteins, motors, transcription factors, and RNA binding proteins, bind pDNA within 15 min of delivery by electroporation (22). Cell defense proteins such as DNA sensors may also bind DNA with these kinetics.
Recent studies from our group demonstrated increased expression of various cytosolic DNA sensors as well as type I IFN after electroporation of control pDNA into different tumor cell lines (23,24). These effects were cell type and pulse protocol specific. However, these observations were made significantly after transfection and therefore did not reveal initial DNA sensing events. In addition, these studies were performed on tumor cell lines, which may have distorted innate immune signaling. Therefore, in this study, we sought to reveal the specific DNA sensors reacting to the presence of plasmid DNA introduced into non-tumor myoblast cells. We found that several putative DNA sensors bound pDNA in the cytosol for up to four h after transfection.
MATERIALS AND METHODS
Cell culture
C2C12 murine skeletal myoblast cells (CRL-1772, American Type Culture Collection, Manassas, VA, USA) in their proliferation phase were cultured as monolayers in Dulbecco's modified Eagle medium (DMEM, Corning, Manassas, VA, USA) supplemented with 5% fetal bovine serum (FBS, Atlanta Biologicals, Flowery Branch, GA, USA) at 37°C in 5% CO2. The cells were routinely tested for mycoplasma infection (LONZA MycoAlert, Basel, Switzerland or e-myco Valid Mycoplasma PCR Detection kit, Intron Biotechnology, Burlington, MA, USA) and were mycoplasma free.
Plasmids and antibodies
A plasmid encoding green fluorescent protein (GFP) driven by the CMV-IE promoter/enhancer, gWizGFP, and gWizBlank, which encodes no transgene, were commercially prepared (Aldevron, Fargo, ND, USA) at a concentration of 2 μg/μl in physiological saline.
The primary antibodies used for pull-down experiments and western blotting were rabbit mouse-reactive anti-DAI/ZBP1 (Thermo Fisher Scientific, Waltham, MA, USA), anti-cyclic AMP response-element binding protein (CREB, Abcam, Cambridge, MA, USA), anti-DDX60 (Abcam), anti-cGAS (Cell Signaling Technology, Danvers, MA, USA), anti-ifi204 (Thermo Fisher Scientific), anti-Ku70 (Thermo Fisher Scientific), anti-DHX9 (Abcam), anti-DEAD box helicase 41 (DDX41; Abcam) and rat mouse-reactive anti-CD4 (eBioscience, San Diego, CA, USA). Goat anti-rabbit IgG H&L (Alexa Fluor 680) and goat anti-rat IgG H&L (Alexa Fluor 680) (both Thermo Fisher Scientific) were used as secondary antibodies. For western blotting, all antibodies were diluted in 5% skim milk. Anti-ds DNA antibody (Abcam) and donkey anti-mouse IgG H&L (Alexa Fluor 488) (Jackson Immunoresearch, Bar Harbor, ME, USA) were used as primary and secondary antibodies, respectively, for the confocal visualization of pDNA. For colocalization studies, anti-DAI/ZBP1 (Thermo Fisher Scientific) and donkey anti-rabbit Cy3 (Jackson Immunoresearch, Bar Harbor, ME, USA) were used as primary and secondary antibodies.
Plasmid DNA transfection
A C2C12 cell suspension of 2.5 × 107 cells/ml in electroporation buffer (23) was mixed with a 2 μg/μl pDNA solution at a 4:1 ratio to a final pDNA concentration of 0.4 μg/μl. The mixture was transferred either between two stainless steel plate electrodes with 2 mm gap or into a cuvette (BTX Harvard Apparatus, Holliston, MA, USA). Transfection was achieved by the application of eight 5 ms pulses at a voltage-to-distance ratio of 600 V/cm and a frequency of 1 Hz (EP1) (25) or six 100 μs pulses at 1300 V/cm at frequency of 4 Hz (EP2) (26). The cells were immediately transferred into the wells of a 6-well culture plate containing FBS to ensure proper cell recovery. After 5 min of incubation, DMEM was added and the plates were incubated at 37°C in 5% CO2 for the time designated in each experiment. Transfections were also performed using Transit LT1 (Mirus Bio LC, Madison, WI, USA) per manufacturer's instructions.
Transfection efficiency
Transfection efficiency was quantified 24 h after transfection as previously described (23).
Cell survival
Cell survival was quantified 6 h after transfection as previously described (23).
Confocal microscopy
Cells were seeded onto 12-well removable chamber slide (Ibidi GmbH, Martinsried, Germany) and transfection was performed 24 h later using EP2. After 15 min, the cells were fixed with 4% paraformaldehyde for 15 min at 37°C. Cell membrane was then labelled with wheat germ agglutinin (WGA) Alexa Fluor 647 (5 μg/ml, Thermo Fisher Scientific). Cells were permeabilized (10 min, 0.1% Tween), blocked for one hour (1% BSA, 0.1% Tween 20, 22.52 mg/ml glycine) and incubated overnight at 4°C with anti-ds DNA antibody (Abcam), followed by incubation with secondary antibody for one hour at room temperature. Nuclei were stained with Hoechst 33258 (1 μg/ml, Thermo Fisher Scientific) for 10 min. Chambers were removed and slides were mounted using Prolong Gold Diamond Antifade Mountant (Thermo Fisher Scientific). Imaging was performed with an LSM 800 confocal microscope (Carl Zeiss, Oberkochen, Germany) with a 63× oil immersion objective and a numerical aperture of 1.4. The lasers providing excitation light at wavelength of 405, 488 and 640 nm were used for visualization of Hoechst 33258, Alexa 488 and Alexa 647, respectively. The emitted light was collected sequentially with Gallium Arsenide Phosphide (GaAsP) detector via a variable dichroic at the following wavelengths: 410–617 nm (Hoechst) where additional 620 short pass filter was used, 400 – 650 nm (Alexa 488) and 645–700 nm (Alexa 647). The collected Z-stacks were then visualized in Imaris software (Bitplane, Zurich, Switzerland).
For colocalization studies, pDNA was stained before transfection with Label IT Nucleic Acid Labeling Kit, MPF488, (MIR7100, Mirus Bio, Madison, USA) according to manufacturer's instructions. Five minutes after the transfection, culture media was added to the wells and slides were incubated for a total of 15 min. Cells were stained with primary anti-DAI/ZBP1 antibody (Thermo Fisher Scientific) overnight and secondary donkey anti-rabbit Cy3 antibody (Jackson Immunoresearch) for one hour, otherwise they were processed as described above with additional laser providing excitation light at wavelength of 570 nm and emission filter 550–570 nm.
Reverse transcription real time PCR
Messenger RNAs were quantified at specific time points after transfection as previously described (23,24).
Plasmid biotinylation, DNA pull-down and slot blot
Plasmid was labeled with photobiotin acetate (Santa Cruz) as described previously (27,28). The final concentration of biotin-labeled gWizGFP was measured by spectrophotometer (NanoVue, GE Healthcare Bio-sciences, Pittsburgh, PA, USA) and adjusted to 2 μg/μL by nuclease-free water.
