Abstract
Infertility is a major health problem affecting ~15% of couples worldwide. Except for cases involving readily detectable chromosome aberrations, confident identification of a causative genetic defect is problematic. Despite the advent of genome sequencing for diagnostic purposes, the preponderance of segregating genetic variants complicates identification of culprit genetic alleles or mutations. Many algorithms have been developed to predict the effects of ‘variants of unknown significance’, typically single nucleotide polymorphisms (SNPs), but these predictions are not sufficiently accurate for clinical action. As part of a project to identify population variants that impact fertility, we have been generating clustered regularly interspaced short palindromic repeats-Cas9 edited mouse models of suspect SNPs in genes that are known to be required for fertility in mice. Here, we present data on a non-synonymous (amino acid altering) SNP (rs140107488) in the meiosis gene Mnd1, which is predicted bioinformatically to be deleterious to protein function. We report that when modeled in mice, this allele (MND1K85M), which is present at an allele frequency of ~ 3% in East Asians, has no discernable effect upon fertility, fecundity or gametogenesis, although it may cause sex skewing of progeny from homozygous males. In sum, assuming the mouse model accurately reflects the impact of this variant in humans, rs140107488 appears to be a benign allele that can be eliminated or de-prioritized in clinical genomic analyses of infertility patients.
Keywords: infertility, meiosis, mouse modeling, CRISPR, variants of unknown significance, recombination
Introduction
Meiosis begins with programmed formation of double stranded breaks (DSBs) induced by the SPO11 protein (Gray and Cohen, 2016). Processing and repair of these DSBs via recombination is crucial for proper pairing and segregation of homologous chromosomes at the first meiotic division. The recombination repair process involves several steps, including resection of the 5′ ends, loading of proteins (including the replication protein A (RPA) complex) that protect the 3′ overhangs, replacement of RPA by the RecA-related recombinases RAD51 and DMC1, then invasion of the single stranded nucleoprotein filament into the homologous chromosome locus to form recombination intermediates called D-loops. Subsequent resolution of the recombination intermediates occurs via non-crossover and crossover recombination. The recombinase loading and D-loop formation depend upon a heterodimer of the meiotic nuclear divisions 1 (MND1) and HOP2 accessory proteins (Bugreev et al., 2014; Chen et al., 2004; Petukhova et al., 2005).
Mnd1 knockout mice of both sexes are sterile, causing an absence of oocytes and postmeiotic spermatids, respectively (Pezza et al., 2014). Most mutant spermatocytes are defective in homologous chromosome synapsis and DSB repair, leading to apoptosis. Nevertheless, 23% of spermatocytes exhibited extensive or complete synapsis and underwent HOP2-dependent DSB repair, although they were deficient in crossovers (chiasmata) that are crucial for normal chromosome disjunction (and thus, aneuploidy prevention) (Pezza et al., 2014).
We have been conducting a project to identify segregating infertility variants in human populations, using a strategy (Singh and Schimenti, 2015) that does not depend upon genome-wide association studies. Our approach begins with a list of genes known to be required for fertility, most of which (~700) have been identified by mutagenesis in mice (Schimenti and Handel, 2018). Next, the genes are scanned for the existence of non-synonymous single nucleotide polymorphisms (nsSNPs) that are predicted computationally to be deleterious to protein function. We focus on nsSNPs rather than those that cause frameshifts or premature stop codons because the latter two are clearly disruptive, whereas predicting the functional impact of nsSNPs depends upon inexact bioinformatic predictions. Third, an amino acid altered by an nsSNP must be conserved between humans and mice, thus allowing for modeling. Fourth, we select nsSNPs that are present at a substantial frequency in at least one population but not exceeding 2% since higher frequency infertility variants are unlikely to persist in populations. We typically use a range of 0.001–2%. Finally, we model the human amino acid change in mice by clustered regularly interspaced short palindromic repeats-Cas9 (CRISPR-Cas9) genome editing. Our ultimate goal is to produce a database of human variants that have been functionally evaluated. We anticipate such a database to be useful for accurately diagnosing genetic causes of infertility in individual patients, and thus to better inform potential clinical interventions or genetic counseling.
