Abstract
The gut microbiome regulates important host metabolic pathways including xenobiotic metabolism and intermediary metabolism, such as the conversion of primary bile acids (BAs) into secondary BAs. The nuclear receptors pregnane X receptor (PXR) and constitutive androstane receptor (CAR) are well-known regulators for xenobiotic biotransformation in liver. However, little is known regarding the potential effects of PXR and CAR on the composition and function of the gut microbiome. To test our hypothesis that activation of PXR and CAR regulates gut microbiota and secondary BA synthesis, 9-week-old male conventional and germ-free mice were orally gavaged with corn oil, PXR agonist PCN (75 mg/kg), or CAR agonist TCPOBOP (3 mg/kg) once daily for 4 days. PCN and TCPOBOP decreased two taxa in the Bifidobacterium genus, which corresponded with decreased gene abundance of the BA-deconjugating enzyme bile salt hydrolase. In liver and small intestinal content of germ-free mice, there was a TCPOBOP-mediated increase in total, primary, and conjugated BAs corresponding with increased Cyp7a1 mRNA. Bifidobacterium, Dorea, Peptociccaceae, Anaeroplasma, and Ruminococcus positively correlated with T-UDCA in LIC, but negatively correlated with T-CDCA in serum. In conclusion, PXR and CAR activation downregulates BA-metabolizing bacteria in the intestine and modulates BA homeostasis in a gut microbiota-dependent manner.
Keywords: bile acid metabolism, CAR, gut-liver axis, gut microbiome, intestine, liver, liver metabolism, nuclear receptors, PXR, 16S rDNA sequencing
Liver is a critical organ for xenobiotic biotransformation and nutrient homeostasis. The nuclear receptors pregnane X receptor (PXR) and constitutive androstane receptor (CAR) are well-known xenobiotic-sensing transcription factors with overlapping target genes involved in the Phase-I and Phase-II xenobiotic metabolism as well as transport (collectively called drug-processing genes [DPGs]) (Cui and Klaassen, 2016; Hernandez et al., 2009; Kliewer et al., 1998; Moore et al., 2003; Petrick and Klaassen, 2007; Wei et al., 2000).
PXR was first identified as a steroid-activating receptor before its function in xenobiotic metabolism was discovered (Kliewer et al., 1998, 2002). PXR is a promiscuous transcription factor that is activated by various xenobiotics, including therapeutic drugs such as dexamethasone, paclitaxel, and rifampicin (Moore et al., 2003), environmental toxicants such as the flame retardant polybrominated diphenyl ethers (PBDEs) (Pacyniak et al., 2007), some pesticides (Lemaire et al., 2006), and herbal supplements such as St. John’s Wort (Moore et al., 2000). PXR is also activated by metabolites produced by the gut microbiome, such as the secondary bile acid (BA), lithocholic acid (LCA), and the tryptophan metabolite indole-3-propionic acid (Staudinger et al., 2001; Venkatesh et al., 2014). Pregnenolone-16α-carbonitrile (PCN) is recognized as a specific PXR ligand in rodents, whereas rifampicin is specific to human PXR activation (Moore et al., 2003).
CAR was identified as a mediator of drug metabolism specifically for altering the sensitivity to barbiturates (Tzameli et al., 2000; Wei et al., 2000). Repressive endogenous ligands for constitutive activity of CAR include androstenol and androstanol (Forman et al., 1998), whereas endogenous activators of CAR are not well characterized. Phenobarbital and LCA can indirectly facilitate the nuclear localization of CAR and upregulation of CAR-target genes (Beilke, Aleksunes, Holland, et al., 2009; Beilke, Aleksunes, Olson, et al., 2009; Swales and Negishi, 2004). A potent and selective CAR activator in mice is 1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene, 3,3′,5,5′-tetrachloro-1,4 bis(pyridyloxy)benzene (TCPOBOP) (Tzameli et al., 2000), whereas 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime (CITCO) is a selective CAR activator in humans (Swales and Negishi, 2004). The flame retardant BDE-47 is an activator of both mouse and human CAR (Sueyoshi et al., 2014).
The gut microbiome is increasingly recognized as a key modifier of host xenobiotic biotransformation through direct and indirect mechanisms (Koppel et al., 2017; Spanogiannopoulos et al., 2016). For direct mechanisms, intestinal bacteria use their own enzymes that are distinct from the host to metabolize ingested compounds, such as dietary components, pharmaceuticals, and environmental chemicals (Koppel et al., 2017; Spanogiannopoulos et al., 2016). These microbial modifications, many of which remain poorly characterized, can either increase or decrease the toxicity or efficacy of drugs. The PharmacoMicrobiomics database has identified over 90 drugs in the literature that are modified by the gut microbiome, but only 28 have identifiable interactions with a specific bacterial species. For example, the first drug-bug interaction was reported by Dr Lindenbaum and Dr Saha for the inactivation of digoxin by Eggerthella lenta (Lindenbaum et al., 1981; Saha et al., 1983). Characterization of these microbial enzymes has led to better understanding of the pharmacokinetics of digoxin as well as improved efficacy and safety (Haiser et al., 2013, 2014; Koppel et al., 2018). Furthermore, microbial β-glucoronidases deconjugate host Phase-II glucuronidation metabolites and modulate the potential toxicity of circulating endogenous and xenobiotic compounds (Little et al., 2018; Pollet et al., 2017; Wallace et al., 2015).
For indirect mechanisms, the gut microbiome can modify host metabolites that are subsequently absorbed into systemic circulation and enter various host organs to interact with host receptors involved in xenobiotic biotransformation (Fu and Cui, 2017; Spanogiannopoulos et al., 2016). The presence of the gut microbiome is necessary in regulating the constitutive expression of many drug processing genes (DPGs), evidenced from altered expression of DPGs in mice lacking a gut microbiome (ie, germ-free [GF] mice) (Bjorkholm et al., 2009; Selwyn, Cheng, et al., 2015; Selwyn, Cui, et al., 2015; Toda, Saito, et al., 2009). For example, the expression of the major Phase-I oxidation enzyme cytochrome P450, family 3, subfamily a, polypeptide 11 (Cyp3a11)—a PXR-target gene and the mouse ortholog to human CYP3A4—was decreased in GF mice compared with conventional (CV) mice. This effect was also observed in CV mice treated with antibiotics and was correlated with decreased bacteria in the feces that produce the PXR-activating BA LCA (Toda et al., 2009a,b). Therefore, modulation of the gut microbiome and the bacterial metabolites can alter xenobiotic biotransformation of the host.
BAs are amphipathic molecules synthesized in liver de novo from cholesterol to primary BAs through two distinct pathways: (1) the classic pathway with the overall rate-limiting BA synthesis enzyme Cyp7a1 and (2) the alternative pathway initiated by Cyp27a1. Primary BAs are conjugated with taurine, glycine, or other cosubstrates and undergo enterohepatic circulation by secretion into the bile and reabsorption from the intestine, facilitating the uptake of dietary fats, steroids, drugs, and lipophilic vitamins (Hofmann, 2009; Li and Chiang, 2014). In the intestine, bacteria further metabolize primary BAs to secondary BAs through deconjugation, oxidation, and epimerization reactions; this, in general, increases the hydrophobicity of the BA pool (Hofmann, 2009; Li and Chiang, 2014; Ridlon et al., 2006). Primary and secondary BAs also include their conjugate forms. In humans, the majority of BAs are conjugated with taurine, glycine, or 3′-phosphoadenosine-5′-phosphosulfate, whereas in mice, the majority of BAs are conjugated with taurine (Thakare et al., 2018). Primary BAs all have a 7-hydroxy substituent (from Cyp7a1), and in humans, the majority of primary BAs are chenodeoxycholic acid (CDCA) and cholic acid (CA). Cholic acid (CA) is metabolized by intestinal bacteria to the secondary BA deoxycholic acid (DCA). CDCA is metabolized to alpha-muricholic acid (αMCA), βMCA, and ursodeoxycholic acid (UDCA) (Hofmann, 2009). UDCA has been found increased in both genders of GF compared with CV mice, suggesting that a host mechanism exists to produce UDCA (Selwyn, Csanaky, et al., 2015; Wahlstrom et al., 2017; Zhang and Klaassen, 2010). There is no known host enzyme for the synthesis of UDCA in mice and humans. Secondary BAs produced from the alternative pathway of BA synthesis include ωMCA, LCA, hyocholic acid (HCA), and hyodeoxycholic (HDCA). These BA biotransformation pathways create diverse sets of BAs with different physiological functions and nuclear receptor affinities (Li and Chiang, 2013).
Several studies have shown the importance of PXR and CAR in BA homeostasis. Pharmacological activation of PXR by PCN represses hepatic Cyp7a1 mRNA and activity in mice (Li and Chiang, 2013, 2014), and endogenous activation of PXR by the secondary BA LCA protects the liver against toxicity (Staudinger et al., 2001). In intestine/enterocytes, PXR activation induces fibroblast growth factor 19 (FGF19; Fgf15 in mice), an intestinally secreted hormone that inhibits BA synthesis by repressing the transcription of Cyp7a1 in liver, and the promoter of FGF19 contains a PXR response element (Li and Chiang, 2013; Wistuba et al., 2007). Activation of CAR has also been shown to repress the expression of CYP7A1 in human hepatocytes (Miao et al., 2006); yet, BAs are not known to serve as ligands of CAR (Li and Chiang, 2013). In a study investigating BA profiles in CV mice following CAR activation, TCPOBOP decreased total BAs in both males and females, largely due to a decrease in the primary BA taurocholic acid (T-CA), and this was associated with an increase in the mRNA expression of Cyp7a1 in liver to restore liver BAs to physiological concentrations (Lickteig et al., 2016). The BA ligase solute carrier family 27 member 5 (Slc27a5/Bal), which regulates BA conjugation, was decreased following TCPOBOP exposure (Cui and Klaassen, 2016), possibly contributes to the improved metabolic health following CAR activation (Dong et al., 2009; Gao et al., 2009).
Transporters are important to the distribution of bile acids throughout the enterohepatic organs. The basolacteral uptake transporter sodium taurocholic transporting polypeptide (Ntcp/Slc10a1) is responsible for the update of the majority of conjugated BAs from the portal blood into hepatocytes, whereas the organic anion transporting polypeptide (Oatp) 1b2 was thought to transport unconjugated BAs into hepatocytes (Csanaky et al., 2011; Klaassen and Aleksunes, 2010). The canalicular bile salt efflux pump (Bsep/Abcb11) is the rate-limiting transporter for BA biliary excretion, whereas on the basolacteral membrane, the solute carriers Slc51a/Ostα and Slc51b/Ostβ, as well as efflux transporter multidrug resistance-associated protein ATP binding cassette subfamily C member 3 (Abcc3/Mrp3) and Abcc4/Mrp4, transport BAs into blood as a compensatory mechanism during cholestasis (Halilbasic et al., 2013). Transporters regulated by PXR and CAR play an important role in the disposition of BAs between different compartments, such as liver, blood, and intestinal lumen. For example, early studies showed that biliary excretion of ouabain was increased by pretreatment with PCN, demonstrating that the biliary clearance could be mediated by pharmacological activation of PXR (Russell and Klaassen, 1972; Thompson and Klaassen, 1995). PXR activators are known to increase the expression of Abcc3/Mrp3 (Cui and Klaassen, 2016; Zelcer et al., 2003); Mrp3 plays a prominent role in protecting the liver from BA overload by lowering the BA concentration in the liver and preventing apoptosis or necrosis (Dawson et al., 2009; Hirohashi et al., 2000; Teng and Piquette-Miller, 2007). TCPOBOP activation of CAR in mice increased Abcc2/Mrp2, Abcc3/Mrp3, and Abcc4/Mrp4. Similar to Mrp3, Mrp4 is a basolateral efflux transporter of BAs, reduced glutathione, and protects the liver from BA-induced injuries during cholestasis (Dawson et al., 2009; Rius et al., 2006). Mrp2 is a canalicular efflux transporter that contributes to the bile flow of sulfated and tetra-hydroxylated BAs (Akita et al., 2001; Nies and Keppler, 2007). Regarding uptake transporters, the promoter of Oatp1a4 has a PXR binding site but was not found to be important in BA uptake, but did modulate secondary BA metabolism in male mice (Cui et al., 2010; Zhang et al., 2013).