After transfection, the cells were incubated for 10, 25, 55 or 235 min, then harvested by centrifugation for 5 min, creating a 15, 30, 60 or 240 min total time, respectively. Cells were then quickly rinsed with PBS and lysed (Cell Lysis Buffer, Cell Signaling Technology). Biotin-labeled DNA was pulled down using Streptavidin Magnetic Beads (Thermo Fisher Scientific) in accordance with manufacturer instructions. After washing with TBS-Tween, the beads were treated with DNase (Ambion DNA-free Kit, Foster City, CA, USA) in accordance with manufacturer instructions. The beads were removed by centrifugation and proteins from the supernatant were transferred to a nitrocellulose membrane using a Bio-Dot SF Microfiltration Apparatus (Bio-Rad) followed by a standard western blot procedure.
Western blotting
For western blotting, C2C12 cells were lysed with Cell Lysis Buffer (Cell Signaling Technology) to prepare whole-cell lysate. Alternatively, nuclei were removed (Nuclei EZ Prep, Sigma-Aldrich, St. Louis, MO, USA) from naïve C2C12 cells to create cytosolic fraction. Western blotting was performed as previously described (24). REVERT Total Protein Stain (LI-COR, Lincoln, NE, USA) was used for normalization according to manufacturer's instructions.
Protein pull-down and PCR to quantify bound plasmid
After transfection, the cells were incubated and lysed as above. The lysates were pre-cleared with Protein A Agarose beads (Cell Signaling Technology) by shaking for one hour at 4°C. Protein was pulled down by Protein A Agarose Beads in accordance with manufacturer's instructions using specific antibodies listed above. The beads were washed three times with lysis buffer and resuspended in molecular grade water for PCR analysis. Bound gWizGFP was quantified by PCR (CFX96 Real Time PCR Detection System, Bio-Rad, Hercules, CA, USA) using custom primers (Integrated DNA Technologies, Coralville, IA, USA) to base pairs 567–751 within the CMV-IE promoter/enhancer region (forward 5′- ATCATATGCCAAGTACGCCC-3′, reverse 5′- TGAGTCAAACCGCTATCCAC-3′) and base pairs 2085–2221 within the GFP open reading frame (forward 5′- CTCTGTGCTATGGTGTTCAATG-3′, reverse 5′- TGTCTTGTAGTTGCCGTCATC-3′) in SYBR Select Master Mix (Applied Biosystems, Foster City, CA , USA). Water was used as no template control. Absolute quantification was performed by comparison to serial dilutions of gWizGFP DNA, then relative quantification was performed by comparison to control sample incubated with anti-CD4 antibody.
siRNA silencing
For siRNA experiments, 1 × 106 cells were seeded to 10 cm Petri dishes 24 h before transfection. Commercially available DHX9, DAI/ZBP1, p204 and control siRNAs (Silencer Select siRNA, Thermo Fischer Scientific) were mixed with Lipofectamine RNAiMAX Transfection Reagent (Thermo Fischer Scientific) following manufacturer's instructions and added to the cells. After 48 h of incubation, the cells were harvested, counted and transfected with pDNA as described above. Values of the experimental groups were normalized to control siRNA values.
Plasmid sequence analysis
The 5757 base pair gWizGFP sequence was analyzed using PROMO version 3 (29,30). Five CREB binding regions within the CMV IE promoter/enhancer were detected (base pairs 454–475, 507–516. 590–599, 776–785, 1035–1045). The non-B DB (31) was used to analyze for Z-DNA forming sequence motifs. A single motif was detected at base pairs 3756–3765. IFI16, the human ortholog of p204, senses dsDNA>60 base pairs long independent of sequence. Therefore, this protein could bind DNA throughout the plasmid (32). DHX9 binds CpG-B DNA (33), which has the general formula purine-purine-CG-pyrimidine-pyrimidine while lacking a poly-G motif (34). gWizGFP contains 28 of these motifs, including 9 that have increased immunostimulatory activity in mouse cells (35,36).
Statistical analysis
Statistical analysis was performed (SigmaPlot, Systat Software, Chicago, IL, USA or GraphPad Software, La Jolla, CA, USA). The data were first tested for normality of distribution with the Shapiro-Wilk test. The differences between the experimental groups were statistically evaluated by two-way analysis of variance followed by a Tukey–Kramer or Dunnett's post-test. A P-value of <0.05 was considered to be statistically significant. A sample size (n) for each experiment represents biological replicates unless otherwise stated.
RESULTS
Initiation of a pro-inflammatory response after plasmid transfection
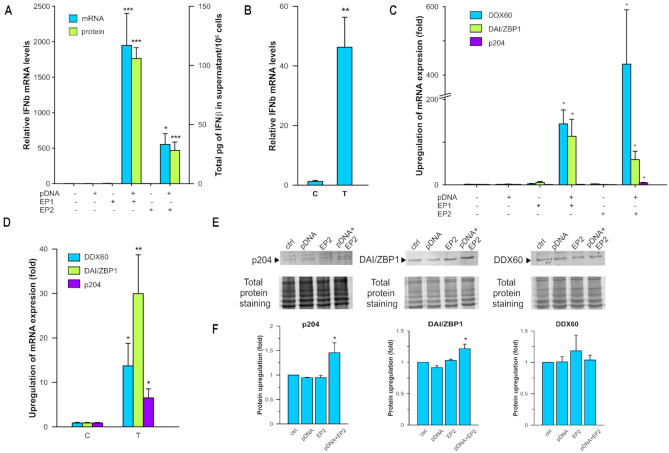
Our previous studies demonstrated that electroporation of control plasmids into tumor cells induced pro-inflammatory signaling in a cell line dependent manner (23,24,37). To investigate the effect of plasmid electroporation on non-tumor cells, we delivered pDNA into C2C12 mouse myoblasts using two different pulse protocols and a chemical delivery method and measured the secretion of a marker of this response, IFN β, by ELISA (PBL Assay Science, Piscataway, NJ, USA). Neither electroporation alone nor addition of pDNA to the cell suspension induced a significant difference from the control (Figure 1A). However, after 4 h, pDNA delivery with EP1, EP2, or chemical delivery significantly (P < 0.001 and P < 0.05, respectively) increased IFN β mRNA levels in comparison to the control. Delivery with EP1 upregulated IFN β mRNA by nearly 1950-fold, significantly higher (P < 0.05) than delivery with EP2, which induced an upregulation of more than 550-fold. Chemical delivery induced a 46-fold upregulation of IFN β mRNA (P < 0.01) after 4 h (Figure 1B).
Figure 1.
Effects of pDNA transfection on IFN β production and expression of putative cytosolic DNA sensors. (A) IFN β mRNA levels 4 h and protein levels 6 h after transfection using electroporation with gWizBlank normalized to control group (n = 3–10). (B) IFN β mRNA levels 4 h after transfection using Transit (T) normalized to control group (C) (n = 6–8). (C) DDX60, DAI/ZBP1 and p204 mRNA levels 4 h after electroporation normalized to control group (n = 3–9). (D) DDX60, DAI/ZBP1 and p204 mRNA levels 4 h after chemical transfection (T) normalized to control group (C) (n = 8). (E) Representative western blots of DDX60, DAI/ZBP1 and p204 proteins 8 h after transfection using EP2. (F) DDX60, DAI/ZBP1 and p204 upregulation 8 h after transfection using EP2 normalized to control group (n = 3 for each protein). *P < 0.05; **P < 0.01; ***P < 0.001 statistical difference from control group.
IFN β protein was coordinately significantly (P < 0.001) upregulated 4 h after pDNA delivery by both pulse protocols. As with the mRNA levels, pDNA delivery with EP1 induced significantly higher levels of IFN β protein (P < 0.001) than EP2. Neither electroporation alone nor addition of pDNA to the cell suspension induced a significant difference from the control in the level of IFN β protein (Figure 1A).
