Abstract
Summary
Human alpha satellite and satellite 2/3 contribute to several percent of the human genome. However, identifying these sequences with traditional algorithms is computationally intensive. Here we develop dna-brnn, a recurrent neural network to learn the sequences of the two classes of centromeric repeats. It achieves high similarity to RepeatMasker and is times faster. Dna-brnn explores a novel application of deep learning and may accelerate the study of the evolution of the two repeat classes.
Availability and implementation
1 Introduction
Eukaryotic centromeres consist of huge arrays of tandem repeats, termed satellite DNA (Garrido-Ramos, 2017). In human, the two largest classes of centromeric satellites are alpha satellite (alphoid) with a 171 bp repeat unit, and satellite II/III (hsat2,3) composed of diverse variations of the ATTCC motif. They are totaled a couple of hundred megabases in length (Schneider et al., 2017). Both alphoid and hsat2,3 can be identified with RepeatMasker (Tarailo-Graovac and Chen, 2009), which is alignment based and uses the TRF tandem repeat finder (Benson, 1999). However, RepeatMasker is inefficient. Annotating a human long-read assembly may take days; annotating high-coverage sequence reads is practically infeasible. In addition, RepeatMasker requires RepBase (Kapitonov and Jurka, 2008), which is not commercially free. This further limits its uses.
We reduce repeat annotation to a classification problem and solve the problem with a recurrent neural network (RNN), which can be thought as an extension to non-profile hidden Markov model but with long-range memory. Because the repeat units of alphoid and hsat2,3 are short, RNN can ‘memorize’ their sequences with a small network and achieve high performance.
2 Materials and methods
2.1 The dna-brnn model
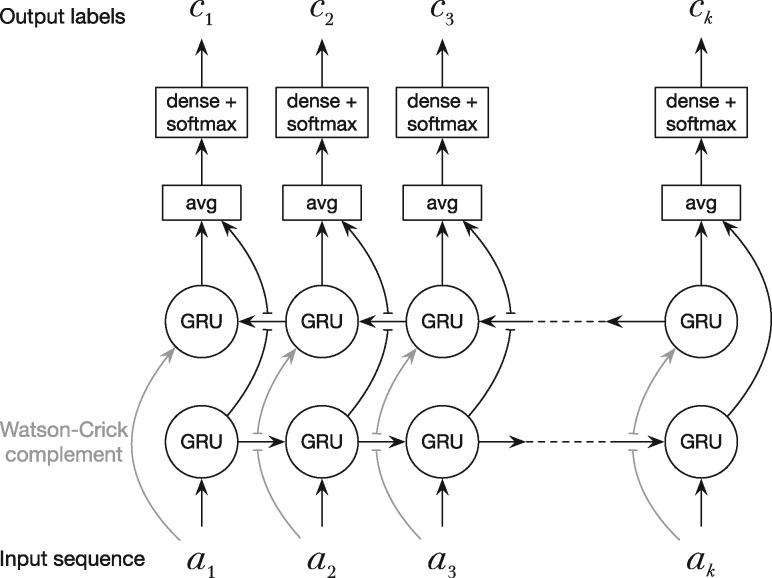
Given m types of non-overlapping features on a DNA sequence, we can label each base with number , where ‘0’ stands for a null-feature. Dna-brnn learns how to label a DNA sequence. Its overall architecture (Fig. 1) is similar to an ordinary bidirected RNN (BRNN), except that dna-brnn feeds the reverse complement sequence to the opposite array of Gated Recurrent Units (GRUs) and that it ties the weights in both directions. Dna-brnn is strand symmetric in that the network output is the same regardless of the input DNA strand. The strand symmetry helps accuracy (Shrikumar et al., 2017). Without weight sharing between the two strands, we will end up with a model with twice as many parameters but 16% worse training cost (averaged in 10 runs).
Fig. 1.
The dna-brnn model. Dna-brnn takes a k-long one-hot encoded DNA sequence as input. It feeds the input and its reverse complement to two GRU arrays running in the opposite directions. At each position, dna-brnn averages the two GRU output vectors, transforms the dimension of the average with a dense layer and applies softmax. The final output is the predicted distribution of labels for each input base. All GRUs in both directions share the same weights
In theory, we can directly apply dna-brnn to arbitrarily long sequences. However, given a single sequence or multiple sequences of variable lengths, it is non-trivial to implement advanced parallelization techniques and without parallelization, the practical performance would be tens of times slower. As a tradeoff, we apply dna-brnn to 150 bp subsequences and discard information in a longer range.