It is known that activation of PXR and CAR can modulate BA homeostasis through the regulation of host BA synthesis enzymes and transporters. However, little is known regarding whether activation of the host nuclear receptors modulates the gut microbiome and the subsequent microbial functions, including BA metabolism. Because primary BAs are metabolized by the gut microbiome into secondary BAs, the host regulation of primary BA synthesis is poorly understood. In addition, little is known regarding the characterization of BA profiles following PXR activation, and there is no comparison of the roles for PXR versus CAR activation on BA homeostasis. Because GF mice have no intestinal bacteria, they do not have secondary BAs, allowing for the investigation of specific regulation of primary BAs by pharmacological interventions. Therefore, the goal of this study was to determine the effect of PXR and CAR activation on gut microbiome composition and the extent that the gut microbiome modulates primary and secondary BA homeostasis.
MATEREIALS AND METHODS
Chemicals
The mouse CAR ligand TCPOBOP (PubChem CID: 5382) and the mouse PXR ligand PCN (PubChem CID: 15032) were purchased from Sigma-Aldrich (St Louis, Missouri). For BA analysis, acetonitrile (PubChem CID: 6342), methanol (PubChem CID: 887), ammonium acetate (PubChem CID: 12432), and acetic acid (PubChem CID: 176; all LC-MS grade) were purchased from Fisher Scientific (Pittsburgh, Pennsylvania). The 56 BA standard compounds (Supplementary Table 1) were purchased from Sigma-Aldrich and Steraloids, Inc (Newport, Rhode Island). Stable isotope-labeled internal standards (IS), cholic acid-2,2,4,4-D4 (CA-D4) (PubChem CID of CA: 221493), lithocholic acid-2,2,4,4-D4 (LCA-D4) (PubChem CID of LCA: 9903), deoxycholic acid-2,2,4,4-D4 (DCA-D4) (PubChem CID of DCA: 222528), glycocholic acid-2,2,4,4-D4 (G-CA-D4) (PubChem CID of GCA: 10140), and glycochenodeoxycholic acid-2,2,4,4-D4 (G-CDCA-D4) (PubChem CID of GCDCA: 12544), were purchased from Cambridge Isotope Laboratories, Inc (Tewksbury, Massachusetts). The purities of nonlabeled standards were above 95%–99%, and the purities of the five isotope-labeled compounds were above 98%.
Animals
C57BL/6J CV mice were purchased from the Jackson Laboratory (Bar Harbor, Maine). Age-matched male GF mice in C57BL/6 background were provided by the University of Washington Gnotobiotic Animal Core Facility (the initial GF breeding colony was established with mice purchased from the National Gnotobiotic Rodent Resource Center [University of North Carolina, Chapel Hill]). All mice were housed according to the Association for Assessment and Accreditation of Laboratory Animal Care International guidelines, and studies were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Washington. The CV mice were exposed to the same diet (laboratory autoclaved rodent diet No. 5010), water (nonacidified autoclaved water), and bedding (autoclaved Enrich-N’Pure) as the GF mice for 4 weeks prior to chemical exposure. All chemical solutions were sterilized using a Steriflip Vacuum-Driven Filtration System with a 0.22 μm Millipore Express Plus Membrane (EMD Millipore, Temecula, California). All gavage needles and syringes were sterilized by autoclave. At 9 weeks of age, CV (n = 5 per group) and GF mice (n = 4–5 per group) were orally gavaged with vehicle (corn oil, 10 ml/kg), PCN (75 mg/kg), or TCPOBOP (3 mg/kg) once daily for 4 days. Doses for activating PXR and CAR with PCN and TCPOBOP, respectively, were selected based on previous studies (Cui and Klaassen, 2016; Li et al., 2016). On the 5th day, livers were collected and immediately frozen in liquid nitrogen. Blood was collected via cardiac puncture, and serum was obtained after centrifugation at 4°C in a MiniCollect blood collection tube (Greiner Bio-One, Monroe, North Carolina). The small and large intestinal contents (SIC and LIC) were flushed using ice-cold PBS containing 10 mM dithiothreitol, and centrifuged at 20 000 × g for 1 h at 4°C to collect pellets. The ileum tissue was also collected and immediately frozen in liquid nitrogen. All samples were stored at −80°C until further analysis.
Bacterial 16S rDNA sequencing
Bacterial DNA was isolated from LIC pellet of CV mice using an OMEGA E.Z.N.A. Stool DNA Kit (OMEGA Biotech, Inc, Norcross, Georgia) per the manufacturer’s protocol. The concentration of DNA was determined using a Qubit fluorometer (Thermo Fisher Scientific, Waltham, Massachusetts). Bacterial 16S rDNA V4 amplicon sequencing was performed using an Illumina HiSeq 2500 sequencing system (250 bp paired-end; n = 3 per group; Beijing Genome Institute Americans Corporation, Cambridge, Massachusetts).
Demultiplexed and quality-filtered FASTQ files were analyzed using various python scripts in QIIME (Caporaso et al., 2010). Briefly, paired-end reads of the same sample were joined using join_paired_ends.py. Joined FASTQ files of all samples were each labeled with a unique identifier and then merged into one FASTA file using split_libraries_fastq.py for downstream analysis. Chimera sequences generated from PCR amplification artifacts were removed using identify_chimeric_seqs.py, and reference-based chimera detection was performed using usearch61 against the Greengenes 13_8 operational taxonomic units (OTUs) database (99_otus.fasta) (http://qiime.org/home_static/dataFiles.html). OTUs were first picked using pick_open_reference_otus.py against the reference database followed by de novo OTU-picking of unaligned sequences, and both forward and reverse strands were considered. OTU tables were then sorted, format-converted, and taxonomy-summarized from level 2 (L2, phylum level) to L7 (species level) using various python scripts in QIIME. The α- and β-diversities were determined using alpha_rarefaction.py and jackknifed_beta_diversity.py, respectively. Predictive functional profiling of 16S rDNA data at the class level (L3) was performed using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) (Langille et al., 2013) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways.
qPCR validation of microbial BA-metabolizing enzymes
The DNA encoding selected microbial enzymes—the BA inducible operons (bai) CD and baiJ—that are known to convert primary BAs to secondary BAs as well as the BA-deconjugation enzyme bile salt hydrolase (bsh) were validated by quantitative polymerase chain reaction (qPCR) using a BioRad CFX384 Real-Time PCR Detection System (Bio-Rad, Hercules, California). For each qPCR reaction, 30 ng of DNA reaction was loaded, and data were normalized to universal bacterial 16S rDNA. The bacterial DNA qPCR primers are shown in Supplementary Table 2.
Bile acid extraction
Bile acids from liver, serum, as well as SIC and LIC pellets were extracted using a similar method as previously described in Zhang and Klaassen (2010) with modifications. Briefly, for liver and intestinal content pellets, 5 μl of ice-cold analytical grade H2O were added per mg of sample, homogenized in a glass Dounce homogenizer until uniform, and 250 μl of homogenate was removed and mixed with 10 μl of the following 5 ISs: CA-d4, DCA-d4, GCA-d4, G-CDCA-d4, and LCA-d4. Samples were then precipitated in acetonitrile with 5% NH4OH and centrifuged to obtain pellets. Pellets were resuspended in methanol, centrifuged, and the supernatant was removed and dried under vacuum. For serum, 50 μl of serum was mixed with 10 μl of IS, precipitated with ice-cold methanol, centrifuged, and the supernatant was dried under vacuum. The precipitates were reconstituted in 100 μl 50% methanol water solution prior to LC-MS.
LC-MS quantification of BAs
LC-MS analysis was performed on a Waters Acquity I-Class UPLC TQS-micro MS (Waters, Milford, Massachusetts) system. Five microliters of each sample were injected for analysis using negative ionization mode. Chromatographic separation was performed on a Waters XSelect HSS T3 column (2.5 µm, 2.1 mm × 150 mm). The flow rate was 0.3 ml/min, auto-sampler temperature was kept at 4°C, and the column compartment was set at 40°C. The mobile phase was composed of Solvents A (5 mM ammonium acetate in H2O with 0.1% acetic acid) and B (acetonitrile with 0.1% acetic acid). After the initial 1 min isocratic elution of 75% A, the percentage of Solvent A progressively decreased to 5% at t = 15 min. The composition of Solvent A was maintained at 5% for 10 min (t = 25 min), and then the percentage of A returned to 75% at t = 25 for 10 min to prepare for the next injection. The total experimental time for each injection was 40 min. The mass spectrometer was equipped with an electrospray ionization (ESI) source. Targeted data acquisition was performed in multiple-reaction-monitoring (MRM) mode. A total of 56 BAs (Supplementary Table 1) and 5 IS MRM transitions were monitored in the negative mode; tauro-α muricholic acid (T-αMCA) and T-βMCA could not be separated by chromatography and were analyzed together as T-α/βMCA. The metabolite identities were confirmed by spiking mixtures of standard compounds. The extracted MRM peaks were integrated using the TargetLynx software (Waters).
RNA isolation and RT-qPCR of host BA-processing genes
Total RNA was isolated from liver and ileum tissue using RNAzol Bee reagent per manufacturer’s protocol (Tel-Test, Inc, Friendswood, Texas). RNA concentration was determined using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Waltham, Massachusetts) at 260 nm. Integrity of total RNA samples was confirmed by gel electrophoresis with visualization of 28S and 18S rRNA bands under ultraviolet light. Reverse transcription was performed using a High Capacity cDNA Synthesis Kit (Applied Biosystems, Foster City, California). The cDNAs were amplified by PCR using SsoAdvanced Universal SYBR Green Supermix in a BioRad CFX384 Real-Time PCR Detection System (Bio-Rad). The qPCR primers targeting the cDNAs of the host BA-processing genes are shown in Supplementary Table 3. Data were normalized to the geometric means of the ddCq values of two housekeeping genes, namely glyceraldehyde 3-phosphate dehydrogenase (Gapdh) and β-actin.
Data analysis
For CV and GF mice exposed to chemicals or vehicle, statistical differences were determined using two-way ANOVA (exposure and enterotype) followed by Tukey’s post hoc test (adjusted p-value < .05) in R. Two-way hierarchical clustering dendrograms of BAs were generated using the native function of R. For CV mice only, differentially regulated taxa were determined using one-way ANOVA followed by Duncan’s post hoc test (p < .05). The R package corrplot was used for Pearson’s correlation and visualization between differentially regulated bacteria at the species level (L7) and the pooled sum of 14 BAs from all four compartments that were statistically significant among exposure groups (p < .05).
RESULTS
Composition and Predicted Functions of Bacteria in the Large Intestine
Overall, following PCN or TCPOBOP, we did not observe an apparent increase in body weight of CV or GF mice. As expected, TCPOBOP-exposed mice had increased liver weight due to increased cell proliferation. To determine the effect of pharmacological activation of PXR and CAR on the composition and predicted functions of the gut microbiome in CV mice, DNA was isolated from LIC and 16S rDNA sequencing was performed as described in the Materials and Methods section. The alpha diversity among corn oil, PCN, and TCPOBOP was determined using the Chao1 index, and no differences were observed in species richness and evenness (Supplementary Figure 1A). The principle coordinates analysis plot of beta diversity as calculated by weighted UniFrac showed that mice of the same exposure had similar taxa abundance and phylogeny (Figure 1A).
