Abstract
Background
MicroRNAs [miRNAs] are key modulators of gene expression in Crohn’s disease [CD] and may drive tissue-specific molecular alterations underlying CD susceptibility. In this study, we analysed differential miRNA expression between CD and healthy subjects across ileal and colonic tissues.
Methods
A cohort of CD and healthy control [HC] subjects was recruited and clinical data collected. Endoscopically quiescent CD [CDq] was defined as inactive or mild by the Simple Endoscopic Score for CD. Total RNA was extracted from endoscopic biopsies taken from the terminal ileum and sigmoid colon. miRNA expression was quantified using NanoString Technologies. Statistical significance was assessed across biopsy site and diagnosis per miRNA, and corrected for multiple testing.
Results
In total, 23 CDq and 38 HC subjects were enrolled; 112 samples were included in the analysis, 51 from the ileum and 61 from the colon. We found 47 miRNAs differentially expressed by biopsy site in healthy tissue. Nine miRNAs were differentially expressed across HC and CDq, accounting for biopsy location. One of these, miR-223-3p, showed age and sex effects. We identified miRNA expression driven by diagnosis targeting genes involved in chemokine and cytokine signalling. miR-31-5p expression was driven by location and may be a biomarker for location subtypes in CD.
Conclusions
We identified differentially expressed miRNAs in healthy ileal and colonic tissues. We discovered spatial miRNA expression patterns in CD and HC, suggesting site-specific regulation in subjects with no or minimal intestinal inflammation. These miRNAs target genes involved in immunoregulatory processes, suggesting a functional, tissue-specific role in CD.
Keywords: Inflammatory bowel disease, miRNA, immune response
1. Introduction
Crohn’s disease [CD] is an inflammatory bowel disease [IBD] resulting in chronic inflammation in the gastrointestinal tract. Whereas the cause of CD is not fully understood, one prevailing hypothesis is that a genetically susceptible host produces an aberrant immune response to commensal microbes in the gut.1 Genome-wide association studies have identified over 200 IBD risk variants important in physiological pathways in this paradigm, such as innate mucosal immunity and cytokine signalling.2–4 Yet, the genetic polymorphisms identified to date only account for approximately one-quarter of the heritability of CD, and these risk variants are unable to explain much of the phenotypic expression of the disease.3,5 This discordance may be explained by examining other non-coding genetic, epigenetic, and environmental contributions to CD. Differential expression of non-coding, regulatory molecules altering expression levels of genes important in CD may therefore provide one mechanism contributing to CD pathogenesis. Pursuant to this, microRNAs [miRNAs], which bind to messenger RNAs [mRNAs] and modulate gene expression,6–8 may play a key regulatory role in the onset and course of IBD. Previous studies in IBD and healthy subjects have found differential expression of miRNAs in blood and tissue.6–13 Some of these miRNAs have also been shown to target mRNAs associated with increased risk of developing CD.14,15 We aimed to investigate the role of miRNAs in CD by studying location-specific differential expression of miRNAs in tissue of inactive or minimally inflamed CD subjects, as well as healthy controls [HC], to reduce the impact of inflammation on miRNA expression.
2. Materials and Methods
2.1. Subject cohort and sample collection
The cohort for this study was recruited at Mount Sinai Hospital in Toronto following approval from the hospital’s research ethics board [REB]. Adult subjects, ages 18–75 years, with [1] a confirmed diagnosis of CD based on endoscopic, histological and radiological evidence, or [2] healthy individuals undergoing routine screening for colorectal cancer showing no macroscopic or microscopic evidence of intestinal inflammation, were enrolled. Clinical and demographic information was collected [Table 1]. The clinical, histological and endoscopic metadata collected by expert IBD endoscopists were confirmed through additional chart review and use of a centralised IBD database. Those with known intercurrent inflammatory disorders including multiple sclerosis [MS], diabetes mellitus, and rheumatoid arthritis, and those with findings of polyps or cancer or any known concurrent infection were excluded. Full ileocolonoscopy with research biopsies acquired from the terminal ileum and sigmoid colon was performed. Endoscopically quiescent disease was defined as inactive or mild by the Simple Endoscopic Score for CD [SES-CD16; total SES-CD ≤11] and biospecimens from these subjects were included in the quiescent Crohn’s disease group [CDq]. Samples from the group undergoing routine screening were included as the healthy control [HC] group. Clinical and demographic characteristics were considered when selecting samples from a larger repository for miRNA expression profiling to reduce possible confounding effects, such as smoking and use of medications. To further minimise erroneous associations, we confirmed that the miRNAs reported as differentially expressed were not influenced by these confounders, and we included those that did have effect in our covariate analysis [such as age]. For some individuals, tissue data at both sites were not available, and they were not included in the paired analysis. The Montreal classification17 was used to define IBD subphenotypes by location of disease.
Table 1.
Demographic and clinical characteristics of study cohort.
| Feature | CDq [n = 23] | HC [n = 38] | p-value |
|---|---|---|---|
| Age at testing [years] | 34 ± 13 | 55 ± 9 | <0.001 |
| Age at diagnosis [years] | 24 ± 12 | N/A | N/A |
| Gender (% [n], male) | 35 [8] | 58 [22] | 0.1 |
| Stricturing (% [n]) | 17 [4] | N/A | N/A |
| Site (% [n]) | N/A | N/A | |
| L1 | 17 [4] | ||
| L2 | 39 [9] | ||
| L3 | 44 [10] | ||
| Treatment (% [n]) | N/A | N/A | |
| Biologic | 22 [5] | ||
| 5ASA | 4 [1] | ||
| Steroid | 0 | ||
| Antibiotic | 0 | ||
| CRP [mg/mL] | 5.1 ± 6.4 | 1.4 ± 1.3 | 0.01 |
p-values for continuous variables and nominal variables are calculated with two-tailed Welch’s t test and Fisher’s exact test, respectively.
CDq, quiescent Crohn’s disease; HC, healthy controls, N/A, not available; 5ASA, 5-aminosalicylates;CRP, C-reactive protein.
2.2. Total RNA extraction
Biopsies were collected at the time of endoscopy and immediately put in RNAlater stabilisation reagent [Qiagen]. Samples were frozen in the RNA stabilising solution and stored at -80°C until extracted. Total RNA was extracted with the miRNeasy kit [Qiagen] as per the manufacturer’s instructions.