Here, we report here on the creation and analysis of a mouse model containing a predicted deleterious nsSNP (rs140107488) in MND1. This SNP is present at an allele frequency of 1.6% in East Asians (meaning that ~ 3.2% are carriers) and 0.14% in South Asians (gnomAD database). We find that mice bearing this SNP are fertile, and show no signs of defective meiosis.
Materials and Methods
Mouse generation and genotyping
The DNA template for making single guide RNA (sgRNA) was generated as described via overlap PCR (Varshney et al., 2015). The guide RNA sequence (all 5′ to 3′) corresponding to Mnd1 was AGCCTCCAACTTGCGCTTCC, embedded within the forward overlap primer GAAATTAATACGACTCACTATAGGAGCCTCCAACTTGCGCTTCCGTTTTAGAGCTAGAAATAGC. This was matched with a universal reverse primer: CAAAATCTCGATCTTTATCGTTCAATTTTATTCCGATCAGGCAATAGTTGAACTTTTTCACCGTGGCTCAGCCACGAAAA.
The DNA template produced by the overlap PCR was reverse-transcribed into RNA using Ambion MEGAshortscript T7 Transcription Kit (cat#AM1354), and purified using Qiagen MinElute columns (cat#28004). For pronuclear injection, the sgRNA (50 ng/μl), single stranded homologous recombination template (ssODN; 50 ng/μl, IDT Ultramer Service) and Cas9 mRNA (25 ng/μl, TriLink) were co-injected into zygotes (F1 hybrids between strains FVB/NJ and B6(Cg)-Tyrc-2J/J) then transferred into the oviducts of pseudopregnant females. The ssODN sequence was as follows: AAAGCACATCTCTTCATAAGGCGGTGACTTACCTGAGAGTTCAGGGCCTCTAACATGCGCTTCCTTGCATGAAGAGCCTTGCTCGGAAAAGCCCAATAGTAATTGGACGTCCCGA. Any founders potentially carrying a copy of the desired alteration were identified by PCR using primers flanking the targeting site, followed by Sanger sequencing. This typically generates a chromatogram reflecting mosaicism in founders, so each founder mouse with an apparent desired allele was mated to a wild-type FVB/NJ mouse, and the offspring genotyped for inheritance of the desired clean allele. One such F1 animal was used to establish the colony of Mnd1K85M mice used in this study.
Crude lysates for PCR were made from small tissue biopsies (tail, toe or ear punches), as described (Truett et al., 2000). Genotyping primers were as follows: 5′-GCGTTGAGCCCAAATAAGAA-3′ and 5′-GTGGATGACGGTATGGTTGA-3′. Cycling conditions were initial denaturation at 95° for 5 min, then 30 cycles of 95° for 30 s, 58° for 30 s, 72° for 30 s, and final elongation at 72° for 5 min. For identification of Mnd1 mutants, the 233 bp amplimer was digested with HaeIII, and run on an agarose gel. The mutant, but not (WT) product was cleaved into two fragments of 133 and 100 bp.
All animal use was conducted under protocol (2004–0038) to J.C.S. and approved by Cornell University’s Institutional Animal Use and Care Committee.
Immunocytochemistry of spermatocyte chromosomes
This was performed essentially as described previously, but summarized as follows (McNairn et al., 2017). To prepare surface spreads, testes were isolated from 8–12-week-old males, detunicated and minced in minimum essential medium. Spermatocytes were hypotonically swollen in 4.5% sucrose solution, lysed in 0.1% Triton X-100, 0.02% sodium dodecyl sulphate and 2% formalin. Slides were blocked for 1 h at room temperature in 5% goat serum in PBS, 0.1% Tween20. Primary antibodies and dilutions used were anti-SYCP3 (1:600, Abcam, Cambridge, MA, USA #ab15093), anti-DMC1 (1:100, Abcam, #ab11054) and anti-MLH1 (1:100, BD Pharmingen, Franklin Lakes, NJ, USA #554073). Immunolabeling was carried out overnight at 4°C. Secondary antibodies used were goat anti-mouse IgG AlexaFluor 488 (1:1000, ThermoFisher Scientific, Waltham, MA, USA #R37120) and goat anti-rabbit IgG AlexaFluor 594 (1:800, ThermoFisher Scientific, Waltham, MA, USA #R37117). Secondary antibodies were incubated for 1 h at room temperature. Foci were quantified using ImageJ with plugins Cell Counter (Kurt De Vos) and Nucleus Counter.