To identify satellites, we assign label ‘1’ to hsat2,3 and label ‘2’ to alphoid. The size of the GRU hidden vector is 32. There are <5000 free parameters in such a model.
2.2 Training and prediction
In training, we randomly sampled 256 subsequences of 150 bp in length and updated the model weights with RMSprop on this minibatch. To reduce overfitting, we randomly dropped out 25% elements in the hidden vectors. We terminated training after processing 250 Mb randomly sampled bases. We generated ten models with different random seeds and manually selected the one with the best accuracy on the validation data.
On prediction, we run the model in each 150 bp long sliding window with 50 bp overlap. In each window, the label with the highest probability is taken as the preliminary prediction. In an overlap between two adjacent windows, the label with higher probability is taken as the prediction. Such a prediction algorithm works well in long arrays of satellites. However, it occasionally identifies satellites of a few bases when there is competing evidence. To address this issue, we propose a post-processing step.
With the previous algorithm, we can predict label ci and its probability pi at each sequence position i. We introduce a score si which is computed as
| (1) |
Here si is usually positive at a predicted satellite base and negative at a non-satellite base. Let be the sum of scores over segment . Intuitively, we say is maximal if it cannot be lengthened or shortened without reducing . Ruzzo and Tompa (1999) gave a rigorous definition of maximal scoring segment (MSS) and a linear algorithm to find all of them. By default, dna-brnn takes an MSS longer than 50 bp as a satellite segment. The use of MSS effectively clusters fragmented satellite predictions and improves the accuracy in practice.
2.3 Training and testing data
The training data come from three sources: chromosome 11, annotated alphoids in the reference genome and the decoy sequences, all for GRCh37. RepeatMasker annotations on GRCh37 were acquired from the UCSC Genome Browser. Repeats on the GRCh37 decoy were obtained by us with RepeatMasker (v4.0.8 with rmblast-2.6.0+ and the human section of RepBase-23.11). RepeatMasker may annotate hsat2,3 as ‘HSATII’, ‘(ATTCC)n’, ‘(GGAAT)n’, ‘(ATTCCATTCC)n’ or other rotations of the ATTCC motif. We combined all such repeats into hsat2,3. We take the RepeatMasker labeling as the ground truth.
For validation, we annotated the GRCh38 decoy sequences (Mallick et al., 2016) with RepeatMasker and used that to tune hyperparameters such as the size of GRU and non-model parameters in Equation (1), and to evaluate the effect of random initialization. For testing, we annotated the CHM1 assembly (AC: GCA_001297185.1) with RepeatMasker as well. Testing data do not overlap training or validation data.
For measuring the speed of RepeatMasker, we used a much smaller repeat database, composed of seven sequences (‘HSATII’, ‘ALR’, ‘ALR_’, ‘ALRa’, ‘ALRa_’, ‘ALRb’ and ‘ALRb_’) extracted from the prepared RepeatMasker database. We used option ‘-frag 300000 -no_is’ as we found this achieves the best performance. The result obtained with a smaller database is slightly different from that with a full database because RepeatMasker resolves overlapping hits differently.
2.4 Implementation
Unlike mainstream deep learning tools which are written in Python and depend on heavy frameworks such as TensorFlow, dna-brnn is implemented in C, on top of the lightweight KANN framework that we developed. KANN implements generic computation graphs. It uses CPU only, supports parallelization and has no external dependencies. This makes dna-brnn easily deployed without requiring special hardware or software settings.
3 Results
Training dna-brnn takes 6.7 wall-clock minutes using 16 CPUs; predicting labels for the full CHM1 assembly takes 56 min. With 16 CPUs, RepeatMasker is 5.3 times as slow in CPU time, but 17 times as slow in real time, likely because it invokes large disk I/O and runs on a single CPU to collate results. Table 1 shows the testing accuracy with different prediction strategies. Applying MSS clustering improves both false negative rate (FNR) and false positive rate (FPR). We use the ‘mss: Y, minLen: 50’ setting in the rest of this section.
Table 1.