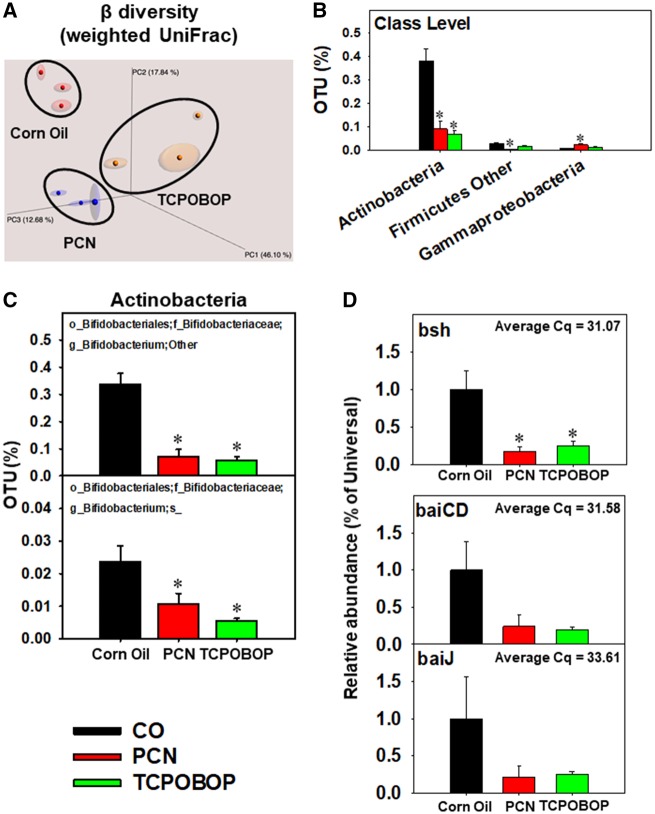
Figure 1.
A, Beta diversity of LIC microbiota in CV mice orally exposed to CO, PCN, or TCPOBOP (n = 3 per group). The 16S rDNA sequencing data were analyzed using QIIME as described in the Materials and Methods section. B, Differentially regulated intestinal bacteria at the class level (L3). C, Differentially regulated bacteria at the species level (L7) from the class Actinobacteria. D, Abundance of intestinal microbial DNA encoding bsh, which is known to be present in certain Bifidobacterium spp. (Ridlon et al., 2006), as well as bile acid inducible operons (bai) CD and J, which are known to be present in certain Clostridia species (Ridlon et al., 2006), from CV mice exposed to corn oil, PCN, or TCPOBOP (n = 5 per group) (qPCR). Asterisks (*) represent statistically significant differences as compared with corn oil-exposed group (one-way ANOVA followed by Duncan’s post hoc test; statistical significance was considered at p < .05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
At the class level, PCN and/or TCPOBOP differentially regulated the percentage of OTU of three taxa: Actinobacteria (decreased by PCN and TCPOBOP), Firmicutes Other (decreased by PCN), and Gammaproteobacteria (increased by PCN) (Figure 1B). Specifically, the PCN- and TCPOBOP-mediated decrease in the relative abundance of Actinobacteria was attributed to decreased Bifidobacterium spp. (Figure 1C). Bifidobacterium spp. are known to express the BA deconjugation enzyme bsh (Ridlon et al., 2006). Consistent with reduced relative abundance of Bifidobacterium spp., qPCR analysis of the LIC DNA showed a decrease in the abundance of bsh genes (Figure 1D). Furthermore, the baiCD and baiJ, which metabolize primary BAs to secondary BAs, tended to decrease, but were not statistically significant (Figs. 2D–G); baiCD and baiJ are known to be present in certain Clostridia species (Ridlon et al., 2006). This data suggests that PCN and TCPOBOP exposure may reduce secondary BA synthesis.
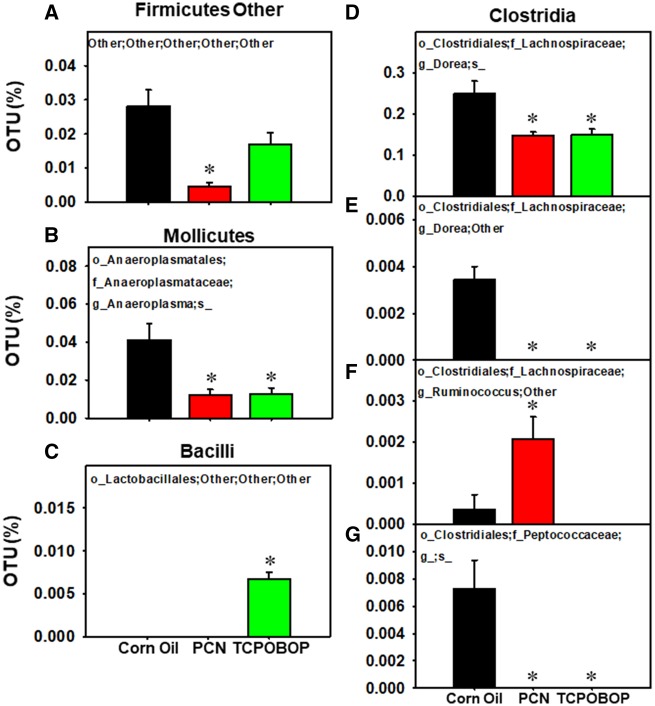
Figure 2.
Differentially regulated intestinal bacteria at the species level (L7) within classes Firmicutes Other (A), Mollicutes (B), Bacilli (C), and Clostridia (D) from LIC of CV mice orally exposed to CO, PCN, or TCPOBOP (n = 3 per group) (16S rDNA sequencing). Data were analyzed using QIIME as described in the Materials and Methods section. Asterisks (*) represent statistically significant differences as compared with corn oil-exposed group (one-way ANOVA followed by Duncan’s post hoc test; statistical significance was considered at p < .05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
As shown in Figure 2, several other taxa were differentially regulated by PCN and/or TCPOBOP exposure. For example, PCN increased the Ruminococcus genus in the Clostridia class (Figure 2F). TCPOBOP increased a taxon of the Lactobacillales order in the Bacilli class, and this taxon was undetected in vehicle and PCN-exposed groups (Figure 2C). Both PCN and TCOBOP reduced the relative abundance of the Actinobacteria class, Anaeroplasma spp. in the Mollicutes class (Figure 2B), two taxa in the Dorea genus (Figs. 2D and 2E), as well as a taxon in the Peptococcaceae family (Figure 2G).
PICRUSt was used to predict the abundance of gene functions of 274 identified gene families that form distinct KEGG pathways. PCN did not affect any of the KEGG pathways of the gut microbiome. However, TCPOBOP increased the gene family counts of three KEGG pathways: mineral absorption, ether lipid metabolism, and phosphotransferase system (Supplementary Figure 1B). The KEGG pathway for secondary bile acid synthesis was present, but not significantly altered by PCN or TCPOBOP exposure.
BAs in Liver, Serum, SIC, and LIC
To determine how oral exposure to PCN and TCPOBOP affects BA homeostasis and to test our hypothesis that the decrease in BA-metabolizing bacteria and BA-metabolizing enzymes (Figs. 1C and 1D) leads to reduced formation of secondary BAs, targeted metabolomics of 56 BAs in serum, liver, SIC, and LIC of CV mice given corn oil, PCN, or TCPOBOP. Individual BAs were classified into seven major categories as described in Supplementary Table 4: total, primary, secondary, conjugated, unconjugated, 6-OH, and 12-OH BAs. Interestingly, in all compartments tested, we detected low but significant levels of total secondary BAs. Because the GF mice were confirmed for the sterile conditions by bacterial culture and 16S rRNA qPCR, the presence of secondary BAs is not likely due to contamination. It is possible that certain BAs that were defined as secondary BAs may undergo host synthesis/repair mechanisms in GF conditions.
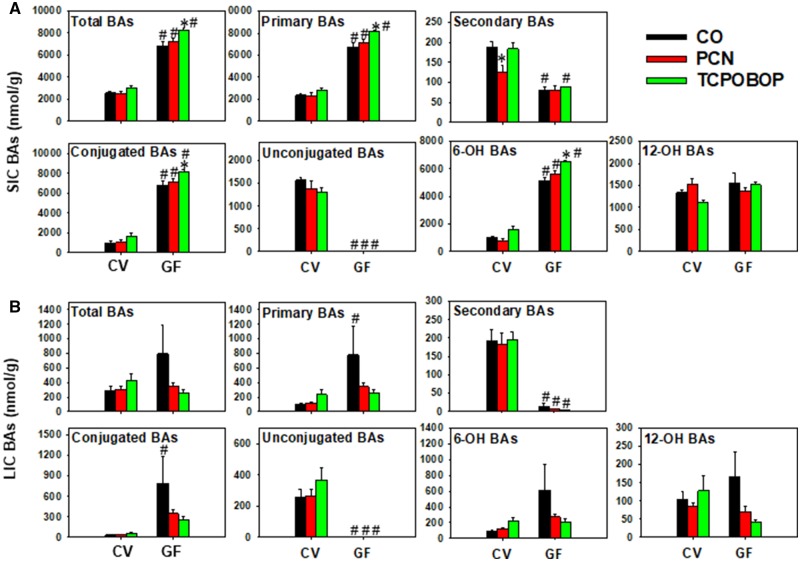
In liver, 21 BAs were above the limit of detection, and 18 of them were differentially regulated by either chemicals or enterotype. Under basal conditions, GF mice had higher total BAs than CV mice, and this was attributed to higher primary, conjugated, and unconjugated BAs (Figure 3A). In addition, there was a basal increase in 6-OH BAs but not 12-OH BAs in the absence of gut microbiota. Following chemical exposure, consistent with an apparent decrease in BA-metabolizing microbial enzymes (bsh, baiCD, and baiJ; Figs. 1C and 1D), both PCN and TCPOBOP decreased total secondary BAs in liver in a microbiota-dependent manner (Figure 3A). PCN and TCPOBOP did not alter BAs from other categories in livers of CV mice. In livers of GF mice, PCN also had minimal effect on any of the seven BA categories. In contrast, lack of gut microbiota augmented the TCPOBOP-mediated hepatic increase in total BAs, which was attributed to an increase in primary, conjugated, and 6-OH BAs, as well as an apparent increase in 12-OH BAs, but not unconjugated BAs (Figure 3A).
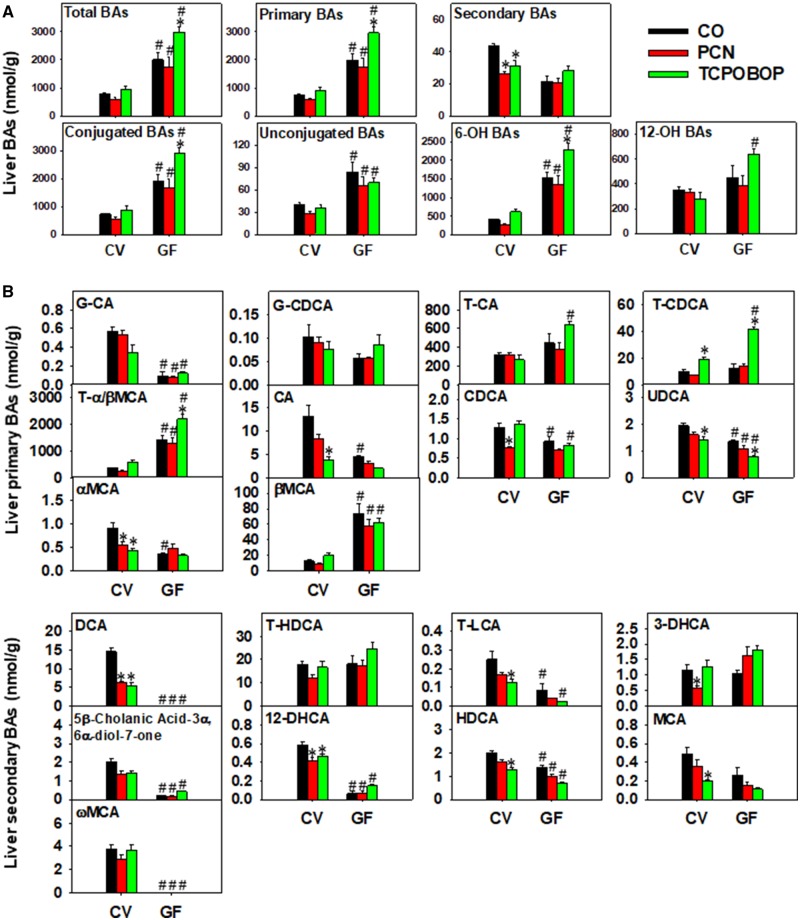
Figure 3.