Sperm counts and histology
For sperm counts, cauda epididymides (both sides) were collected from 8-week-old males, minced in PBS, then incubated at 37°C for 10 min to allow sperm to swim out. Sperm solutions were diluted and counted on a hemocytometer.
For histological sections, testes were fixed in Bouin’s for 24 h, washed in 70% ethanol for 24 h, then embedded in paraffin. Sections were made at 6 μM thickness. Slides were stained with hematoxylin and eosin (H&E).
Statistical analysis
All plotting of data and Student’s t-test calculations were performed using Prism 7 (GraphPad Software, San Diego, CA, USA.).
Results
Selection of the MND1K85M, encoded by SNP variant rs140107488, as a potential infertility allele
In our ongoing project to identify and functionally validate segregating infertility alleles in human populations by mouse modeling, we select candidate nsSNPs in genes that are known to be required for fertility in mice, and which are computationally predicted to be pathogenic. rs140107488 encodes a lysine-to-methionine change at amino acid (AA) position 85 of human and mouse MND1. The human and mouse proteins are both 205 amino acids in length, being 90% identical and 94% similar. The SIFT, PolyPhen-2 and Mutation Assessor algorithms, which predict deleteriousness of an nsSNP based on physico-chemical properties of the substituted AA, potential structural impact and evolutionary conservation (Adzhubei et al., 2013; Kumar et al., 2009; Reva et al., 2007), all predict this variant to be highly deleterious to protein function. The SIFT, PolyPhen-2 and Mutation Assessor algorithm scores for rs140107488 are, respectively, 0 (the ‘worst’ possible), 0.963 (‘probably damaging’) and 0.933 (‘high’ likelihood of being damaging). Given these predictions, in conjunction with the knowledge that MND1 is essential for meiosis and exists as a significant minor allele in East Asians, we selected it to model in mice and assess the consequences in vivo.
Mnd1K85M/K85M mice are fertile and produce normal numbers of sperm
To generate mice modeling the human MND1K85M allele, CRISPR/Cas9-mediated genome editing in single-celled zygotes was performed. An ssODN was used as a repair template to introduce a nucleotide change into codon 85 via homologous recombination, causing this codon to encode methionine instead of lysine (Fig. 1A). Other silent changes were also introduced to facilitate genotyping (creation of a novel restriction enzyme site) and to reduce deletion mutations (mutation of the protospacer adjacent motif (PAM) site required for Cas9:sgRNA recognition) (Singh et al., 2015). A founder mouse transmitting the correct mutation was identified (Fig. 1B), the allele was backcrossed into FVB/NJ for at least two generations, then intercrossed to generate homozygous mice for phenotypic analysis.
Figure 1.