2.3. miRNA expression profiling of ileal and colonic samples
Total RNA [100 ng] that passed quality control for each sample was randomly allocated for miRNA profiling by Nanostring [Nanostring Technologies, Seattle, WA].18 The samples included in this study were profiled as part of a larger cohort of patients comprising 22 batches altogether, with 12 samples per batch. Samples were randomised to minimise possible biasing effects due to phenotype, location of biopsy, gender, and age, and the miRNA expression profiling procedure was done as per manufacturer’s protocols [Nanostring Technologies, Seattle, WA]. Briefly, as per the Nanostring user manual [user manual found online 19], library preparation for small RNA expression profiling by Nanostring involves ligation of transcript tags onto miRNAs, using the nCounter miRNA Sample Preparation Kit [Nanostring Technologies]. The codeset used for this study was Nanostring’s Human v3 miRNA Panel containing 798 miRNA probes, after removing controls. After the ligation reaction, the miRNA tagged material was hybridised overnight with capture probes and reporter probes carrying fluorescent barcodes. These target-specific, barcoded reporter probe and capture probe complexes were purified and immobilised on a cartridge where the individual barcodes [each corresponding to a specific transcript] were counted on the nCounter Digital Analyzer [Nanostring Technologies]. The codeset used in this study included six positive spike-in controls per 12-strip tube. Raw counts were normalised using internal positive controls, as well as housekeeping genes provided in the manufacturer’s codeset, using the nSolver v2.5 software according to accompanying manuals.20 Background noise was assessed using eight negative control probes as described in the nSolver software guidelines.20 The NanoString miRNA expression assay was performed at the Princess Margaret Genomics Centre [Toronto, ON, Canada].
2.4. Differential miRNA expression analysis
Normalised tissue expression was filtered for low abundance miRNAs, where only miRNAs with counts across all cohort samples with a 70th percentile greater than 2 were included in the analysis. This resulted in 582 out of the original 798 miRNAs being included in the miRNA expression dataset in subsequent analysis [Supplementary Table 1, available as Supplementary data at ECCO-JCC online]. A log2 transformation was applied to normalised data, and corrected for technical batch effects using the package ComBat21 in R.3.1.1.
Since more than one biopsy location was included for each individual in this analysis, the differential miRNA expression between CDq and HC while accounting for location was analysed using a linear mixed-effects [LME] model across all miRNAs, using the package nlme in R.3.1.1. In this model, diagnosis and location were fixed effects [as factors] and Sample ID was a random effect: lme = lme[miRNA~Diagnosis + Location, rand = ~1|Sample, na.action = na.exclude]. Age [as numeric] and gender [as factor] were included as covariates in the lme model when assessing their effects on differential expression. Non-parametric Wilcoxon tests assessed differential miRNA expression in matched subject samples across sites. Non-parametric Kruskal-Wallis tests assessed differential miRNA expression across phenotypes for all samples within each biopsy site. For analysis across multiple miRNAs, raw p-values were corrected for multiple testing by the Benjamini-Hochberg false-discovery rate [FDR] method].22
2.5. Prediction of miRNA targets
Two experimentally supported online databases of miRNA-mRNA pairs were used to identify validated targets of our identified differentially expressed miRNAs: miRTarBase Release 6.023 and Diana Tools TarBase version 7.0.24 In a secondary in silico analysis, bioinformatically predicted targets were identified using miRWalk version 2.025 selecting four predictive algorithms, namely miRanda, miRWalk, RNA22, and TargetScan.
3. Results
3.1. Subject demographic and clinical characteristics
A total of 61 participants were included in this analysis; 23 CDq [eight male, 15 female] and 38 HC [22 male, 16 female]. Clinical and demographic details are summarised in Table 1. HC subjects were significantly older than those in the CDq group [p <0.001, two-tailed Welch’s t test]. The majority of the CDq group had ileocolonic disease [L3, n = 10], followed by isolated colonic disease [L2, n = 9], and finally isolated ileal disease [L1, n = 4]. In total, 112 samples were included in our analyses, with 51 samples from the terminal ileum and 61 samples from the sigmoid colon. Two CDq individuals and eight HC individuals had expression data from only one location and thus were not included in the paired analyses. The breakdown by location and diagnosis, as well as indication of number of paired samples for each diagnostic group, are summarised in Table 2.
Table 2.
Number of endoscopic biopsies analysed for each location and phenotype.
| CDq | HC | |
|---|---|---|
| Terminal ileum | 21 samples | 30 samples |
| Sigmoid colon | 23 samples | 38 samples |
| Matched terminal ileum and sigmoid colon samples | 21 pairs | 30 pairs |
miRNAimmunoregulatory mRNA targets.
CDq, quiescent Crohn’s disease; HC, healthy controls.
3.2. Differential expression of miRNAs in colonic and ileal tissue
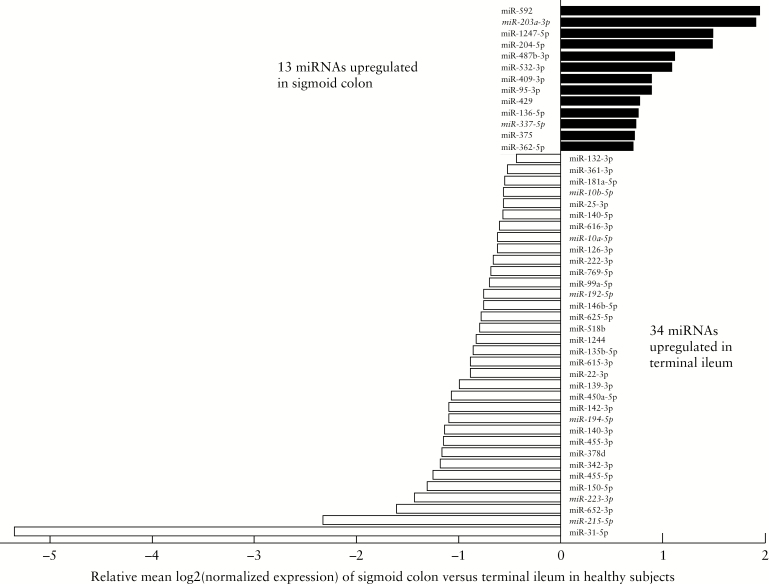
miRNA expression in colonic and ileal samples from HC subjects using Nanostring [see Methods] was measured. Non-parametric analysis of miRNA expression in matched samples across the two biopsy sites in each individual in the HC group was performed, giving a baseline spatial expression pattern in healthy intestinal tissue; 47 miRNAs were differentially expressed by site of biopsy after FDR correction, with 13 upregulated in the sigmoid colon and 34 upregulated in the terminal ileum [Figure 1, FDRp <0.05]. Boxplots with log2 normalised expression values of all 47 differentially expressed miRNAs in the sigmoid colon versus ileum are shown in Supplementary Figure 1, available as Supplementary data at ECCO-JCC online.