The increase in IFN β production might be attributed to the binding and activation of endosomal or cytosolic DNA sensors. We therefore quantified changes in the mRNA levels for several putative DNA sensors. The mRNAs of retinoic acid inducible gene I (RIG-I) and toll-like receptor 9 (TLR9) were not detected in any group in these cells. The mRNAs of several sensors, DEAD box polypeptide 41 (DDX41), DEAH box helicase 9 (DHX9), 70 kDa subunit of Ku antigen (Ku70) and cGAS, were detected in the cells but not upregulated in any group (data not shown). However, the mRNA of the DEAD box helicase 60 (DDX60) protein was comparably upregulated after pDNA delivery with EP1 and EP2, by ∼140-fold and 430-fold respectively. Interestingly, pDNA delivery using EP2 induced a small but significant 5-fold increase in the mRNA of interferon activated gene 204 protein (p204) not observed in other groups. Finally, the mRNA of DAI/ZBP1 was upregulated by ∼115-fold after pDNA delivery with EP1, which was significantly higher (P < 0.05) than the 60-fold upregulation after delivery with EP2 (Figure 1C). Chemical delivery induced an approximately 14-fold upregulation of DAI/ZBP1 mRNA (P < 0.05), an approximately 30-fold upregulation of DDX60 mRNA (P < 0.01), and a >6-fold upregulation of p204 mRNA (P < 0.05) after 4 h (Figure 1D). As with IFN β, protein levels 8 h after pDNA transfection were coordinately upregulated 1.2-fold for DAI/ZBP1 and nearly 1.5-fold for p204, but, surprisingly, no significant upregulation was detected for DDX60 (Figure 1E, F).
Variation in cytotoxicity between pulse protocols with similar transfection efficiencies
The transfection efficiency of ∼20% of the cells was similar between the two pulse protocols (Figure 2A). Delivery using EP1 produced a significantly higher median fluorescence intensity, suggesting that a greater plasmid copy number reached the nucleus and was expressed in this group.
Figure 2.
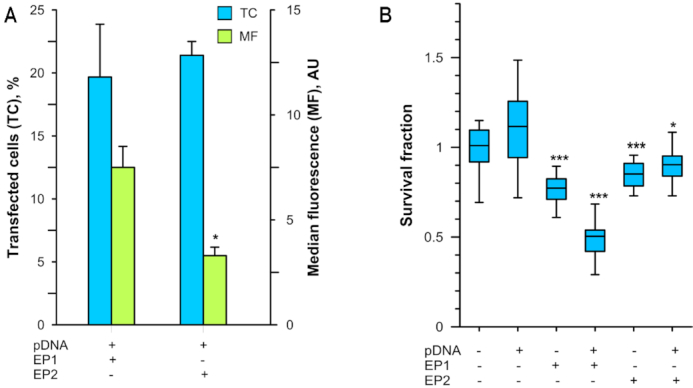
Effects of different pulse protocols on transfection efficiency and cell survival. (A) The fraction of transfected cells (blue) and median fluorescence (green) were determined by flow cytometry 24 h after gWizGFP delivery (EP1, n = 10; EP2, n = 6). (B) Survival fraction determined by Presto Blue assay 6 h after gWizBlank delivery normalized to control group (n = 24 technical replicates, n = 2 biological replicates). *P < 0.05; ***P < 0.001 statistical difference from control group.
Cell survival was also pulse protocol dependent. No significant difference in survival was found between pulse protocols in the absence of pDNA, although the viability for each was statistically different from the control (Figure 2B). Plasmid DNA delivery using EP1 reduced survival to 48% after 6 h, while pDNA delivery with EP2 maintained a 90% cell survival. Since we were attempting to reveal the early events of DNA sensing, we chose EP2 for our further experiments as a less damaging to the cells.
Kinetics of mRNA upregulation
Several mRNAs were upregulated after transfection. PCR or reverse transcription PCR of RNA from cells transfected with gWizGFP was now employed to more precisely assess the dynamic changes in the mRNA levels of the biomarker IFN β, the transgene GFP, and the upregulated DNA sensors. IFN β mRNA levels increased significantly within 60 min of transfection (Figure 3A). This preceded detectable transgene expression; an increase GFP mRNA levels was detected after 2 h that continued up to 24 h (Figure 3B). A significant increase in DDX60 mRNAs was detected at 2 h, while p204 was significantly increased at 5 h after transfection. DAI/ZBP1 mRNA was significantly upregulated from 5 to 24 h (Figure 3C). Peak levels did not necessarily mirror those after transfection with gWizBlank (Figure 1A, C); we previously noted plasmid-dependent variations in DNA sensor mRNA induction (23).
Figure 3.
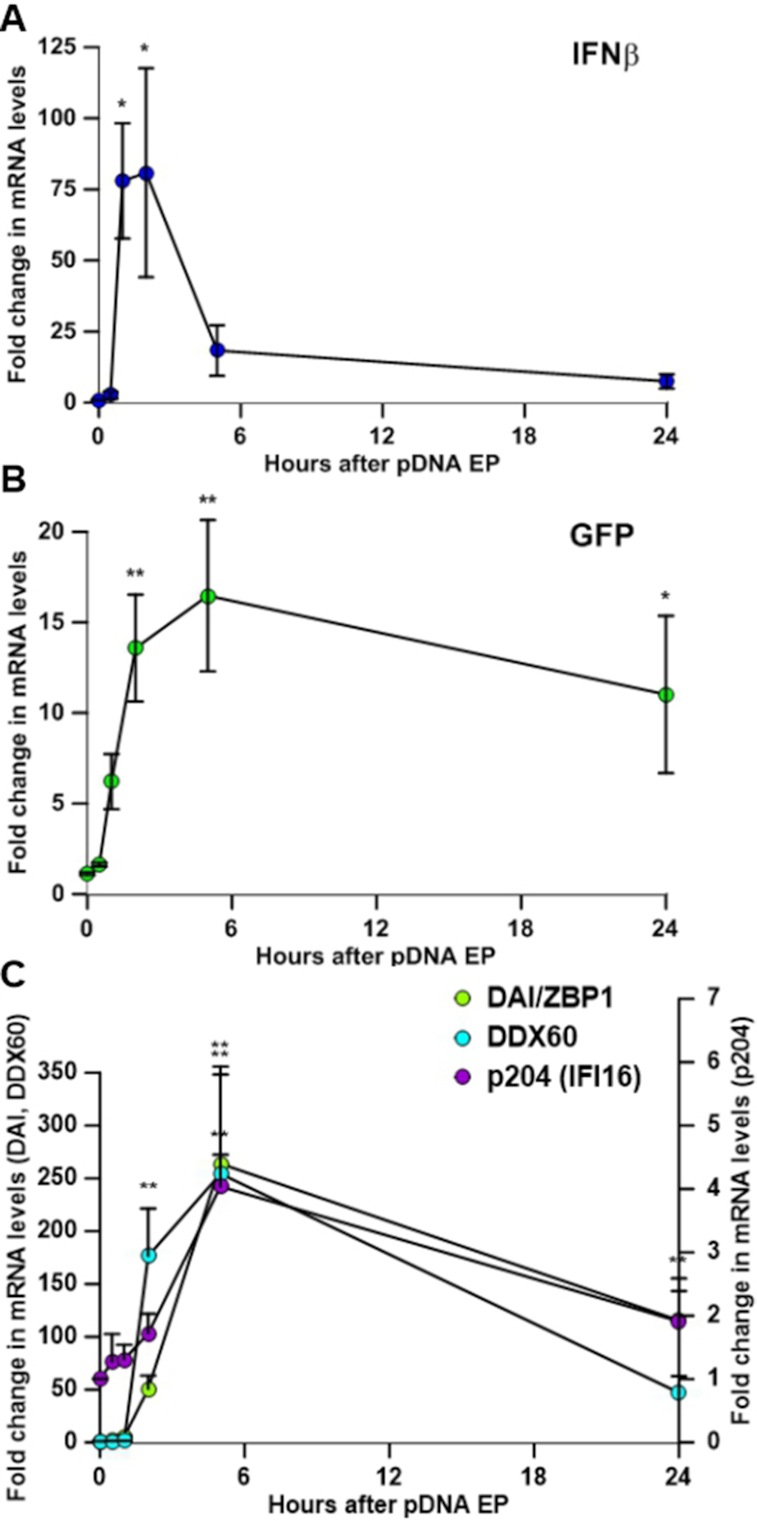
Kinetics of mRNA expression. (A) IFN β, (B) GFP, (C) DAI/ZBP1, DDX60, and p204 mRNA levels at 0, 0.5, 1, 5 and 24 h after transfection (n = 5–8). *P < 0.05; **P < 0.01 statistical difference from time 0.