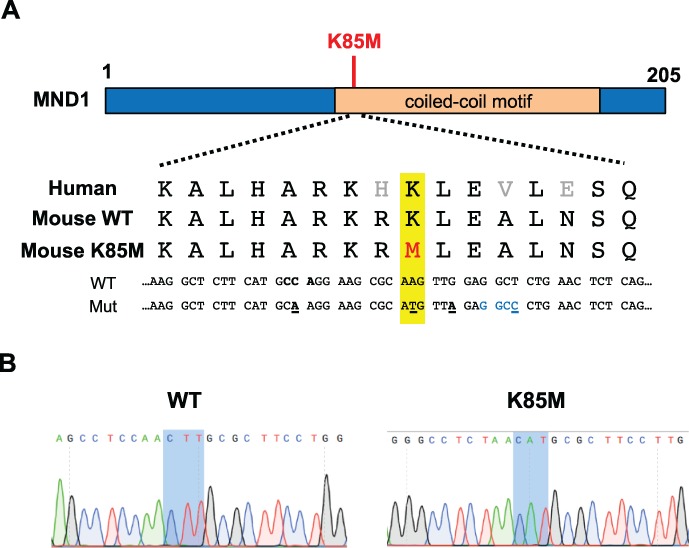
CRISPR/Cas9 genome editing strategy to generate mice Mnd1K85M. (A) Schematic of the human meiotic nuclear divisions 1 (MND1) protein and amino acid and nucleotide sequences surrounding the target single nucleotide polymorphism (SNP) in the human and mouse genomes. Point changes introduced by the single stranded homologous recombination (ssODN/HR) template are indicated as follows: yellow highlighted area contains the altered amino acid (M) in red, and the nucleotide that was mutated text/(underlined) to cause the amino acid change that models the human SNP variant; black text/underline = silent nucleotide changes to prevent Cas9 re-cleavage, including the PAM site (also bolded); blue text = HaeIII restriction enzyme site used for genotyping. K85M, MND1K85M. (B) Chromatograms of wild-type (WT) and Mnd1K85M/K85M mice (sequence is reverse complement of the sequences above).
Homozygous mutant mice were normal in gross appearance, consistent with the lack of somatic phenotypes reported for null mice (Pezza et al., 2014). To determine if the Mnd1K85M allele impacts fertility, homozygotes and wild-type (WT; +/+ or heterozygotes from the same genetic background) adults between 2 and 7 months of age were bred to WT mates, and litter sizes were recorded. The average litter sizes produced by Mnd1K85M/K85M males and females (Fig. 2A) were 8.2 (n = 123 pups) and 8.6 (n = 95 pups), respectively, compared to 8.3 for WT intercrosses (n = 58 pups). Thus, fecundity of mutants was normal (P = 0.78 and 0.93 for females and males, respectively by Student’s t-test). Interestingly, however, male (but not female) homozygotes produced more male (82) than female (41) offspring (P = 0.000073 by Student’s t-test), a 2:1 ratio (Fig. 2B). The basis for this is unknown.
Figure 2.
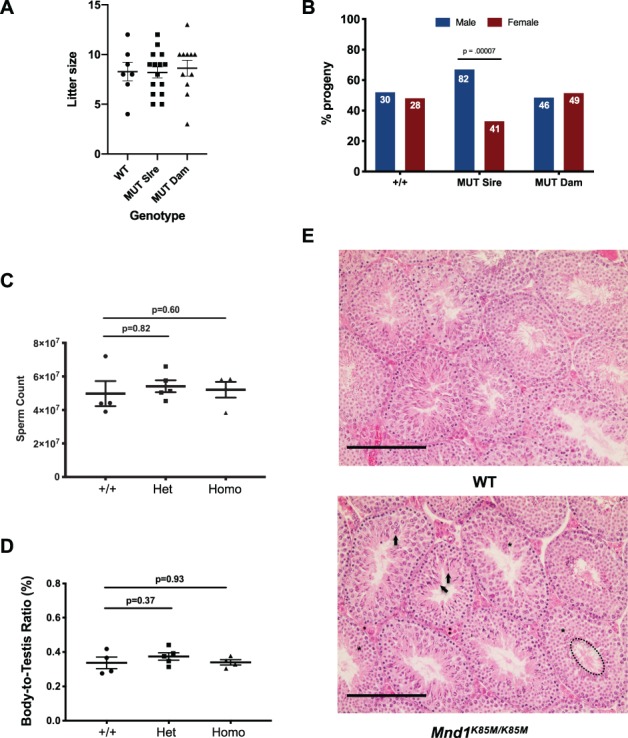
Mnd1K85M/K85M males have normal sperm counts and spermatogenesis, but exhibit sex distortion. (A) Litter sizes from matings of indicated genotypes of males or females to WT mates. WT mice were intercrossed, so no sex is indicated. MUT = Mnd1K85M/K85M. Each point represents a litter (males and females combined), and horizontal bars mark the average and SEM. (B) Progeny separated by sex of WT and Mnd1K85M/K85M (MUT) mice mated to WT animals. Note that mutant males (sires) produce a 2:1 ratio of male:female offspring. The P value is from Student’s t-test. (C) Total sperm quantified from both epididymides of each animal. N = 3–5 per genotype. P values are from Student’s t-test. Het: heterozygous, Homo: homozygous. (D) Relative body:testis masses. N = 4–5 per genotype. P values are from Student’s t-test. (E) Testis histology. Hematoxylin and eosin stained testis cross-sections of 8-week-old WT and mutant males. Highlighted in selected seminiferous tubules of the mutant sample are normal examples of elongated spermatids (arrows), round spermatids (asterisks) and flagellated, spermiated spermatozoa (dashed ellipse in lower right tubule). Size bar = 75 μM.