Figure 1.
Relative expression of significantly differentially expressed miRNAs across paired samples in the sigmoid colon [black] and terminal ileum [white] in healthy controls. Relative expression levels on X-axis are based on comparison with expression in the terminal ileum. MiRNAs italicised and underlined refer to those also differentially expressed by diagnosis accounting for location of biopsy [Figure 2]. Statistical significance obtained by Wilcoxon signed-rank test, corrected for multiple testing at FDRp <0.05.
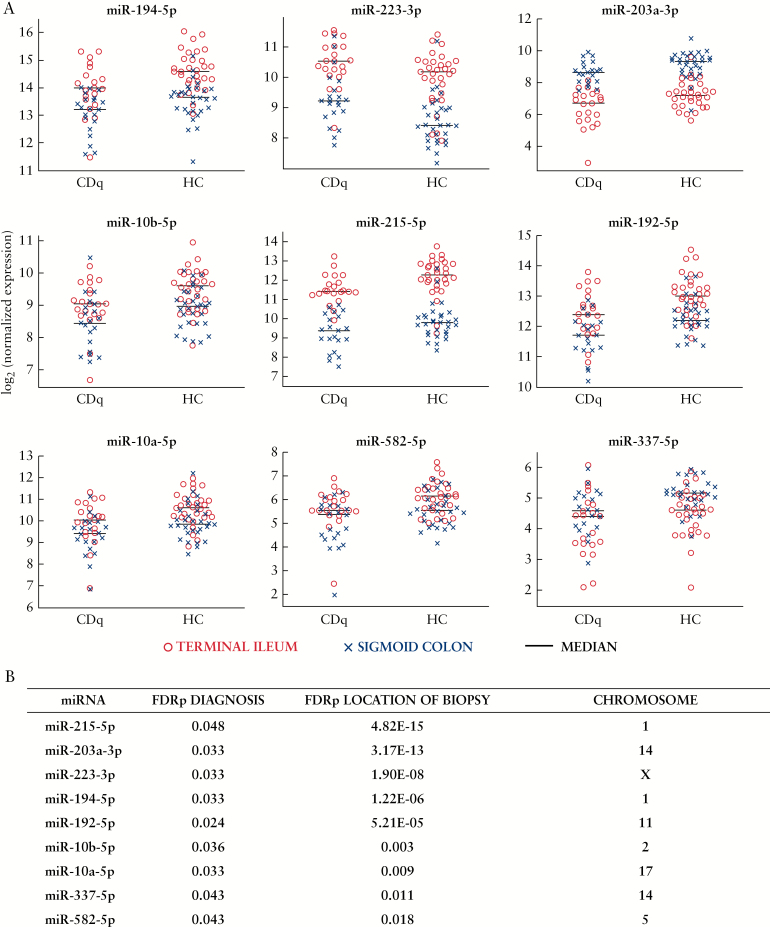
We then investigated the differential expression of miRNAs driven both by location and phenotype, using a linear mixed-effects model for matched colonic and ileal samples in CDq and HC individuals. We found nine miRNAs that were differentially expressed after FDR correction across HC and CDq accounting for the location of the biopsy: miR-215-5p, miR-203a-3p, miR-223-3p, miR-194-5p, miR-192-5p, miR-10b-5p, miR-10a-5p, miR-337-5p, miR-582-5p [Figure 2]. For the majority of differentially expressed miRNAs, the FDR-adjusted p-values for location are orders of magnitude larger than those for diagnosis [Figure 2b], suggesting that the site of the biopsy is the main driver of differential expression in our model. There were no significant interaction effects observed between diagnosis and location of biopsy in our model [Supplementary Table 2, available as Supplementary data at ECCO-JCC online].
Figure 2.
Nine differentially expressed miRNAs by diagnosis accounting for location of biopsy. A: Beeswarm plots of differentially expressed miRNAs by location and phenotype from lme model for matched samples; n = 51 total pairs from terminal ileum [red o] and sigmoid colon [blue x]. CDq, quiescent Crohn’s disease [n = 21]; HC, healthy controls [n = 30]. B: FDR-adjusted p-values from the lme model of differential expression of miRNAs across diagnosis [FDRp Diagnosis] and location [FDRp Location of Biopsy]. Chromosome location for miRNAs was obtained from miRBase release 21 [http://www.mirbase.org].
To explore the possible function of this subset of miRNAs in CD susceptibility, we looked at both bioinformatically predicted and experimentally validated mRNA targets using publicly available software miRWalk, miRTarBase, and Diana-TarBase [see Methods; Table 3].23–25 We found that these co-expressed miRNAs targeted several genes such as NOD2, TLR4, and IL6ST [Table 3], involved in immunoregulatory processes important in CD pathogenesis2–4 including the innate immune response to pathogens as well as cytokine signalling.
Table 3.
Immune-related mRNA targets with relevance in CD of differentially expressed miRNAs driven by location and diagnosis.
| miRNA | Immunoregulatory mRNA targets | |||||
|---|---|---|---|---|---|---|
| miR-10a-5p | LILRA2 | IL6R* | IFNAR2* | |||
| miR-10b-5p | LILRA2 | IL6R* | IFNAR2* | |||
| miR-192-5p | IL6R | IL6ST | NOD2 | IRAK1 | TRAF6* | |
| miR-194-5p | SOCS2 | IL6ST | IL6R* | IL21* | ||
| miR-203a-3p | IL6 | IL6ST | NOD2* | TLR4* | TNF | SOCS3 |
| miR-215-5p | IL6R | IL6ST | NOD2 | IRAK1 | TRAF6* | |
| miR-223-3p | IL6 | IL6ST | NLRP3 | IFNAR1* | ||
| miR-337-5p | NOD2* | |||||
| miR-582-5p | IL17RD | TGFBR2 | TLR2* |
The mRNA targets with an asterisk [*] are bioinformatically predicted from miRWalk 2.0; otherwise, no asterisk refers to targets which are experimentally validated in either miRTarBase or TarBase.