A, LC-MS quantification of total BAs, primary BAs, secondary BAs, conjugated BAs, unconjugated BAs, 6-OH BAs, and 12-OH BAs in livers of CV and GF mice exposed to corn oil, PCN, or TCPOBOP (n = 4–5 per group) as described in the Materials and Methods section. B, Individual differentially regulated primary BAs (top panels) and secondary BAs (bottom panels) in livers of CV and GF mice exposed to corn oil, PCN, and TCPOBOP. Asterisks (*) represent statistically significant differences as compared with corn oil-exposed group of the same enterotype; pounds (#) represent statistically significant differences between CV and GF mice exposed to the same chemical (two-way ANOVA followed by Tukey’s post hoc test; statistical significance was considered at adjusted p-value < .05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A two-way hierarchical clustering dendrogram of the relative concentrations of individual BAs in livers of CV and GF mice is shown in Supplementary Figure 2, and examples of individual differentially regulated BAs are shown in Figure 3B. Under basal conditions, a total of two BAs (βMCA and T-α/βMCA) were higher and 11 BAs (UDCA, HDCA, CDCA, T-LCA, 12-DHCA, G-CA, T-DCA, ωMCA, 5β-cholanic acid-3α, 6α-diol-7-one, T-CA, and T-CDCA) were lower in livers of GF mice compared with CV mice. In livers of CV mice, PCN-reduced CDCA, αMCA, 12-DHCA, T-DCA, and 5β-cholanic acid-3α, 6α-diol-7-one, all in a gut microbiota-dependent manner; whereas TCPOBOP-reduced UDCA, HDCA, MCA, T-LCA, αMCA, 12-DHCA, T-DCA, 5β-cholanic acid-3α, 6α-diol-7-one, and CA in a gut microbiota-dependent manner except for UDCA. Conversely, TCPOBOP increased T-CDCA in livers of CV mice, and such increase was further augmented in livers of GF mice. In addition, lack of gut microbiota potentiated the TCPOBOP-mediated increase in T-α/βMCA. T-HDCA was not altered by chemical exposure or lack of microbiota, but was a major contributor to secondary BAs in GF mice.
In serum, 13 BAs were above the LOD, and 8 were differentially regulated by either chemicals or enterotype. As shown in Supplementary Figure 3A, control GF mice had higher primary, conjugated, and 6-OH BAs in serum than control CV mice. In serum of GF mice, PCN appeared to lower the serum primary, conjugated and 6-OH BAs, whereas TCPOBOP did not have a major effect on these BAs.
The regulation of individual BAs in serum of CV and GF mice is shown in Supplementary Figure 3B. Under basal conditions, two BAs (ie, the secondary BAs DCA and T-DCA) were decreased, whereas three BAs (T-α/βMCA, T-HDCA, and T-UDCA) were increased in serum of GF mice relative to CV mice. PCN and TCPOBOP in general did not alter these individual BAs in serum, except for a marked increase in T-CDCA by TCPOBP in serum of GF mice. Serum of PCN-exposed GF mice had lower T-DCA and CA as compared with serum of PCN-exposed CV mice. Serum of TCPOBOP-exposed CV mice also had lower T-DCA as compared with serum of TCPOBOP-exposed CV mice, but had higher T-α/βMCA, T-CA, T-HDCA, and T-UDCA. These data are summarized as a two-way hierarchical clustering dendrogram of the relative concentrations of individual BAs as shown in Supplementary Figure 3C.
For intestinal content, a systematic comparison between BA profiles in SIC and LIC of CV and GF mice exposed to corn oil, PCN, or TCPOBPOP was performed as shown in Figures 5 and 6 and Supplementary Figures 4 and 5.
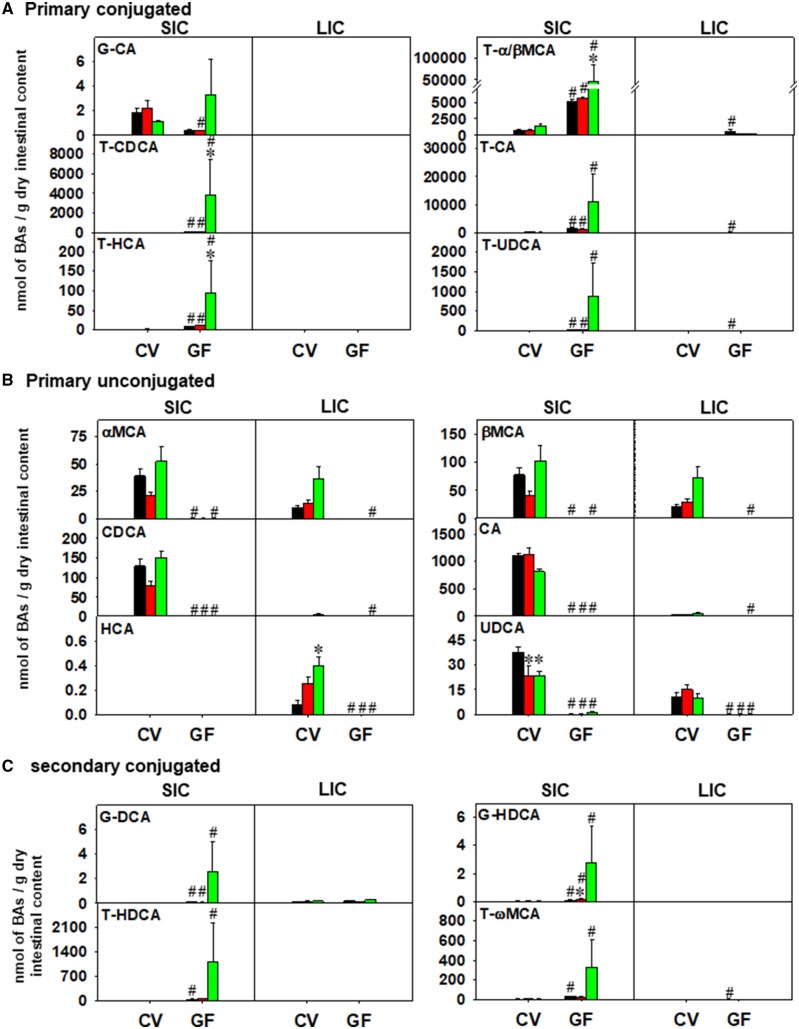
Figure 5.
LC-MS quantification of primary conjugated BAs (A), primary unconjugated BAs (B), and secondary conjugated BAs (C) in SIC and LIC of CV and GF mice exposed to corn oil, PCN, or TCPOBOP (n = 4–5 per group) as described in the Materials and Methods section. Asterisks (*) represent statistically significant differences as compared with corn oil-exposed group of the same enterotype; pounds (#) represent statistically significant differences between CV and GF mice exposed to the same chemical (two-way ANOVA followed by Tukey’s post hoc test; statistical significance was considered at adjusted p-value < .05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Figure 6.
Individual differentially regulated BAs in SIC and LIC of CV and GF mice exposed to corn oil, PCN, or TCPOBOP (n = 4–5 per group) as described in the Materials and Methods section. Asterisks (*) represent statistically significant differences as compared with corn oil-exposed group of the same enterotype; pounds (#) represent statistically significant differences between CV and GF mice exposed to the same chemical (two-way ANOVA followed by Tukey’s post hoc test; statistical significance was considered at adjusted p-value < .05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
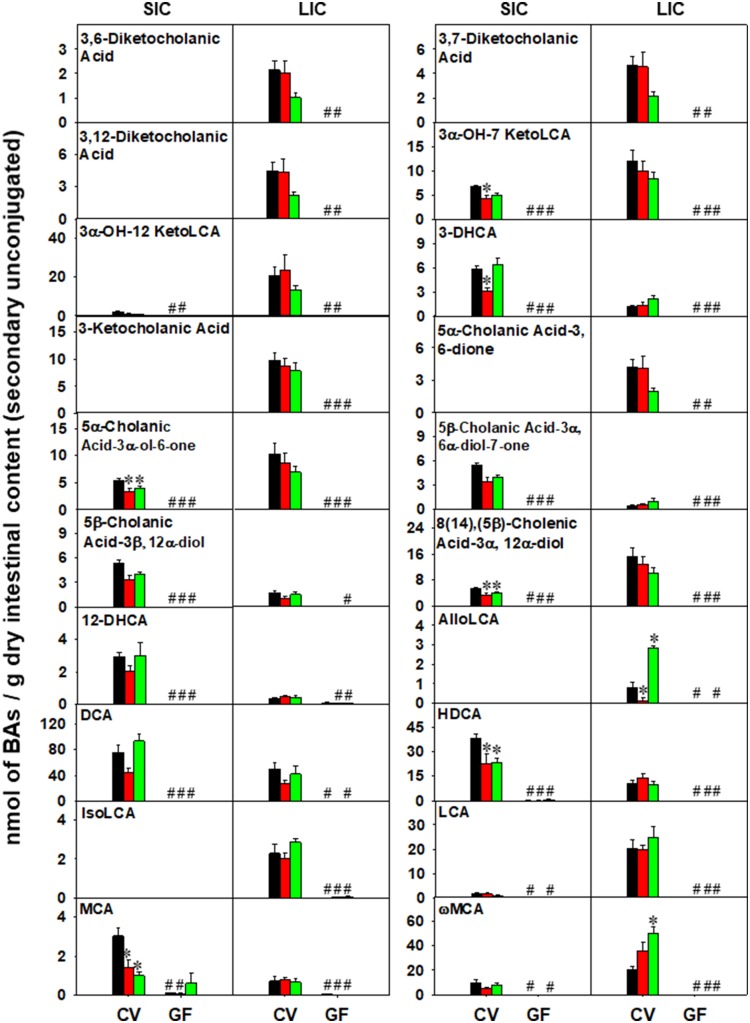
In SIC, 32 BAs were detected above the LOD, and they were all differentially regulated by either chemical exposure or enterotype. As shown in Figure 4A, control GF mice, as expected, had higher total BAs compared with CV mice, and this was attributed to primary BAs, whereas secondary BAs were decreased. The basal levels of conjugated BAs were higher and unconjugated BAs were lower in GF control compared with CV control mice. BAs from the alternative pathway (6-OH BAs) but not the classic pathway (12-OH BAs) were preferably increased in the absence of gut microbiota. The increase in total, primary, conjugated, and 6-OH BAs and the decrease in unconjugated BAs was maintained in GF mice following PCN and TCPOBOP exposure. Interestingly, there was a decrease in secondary BAs by PCN exposure. In contrast, lack of gut microbiome potentiated the TCPOBOP-mediated increase in total, primary, conjugated, and 6-OH BAs in SIC. In LIC, 35 BAs were detected above the LOD. As shown in Figure 4B, control GF mice had higher primary and conjugated BAs than control CV mice, but there was no statistical increase in total BAs (Figure 5A). The basal levels of secondary and unconjugated BAs were markedly lower in GF mice, and the decreased levels of secondary and unconjugated BAs was remained significant following PCN and TCPOBOP exposure. There were no PCN- or TCPOBOP-mediated differences among the BA categories listed in Figure 4B.
Figure 4.