Direct detection of cytoplasmic pDNA
Previous studies demonstrated that electroporated pDNA enters the cytosol or cytoplasm within 10 min (19–21) and transcription factors can bind to it within 15 min (38) after transfection. To confirm the presence of pDNA in the cytoplasm of C2C12 cells, we performed confocal imaging 15 min after transfection. Cells in the control group and electroporated cells demonstrated basal level of staining which could be attributed to the presence of mitochondrial DNA. Plasmid DNA aggregates were detected extracellularly in cells suspended in pDNA. However, increased DNA staining was detected only in the transfected cells (Figure 4), corroborating the reports that pDNA enters the cytoplasm within minutes of transfection (19–21). Based on these data, we chose this time point as a starting point to test for early DNA sensor binding events.
Figure 4.
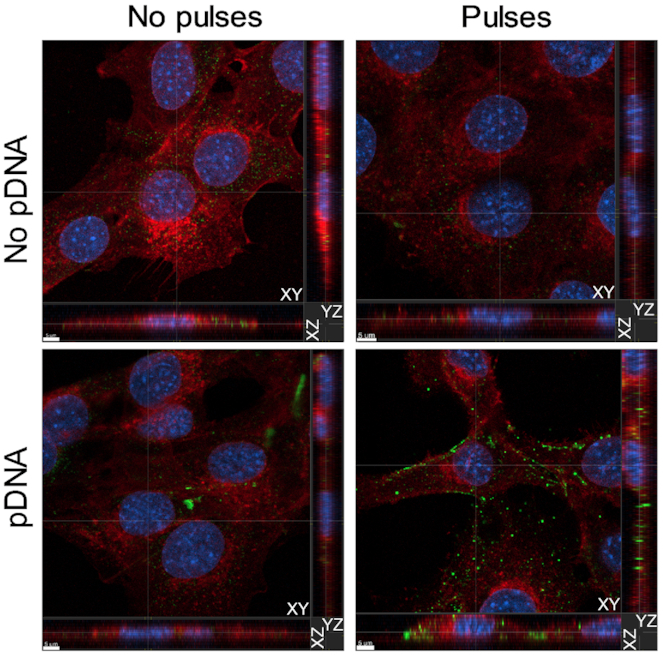
Plasmid DNA enters the cytoplasm of C2C12 cells within 15 min of transfection. Representative confocal images from the Z-stacks demonstrate the presence of pDNA in the cytoplasm of C2C12 cells 15 min after electroporation with EP2. Plasmid DNA was detected using anti-ds DNA antibody (green), while membrane structures were labeled with WGA (red). Cell nuclei were labeled with Hoescht 33258 (blue). Scale bar equals 5 μm.
DNA sensor proteins present at basal conditions
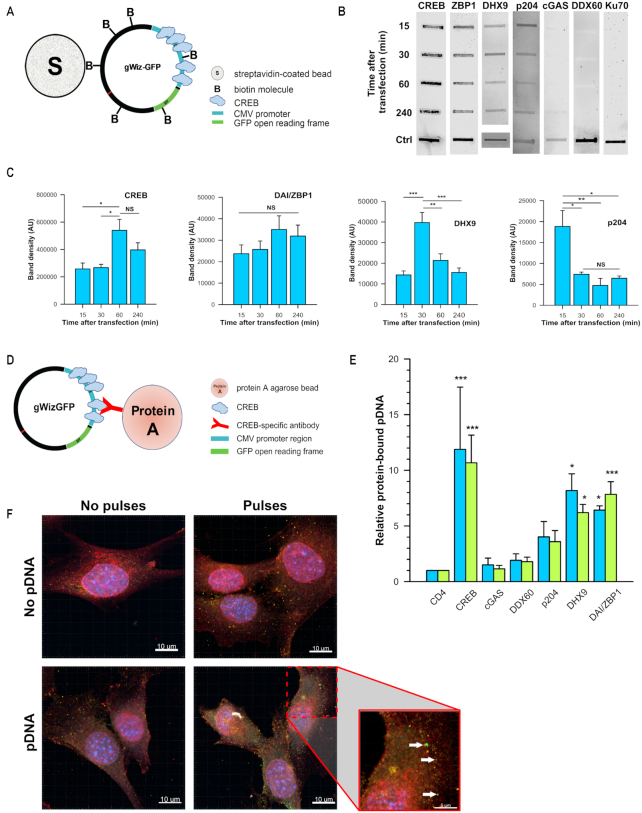
A protein must be present in cytoplasm in order to bind DNA upon its entry into the cell. Therefore, we tested the cytosolic fraction of lysates obtained from control C2C12 cells for the presence of the putative DNA sensors correlating to the mRNAs detected by RT-PCR. We also tested for the presence of CREB, which is known to bind the CMV-IE promoter/enhancer region (22). With the exception of DDX41, all of these proteins were detected in the cytosol of control cells (Supplementary Figure S1).
Protein binding to cytoplasmic pDNA after transfection
Several putative DNA sensors were detected in control cells. It was therefore possible that these proteins functioned in pDNA binding. We transfected cells with biotinylated gWizGFP, pulled down the pDNA using streptavidin-coated beads, and tested for the presence of CD4 (as a negative control since this protein is not present in the cytoplasm nor is it known to bind DNA), CREB (as a positive control), DAI/ZBP1, DDX60, p204, cGAS, DHX9 or Ku70. Whole-cell lysate of naïve C2C12 cells served as additional positive control (Figure 5A). No bands were detected in the samples probed with anti-CD4 antibody, which suggested the absence of non-specific fluorescence signal (data not shown). Intense bands were detected at each time point on the membranes probed against CREB, peaking at 60 min after transfection. DDX60 and Ku70 proteins were not detected in pull-downs at any tested time point, although each of these proteins was detected in a positive control sample, indicating properly performed immunoblotting. A faint band was visible at the four hour time point on the membrane probed against cGAS protein, although this band was not quantifiable by the software. Protein bands were detected on the membrane probed against DAI/ZBP1, DHX9 and p204 at each time point (Figure 5B). DHX9 and p204 binding peaked at 30 and 15 min after transfection, respectively, with the further decrease, while no significant difference in band intensity was found for DAI/ZBP1 at any time point (Figure 5C).