CD, Crohn’s disease.
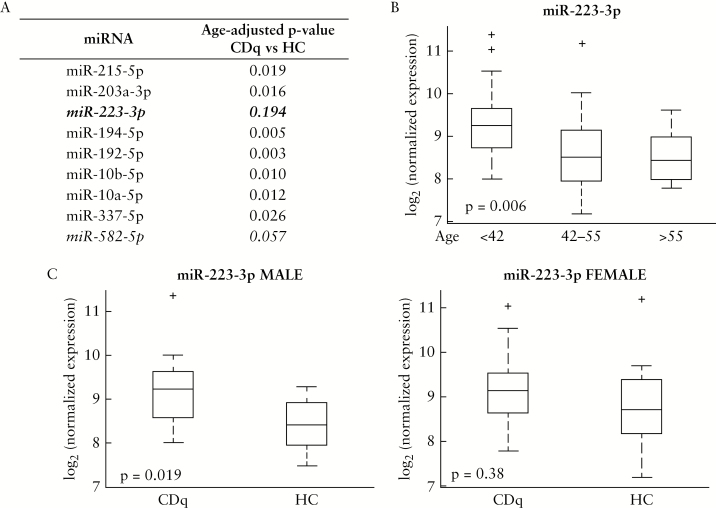
3.3. Age and sex effects on differential expression of miR-223-3p
Differential expression of miR-223-3p has been associated with age and age-related changes in cellular composition.26–28 The CD patients in this study were significantly younger than our control group [Table 1, p <0.001], and we found that miR-223-3p was upregulated in in CDq versus HC [Figure 2]. We therefore repeated our differential expression analysis, using the lme model including age as a covariate, for the nine differentially expressed miRNAs summarised in Figure 2.
We found that the raw p-value for the differential expression of miR-223-3p was no longer significant when accounting for the age disparity in our outcome groups [Figure 3A], and found an inverse association between the expression of miR-223-3p in the sigmoid colon and subject age [Figure 3B, p = 0.006, Kruskal-Wallis test, Dunn’s post-hoc; terminal ileum results not significant at p = 0.1 and shown in Supplementary Figure 2A, available as Supplementary data at ECCO-JCC online].
Figure 3.
Sex and age effects on miR-223 expression. A: Table of results from lme model for age-corrected differential expression across diagnosis accounting for location for each miRNA in Figure 2. B: Association of miR-223-3p expression in all sigmoid colon samples [CDq and HC] with age tertiles. p = 0.006 three-way Kruskal-Wallis test, Dunn’s post-hoc for statistically significant pairwise values [group 1:2, p = 0.017; group 1:3, p = 0.014]. C: Boxplots of differential expression across diagnosis [CDq versus HC] in the sigmoid colon stratified by sex. miR-223-3p expression significantly upregulated in CDq males [n = 8] versus HC males [n = 22], p = 0.019 [left panel]. No significant difference in CDq females [n = 15] versus HC females [n = 16] in miR-223-3p expression in sigmoid colon [right panel]. Boxplots of terminal ileum expression in Supplementary Figure 2B, available at ECCO-JCC online. CDq, quiescent Crohn’s disease; HC, healthy controls.
In addition to association of age with the expression of miR-223-3p, sex may also be important to consider since the gene encoding human miR-223 is located on the X chromosome as shown in Figure 2B; miRBase release 21, [http://www.mirbase.org]. To explore this possibility, the cohort was stratified by sex and expression of miR-223-3p at each biopsy site analysed across phenotypes. We found miR-223-3p expressed in the sigmoid colon was significantly upregulated in CDq males compared with HC males [Figure 3C, p = 0.019, Kruskal-Wallis test], but not CDq females compared with HC females [Figure 3C, p = 0.38, Kruskal-Wallis test]. No differences were seen in the terminal ileum for either males or females [Supplementary Figure 2B, available as Supplementary data at ECCO-JCC online]. As a supplementary analysis, we also adjusted the sigmoid colon CDq male vs HC male result accounting for age in a linear model; our result remained significant with a p-value = 0.012. Although interpretation of this result is beyond the scope of this study, these preliminary data suggest that genomic context may be important in interpreting the mechanistic effects of differential expression of miR-223-3p, since its location on the X chromosome might influence sex-specific immune responses.
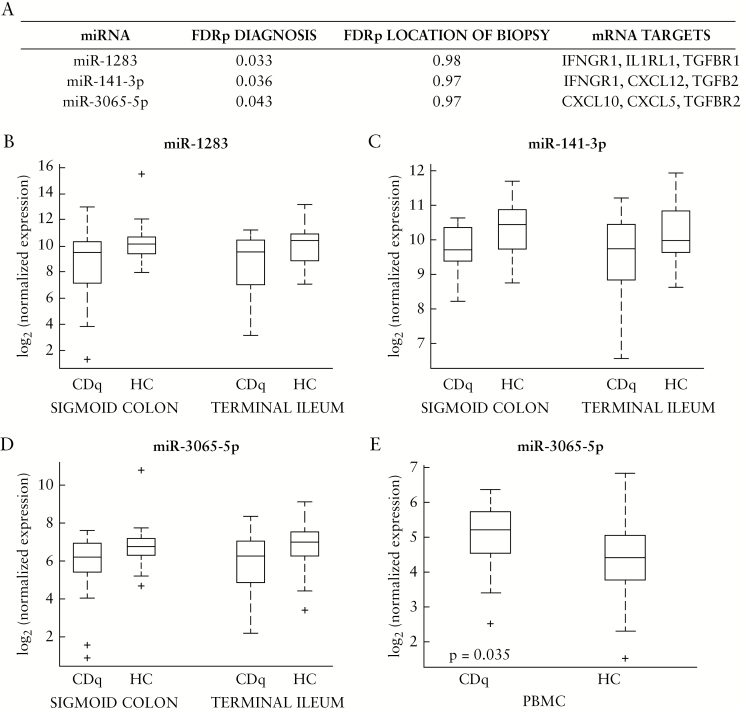
3.4. Differential expression of miRNAs by diagnosis not affected by biopsy location
We then examined the differential expression of miRNAs driven by diagnostic phenotype [CDq versus HC], regardless of the location of the biopsy. To this end, we looked at the results from our linear mixed-effect model and chose the top three miRNAs that were significantly differentially expressed after FDR correction by diagnosis, but were not differentially expressed by location: miR-1283, miR-141-3p, miR-3065-5p [Figure 4A–D, FDRp <0.05 for diagnosis, FDRp = 0.97 or 0.98 for location, Supplementary Table 1]. All were downregulated in CDq versus HC. We constrained the miRNA target analysis to only experimentally validated databases,23,24 and found that several targets of these miRNAs are involved in chemokine and cytokine signalling, such as IFNGR1, IFNAR1, and CXCL5 [Figure 4].