A, LC-MS quantification of total BAs, primary BAs, secondary BAs, conjugated BAs, unconjugated BAs, 6-OH BAs, and 12-OH BAs in SIC of CV and GF mice exposed to corn oil, PCN, or TCPOBOP (n = 4–5 per group) as described in the Materials and Methods section. B, Individual differentially regulated primary BAs (top panels) and secondary BAs (bottom panels) in SIC of CV and GF mice exposed to corn oil, PCN, and TCPOBOP. Asterisks (*) represent statistically significant differences as compared with corn oil-exposed group of the same enterotype; pounds (#) represent statistically significant differences between CV and GF mice exposed to the same chemical (two-way ANOVA followed by Tukey’s post hoc test; statistical significance was considered at adjusted p-value < .05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Regarding the regulation of individual BAs in SIC of CV and GF mice, as shown in Figure 5A and Supplementary Figure 4, under basal conditions, a total of 17 BAs (UDCA, HDCA, 5β-Cholanic Acid-3α, 6α-diol-7-one, CA, βMCA, αMCA, CDCA, DCA, G-HDCA, G-DCA, T-HDCA, T-UDCA, T-α/βMCA, T-HCA, T-CA, T-ωMCA, and T-CDCA) were increased, and ten BAs (8(14),(5β)-cholenic acid-3α, 12α-diol, 3α-OH-7 KetoLCA, 5α-cholanic acid-3α-ol-6-one, 12-DHCA, ωMCA, 3-DHCA, LCA, MCA, 3α-OH-12 KetoLCA, and T-CDCA) were decreased in SIC of GF mice. In livers of CV mice, PCN decreased UDCA, HDCA, 8(14),(5β)-cholenic acid-3α, 12α-diol, 3α-OH-7 KetoLCA, 5α-cholanic acid-3α-ol-6-one, and MCA. TCPOBOP also decreased MCA, but increased UDCA, HDCA, 8(14),(5β)-cholenic acid-3α, 12α-diol, 5α-cholanic acid-3α-ol-6-one in SIC of CV mice. In SIC of GF mice, lack of gut microbiota potentiated the TCPOBOP-mediated increase in T-α/βMCA, T-HCA, and T-CDCA and the PCN-mediated increase in G-HDCA.
Regarding the regulation of individual BAs in LIC of CV and GF mice, as shown in Figure 6 and Supplementary Figure 5, the lack of gut microbiota had a greater regulatory effect on BA levels in LIC than did chemical exposure. In LIC of control GF mice, a total of four BAs (T-α/βMCA, T-ωMCA, T-UDCA, and T-CA) were increased and 20 BAs were decreased (3,12-diketocholanic acid, 3,7-diketocholanic acid, 5α-cholanic acid-3, 6-dione, 3,6-diketocholanic acid, 3α-OH-12 KetoLCA, 3-ketocholanic acid, 3α-OH-7 KetoLCA, 5α-cholanic acid-3α-ol-6-one, 8(14),(5β)-cholenic acid-3α, 12α-diol, HDCA, UDCA, MCA, DCA, LCA, IsoLCA, 3-DHCA, 5β-cholanic acid-3α, 6α-diol-7-one, ωMCA, HCA, and AlloLCA). In LIC of CV mice, PCN-reduced AlloLCA, whereas TCPOBOP increased AlloLCA, HCA, and ωMCA in a gut microbiome-dependent manner. Neither PCN nor TCPOBOP altered LIC BAs in GF conditions.
Pearson Correlation Analysis of LIC Bacteria and BAs
To determine the relationship between BAs and gut microbiota, BAs that were differentially regulated in serum (Supplementary Figure 6A) and LIC (Supplementary Figure 6B) were correlated against the 11 differentially regulated bacteria in LIC. In serum, the primary conjugated BAs T-α/βMCA and T-CDCA were negatively correlated with seven taxa (two Bifidobacterium spp., two Dorea spp., Peptociccaceae, Anaeroplasma spp., and Ruminococcus). In contrast, the majority of these taxa (except for Ruminococcus) were positive correlated with the secondary unconjugated BA DCA. The taxa Lactobacillales, Bradyrhizobiaceae, and Anaerotruncus spp. were positively correlated with T-α/βMCA and T-CDCA but negatively correlated with DCA. In LIC, Bifidobacterium spp., Dorea spp., Peptociccacea, and Anaeroplasma spp. were positively correlated with the conjugated primary BA T-UDCA, but negatively correlated with the unconjugated BAs (HCA, αMCA, βMCA, AlloLCA, ωMCA, 3-DHCA). Conversely, Lactobacillales, Bradyrhizobiaceae, and Anaerotruncus spp. were positively correlated with these unconjugated BAs, but negatively correlated with T-UDCA.
Hepatic Gene Expression
Xenobiotic receptor-target genes
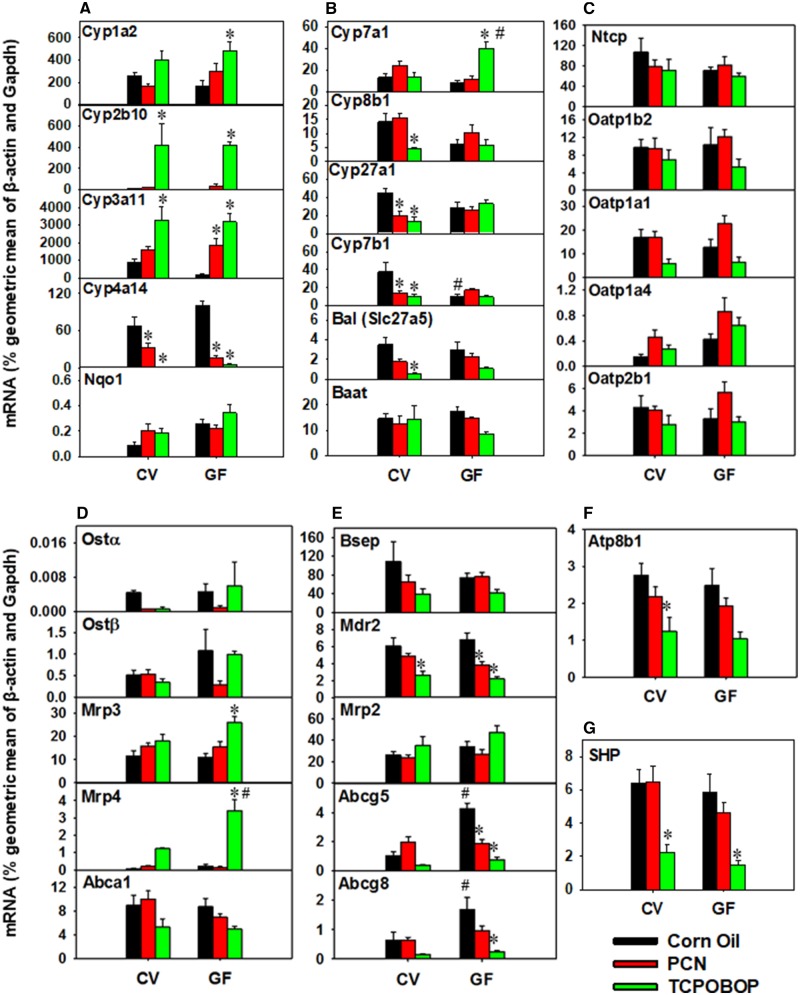
The mRNA expression of five prototypical target genes of major xenobiotic-sensing transcription factors were determined to confirm pharmacological activation of PXR by PCN or CAR by TCPOBOP (Figure 7A). Regarding PXR activation, PCN increased the expression of Cyp3a11 in GF mice and tended to increase expression in CV mice, suggesting that lack of gut microbiota potentiates PXR activation, a similar observation as we reported previously (Cui and Klaassen, 2016). CAR activation by TCPOBOP increased the expression of Cpy2b10 as expected (Cui and Klaassen, 2016). Lack of gut microbiota sensitized the TCPOBOP-mediated increase in the expression of Cyp1a2, which has been reported to carry a CAR binding motif in the promoter region (Yoshinari et al., 2010). Both PXR and CAR activation decreased the peroxisome proliferator-activated receptor α (PPARα) target gene Cyp4a14, and this was consistent with our previous reports (Cui and Klaassen, 2016; Li et al., 2016). No changes in gene expression were observed in CV or GF mice for the nuclear factor, erythroid derived 2, like 2 (Nfe2l2/Nrf2) target gene NAD(P)H dehydrogenase, quinone 1 (Nqo1).
Figure 7.
RT-qPCR quantification of mRNAs in livers of CV and GF mice exposed to corn oil, PCN, and TCPOBOP (n = 4–5 per group) for prototypical PXR- and CAR-regulated target genes (A), primary BA synthesis (B), hepatic BA uptake transporters (C), basolateral efflux transporters for BAs and cholesterol (D), canalicular efflux transporters for BAs, lipids, and cholesterol (E and F), and the repressive nuclear receptor Shp (G). The mRNA was calculated by dividing the ddCq for the gene of interest by the ddCq for the geometric mean of β-actin and Gapdh. Asterisks (*) represent statistically significant differences as compared with corn oil-exposed group of the same enterotype; pounds (#) represent statistically significant differences between CV and GF mice exposed to the same chemical (two-way ANOVA followed by Tukey’s post hoc test with adjusted p-value < .05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Hepatic BA synthesis genes
The mRNA expression for six major BA-processing genes was determined (Figure 7B). Interestingly, for the classic pathway of BA synthesis, lack of gut microbiota potentiated the TCPOBOP-mediated increase in the mRNA of the rate-limiting BA-synthesizing enzyme Cyp7a1 in liver, and this corresponded with an increase in total and primary BAs in liver of GF mice but not in CV mice (Figure 3A). In livers of CV mice, TCPOBOP decreased the expression of Cyp8b1, which produces CA and determines the ratio between CA (classic pathway) and CDCA (alternative pathway). For the alternative pathway of BA synthesis, in CV mice, PCN and TCPOBOP exposure decreased the expression of the rate-limiting enzyme Cyp27a1 and the downstream enzyme Cyp7b1. Regarding BA conjugation, TCPOBOP decreased Slc27a5/Bal in a gut microbiome-dependent manner; however, expression of the BA-amino acid conjugating enzyme BA-CoA: amino acid N-acyltransferase Baat remained unchanged.
Hepatic basolateral uptake transporters
In general, neither chemical exposure nor enterotype altered the mRNA expression of hepatic basolateral uptake transporters examined (Figure 7C). TCPOBOP exposure tended to decrease the expression of several transporters (Oatp1a1, 1b2, 2b1), but statistical significance was not achieved.
Hepatic basolateral efflux transporters
PCN and TCPOBOP had no effect on basolateral efflux transporters examined in the livers of CV mice (Figure 7D). However, lack of gut microbiota potentiated the TCPOBOP-mediated increase in the multidrug resistance-associated protein ATP-binding cassette, sub-family C (CFTR/MRP), member 3 (Abcc3/Mrp3), and Abcc4/Mrp4.
Hepatic canalicular efflux transporters for BAs, lipids, and cholesterol
For BA canalicular efflux transporters in CV and GF mice, TCPOBOP exposure decreased the expression of the phospholipid transporter Abcb4/Mdr2, whereas lack of gut microbiota and PCN downregulated mRNA expression of Mdr2 (Figure 7E). In livers of GF mice, the basal expression of the cholesterol efflux dimer Abcg5/g8 was higher than in livers of CV mice in control conditions, but PCN and TCPOBOP exposure decreased their expression toward CV levels. In liver of CV mice, the mRNA of the canalicular membrane transporter ATPase, class I, type 8B, member 1 (Atp8b1), which translocates phosphatidylserine and phosphatidylethanolamine from the outer to the inner leaflet bilayer (Klaassen and Aleksunes, 2010), was decreased by TCPOBOP exposure and tended to decrease by TCPOBOP in GF mice (Figure 7F).