Evaluation of dna-brnn accuracy
| Alphoid |
hsat2,3 |
|||
|---|---|---|---|---|
| Setting | FNR (%) | FPR | FNR (%) | FPR |
| mss: N, minLen: 0 | 0.42 | 1/9952 | 0.42 | 1/4086 |
| mss: N, minLen: 50 | 0.59 | 1/28 908 | 0.68 | 1/4639 |
| mss: Y, minLen: 50 | 0.33 | 1/44 095 | 0.30 | 1/4370 |
| mss: Y, minLen: 200 | 0.36 | 1/60 078 | 0.50 | 1/6010 |
| mss: Y, minLen: 500 | 0.48 | 1/69 825 | 0.85 | 1/10 505 |
Note: RepeatMasker annotations on the CHM1 assembly (3.0 Gb in total, including 55 Mb alphoid and 50 Mb hsat2,3) are taken as the ground truth. ‘mss’: whether to cluster predictions with MSSs. ‘minLen’: minimum satellite length. ‘FNR’: false negative rate, the fraction of RepeatMasker annotated bases being missed by dna-brnn. ‘FPR’: false positive rate, the fraction of non-satellite bases being predicted as satellite by dna-brnn. A format ‘’ in the table implies one false positive prediction per x-bp.
Dna-brnn takes ∼1.5 days on 16 threads to process whole-genome short or long reads sequenced to 30-fold coverage. For the NA24385 CCS dataset (Wenger et al., 2019), 2.91% of bases are alphoid and 2.56% are hsat2,3. If we assume the human genome is 3 Gb in size, these two classes of satellites amount to 164 Mb per haploid genome. The CHM1 assembly contains 105 Mb hsat2,3 and alphoid, though 70% of them are in short contigs isolated from non-repetitive regions. In the reference genome GRCh37, both classes are significantly depleted (<0.3% of the genome). GRCh38 includes computationally generated alphoids but still lacks hsat2,3 (<0.1%). Partly due to this, 82% of human novel sequences found by Sherman et al. (2019) are hsat2,3. There are significantly less novel sequences in euchromatin.
We have also trained dna-brnn to identify the Alu repeats to high accuracy. Learning beta satellites, another class of centromeric repeats, is harder. We can only achieve moderate accuracy with larger hidden layers. Dna-brnn fails to learn the L1 repeats, which are longer, more divergent and more fragmented. We are not sure if this is caused by the limited capacity of dna-brnn or by innate ambiguity in the RepeatMasker annotation.
4 Conclusion
Dna-brnn is a fast and handy tool to annotate centromeric satellites on high-throughput sequence data and may help biologists to understand the evolution of these repeats. Dna-brnn is also a general approach to modeling DNA sequences. It can potentially learn other sequence features and can be easily adapted to different types of sequence classification problems.
Acknowledgements
We thank the second anonymous reviewer for pointing out an issue with our running RepeatMasker, which led to unfair performance comparison in an earlier version of this manuscript.
Funding
This work was supported by NHGRI R01-HG010040.
Conflict of Interest: none declared.
References
- Benson G. (1999) Tandem repeats finder: a program to analyze dna sequences. Nucleic Acids Res., 27, 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Ramos M.A. (2017) Satellite DNA: an evolving topic. Genes (Basel), 8, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov V.V., Jurka J. (2008) A universal classification of eukaryotic transposable elements implemented in Repbase. Nat. Rev. Genet., 9, 411–412. [DOI] [PubMed] [Google Scholar]
- Mallick S. et al. (2016) The Simons genome diversity project: 300 genomes from 142 diverse populations. Nature, 538, 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzo W.L., Tompa M. (1999) A linear time algorithm for finding all maximal scoring subsequences In: Proceedings of the Seventh International Conference on Intelligent Systems for Molecular Biology, August 6–10, 1999, Heidelberg, Germany, pp. 234–241. [PubMed] [Google Scholar]
- Schneider V.A. et al. (2017) Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res., 27, 849–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman R.M. et al. (2019) Assembly of a pan-genome from deep sequencing of 910 humans of African descent. Nat. Genet, 51, 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrikumar A. et al. (2017) Reverse-complement parameter sharing improves deep learning models for genomics. doi: 10.1101/103663.
- Tarailo-Graovac M., Chen N. (2009) Using repeatmasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics, 25, 4.10.1–4.10.14. [DOI] [PubMed] [Google Scholar]
- Wenger A.M. et al. (2019) Highly-accurate long-read sequencing improves variant detection and assembly of a human genome. doi: 10.1101/519025. [DOI] [PMC free article] [PubMed]