Figure 4.
Top three miRNAs differentially expressed by diagnosis but not the location of biopsy. A: Summary of FDR-adjusted p-values for top three differentially expressed miRNAS by phenotype and not location. Full list in Supplementary Table 1, available at ECCO-JCC online, FDRp Diagnosis <0.05, FDRp Location >0.05. Predicted targets are from experimentally validated databases, miRTarBase and TarBase. B–D: Boxplots of miRNAs in 4A in the sigmoid colon and terminal ileum, showing differential expression across CDq and HC within each site. E: Boxplot of CDq versus HC differential expression of miR-3065-5p in the PBMCs of the same cohort of patients from another study in our group; n = 61, p = 0.035, Kruskal-Wallis test. miR-1283 and miR-141-3p PBMC differential expression not significant [p >0.05, Kruskal-Wallis test]; boxplots in Supplementary Figure 3, available at ECCO-JCC online. CDq, quiescent Crohn’s disease; HC, healthy controls; PBMCs, peripheral blood mononuclear cells.
In a separate study,29,30 our group analysed miRNA differential expression in the peripheral blood mononuclear cells [PBMCs] of the same cohort of subjects. Since the differential expression for miR-1283, miR-141-3p, and miR-3065-5p in tissue is not driven by location, we used the PBMC data for a targeted analysis, to see if similar differential expression of these three miRNAs was seen in the peripheral blood as a systemic manifestation in these same individuals [n = 61]. We found that miR-3065-5p was also differentially expressed in the blood, although with inverse directionality to the expression in tissue [Figure 4E, p <0.05, Kruskal-Wallis test; see Supplementary Figure 3, available as Supplementary data at ECCO-JCC online for miR-1283 and miR-141-3p PBMC expression, p >0.05].
3.5. miR-31-5p is most differentially expressed by location but not affected by diagnosis
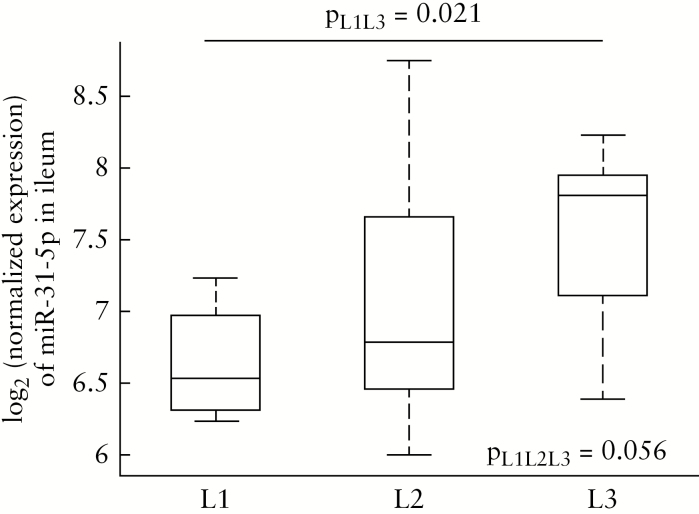
miR-31-5p was the most differentially expressed miRNA driven by location in our mixed-effects model, and demonstrated no differential expression by diagnosis [FDRp = 9.96E-19 for location, FDRp = 0.99 for diagnosis, Supplementary Table 1]. miR-31-5p was significantly upregulated in the ileum of healthy subjects compared with the sigmoid colon [Figure 1; Supplementary Figure 1, available as Supplementary data at ECCO-JCC online]. Directly comparing CDq and HC expression in the ileum with a non-parametric test confirmed there is no discernable effect of diagnosis on miR-31-5p differential expression with the site of biopsy [Supplementary Figure 4, available as Supplementary data at ECCO-JCC online]. Since biopsy location was the main driver of differential expression of miR-31-5p in our data, we investigated whether miR-31-5p was also associated with the site of disease. We stratified our CDq cohort based on the Montreal classification for disease location,17 and analysed the expression of miR-31-5p in the ileum [where the expression was significantly higher and provided a stronger signal to analyse], comparing those with ileal involvement [L1, n = 4], colonic involvement [L2, n = 8], and ileocolonic involvement [L3, n = 9]. Performing a three-way non-parametric test, we found the differential expression across the subtypes to be just above our significance threshold at p = 0.056 [Figure 5]. The small sample size in each group after stratification likely rendered our analysis statistically underpowered. The L1 and L3 groups exhibited the greatest differential expression [Figure 5], and when these two subtypes were directly compared using a non-parametric Kruskal-Wallis test, ileal-only CD was significantly downregulated compared with ileocolonic CD [Figure 5, p = 0.021]. Although this is a small sample size that necessitates a larger cohort for replication, the expression of miR-31-5p may be a biomarker for disease location subtypes in CD, and may regulate mechanisms underlying the location and extent of disease.
Figure 5.
Boxplots of miR-31-5p expression in the terminal ileum for patients with ileal [n = 4, L1], colonic [n = 8, L2], and ileocolonic [n = 9, L3] involvement; p = 0.056, three-way Kruskal-Wallis test; p = 0.021, L1 versus L3, KruskalWallis test.
4. Discussion
This study examined the quiescent intestinal mucosa of individuals with CD as well as that of HC at two biopsy sites [n = 112 total samples], revealing molecular alterations potentially underlying disease susceptibility. Our investigation focused on the differential expression of miRNAs, which play a major role in regulating levels of gene expression through mRNA degradation or translation repression.31–33 Our analysis highlighted miRNAs that were differentially expressed by location, by diagnosis, and concurrently by both. We started by examining differential expression across the gut of 30 healthy subjects with paired colonic and ileal biopsies, and found 13 miRNAs upregulated in the sigmoid colon and 34 miRNAs upregulated in the terminal ileum [Figure 1, Supplementary Figure 1]. Previous work by Wu et al.10 used microarray profiling to assess the region-specific miRNA expression in the gut of six healthy subjects undergoing routine screening, and found 20 miRNAs differentially expressed by location, five of these overlapping with our results: miR-31, miR-215, miR-126, miR-22, and miR-140 [Figure 1]. Our larger dataset of paired samples from HC subjects was better powered to identify a greater subset of spatially differentially expressed miRNAs, serving as a baseline for expression in the normal gut.