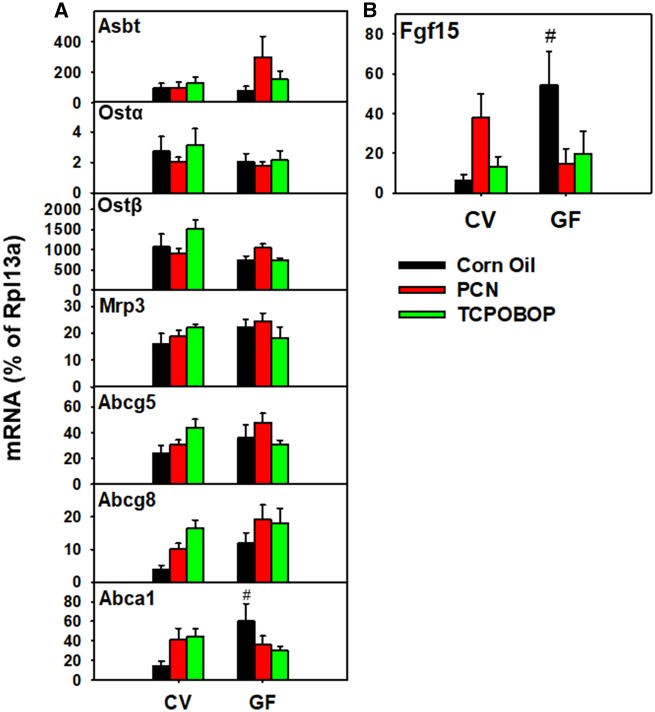
Hepatic Small Heterodimer Partner and Ileal BA-Processing Gene Expression
Hepatic expression of the repressive nuclear hormone nuclear receptor subfamily 0, group B, member 2 (Nr0b2/small heterodimer partner [SHP]) was decreased by TCPOBOP exposure in both CV and GF mice, but not by PCN exposure (Figure 7G). Overall, there were no significant PCN- or TCPOBOP-mediated effects on the expression of BA transporters in CV or GF mice (Figure 8). The mRNA expression of the cholesterol efflux transporter Abca1 was higher in vehicle-exposed GF mice than CV, and the enterotype effect was attenuated by PCN and TCPOBOP in GF mice (Figure 8A). Similarly, the BA signaling hormone Fgf15 was increase in GF corn oil mice compared with CV corn oil mice, but the enterotype difference was attenuated following exposure to PCN or TCPOBOP (Figure 8B).
Figure 8.
RT-qPCR quantification of mRNAs in ileum of CV and GF mice exposed to corn oil, PCN, and TCPOBOP (n = 4–5 per group) for intestinal BA transporters (A) and the murine bile acid synthesis regulating hormone Fgf15 (B). The mRNA was calculated by dividing the ddCq for the gene of interest by the ddCq for the geometric mean of β-actin and Gapdh. Asterisks (*) represent statistically significant differences as compared with corn oil-exposed group of the same enterotype; pounds (#) represent statistically significant differences between CV and GF mice exposed to the same chemical (two-way ANOVA followed by Tukey’s post hoc test with adjusted p-value < .05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Hepatic Muricholic Acid Synthesis Genes
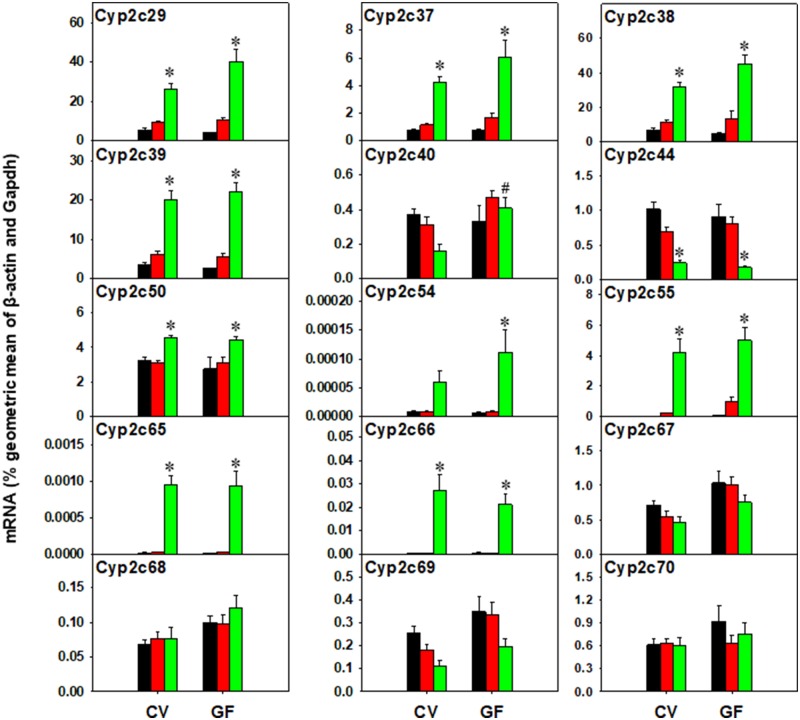
Recently, it was shown that in mice, Cyp2c70 is responsible for the species difference in mice regarding the synthesis of αMCA and βMCA from CDCA and UDCA, (Takahashi et al., 2016), but not Cyp3a11 (Wahlstrom et al., 2017). We speculate that other Cyp2c members may also contribute to muricholic acid-synthesis in liver. Therefore, the mRNA of Cyp2c70 and other Cyp2c members was quantified by RT-qPCR as shown in Figure 9.
Figure 9.
RT-qPCR quantification of mRNAs of Cyp2c genes in liver of CV and GF mice exposed to corn oil, PCN, and TCPOBOP (n = 4–5 per group). The mRNA was calculated by dividing the ddCq for the gene of interest by the ddCq for the housekeeping gene Rpl13a. Asterisks (*) represent statistically significant differences as compared with corn oil-exposed group of the same enterotype; pounds (#) represent statistically significant differences between CV and GF mice exposed to the same chemical (two-way ANOVA followed by Tukey’s post hoc test with adjusted p-value < .05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
There were no differences in basal expression of Cyp2c genes between CV and GF mice, and PCN did not alter the Cyp2c gene expression in CV or GF mice. In contrast, TCPOBOP increased the mRNAs of Cyp2c29, 2c37, 2c38, 2c39, Cyp2c50, 2c55, 2c65, and 2c66 in both CV and GF mice. The TCPOBOP-mediated increase in the expression of Cyp2c54 was potentiated in livers of GF mice. Conversely, Cyp2c44 mRNA was decreased by TCPOBOP in both CV and GF mice. The expression of Cyp2c67, 2c68, 2c69, and 2c70 was not altered by any exposure conditions.
DISCUSSION
Taken together, this study used 16S rDNA sequencing, BA targeted metabolomics, bioinformatics, and RT-qPCR analysis, to determine the gut microbiota-dependent regulation of BA homeostasis in response to pharmacological activation of PXR or CAR in mice. Although there is a breadth of research demonstrating how PXR and CAR activators and repressors modulate host gene expression in the liver, there is limited information regarding how bacteria in the gut are regulated by host receptor activation. This study provides evidence of the effect of pharmacological activation of the host xenobiotic sensing nuclear receptors PXR and CAR on the gut microbiome composition and functions with a primary focus on bile acid homeostasis. Prior to our work, Dr Curtis Klaassen’s laboratory has systematically characterized the effect of pharmacological activation of PXR, aryl hydrocarbon receptor (AhR), and peroxisome proliferator-activated receptor α (PPARα) on bile acid homeostasis and the regulation of host bile acid processing genes in CV mice (Csanaky et al., 2018; Lickteig et al., 2016; Zhang et al., 2017, 2018). As a follow-up, this study adds the knowledge to this field by (1) examining the effect of pharmacological activation of PXR and CAR in both CV and GF mice; and (2) unveiling the regulation of gut microbiota by these receptors. The rationale for using GF mice was that they allowed for the investigation of primary BA regulation independently from secondary BAs. Investigation of the bacterial composition of LIC enabled the identification of taxonomic shift associated with host xenobiotic-sensing nuclear receptor activation and BA changes. Targeted metabolomics of BAs in four major compartments (liver, serum, SIC, and LIC) unveiled gut microbiota-dependent and xenobiotic-mediated effects on BA homeostasis.
Regarding PXR, several studies have established a role between PXR activation, anti-inflammation, and the microbiome. For example, the microbial metabolite indole-3-propionic acid is a PXR activator in intestine that promotes PXR to interact with Toll-like receptor 4 to protect the epithelial barrier integrity and reduce inflammation (Venkatesh et al., 2014). PXR activation is also known to suppress inflammatory responses in hepatocytes, and PXR activation was found to be lower in patients with inflammatory bowel disease (Sun et al., 2015; Wallace et al., 2010). In this study, we demonstrated that pharmacological activation of PXR reduced the relative abundance of Bifidobacterium spp., which have been shown to protect the intestinal epithelial barrier in inflammatory models (Figure 1 and summarized in Figure 10) (Guo et al., 2017; Wang et al., 2014). Previously, it was reported that PXR activation by the cholesterol-lowering drugs pravastatin and atorvastatin also decreased the abundance of Bifidobacterium spp. in a female mouse model (Caparros-Martin et al., 2017). Our study using PCN as a prototypical PXR activator showed consistent findings with the previous report using statins as PXR activators. Together, these studies suggest that the anti-inflammatory properties of PXR may decrease the necessity for immune-modulation by Bifidobacterium spp. in the gut. For other bacteria, Ruminococcus spp., which was increased by pharmacological activation of PXR (Figure 2 and summarized in Figure 10), is considered a beneficial taxon that facilitates the digestion of complex carbohydrates from high fiber food and has been negatively associated with diabetes and colon cancer (Lagier et al., 2012; Ze et al., 2012). PCN also decreased a nonspecific taxon in the Firmicutes phylum, which contains the class Clostridiales and are capable of producing toxic secondary BAs (Ridlon et al., 2006). Therefore, the benefits of host PXR activation may act in part through increasing the percentage of beneficial bacteria in the gut.
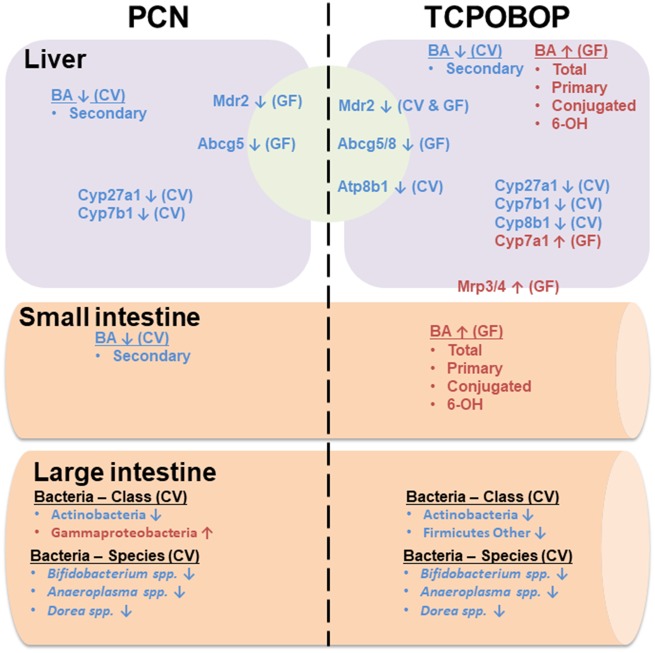
Figure 10.
A diagram illustrating the major findings of this study. Briefly, (1) PCN and TCPOBOP exposure decreased Bifidobacterium spp., Anaeroplasma spp., and Dorea spp.; (2) lack of gut microbiota potentiated an increase in total, primary, conjugated and 6-OH BAs in SIC and liver, whereas secondary BAs were increased in CV mice by PCN exposure; and (3) expression of the genes in the alternative BA synthesis were decreased by PCN and TCPOBOP exposure in CV and GF mice, whereas Cyp7a1 was increased in GF mice by TCPOBOP.