To determine if there were potential subclinical effects in sperm production or spermatogenesis in mutants, we performed gross and histological analyses of testes and sperm. Eight-week-old Mnd1K85M/K85M males had testis weights and epididymal sperm numbers that were similar to WT (Fig. 2C and D). Consistent with these observations, testis histological cross-sections appeared normal in the mutant; all spermatogenic stages were present, and no notable pathologies or abnormal cellular phenotypes were observed (Fig. 2E).
Meiosis and recombination appears to be normal in Mnd1K85M/K85M mice
MND1 is an important binding partner of HOP2, and this complex stabilizes DMC1 and RAD51 binding to ssDNA to promote strand invasion into, and recombination with, the homologous chromosome (Tourtellotte et al., 1999). To assess if MND1K85M affects DMC1 recruitment and displacement, spermatocyte chromosome surface spreads were immunolabeled with DMC1 and the synaptonemal complex axial element protein SYCP3 (Fig. 3A). In response to meiotically programmed DSBs, DMC1 loading was observed in foci that were present at normal levels (compared to WT; ~ 200 foci/nucleus) in leptotene spermatocytes (cells in which homologous chromosome pairing and synapsis has yet to occur). Focus numbers also disappeared normally as meiotic prophase I progressed into pachynema (when chromosome synapsis is complete), indicating that homologous recombination-mediated DSB repair was occurring without defects in the mutant (Fig. 3A). Unlike Mnd1 null spermatocytes, we did not observe defects in chromosome synapsis (Pezza et al., 2014). To assess crossing-over, late pachytene spreads were immunolabeled with the chiasmata marker MLH1. In contrast to null spermatocytes that have decreased crossing-over in surviving spermatocytes (Pezza et al., 2014), there was no significant difference in MLH1 focus numbers between WT and Mnd1K85M/K85M (P = 0.13) (Fig. 3B), indicating a normal level of crossing over. Aberrations in crossover number can lead to aneuploidy and death of offspring, typically resulting in reduced litter sizes, which is not the case with Mnd1K85M mutants, as indicated above.
Figure 3.
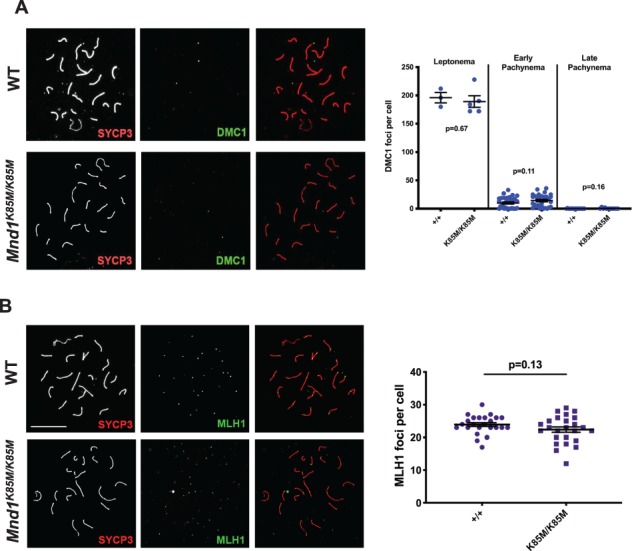
Cytological data indicating that Mnd1K85M/K85M is not defective in meiotic recombination. (A) Late pachytene spermatocyte chromosome surface spreads immunolabeled with synaptonemal complex axial element protein (SYCP3) and DMC1 (merged in rightmost subpanels). To the right of each panel are plots of DMC1 focus counts at indicated substages. N = 2 per genotype. P values are from Student’s t-test. (B) Spreads immunolabeled with MLH1 (a chiasmata marker protein) and SYCP3 (merged in rightmost subpanels). Size bar in WT panel of B = 20 μm, and applies to all panels. To the right of each panel are plots of the MLH1 focus counts at indicated substages. N = 3 per genotype. DMC1: dosage suppressor of mck1 homolog.