We then accounted for diagnosis [either CDq or HC] in our analysis, and found a panel of nine miRNAs whose expression pattern was driven by both diagnosis and site of biopsy: miR-215-5p, miR-203a-3p, miR-223-3p, miR-194-5p, miR-192-5p, miR-10b-5p, miR-10a-5p, miR-337-5p, miR-582-5p. All but one of these, miR-582-5p, are also differentially expressed by location in healthy tissue after correction for multiple testing. It is possible that tissue-driven expression of miR-582-5p is related to the site of the disease, which could explain it not being differentially expressed in healthy colon versus ileum. Investigation of this hypothesis would require a cohort with paired samples that has a better distribution of disease locations to explore further. We also probed the possible function of these dysregulated miRNAs in CD, by examining their mRNA targets using publicly available bioinformatics software [see Methods]. We found that our panel of miRNAs targeted several genes important in regulating the immune response via cytokine signalling and/or pathogen recognition, including NOD2, IL6ST, IL6, and TLR4. These have been linked to CD in genetic studies, suggesting that alteration of gene expression levels by these miRNAs in pathways is relevant in CD pathogenesis.2–4 Confirmation of this hypothesis would require quantifying the mRNA levels in our cohort to confirm target suppression, which is currently under way. Future work including a validation cohort to confirm these differentially expressed miRNAs and miRNA-mRNA interactions is also warranted. Furthermore, one drawback of this study is that the mucosal biopsies include a heterogeneous population of cells. Therefore experiments quantifying the cellular sources of the miRNA differential expression are necessary to better characterise the immunoregulatory role of these miRNAs.
Furthermore, miR-223, which showed an age-related effect in our study, has previously been shown to suppress NLRP3,34,35 an activator of NF-kappaB signalling and a key component in the inflammatory response. A recent study by Neudecker et al.36 showed that synthetic mimics of miR-223, delivered via nanoparticles, reduced levels of NLRP3 and inflammation in a murine model of experimental colitis, suggesting the possibility of miRNA-based therapeutics targeting inflammation in the gut. Whereas there is a known role for miR-223 in inflammation and innate immunity36–40 which may still contribute to its differential expression in our cohort, our finding is interesting in the context of some previous human studies in IBD which have shown differential expression of miR-223-3p across phenotypes; but the ages were also significantly different between patient and control groups, and this did not seem to be accounted for in those analyses.6,10,12,41 For example, a recent study suggested upregulation of miR-223 as a biomarker of IBD activity41; however a two-tailed t test we performed on the published demographics of the cohort shows that the CD group [n = 50, age 27.4 ± 9.1 years] was significantly younger [p <0.0001] than the controls [n = 50, age 38.2 ± 10.1 years]. miR-223 could potentially monitor disease activity as proposed therein, but accounting for differences in ages between groups may be of particular importance when evaluating its utility as a biomarker, since our results [Figure 3A, B] as well as others’ studies on ageing demonstrate that it is associated with age and age-related changes in cellular composition.26–28
A search on miRBase.org at the time of writing [miRBase Release 21] for miRNAs located on the X chromosome shows that the human X chromosome is highly enriched for miRNAs compared with the Y-chromosome, encoding 118 miRNAs compared with only two Y-located miRNAs. Several molecular mechanisms have been proposed to support the contribution of the X chromosome in sex-specific immune responses, including X chromosome-encoded miRNA expression and function.42–44 Previous studies have identified X-linked miRNAs associated with sex differences in disease occurrence and development.45–48 Since miR-223 is located on the X chromosome, we investigated the potential role of sex on its differential expression in our cohort. After stratifying the cohort by sex, the males in our group demonstrated differential expression of miR-223 in colonic biopsies across phenotypes [p = 0.019], but the females showed no significant difference [p = 0.38]. Interpretation of this result in context of X-linked miRNAs and CD risk is beyond the scope of this study, but further investigation may be warranted given the current literature and hypotheses relating miRNAs located on the X chromosome to immune-mediated disease susceptibility.42–48
We then also evaluated differential miRNA expression driven primarily by diagnosis [CDq or HC], where the site of biopsy had no effect. We selected the top three miRNAs from this analysis: miR-1283, miR-141-3p, and miR-3065-5p, all downregulated in CDq versus HC. We constrained the mRNA target prediction to those in the experimentally validated databases miRTarBase23 and TarBase,24 and found these miRNAs target genes involved in cytokine/chemokine signalling key in regulating mucosal immunity, such as CXCL10, IFNGR1, and IL1RL1. Since location was not driving the differential expression, we also looked at possible systemic manifestation using the peripheral blood of these patients from another study in our group.29,30 In a targeted analysis of these miRNAs, we found that miR-3065-5p was also differentially expressed in the blood with inverse directionality [CDq upregulated compared with HC]. An inverse relationship between tissue and miRNAs in circulation has been reported in previous studies in other diseases, such as various cancers49–52 and abdominal aortic aneurysm.53 It is unclear what the physiologically relevant mechanism[s] behind inverse tissue and blood miRNA expression are, but the hypothesis of miRNA trafficking has been suggested,49,50,54 where specific miRNAs are packaged in exosomes and secreted from donor cells and transferred to recipient cells, thus increasing or decreasing miRNA expression levels in blood or tissue, respectively.55 This hypothesis is interesting in the context of our study, where some targets of miR-3065-5p include chemokines which are known to be important during lymphocyte homing from blood to tissue in the immune response.56
Finally, miR-31-5p was the most differentially expressed miRNA in our cohort driven by location, but diagnosis had no effect in its expression levels [Figure 1; Supplementary Table 1 and Supplementary Figure 4]. Interestingly, miR-31-5p has been proposed by Sheikh et al. as a differentiator of two clinically distinct forms of CD, namely ‘ileum-like’ and ‘colon-like’ CD.57 Since miR-31-5p may play a role in regional expression of clinical phenotypes in CD, we stratified the terminal ileum biopsies by the location of disease, and found that miR-31-5p was differentially expressed in ileal-only disease compared with ileocolonic disease [Figure 5]. Our sample size after stratification was very small, and therefore replication is required in a larger cohort to confirm this result. Our result adds to the current evidence and encourages future investigation of mechanisms driving miR-31-5p association with regional clinical subtypes, such as examining differentially expressed gene targets of miR-31-5p in healthy colon versus ileum. As a supplementary, preliminary exploration, we referred to the study by Peloquin et al.58 which reported differential expression of IBD risk genes in healthy colon and terminal ileum of control subjects. We performed miR-31-5p bioinformatic target prediction of experimentally validated targets in miRWalk 2.0, as described in Methods here. We then cross-referenced these miR-31-5p targets from miRWalk with the location-driven differentially expressed genes known from Peloquin et al. 2016,58 using the methods described in our previous [PBMC-derived] miRNA study.59 From this, we found miR-31-5p targets PPIL2 [peptidylprolyl isomerase like 2], a member of the cyclophilin family. Furthermore, our miRNA expression has inverse directionality to the reported mRNA expression in the terminal ileum, which suggests biological relevance. This exploratory analysis and its interpretation are beyond our context of a potential miRNA biomarker for location subtypes [our main emphasis here], but this is an interesting avenue for future work probing mechanisms of miRNA-mediated pathogenesis of clinical subtypes of CD.