There are limited studies examining the interaction between CAR activation and the gut microbiome. The mRNA of CAR was found higher in livers of mice lacking a gut microbiome (Lundin et al., 2008). This study showed that pharmacological activation of CAR increased the Lactobacillales order, which is known to decompose plant and milk products through carbohydrate fermentation, producing lactic acid as the major metabolic product (Figure 2) (Kandler, 1983). Aneroplasma spp. was decreased by both CAR and PXR activation and is associated with fiber digestibility (Niu et al., 2015); Dorea spp., which colonizes mucosal regions where sialic acid is abundant, was also decreased by both PXR and CAR and has been shown to an increase in IBS patients, suggesting that Dorea spp. may not act positively on the host (Leclercq et al., 2014; Rajilic-Stojanovic et al., 2011; Tailford et al., 2015). CAR activation also decreased Bifidobacterium spp., which has been shown to protect the intestinal epithelial barrier in inflammatory models of postinfectious irritable bowel syndrome model in mice and in the human epithelial colorectal adenocarcinoma Caco-2 cell line (Guo et al., 2017; Wang et al., 2014). Activation of CAR has long been known to reduce blood-glucose and improve insulin sensitivity in diabetic patients (Lahtela et al., 1985; Sotaniemi and Karvonen, 1989) as well as suppress the expression of gluconeogenic genes in rat primary hepatocytes (Argaud et al., 1991) and mouse liver (Ueda et al., 2002). Later, in both leptin-deficient ob/ob mice and wild-type mice fed a high-fat diet, it was shown that CAR agonists decreased fasting serum-glucose levels and hepatic glucose production to decrease hyperglycemia and improve insulin sensitivity (Dong et al., 2009; Gao et al., 2009). However, it was also shown in a choline-deficient nonalcoholic steatohepatitis (NASH) mouse model that CAR activation caused lipid peroxidation and increased fibrosis (Yamazaki et al., 2007). Certain compounds from the gut microbiome, such as BAs and short-chain fatty acids from fiber, act as signaling molecules to host organs to regulate metabolic health, including obesity, type II diabetes, nonalcoholic fatty liver disease, and NASH (Makki et al., 2018; Schroeder and Backhed, 2016). More research is needed to understand how CAR may interact with the gut microbiome to improve metabolic health.
This study found that several bacterial genes involved in secondary BA synthesis were decreased or appeared to be downregulated after PXR or CAR activation (Figure 1). In particular, the PCN- and TCPOBOP-mediated decrease in Bifidobacterium spp. was associated with a decrease in the DNA encoding the bacterial BA deconjugation enzyme gene bsh. Because Bifidobacterium is known to carry bsh activity, the decrease in this taxa explains the decrease in the DNA encoding the bsh enzyme (Jarocki et al., 2014). In addition, there was an apparent decrease in the DNA encoding baiCD and baiJ, which are responsible for secondary BA synthesis. This corresponded with a decrease in total secondary BAs in liver following PCN and TCPOBOP exposure. Conjugated BAs are considered less toxic than unconjugated BAs and do not undergo passive diffusion due to increased size and ionization in the small intestine (Hofmann, 2009). However, overall, the decreased bsh genes did not significantly change the pool of conjugated or unconjugated BA in any compartment of CV mice, suggesting that the expression of bsh genes was maintained despite a decrease in gene number or additional genes may act similarly to bsh, but they have not been identified. Because the total BAs and primary BAs did not change due to treatment in CV mice, it is possible that PXR and CAR activation diminished the synthesis and/or increased the excretion of secondary BAs.
There is no evidence demonstrating that BAs can directly function as ligands of CAR, but the CAR activators phenobarbital and TCPOBOP have been shown to repress Cyp7a1 and may protect against BA toxicity similar to the functions of farnesoid X receptor (FXR/Nr1h4) and PXR (Guo et al., 2003; Li and Chiang, 2014; Miao et al., 2006). Following exposure to TCPOBOP, there was no change in the expression of Cyp7a1 in CV mice. However, there was a marked increase in GF mice that was potentiated by the lack of gut microbiome. This corresponded with an increase in total, primary, conjugated and 6-OH BAs in liver and SIC of GF mice exposed to TCPOBOP (summarized in Figure 10). Very little is known regarding the molecular mechanisms underlying the potentiated CAR ligand-mediated increase in Cyp7a1 mRNA in germ free conditions. It has been shown that during a lithogenic diet CAR increases Cyp7a1 expression to promote the metabolism of cholesterol into bile acids, as a compensatory mechanism (Cheng et al., 2017). It is known that livers of GF mice have higher cholesterol content (Mistry et al., 2017), thus it is possible that germ free conditions triggered the same signaling pathway for Cyp7a1 mRNA upregulation. Dr Curtis Omiecinski’s laboratory recently demonstrated identified a positive CAR-DNA binding site with over 30-fold of enrichment near the promoter of mouse Cyp7a1 gene following TCPOBOP exposure (Niu et al., 2018) (https://genome.ucsc.edu/cgi-bin/hgTracks? hgS_doOtherUser=submit&hgS_otherUserName=nxb939&hgS_otherUserSessionName=ChIP_exo_CAR). In summary, these observations from the literature and our data suggest that the increase in Cyp7a1 mRNA by CAR activation is sensitized by cholesterol overload and likely a result of an increase in direct CAR-binding.
Regarding individual BAs, the TCPOBOP-mediated, gut microbiome-dependent increase in BAs in liver is attributable to T-α/βMCA and T-CDCA as well as βMCA and T-CA, whereas the increase in SIC this was attributed to T-α/βMCA, T-CDCA, and T-HCA, as well as T-UDCA, T-CA, and T-ωMCA. The majority of these BAs are derivatives of CDCA an alternative pathway BA, which is converted to α/βMCA in mice by Cyp2c (Takahashi et al., 2016). To note, βMCA is an antagonist of FXR (Sayin et al., 2013). Because activation of FXR inhibits the expression of Cyp7a1, it is possible that potent CAR activation increases muricholic acids, which in turn blocks the FXR-mediated inhibition of BA synthesis. In a mouse bile duct ligation model, total BAs in serum increased less in CAR-null mice than PXR-null mice; in particular, this was due to a lesser increase in T-CA, T-CDCA, and T-βMCA, suggesting that constitutive activation of CAR is important in the synthesis of these BAs during conditions of toxicity (Stedman et al., 2005). Despite several studies showing indirect associations between CAR and FXR, there are no known direct interactions such that CAR and FXR co-regulate BA homeostasis (Beilke, Aleksunes, Olson, et al. 2009; Guo et al., 2003; Saini et al., 2004; Stedman et al., 2005). The present research shows that lack of gut microbiome potentiates BA synthesis following CAR activation and that the regulation of BA homeostasis by activation of CAR in mice may be microbiome-dependent.
In a study investigating the basal regulation of BAs between CV and GF mice, in liver, Selwyn et al. found increased total, primary, and conjugated BA species in GF mice compared with CV mice (Selwyn, Csanaky, et al., 2015), which is comparable with this study, and this was most likely attributable to an increase in T-α/βMCA (Figure 3). Under basal conditions, the concentrations of total, primary, conjugated, and 6-OH BAs were higher in GF compared with CV mice in multiple bio-compartments (liver, SIC, and LIC); in addition, unconjugated BAs were also higher in the liver of GF mice. It has been suggested that the increased BA pool in GF mice may be due to increased reabsorption of BAs and decreased excretion in the feces (Gustafsson et al., 1957; Riottot and Sacquet, 1985; Sayin et al., 2013). Consistent with Sayin et al., muricholic acids (T-α/βMCA and βMCA), which are derivatives of CDCA, were also increased in GF mice (Figs. 3 and 5, andSupplementary Figure 3), but was not previously reported in Swiss Webster mice (Sayin et al., 2013) and could reflect a strain difference with C57BL/6J mice. The increased BA pool may also be contributed by FXR antagonism due to increased T-α/βMCAs, which are known FXR antagonists (Sayin et al., 2013). FXR antagonism in intestine may subsequently suppress the intestinal Fgf15-mediated downregulation of the hepatic bile acid synthetic enzyme Cyp7a1. The lack of this FXR-Fgf15-Cyp7a1 negative feedback loop may explain the higher basal levels of bile acids in GF mice. The increased T-α/βMCAs may be due to decreased expression of Cyp8b1, which affects the ratio of CA (12-OH) to CDCA (6-OH). Alternatively, bacteria could preferentially metabolize muricholic acids over CA, resulting in the large increase in GF mice. Furthermore, the increased BA pool in GF mice may be due to the FXR antagonistic activity of T-α/βMCA in liver. T-α/βMCA had the highest concentration in GF mice in this study; following TCPOBOP in GF mice, both T-α/βMCA and Cyp7b1 increased, which is consistent with known FXR antagonism by T-βMCA.
Sayin et al. (2013) quantified 16 bile acids between CV and GF mice, among which six were secondary bile acids (DCA, LCA, ωMCA, and their taurine conjugates). In their GF mice, these secondary bile acids were not detected in liver, small intestine, or large intestine, and DCA and ωMCA were minimally detected in serum (Sayin et al., 2013). Quantification of DCA, LCA, ωMCA, and their taurine conjugates in this study, matched Sayin et al., with the exception of T-ωMCA in SIC. It was highly abundant in SIC, especially following exposure to TCPOBOP, suggesting that there may be an unknown host mechanism be capable of producing T-ωMCA that could be CAR-dependent. Other BAs traditionally considered secondary BAs monitored in this study were identified in GF mice. In particular, T-HDCA has similar concentration levels in liver between CV and GF mice and was markedly increased in GF SIC following TCPOBOP exposure, suggesting that there is a host mechanism for T-HDCA production that is CAR-dependent. Indeed, HDCA was previously identified in GF pigs (Haslewood, 1971). Overall, the quantification of some traditional secondary BAs in GF mice suggests that the gut microbiota may mask host synthesis of these BAs.
In a previous study, CAR was activated by TCPOBOP (3 mg/kg) in 12- to 15-week-old CV male mice for 4 days by intraperitoneal (i.p.) route and BAs were quantified in serum and liver and BA synthesis genes in liver and ileum were measured by qPCR (Lickteig et al., 2016). In serum, Lickteig et al. observed increased T-α/βMCA, T-UDCA, T-HDCA, and αMCA as well as decreased T-DCA and DCA following TCPOBOP exposure; whereas this study found no changes in BA categories in serum (Supplementary Figure 2). In liver, Lickteig et al. found that TCPOBOP exposure decreased total, primary, secondary, conjugated, unconjugated, and 12-OH BAs; regarding specific BAs, the primary BA T-CA was markedly decreased whereas the secondary BAs T-DCA, DCA, and T-LCA were decreased. This study only found a decrease in livers of CV mice of secondary BAs due to the minor BAs HDCA, MCA, and T-LCA, whereas T-DCA increased. Comparison of the BA concentrations between the two studies suggests that the route of exposure (i.p. vs oral) or age can influence individual BA species in serum. In addition, because the studies were performed at two different facilities, the different housing conditions (food, bedding, water, etc.) could have led to differing responses to CAR activation and microbiome composition. There were also different responses seen in hepatic BA synthesis gene expressions. Lickteig et al. found increased hepatic expression of Cyp7a1 and Cyp27a1 and decreased Cyp7b1 and Cyp8b1, which could be a compensatory response to decreased concentrations of BAs in liver. Whereas this study found no change in Cyp7a1 and decreased Cyp27a1, Cyp7b1, Cyp8b1, which could explain why total BAs were unchanged whereas the concentration of CA, CDCA, and their derivatives tended to decrease. Overall, both studies have demonstrated that CAR is an important regulator of BA homeostasis.
Pearson correlation was used to identify relationships between the differentially regulated taxa and BAs in serum and LIC (Figure 6). Bifidobacterium spp. and Lactobacillus spp. express the BA deconjugating enzyme bsh (Ridlon et al., 2006), so they were anticipated to be positively correlated with deconjugated BAs. In serum, Bifidobacterium spp. was positively correlated with deconjugated BAs; however, in LIC, Bifidobacterium spp. was positively correlated with conjugated BAs. It is possible that the deconjugation reactions by Bifidobacterium spp. are BA-specific. Consistent with the literature, Dorea was positively correlated with DCA in serum (Martin et al., 2018); Ruminococcus was also reported to be positively associated with deconjugation of TCA (Martin et al., 2018). The BA metabolism of Bradyrhizobiaceae is not known, however Gram-negatives are thought to have a higher BA tolerance than Gram-positive bacteria (Ridlon et al., 2016). The unknown taxa in the phylum Firmicutes may have a competitive advantage following changes in BA homeostasis as rodents fed BAs or high-fat diets tend to have increased Firmicutes (Hildebrandt et al., 2009; Ridlon et al., 2016). More research is needed to understand the specificity of BA-bacteria interactions and the selectivity of bacterial BA metabolizing enzymes.