Discussion
In summary, our studies indicate that MND1K85M is a benign allele that has no discernable effect upon fertility. Homozygous mutants of both sexes were normal in both reproductive and molecular phenotypes. By eliminating this as a candidate infertility allele, these finding may be useful in prioritizing candidate genetic variants that may underlie reproductive infertility in people bearing this SNP. While it is conceivable that the allele is pathogenic in humans but not mice, the consequences are likely to be subtle or subclinical, especially since the allele is present at a relatively high frequency in East Asians. One remaining question is the unusual sex distortion ratio exhibited by allele transmission from homozygous males. At present, it is unclear whether this is a consequence of the mutant allele itself, a statistical rarity, or perhaps an unrelated closely linked allele. Further studies of this phenomenon are under way.
Acknowledgements
We thank R. Munroe and C. Abratte of Cornell’s transgenic facility for generating the edited mice.
Authors’ roles
T.T. conducted most of the experiments, wrote a draft of the text and figures and supervised J.M., who contributed to the genotyping and immunohistochemistry analyses. J.S. supervised the work and the aforementioned students, and prepared the manuscript.
Funding
National Institutes of Health (grant R01 HD082568 to J.C.S.); NY State Stem Cell Program (NYSTEM) (contract CO29155 to J.C.S.). J.M. was a student in the Molecular Biology and Genetics Research Experience for Undergraduate program, which was supported by the National Science Foundation (DBI1659534), the Department of Molecular Biology and Genetics, the Weill Institute of Cell and Molecular Biology and the Division of Nutritional Sciences at Cornell University.
Conflict of interest
None declared.
References
- Bugreev DV, Huang F, Mazina OM, Pezza RJ, Voloshin ON, Camerini-Otero RD, Mazin AV. HOP2-MND1 modulates RAD51 binding to nucleotides and DNA. Nat Commun 2014;5:4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-K, Leng C-H, Olivares H, Lee M-H, Chang Y-C, Kung W-M, Ti S-C, Lo Y-H, Wang AH-J, Chang C-S et al. Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc Natl Acad Sci U S A 2004;101:10572–10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Cohen PE. Control of meiotic crossovers: from double-strand break formation to designation. Annu Rev Genet 2016;50:175–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNairn AJ, Rinaldi VD, Schimenti JC. Repair of meiotic DNA breaks and homolog pairing in mouse meiosis requires a minichromosome maintenance (MCM) paralog. Genetics 2017;205:529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova GV, Pezza RJ, Vanevski F, Ploquin M, Masson J-Y, Camerini-Otero RD. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat Struct Mol Biol 2005;12:449–453. [DOI] [PubMed] [Google Scholar]
- Pezza RJ, Voloshin ON, Volodin AA, Boateng KA, Bellani MA, Mazin AV, Camerini-Otero RD. The dual role of HOP2 in mammalian meiotic homologous recombination. Nucleic Acids Res 2014;42:2346–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Schimenti JC, Bolcun-Filas E. A mouse geneticist’s practical guide to CRISPR applications. Genetics 2015;199:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourtellotte WG, Nagarajan R, Auyeung A, Mueller C, Milbrandt J. Infertility associated with incomplete spermatogenic arrest and oligozoospermia in Egr4-deficient mice. Development 1999;126:5061–5071. [DOI] [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 2000;29:52–54. [DOI] [PubMed] [Google Scholar]
- Varshney GK, Pei W, LaFave MC, Idol J, Xu L, Gallardo V, Carrington B, Bishop K, Jones M, Li M et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res 2015;25:1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]