In conclusion, we found a panel of differentially expressed miRNAs across ileal and colonic endoscopic biopsies in the healthy gut. We further identified subsets of tissue-derived miRNA signatures differentially expressed by location of biopsy, disease phenotype of the individual, and both location and disease, in the absence of active endoscopic inflammation. Through integration of predicted and validated mRNA targets that have been associated with CD pathogenesis through genetic studies and relevant pathways, our findings advance the understanding of tissue-specific molecular alterations underlying CD susceptibility.
Funding
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases [NIDDK] grant 5U01DK062423-15.
Conflict of Interest
The authors have no conflicting financial interests.
Author Contributions
AM: study design and planning; acquisition, analysis, and interpretation of data; manuscript drafting. OBK: study design and planning; clinical data collection and analysis; critical revision of the manuscript for important intellectual content. MIS: study design and planning; critical revision of the manuscript for important intellectual content. BK: study design and planning; acquisition and accessing of data; critical revision of the manuscript for important intellectual content. MSS: study concept and design; planning of the study; acquisition, analysis, and interpretation of data; critical revision of the manuscript for important intellectual content. Conference presentation: part of this work was presented as a poster at Digestive Disease Week, Chicago, IL, May 2017.
Supplementary Material
Acknowledgments
We would like to acknowledge the National Institute of Diabetes and Digestive and Kidney Diseases IBD Genetics Consortium [NIDDK IBDGC] for their support and critical review of this work.
References
- 1. Sartor RB. Mechanisms of disease: pathogenesis of Crohn’s disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol 2006;3:390–407. [DOI] [PubMed] [Google Scholar]
- 2. Jostins L, Ripke S, Weersma RK, et al. ; International IBD Genetics Consortium [IIBDGC] Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011;474:307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ellinghaus D, Jostins L, Spain SL, et al. ; International IBD Genetics Consortium [IIBDGC]; International Genetics of Ankylosing Spondylitis Consortium [IGAS]; International PSC Study Group [IPSCSG]; Genetic Analysis of Psoriasis Consortium [GAPC]; Psoriasis Association Genetics Extension [PAGE] Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet 2016;48:510–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cleynen I, Boucher G, Jostins L, et al. ; International Inflammatory Bowel Disease Genetics Consortium Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet 2016;387:156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fasseu M, Treton X, Guichard C, et al. . Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS One 2010, Oct 5. doi: 10.1371/journal.pone.0013160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kalla R, Ventham NT, Kennedy NA, et al. . MicroRNAs: new players in IBD. Gut 2015;64:504–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paraskevi A, Theodoropoulos G, Papaconstantinou I, Mantzaris G, Nikiteas N, Gazouli M. Circulating microRNA in inflammatory bowel disease. J Crohns Colitis 2012;6:900–4. [DOI] [PubMed] [Google Scholar]
- 9. Wu F, Guo NJ, Tian H, et al. . Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis 2011;17:241–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu F, Zhang S, Dassopoulos T, et al. . Identification of microRNAs associated with ileal and colonic Crohn’s disease. Inflamm Bowel Dis 2010;16:1729–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu F, Zikusoka M, Trindade A, et al. . MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology 2008;135:1624–35.e24. [DOI] [PubMed] [Google Scholar]
- 12. Peck BC, Weiser M, Lee SE, et al. . MicroRNAs classify different disease behavior phenotypes of Crohn’s disease and may have prognostic utility. Inflamm Bowel Dis 2015;21:2178–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin J, Cao Q, Zhang J, et al. . MicroRNA expression patterns in indeterminate inflammatory bowel disease. Mod Pathol 2013;26:148–54. [DOI] [PubMed] [Google Scholar]
- 14. Brest P, Lapaquette P, Souidi M, et al. . A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet 2011;43:242–5. [DOI] [PubMed] [Google Scholar]
- 15. Chapman CG, Pekow J. The emerging role of miRNAs in inflammatory bowel disease: a review. Therap Adv Gastroenterol 2015;8:4–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Daperno M, D’Haens G, Van Assche G, et al. . Development and validation of a new, simplified endoscopic activity score for Crohn’s disease: the SES-CD. Gastrointest Endosc 2004;60:505–12. [DOI] [PubMed] [Google Scholar]
- 17. Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 2006;55:749–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geiss GK, Bumgarner RE, Birditt B, et al. . Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 2008;26:317–25. [DOI] [PubMed] [Google Scholar]
- 19.nanoString. nCounter® miRNA Expression Assay User Manual. NanoString Technologies, Inc., Seattle, Washington. 2018. https://www.nanostring.com/download_file/view/305/3781. Accessed April 30, 2019. [Google Scholar]
- 20.nanoString. nSolver™ Analysis Software. NanoString Technologies, Inc., Seattle, Washington. https://www.nanostring.com/products/analysis-software/nsolver [Google Scholar]
- 21. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–27. [DOI] [PubMed] [Google Scholar]
- 22. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995;57: 289–300. [Google Scholar]
- 23. Chou CH, Chang NW, Shrestha S, et al. . miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res 2016;44:D239–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vlachos IS, Paraskevopoulou MD, Karagkouni D, et al. . DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res 2015;43:D153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dweep H, Sticht C, Pandey P, Gretz N. miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 2011;44:839–47. [DOI] [PubMed] [Google Scholar]