Figure 5.
Protein immunoprecipitation and pDNA pull-down. (A) Schematic representation of biotinylated pDNA pull-down. (B) Representative membranes demonstrating proteins’ binding to pDNA detected by western blotting. Whole-cell C2C12 lysate serves as positive control (bottom line). (C) Time course of CREB, DAI/ZBP1, DHX9 and p204 binding to pDNA (n = 3 for each protein). All time points represent time after gWizGFP transfection. (D) Schematic representation of protein immunoprecipitation. (E) gWizGFP plasmid copy number normalized to CD4 group (n = 3 for CREB, DHX9 and cGAS groups, n = 6 for the other groups). No statistical difference was observed within any group whether the CMV promoter (blue) or the GFP open reading frame (green) was targeted. *P < 0.05; **P < 0.01; ***P < 0.001 statistical difference from control group. (F) Representative confocal microscopy images demonstrating colocalization of DAI/ZBP1 and pDNA. Nuclei were counterstained with Hoechst 33258 (blue) and membranes with WGA (red). Scale bar equals 10 μm. Inset: White arrows indicate the points of colocalization of DAI/ZPB1 (orange) and pDNA (green). Scale bar equals 5 μm.
We next performed protein immunoprecipitations (Figure 5D) to confirm DNA sensor binding using specific antibodies against DAI/ZBP1, DHX9 and p204, which were found to bind biotinylated plasmid. We also included DDX60 and cGAS in the tested panel, since DDX60 mRNA was upregulated after transfection, and cGAS is considered to be essential for the induction of a type I IFN response to cytosolic DNA (39). An antibody against CD4 was applied to serve as negative control, while an antibody against CREB served as positive control. After protein immunoprecipitation, bound pDNA was quantified by PCR. Fifteen minutes after transfection, the relative amounts of pDNA pulled down with the antibodies against cGAS and DDX60 did not differ from the negative control, while the antibodies against DHX9 and DAI/ZPB1 proteins pulled down ∼8-fold more pDNA copies (Figure 5E). Surprisingly, the amount of pDNA pulled down with the antibody against p204 was not significantly different from the negative control. No difference was observed in the quantification of the two targeted pDNA regions, implying that the pDNA molecules were intact. Plasmid DNA colocalization with DAI/ZBP1, which is a rare event, was confirmed by confocal microscopy at 15 min after transfection (Figure 5F).
siRNA
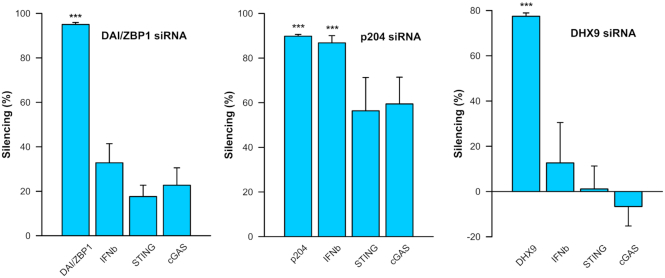
The mRNA and protein upregulation coupled with the confirmation of direct pDNA binding implied that specific sensors might play a part in the activation of the DNA sensor pathway. We used siRNA silencing to deduce the relative contributions of DAI/ZBP1, p204 and DHX9. Since the levels of IFN β and DDX60 were very low in control cells, we were unable to test these mRNAs reliably. Exposure of C2C12 cells to control siRNA alone did not significantly modulate any of the targets from background levels at 48 h (data not shown). Nevertheless, expression levels were normalized to control siRNA to exclude the effects of its presence in the cells.
Incubation of C2C12 cells with DAI/ZBP1, DHX9 or p204 siRNAs led to significant downregulation of the corresponding mRNAs with or without pDNA transfection. To investigate the importance of these sensors in the DNA sensing pathway, we tested the level of IFN β mRNA after pDNA electroporation of C2C12 cell treated with these siRNAs. No significant changes in IFN β expression were detected after treatment the cells with control siRNAs, while IFN β mRNA levels were downregulated after incubation with p204 siRNA by 87%. DAI/ZBP1 or DHX9 siRNA incubation did not affect IFN β mRNA levels. No siRNA tested in this study effected the mRNA levels of cGAS or STING after pDNA transfection (Figure 6).
Figure 6.
Effect of DAIZBP1, p204 and DHX9 silencing on the mRNA levels of IFN β, STING and cGAS. Levels of mRNA of cytosolic DNA sensors (DAI/ZBP1, p204 and DHX9), IFN β, STING and cGAS 4 h after transfection with pDNA in cells treated for 48 h with siRNAs as noted. Levels of mRNA were normalised to the mRNA levels in control cells treated with scrambled siRNA (n = 3 for p204 and n = 4 for DAI/ZBP1 and DHX9). ***P < 0.001 statistical difference from the same groups in cells treated with control, scrambled siRNA.
DISCUSSION
Cytosolic nucleic acid sensing may be a key component of innate immune sensing. Several proteins have been proposed as cytosolic DNA sensors over the last decade, but the detailed mechanisms of their actions have not been clarified. In this study, we sought to reveal early events upon pDNA entrance into the cell. After testing several candidate proteins, we identified DAI/ZBP1, p204, and DHX9 as potential early pDNA binding proteins (Figure 5). This conclusion is based on several findings: first, the levels of DAI/ZBP1 and p204 mRNAs and proteins were upregulated after pDNA transfection; second, these proteins were present in the cell cytoplasm at a basal level; and third, we directly demonstrated binding of these proteins to pDNA after transfection. Although DHX9 mRNA and protein were not upregulated, the basal level present in the cells was clearly adequate for the detection of pDNA binding. Taken together, these findings allow us to hypothesize that since these proteins bind pDNA soon after cell entry, they may be involved in the early events in DNA sensing.
Plasmid DNA can be delivered into cells by both physical and chemical methods. Since we sought to elucidate the immediate events after pDNA entrance into the cell, we needed a tool that could provide fast and effective pDNA transfection with a reliable time scale as to when the pDNA becomes available to intracellular proteins. Works from different groups demonstrated that pDNA is detectable in the cytosol or cytoplasm within 10 min of electroporation (19–21), which gave a good reference point for our study. Furthermore, this method mitigates the need for viral or chemical agents that could confound the source of sensor activation. To test the applicability of electroporation for our study, we performed confocal imaging of C2C12 cells 15 min after transfection. Transfected cells demonstrated an increased level of staining for cytoplasmic double-strand DNA, confirming these earlier reports.
We observed three major differences between two electroporation protocols that produced similar levels of transfection. First, the median fluorescence of transgene expression was significantly different (Figure 2A), implying that EP1 delivered a greater plasmid copy number to the nucleus, producing higher transgene expression. However, this does not account for plasmids reaching the cell's cytoplasm but unable to achieve nuclear entry for expression. Second, the protocols produced significant differences in cell viability (Figure 2B). Finally, these two protocols induced significantly different IFN β and DDX60 mRNA and protein upregulation (Figure 1). We previously noted that transfection efficiency was unrelated to the upregulation of mRNAs in this pathway (23). The protocol producing higher mRNA and protein levels was also the protocol producing increased cell death. Therefore, these events may be related.