- 26. Teteloshvili N, Kluiver J, van der Geest KS, et al. . Age-associated differences in MiRNA signatures are restricted to CD45RO Negative T Cells and are associated with changes in the cellular composition, activation and cellular ageing. PLoS One 2015;10:e0137556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kumar S, Vijayan M, Bhatti JS, Reddy PH. MicroRNAs as peripheral biomarkers in aging and age-related diseases. Prog Mol Biol Transl Sci 2017;146:47–94. [DOI] [PubMed] [Google Scholar]
- 28. Dimmeler S, Nicotera P. MicroRNAs in age-related diseases. EMBO Mol Med 2013;5:180–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mohammadi A, Kelly O, Kabakchiev B, et al. . Su1786 a PBMC-derived microRNA signature differentiates colonic Crohn’s disease from ulcerative colitis. Gastroenterology 2016;150:S550–1. [Google Scholar]
- 30. Mohammadi A, Kelly O, Kabakchiev B, et al. . Tu1928 a peripheral blood derived microRNA signature reveals molecular differences in patients with endoscopically quiescent Crohn’s disease versus healthy controls. Gastroenterology 2016;150: S980. [Google Scholar]
- 31. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev 2004;18:504–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol 2005;3:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bauernfeind F, Rieger A, Schildberg FA, Knolle PA, Schmid-Burgk JL, Hornung V. NLRP3 inflammasome activity is negatively controlled by miR-223. J Immunol 2012;189:4175–81. [DOI] [PubMed] [Google Scholar]
- 35. Haneklaus M, Gerlic M, Kurowska-Stolarska M, et al. . Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1β production. J Immunol 2012;189:3795–9. [DOI] [PubMed] [Google Scholar]
- 36. Neudecker V, Haneklaus M, Jensen O, et al. . Myeloid-derived miR-223 regulates intestinal inflammation via repression of the NLRP3 inflammasome. J Exp Med 2017;214:1737–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnnidis JB, Harris MH, Wheeler RT, et al. . Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008;451:1125–9. [DOI] [PubMed] [Google Scholar]
- 38. Ha TY. The role of MicroRNAs in regulatory T cells and in the immune response. Immune Netw 2011;11:11–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fazi F, Rosa A, Fatica A, et al. . A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell 2005;123:819–31. [DOI] [PubMed] [Google Scholar]
- 40. Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol 2008;9:839–45. [DOI] [PubMed] [Google Scholar]
- 41. Wang H, Zhang S, Yu Q, et al. . Circulating MicroRNA223 is a new biomarker for inflammatory bowel disease. Medicine 2016;95:e2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dai R, Ahmed SA. Sexual dimorphism of miRNA expression: a new perspective in understanding the sex bias of autoimmune diseases. Ther Clin Risk Manag 2014;10:151–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Libert C, Dejager L, Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nat Rev Immunol 2010;10:594–604. [DOI] [PubMed] [Google Scholar]
- 44. Fish EN. The X-files in immunity: sex-based differences predispose immune responses. Nat Rev Immunol 2008;8:737–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pinheiro I, Dejager L, Libert C. X-chromosome-located microRNAs in immunity: might they explain male/female differences? The X chromosome-genomic context may affect X-located miRNAs and downstream signaling, thereby contributing to the enhanced immune response of females. Bioessays 2011;33:791–802. [DOI] [PubMed] [Google Scholar]
- 46. Khalifa O, Pers Y-M, Ferreira R, et al. . X-Llnked miRNAs associated with gender differences in rheumatoid arthritis. Int J Mol Sci 2016, Nov 8. pii: E1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sharma S, Eghbali M. Influence of sex differences on microRNA gene regulation in disease. Biol Sex Differ 2014;5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guo L, Zhang Q, Ma X, Wang J, Liang T. miRNA and mRNA expression analysis reveals potential sex-biased miRNA expression. Sci Rep 2017;7:39812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tanaka M, Oikawa K, Takanashi M, et al. . Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS One 2009;4:e5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Plieskatt J, Rinaldi G, Feng Y, et al. . A microRNA profile associated with Opisthorchis viverrini-induced cholangiocarcinoma in tissue and plasma. BMC Cancer 2015;15:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guo L, Zhao Y, Yang S, Cai M, Wu Q, Chen F. Genome-wide screen for aberrantly expressed miRNAs reveals miRNA profile signature in breast cancer. Mol Biol Rep 2013;40:2175–86. [DOI] [PubMed] [Google Scholar]
- 52. Torres A, Torres K, Pesci A, et al. . Deregulation of miR-100, miR-99a and miR-199b in tissues and plasma coexists with increased expression of mTOR kinase in endometrioid endometrial carcinoma. BMC Cancer 2012;12:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kin K, Miyagawa S, Fukushima S, et al. . Tissue- and plasma-specific MicroRNA signatures for atherosclerotic abdominal aortic aneurysm. J Am Heart Assoc 2012;1:e000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ohyashiki K, Umezu T, Yoshizawa S, et al. . Clinical impact of down-regulated plasma miR-92a levels in non-Hodgkin’s lymphoma. PLoS One 2011;6:e16408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654–9. [DOI] [PubMed] [Google Scholar]
- 56. Stein JV, Nombela-Arrieta C. Chemokine control of lymphocyte trafficking: a general overview. Immunology 2005;116:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sheikh S, Furey T, Keith B, et al. . P-308 identification of miR-31 as a molecular stratifier of clinical Crohn’s disease phenotypes. Inflam Bowel Dis 2017; 23: S98–9. [Google Scholar]
- 58. Peloquin JM, Goel G, Kong L, et al. . Characterization of candidate genes in inflammatory bowel disease-associated risk loci. JCI Insight 2016;1:e87899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mohammadi A, Kelly OB, Filice M, Kabakchiev B, Smith MI, Silverberg MS. Differential expression of microRNAs in peripheral blood mononuclear cells identifies autophagy and TGF-beta-related signatures aberrantly expressed in inflammatory bowel disease. J Crohns Colitis 2018;12:568–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.