These consequences of DNA electroporation may be a reflection of the mechanism by which DNA enters the cell. Similar to cationic lipids and polymers (40), endocytosis-like mechanisms are necessary for transgene expression after electroporation (20,41), although pDNA may enter by other mechanisms. After transfection, pDNA may reach the cytosol via defective endocytosis, errors during vesicle formation or transitions, or endosomal escape to bind and activate cytosolic DNA sensors. These effects may vary by cell type based on membrane composition (41), cell-specific endocytotic pathways (42), the speed of the specific endocytotic pathway (43), or the level of cytosolic nucleases (40). Variations in the parameters between protocols such as pulse number, length, applied field may influence the efficiency of pDNA entry or of endocytotic processing.
In previous studies, we demonstrated an increase in type I IFN and pro-inflammatory cytokine production after pDNA delivery, indicators of DNA detection in the cytosol, (23,24,37,44). We also observed an upregulation of several cytosolic DNA sensing proteins. However, our studies were conducted on various tumor cell lines, which could have impaired physiology as well as modulated DNA sensing mechanisms. Thus, we decided to clarify early cytosolic DNA sensing events in non-tumor cells.
In the present study, the increase in IFN β production confirmed that the sensing events observed in tumor cells also take place in C2C12 myoblasts. We found a dramatic increase in the mRNA levels of DDX60 and DAI/ZBP1 and a small but significant increase in p204 mRNA after transfection. An incremental, but significant increase in DAI/ZBP1 and p204 protein levels was also detected. Despite a >400-fold increase in DDX60 mRNA level after delivery with EP2, this upregulation was not reflected in an increased protein level at the specific time point measured (45). Interestingly, these were the same DNA sensors upregulated in tumor cells, although to differing extents (23,24). We confirmed these observations using another method of non-viral delivery, a non-liposomal formulation comprised of a lipid and a protein/polyamine mixture (Figure 1B, D). These results parallel previous observations of IFN β and/or DNA sensor mRNA or protein upregulation after liposomal DNA delivery (7,14,39,46–48) in a broad range of cell types (4). Overall, these findings suggest the existence of a general activation pattern in both tumor and non-tumor cells.
Reasonably, the first step in the cascade of events that comprise DNA sensing in the cytosol is the binding of a sensing protein or proteins to DNA. To accomplish that, the protein should be immediately available in the cytoplasm of the cell. We therefore tested control cells for the presence of proteins associated with detectable mRNAs: DAI/ZPB1, DDX60, p204, cGAS, DHX9, Ku70 and DDX41. CREB was included in the tested panel for its known ability to bind pDNA (22), which made it a useful positive control for our study. With the exception of DDX41, these proteins were detected in the cytoplasmic fraction of C2C12 cells (Supplementary Figure S1), potentially in adequate abundance to initiate a signaling cascade culminating in IFN β mRNA upregulation, which occurs before upregulation any of the DNA sensors tested (Figure 3).
To determine the proteins that initially bind pDNA, we performed pDNA pull downs from transfected cells. Since the binding process could potentially be a dynamic event, we tested several time points, specifically 15 min, 30 min, 1 h and 4 h after transfection. The positive control, CREB, was bound to pDNA at each time point, comparable to the results reported for CREB binding to the CMV-IE promotor/enhancer by Dean's group (22). Among the tested proteins, we found that DAI/ZBP1, DHX9 and p204 bound pDNA at each time point, while we could not detect DDX60 or Ku70 binding (Figure 5). Both p204 and DHX9 demonstrated fluctuating binding levels, peaking at 15 and 30 min, respectively, with decreased binding with time. Conversely, DAI/ZBP1 binding did not change significantly over the tested time course.
The differences in detection levels and kinetics could potentially be explained by the widely varying number of putative consensus binding sites in gWizGFP for the different proteins. Plasmid DNA sequence analysis revealed five binding regions for CREB within CMV-IE promoter/enhancer (29,30). A single Z-DNA motif (31) might be potentially bound by DAI/ZBP1, which prefers longer DNA such as plasmids (49). Up to 28 CpG-B motifs, potentially bound by DHX9 (33–36) were detected, while p204 may bind any long double stranded DNA without sequence specificity (32). Plausibly, the single binding site for DAI/ZBP1 may become almost immediately occupied when the plasmid reaches the cytoplasm, while the more abundant occupancy opportunities for CREB and DHX9 take some time to be filled. The decreasing levels of p204 and DHX9 binding may be due to binding by an unknown protein, forming a complex that shields the antibody epitope, or these proteins may be released from the plasmid after the activation of the signaling cascade.
To confirm pDNA binding, we performed protein immunoprecipitations after transfection. Anti-CREB, anti-DAI/ZBP1, and anti-DHX9 antibodies pulled down a significant amount of pDNA, confirming protein-DNA binding. Surprisingly, the amount of pDNA pulled down by anti-p204 antibody did not significantly differ from the negative control. This discrepancy in p204 binding detection between the two methods could be due to the previously observed variations in binding affinity of p204 to pDNA based on the salt concentration in the assay (32). Indeed, different reagents were used in the pDNA and protein pull down assays. Another interesting observation from the protein immunoprecipitation assay was that anti-CREB, anti-DAI/ZBP1 and anti-DHX9 antibodies pulled down pDNA with similar efficiency regardless of the number of putative binding sites. With the protein immunoprecipitations, only one pDNA copy could be pulled down no matter how many protein-binding sites were occupied.
In addition to CREB, nearly 500 proteins were demonstrated to bind pDNA 15 min after entry into the cytosol (22). However, we confirmed pDNA binding for only CREB and DHX9 (ATP-dependent RNA helicase A). The discrepancies between the results of these studies could be explained by the difference in cell types, pDNA concentration, or other experimental conditions. Badding et al. used a human tumor cell line and electroporated attached cells, while we used a non-tumor murine cell line and electroporated cells in suspension. Another source of discrepancy could be formaldehyde cross-linking, which, as the authors stated, can modify the mass of tryptic peptides, leading to misinterpretation of mass-spectrometry data (22).
Although cytosolic DDX60 protein was abundant in this study, we did not detect pDNA binding by this protein. However, the upregulation of the DDX60 mRNA 4 h after the transfection implicates its participation in the cascade of events initiated by the presence of pDNA in the cytosol. DDX60 was previously demonstrated to associate with RIG-I protein after vesicular stomatitis virus infection, but not in its absence (50). Hence, one could speculate that DDX60 plays a redundant role in cytosolic DNA sensing to ensure accurate and thorough detection of invading nucleic acids. Interestingly, DHX9, which bound pDNA in our study, belongs to the family of RIG-like helicases, supporting the idea of DDX60 role as an auxiliary protein in pDNA sensing. Further studies are required to clarify the details of DDX60 function.
DAI/ZBP1 was originally described as a tumor-associated protein with unknown function specifically recognizing Z-DNA (51). It was later reported as a cytosolic double-strand DNA sensor activating IRF3 and inducing type I IFN expression (7), although redundant sensors were later described (8,13,49). Recently, several groups (52–55) identified DAI/ZBP1 as an innate sensor of influenza A virus and demonstrated Z-RNA binding and colocalization with viral ribonucleoprotein complexes formed by influenza A virus. In our study, we confirmed DAI/ZPB1 bound pDNA by both plasmid and protein pull-down assays and by confocal microscopy after transfection.
DHX9 is a DExD/H-box helicase that was initially shown to play a crucial role in HIV replication (56), further described as a central factor in replication of foot-and-mouth disease virus (57) and influenza A virus (58). Later it was identified as a sensor for both CpG DNA and double-strand RNA in the cell cytoplasm (33,59) and a potentially important protein for viral replication and anti-viral immunity. We confirmed that DHX9 bound throughout the four hour period tested, peaking at 30 min.
p204 belongs to the interferon-inducible p200 family proteins, known as PYHIN or HIN-200 proteins. Initially, it was studied for its antiproliferative role (60) and for the regulation of cell differentiation, in particular, during the differentiation of C2C12 myoblasts into skeletal muscle type myotubes (61). Later it was shown, together with its ortholog IFI16, to be the first PYHIN protein, involved in IFN β induction, and therefore, having a role in cytosolic DNA sensing (46). We demonstrated that p204 bound pDNA, for minimally 4 h, peaking at 15 min.
Silencing experiments using siRNA implied that p204, but not DAI/ZBP1 or DHX9, partially modulated IFN β mRNA expression (Figure 6). This confirmed studies by Unterholzner et al., who demonstrated that IFI16 or p204 siRNA inhibited DNA-induced IFN β (46). Silencing of these mRNAs did not modulate cGAS or STING mRNA levels, however. We also confirmed the interferon inducibility of DAI/ZBP1 (7) and p204 (46), which may explain the subsequent increases in mRNA levels of these proteins (Figure 3) by a positive feedback loop.
In summary, our results indicate that DAI/ZBP1, p204 and DHX9 directly bind cytosolic plasmid DNA after transfection. Silencing of the DNA sensor p204 modulated IFN β expression, indicating it acts as an early cytosolic DNA sensor in C2C12 mouse myoblasts. However, we were unable to demonstrate a function in DNA sensing by DAI/ZBP1 or DHX9 in our model. This does not rule out these proteins functioning in DNA sensing in other cell types and other physiological conditions. Furthermore, alternative DNA sensors may be required simultaneously or at other time points to elicit the observed pro-inflammatory response.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute of the National Institutes of Health [R01CA196796 to L.H. and M.C.]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Funding for open access charge: NIH R01CA196796 (NCI).
Conflict of interest statement. None declared.
REFERENCES
- 1. Takaoka A., Taniguchi T.. Cytosolic DNA recognition for triggering innate immune responses. Adv. Drug. Deliv. Rev. 2008; 60:847–857. [DOI] [PubMed] [Google Scholar]
- 2. McNab F., Mayer-Barber K., Sher A., Wack A., O’Garra A.. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015; 15:87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kawai T., Akira S.. Innate immune recognition of viral infection. Nat. Immunol. 2006; 7:131–137. [DOI] [PubMed] [Google Scholar]
- 4. Stetson D.B., Medzhitov R.. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006; 24:93–103. [DOI] [PubMed] [Google Scholar]
- 5. Ishii K.J., Coban C., Kato H., Takahashi K., Torii Y., Takeshita F., Ludwig H., Sutter G., Suzuki K., Hemmi H. et al.. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. 2006; 7:40–48. [DOI] [PubMed] [Google Scholar]
- 6. Desmet C.J., Ishii K.J.. Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nat. Rev. Immunol. 2012; 12:479–491. [DOI] [PubMed] [Google Scholar]
- 7. Takaoka A., Wang Z., Choi M.K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K. et al.. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007; 448:501–505. [DOI] [PubMed] [Google Scholar]
- 8. Dempsey A., Bowie A.G.. Innate immune recognition of DNA: a recent history. Virology. 2015; 479–480:146–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Unterholzner L. The interferon response to intracellular DNA: why so many receptors. Immunobiology. 2013; 218:1312–1321. [DOI] [PubMed] [Google Scholar]
- 10. Paludan S.R., Bowie A.G.. Immune sensing of DNA. Immunity. 2013; 38:870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thaiss C.A., Levy M., Itav S., Elinav E.. Integration of Innate Immune Signaling. Trends Immunol. 2016; 37:84–101. [DOI] [PubMed] [Google Scholar]
- 12. Chen Q., Sun L., Chen Z.J.. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016; 17:1142–1149. [DOI] [PubMed] [Google Scholar]
- 13. Yoh S.M., Schneider M., Seifried J., Soonthornvacharin S., Akleh R.E., Olivieri K.C., De Jesus P.D., Ruan C., de Castro E., Ruiz P.A. et al.. PQBP1 Is a proximal sensor of the cGAS-dependent innate response to HIV-1. Cell. 2015; 161:1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jonsson K.L., Laustsen A., Krapp C., Skipper K.A., Thavachelvam K., Hotter D., Egedal J.H., Kjolby M., Mohammadi P., Prabakaran T. et al.. IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat. Commun. 2017; 8:14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan Y.K., Gack M.U.. Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 2016; 14:360–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kawakami S., Higuchi Y., Hashida M.. Nonviral approaches for targeted delivery of plasmid DNA and oligonucleotide. J. Pharm. Sci. 2008; 97:726–745. [DOI] [PubMed] [Google Scholar]
- 17. Neumann E., Schaefer-Ridder M., Wang Y., Hofschneider P.H.. Gene transfer into mouse lyoma cells by electroporation in high electric fields. EMBO J. 1982; 1:841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stewart M.P., Sharei A., Ding X., Sahay G., Langer R., Jensen K.F.. In vitro and ex vivo strategies for intracellular delivery. Nature. 2016; 538:183–192. [DOI] [PubMed] [Google Scholar]
- 19. Golzio M., Teissie J., Rols M.P.. Cell synchronization effect on mammalian cell permeabilization and gene delivery by electric field. Biochim. Biophys. Acta. 2002; 1563:23–28. [DOI] [PubMed] [Google Scholar]
- 20. Wu M., Yuan F.. Membrane binding of plasmid DNA and endocytic pathways are involved in electrotransfection of mammalian cells. PLoS One. 2011; 6:e20923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rosazza C., Buntz A., Riess T., Woll D., Zumbusch A., Rols M.P.. Intracellular tracking of single-plasmid DNA particles after delivery by electroporation. Mol. Ther. 2013; 21:2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Badding M.A., Lapek J.D., Friedman A.E., Dean D.A.. Proteomic and functional analyses of protein-DNA complexes during gene transfer. Mol. Ther. 2013; 21:775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Znidar K., Bosnjak M., Cemazar M., Heller L.C.. Cytosolic DNA Sensor Upregulation Accompanies DNA Electrotransfer in B16.F10 Melanoma Cells. Mol. Ther. Nucleic Acids. 2016; 5:e322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Znidar K., Bosnjak M., Semenova N., Pakhomova O., Heller L., Cemazar M.. Tumor cell death after electrotransfer of plasmid DNA is associated with cytosolic DNA sensor upregulation. Oncotarget. 2018; 9:18665–18681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cemazar M., Sersa G., Wilson J., Tozer G.M., Hart S.L., Grosel A., Dachs G.U.. Effective gene transfer to solid tumors using different nonviral gene delivery techniques: electroporation, liposomes, and integrin-targeted vector. Cancer Gene Ther. 2002; 9:399–406. [DOI] [PubMed] [Google Scholar]
- 26. Heller R., Jaroszeski M., Atkin A., Moradpour D., Gilbert R., Wands J., Nicolau C.. In vivo gene electroinjection and expression in rat liver. FEBS Lett. 1996; 389:225–228. [DOI] [PubMed] [Google Scholar]
- 27. Forster A.C., McInnes J.L., Skingle D.C., Symons R.H.. Non-radioactive hybridization probes prepared by the chemical labelling of DNA and RNA with a novel reagent, photobiotin. Nucleic Acids Res. 1985; 13:745–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McInnes J.L., Forster A.C., Skingle D.C., Symons R.H.. Preparation and uses of photobiotin. Methods Enzymol. 1990; 184:588–600. [DOI] [PubMed] [Google Scholar]
- 29. Messeguer X., Escudero R., Farre D., Nunez O., Martinez J., Alba M.M.. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002; 18:333–334. [DOI] [PubMed] [Google Scholar]
- 30. Farre D., Roset R., Huerta M., Adsuara J.E., Rosello L., Alba M.M., Messeguer X.. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003; 31:3651–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cer R.Z., Donohue D.E., Mudunuri U.S., Temiz N.A., Loss M.A., Starner N.J., Halusa G.N., Volfovsky N., Yi M., Luke B.T. et al.. Non-B DB v2.0: a database of predicted non-B DNA-forming motifs and its associated tools. Nucleic Acids Res. 2013; 41:D94–D100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morrone S.R., Wang T., Constantoulakis L.M., Hooy R.M., Delannoy M.J., Sohn J.. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E62–E71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim T., Pazhoor S., Bao M., Zhang Z., Hanabuchi S., Facchinetti V., Bover L., Plumas J., Chaperot L., Qin J. et al.. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:15181–15186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krieg A.M. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 2002; 20:709–760. [DOI] [PubMed] [Google Scholar]
- 35. Krieg A.M., Yi A.K., Matson S., Waldschmidt T.J., Bishop G.A., Teasdale R., Koretzky G.A., Klinman D.M.. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995; 374:546–549. [DOI] [PubMed] [Google Scholar]
- 36. Yu D., Kandimalla E.R., Zhao Q., Bhagat L., Cong Y., Agrawal S.. Requirement of nucleobase proximal to CpG dinucleotide for immunostimulatory activity of synthetic CpG DNA. Bioorg. Med. Chem. 2003; 11:459–464. [DOI] [PubMed] [Google Scholar]
- 37. Bosnjak M., Jesenko T., Kamensek U., Sersa G., Lavrencak J., Heller L., Cemazar M.. Electrotransfer of different control plasmids elicits different antitumor effectiveness in B16.F10 melanoma. Cancers. 2018; 10:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Badding M.A., Vaughan E.E., Dean D.A.. Transcription factor plasmid binding modulates microtubule interactions and intracellular trafficking during gene transfer. Gene Ther. 2012; 19:338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun L., Wu J., Du F., Chen X., Chen Z.J.. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013; 339:786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cervia L.D., Chang C.C., Wang L., Yuan F.. Distinct effects of endosomal escape and inhibition of endosomal trafficking on gene delivery via electrotransfection. PLoS One. 2017; 12:e0171699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rosazza C., Phez E., Escoffre J.M., Cezanne L., Zumbusch A., Rols M.P.. Cholesterol implications in plasmid DNA electrotransfer: Evidence for the involvement of endocytotic pathways. Int. J. Pharm. 2012; 423:134–143. [DOI] [PubMed] [Google Scholar]
- 42. Mao M., Wang L., Chang C.C., Rothenberg K.E., Huang J., Wang Y., Hoffman B.D., Liton P.B., Yuan F.. Involvement of a Rac1-Dependent Macropinocytosis Pathway in Plasmid DNA Delivery by Electrotransfection. Mol. Ther. 2017; 25:803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chang C.C., Wu M., Yuan F.. Role of specific endocytic pathways in electrotransfection of cells. Mol. Ther. Methods Clin. Dev. 2014; 1:14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Heller L.C., Cruz Y.L., Ferraro B., Yang H., Heller R.. Plasmid injection and application of electric pulses alter endogenous mRNA and protein expression in B16.F10 mouse melanomas. Cancer Gene Ther. 2010; 17:864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vogel C., Marcotte E.M.. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012; 13:227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S., Sirois C.M., Jin T., Latz E., Xiao T.S. et al.. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010; 11:997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Z., Yuan B., Bao M., Lu N., Kim T., Liu Y.J.. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011; 12:959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li T., Diner B.A., Chen J., Cristea I.M.. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:10558–10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Z., Choi M.K., Ban T., Yanai H., Negishi H., Lu Y., Tamura T., Takaoka A., Nishikura K., Taniguchi T.. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:5477–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miyashita M., Oshiumi H., Matsumoto M., Seya T.. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol. Cell Biol. 2011; 31:3802–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schwartz T., Behlke J., Lowenhaupt K., Heinemann U., Rich A.. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 2001; 8:761–765. [DOI] [PubMed] [Google Scholar]
- 52. Thapa R.J., Ingram J.P., Ragan K.B., Nogusa S., Boyd D.F., Benitez A.A., Sridharan H., Kosoff R., Shubina M., Landsteiner V.J. et al.. DAI Senses Influenza A Virus Genomic RNA and Activates RIPK3-Dependent Cell Death. Cell Host Microbe. 2016; 20:674–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Maelfait J., Liverpool L., Bridgeman A., Ragan K.B., Upton J.W., Rehwinkel J.. Sensing of viral and endogenous RNA by ZBP1/DAI induces necroptosis. EMBO J. 2017; 36:2529–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kuriakose T., Man S.M., Subbarao Malireddi R.K., Karki R., Kesavardhana S., Place D.E., Neale G., Vogel P., Kanneganti T.D.. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 2016; 1:aag2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kesavardhana S., Kuriakose T., Guy C.S., Samir P., Malireddi R.K.S., Mishra A., Kanneganti T.D.. ZBP1/DAI ubiquitination and sensing of influenza vRNPs activate programmed cell death. J. Exp. Med. 2017; 214:2217–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li J., Tang H., Mullen T.M., Westberg C., Reddy T.R., Rose D.W., Wong-Staal F.. A role for RNA helicase A in post-transcriptional regulation of HIV type 1. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:709–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lawrence P., Rieder E.. Identification of RNA helicase A as a new host factor in the replication cycle of foot-and-mouth disease virus. J. Virol. 2009; 83:11356–11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lin L., Li Y., Pyo H.M., Lu X., Raman S.N., Liu Q., Brown E.G., Zhou Y.. Identification of RNA helicase A as a cellular factor that interacts with influenza A virus NS1 protein and its role in the virus life cycle. J. Virol. 2012; 86:1942–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang Z., Yuan B., Lu N., Facchinetti V., Liu Y.J.. DHX9 pairs with IPS-1 to sense double-stranded RNA in myeloid dendritic cells. J. Immunol. 2011; 187:4501–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gribaudo G., Ravaglia S., Caliendo A., Cavallo R., Gariglio M., Martinotti M.G., Landolfo S.. Interferons inhibit onset of murine cytomegalovirus immediate-early gene transcription. Virology. 1993; 197:303–311. [DOI] [PubMed] [Google Scholar]
- 61. Liu C., Wang H., Zhao Z., Yu S., Lu Y.B., Meyer J., Chatterjee G., Deschamps S., Roe B.A., Lengyel P.. MyoD-dependent induction during myoblast differentiation of p204, a protein also inducible by interferon. Mol. Cell Biol. 2000; 20:7024–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.