CAR activation by TCPOBOP decreased the cholesterol canalicular efflux transporters Abcg5 and Abcg8 in livers of GF mice but not CV mice. The decreased hepatic expression of these transporters predicts a build-up of cholesterol and oxysterols in hepatocytes in the absence of the gut microbiome. The liver X receptors (LXRs) coordinately regulate sterol catabolism, storage, efflux, and elimination, including Abcg5 and Abcg8 (Berge et al., 2000; Repa et al., 2002), and mice lacking either gene accumulate cholesterol in the liver (Yu et al., 2002). CAR and LXR have distinct functions for coordinating a response to xenobiotics and promoting lipogenesis. In vivo CAR activation inhibited the expression of LXR target genes and LXR-induced lipogenesis, demonstrating the presence of a crosstalk between CAR and LXR (Gnerre et al., 2005). Therefore, it is plausible that CAR activation inhibited LXR-regulated genes, such as Abcg5 and Abcg8. In addition, both LXR and CAR interact with peroxisome proliferator-activated receptors (PPARs), and these interactions have been associated with atherosclerosis and obesity (Xu et al., 2018). Understanding the interactions between CAR, LXR, and PPARs may reveal new therapeutic targets for metabolic diseases and provide insight for the evaluation of BA-dependent toxicological responses. Similarly, in this study, SHP was decreased by CAR activation. It is known that SHP can interact with CAR to inhibit CAR-target gene expression; however, more research is needed to identify co-regulatory networks between SHP, CAR, and BA synthesis (Bae et al., 2004; Li and Chiang, 2013).
This study investigated how pharmacological activation of PXR and CAR can modulate the gut microbiome as well as primary and secondary BA homeostasis. The gut microbiome was sequenced from the LIC by 16S rDNA. Because this method uses primers, the identified taxa may be a biased representation of the DNA that was amplified during library preparation. In addition, resolution of 16S rDNA data was limited to the mapping database and therefore PXR or CAR activation could not reliably be associated with bacteria at the species level. Furthermore, there was weak evidence correlating the significantly differentiated bacteria with altered BAs. Active BA uptake occurs in the ileum, but some BAs are passively absorbed in the colon. In this study, we did not sequence the bacterial DNA from the small intestine. However, two major reactions occur in the large intestine, namely 7-dehyoxylation and epimerization reactions of hydroxyl moieties (Hofmann, 2009), and we were able to capture the bacteria performing these important BA modifications. Whereas we measured BAs in four compartments (blood, liver, SIC, and LIC), we did not quantify BAs in the gall bladder or other peripheral tissues that may also be affected by BA signaling, such as brown adipose tissue (Broeders et al., 2015). Overall, this study showed that PXR and CAR activation altered the gut microbiome composition. Lack of gut microbiota potentiated an increase in total, primary, conjugated, and 6-OH BAs in SIC and liver following TCPOBOP exposure; in CV mice, PCN exposure decreased secondary BAs in liver and SIC, whereas TCPOBOP only decreased secondary BAs in liver (summarized in Figure 10). In addition, because the turnover rate for bile acids is about 5% per day (Chiang, 2013), a new steady-state for bile acid homeostasis following PXR or CAR activation was not achieved; although the microbiome can change rapidly following a dietary exposure (David et al., 2014), the lack of a new BA steady-state will affect the gut microbiota composition, particularly bile acid metabolizing bacteria. Therefore, the results should be interpreted as the effect of an acute exposure activating these nuclear receptors on BAs and gut microbiota. Expression of genes in the alternative pathway of BA synthesis was decreased by PCN and TCPOBOP exposure in CV and GF mice, whereas Cyp7a1 mRNA was increased in GF mice by TCPOBOP. Future studies should focus on the metabolic and mechanistic effects of BAs modulated by PXR and CAR activation to identify new therapeutic methods to treat metabolic diseases.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank members of the Cui Lab and Dr Curtis Klaassen for reviewing the manuscript.
FUNDING
National Institute of Health (T32 ES007032-39, GM111381); University of Washington Start-Up Funds from University of Washington Center for Exposures, Diseases, Genomics, and Environment (P30 ES007033) and the Sheldon Murphy Endowment.
REFERENCES
- Akita H., Suzuki H., Ito K., Kinoshita S., Sato N., Takikawa H., Sugiyama Y. (2001). Characterization of bile acid transport mediated by multidrug resistance associated protein 2 and bile salt export pump. Biochim. Biophys. Acta 1511, 7–16. [DOI] [PubMed] [Google Scholar]
- Argaud D., Halimi S., Catelloni F., Leverve X. M. (1991). Inhibition of gluconeogenesis in isolated rat hepatocytes after chronic treatment with phenobarbital. Biochem. J. 280, 663–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae Y., Kemper J. K., Kemper B. (2004). Repression of CAR-mediated transactivation of CYP2B genes by the orphan nuclear receptor, short heterodimer partner (SHP). DNA Cell Biol. 23, 81–91. [DOI] [PubMed] [Google Scholar]
- Beilke L. D., Aleksunes L. M., Holland R. D., Besselsen D. G., Beger R. D., Klaassen C. D., Cherrington N. J. (2009). Constitutive androstane receptor-mediated changes in bile acid composition contributes to hepatoprotection from lithocholic acid-induced liver injury in mice. Drug Metab. Dispos. 37, 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilke L. D., Aleksunes L. M., Olson E. R., Besselsen D. G., Klaassen C. D., Dvorak K., Cherrington N. J. (2009). Decreased apoptosis during CAR-mediated hepatoprotection against lithocholic acid-induced liver injury in mice. Toxicol. Lett. 188, 38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berge K. E., Tian H., Graf G. A., Yu L., Grishin N. V., Schultz J., Kwiterovich P., Shan B., Barnes R., Hobbs H. H. (2000). Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science 290, 1771–1775. [DOI] [PubMed] [Google Scholar]
- Bjorkholm B., Bok C. M., Lundin A., Rafter J., Hibberd M. L., Pettersson S. (2009). Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One 4, e6958.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeders E. P., Nascimento E. B., Havekes B., Brans B., Roumans K. H., Tailleux A., Schaart G., Kouach M., Charton J., Deprez B., et al. (2015). The bile acid chenodeoxycholic acid increases human brown adipose tissue activity. Cell Metab. 22, 418–426. [DOI] [PubMed] [Google Scholar]
- Caparros-Martin J. A., Lareu R. R., Ramsay J. P., Peplies J., Reen F. J., Headlam H. A., Ward N. C., Croft K. D., Newsholme P., Hughes J. D., et al. (2017). Statin therapy causes gut dysbiosis in mice through a PXR-dependent mechanism. Microbiome 5, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Pena A. G., Goodrich J. K., Gordon J. I., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S., Zou M., Liu Q., Kuang J., Shen J., Pu S., Chen L., Li H., Wu T., Li R., et al. (2017). Activation of constitutive androstane receptor prevents cholesterol gallstone formation. Am. J. Pathol. 187, 808–818. [DOI] [PubMed] [Google Scholar]
- Chiang J. Y. (2013). Bile acid metabolism and signaling. Compr. Physiol. 3, 1191–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csanaky I. L., Lickteig A. J., Klaassen C. D. (2018). Aryl hydrocarbon receptor (AhR) mediated short-term effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) on bile acid homeostasis in mice. Toxicol. Appl. Pharmacol. 3E43, 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csanaky I. L., Lu H., Zhang Y., Ogura K., Choudhuri S., Klaassen C. D. (2011). Organic anion-transporting polypeptide 1b2 (Oatp1b2) is important for the hepatic uptake of unconjugated bile acids: Studies in Oatp1b2-null mice. Hepatology 53, 272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J. Y., Gunewardena S. S., Rockwell C. E., Klaassen C. D. (2010). ChIPing the cistrome of PXR in mouse liver. Nucleic Acids Res. 38, 7943–7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J. Y., Klaassen C. D. (2016). RNA-Seq reveals common and unique PXR- and CAR-target gene signatures in the mouse liver transcriptome. Biochim. Biophys. Acta 1859, 1198–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L. A., Maurice C. F., Carmody R. N., Gootenberg D. B., Button J. E., Wolfe B. E., Ling A. V., Devlin A. S., Varma Y., Fischbach M. A., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson P. A., Lan T., Rao A. (2009). Bile acid transporters. J. Lipid Res. 50, 2340–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B., Saha P. K., Huang W., Chen W., Abu-Elheiga L. A., Wakil S. J., Stevens R. D., Ilkayeva O., Newgard C. B., Chan L., et al. (2009). Activation of nuclear receptor CAR ameliorates diabetes and fatty liver disease. Proc. Natl. Acad. Sci. U.S.A. 106, 18831–18836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman B. M., Tzameli I., Choi H. S., Chen J., Simha D., Seol W., Evans R. M., Moore D. D. (1998). Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature 395, 612–615. [DOI] [PubMed] [Google Scholar]
- Fu Z. D., Cui J. Y. (2017). Remote sensing between liver and intestine: Importance of microbial metabolites. Curr. Pharmacol. Rep. 3, 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., He J., Zhai Y., Wada T., Xie W. (2009). The constitutive androstane receptor is an anti-obesity nuclear receptor that improves insulin sensitivity. J. Biol. Chem. 284, 25984–25992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre C., Schuster G. U., Roth A., Handschin C., Johansson L., Looser R., Parini P., Podvinec M., Robertsson K., Gustafsson J. A., et al. (2005). LXR deficiency and cholesterol feeding affect the expression and phenobarbital-mediated induction of cytochromes P450 in mouse liver. J. Lipid Res. 46, 1633–1642. [DOI] [PubMed] [Google Scholar]
- Guo S., Gillingham T., Guo Y., Meng D., Zhu W., Walker W. A., Ganguli K. (2017). Secretions of Bifidobacterium infantis and Lactobacillus acidophilus protect intestinal epithelial barrier function. J. Pediatr. Gastroenterol. Nutr. 64, 404–412. [DOI] [PubMed] [Google Scholar]
- Guo G. L., Lambert G., Negishi M., Ward J. M., Brewer H. B. Jr, Kliewer S. A., Gonzalez F. J., Sinal C. J. (2003). Complementary roles of farnesoid X receptor, pregnane X receptor, and constitutive androstane receptor in protection against bile acid toxicity. J. Biol. Chem. 278, 45062–45071. [DOI] [PubMed] [Google Scholar]
- Gustafsson B. E., Bergstrom S., Lindstedt S., Norman A. (1957). Turnover and nature of fecal bile acids in germfree and infected rats fed cholic acid-24-14C; bile acids and steroids 41. Proc. Soc. Exp. Biol. Med. 94, 467–471. [DOI] [PubMed] [Google Scholar]
- Haiser H. J., Gootenberg D. B., Chatman K., Sirasani G., Balskus E. P., Turnbaugh P. J. (2013). Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser H. J., Seim K. L., Balskus E. P., Turnbaugh P. J. (2014). Mechanistic insight into digoxin inactivation by Eggerthella lenta augments our understanding of its pharmacokinetics. Gut Microbes 5, 233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halilbasic E., Claudel T., Trauner M. (2013). Bile acid transporters and regulatory nuclear receptors in the liver and beyond. J. Hepatol. 58, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslewood G. A. (1971). Bile salts of germ-free domestic fowl and pigs. Biochem. J. 123, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez J. P., Mota L. C., Baldwin W. S. (2009). Activation of CAR and PXR by dietary, environmental and occupational chemicals alters drug metabolism, intermediary metabolism, and cell proliferation. Curr. Pharmacogenomics Person. Med. 7, 81–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt M. A., Hoffmann C., Sherrill-Mix S. A., Keilbaugh S. A., Hamady M., Chen Y. Y., Knight R., Ahima R. S., Bushman F., Wu G. D. (2009). High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 137, 1716–1724, e1711–e1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirohashi T., Suzuki H., Takikawa H., Sugiyama Y. (2000). ATP-dependent transport of bile salts by rat multidrug resistance-associated protein 3 (Mrp3). J. Biol. Chem. 275, 2905–2910. [DOI] [PubMed] [Google Scholar]
- Hofmann A. F. (2009). The enterohepatic circulation of bile acids in mammals: Form and functions. Front. Biosci. (Landmark Ed.) 14, 2584–2598. [DOI] [PubMed] [Google Scholar]
- Jarocki P., Podleśny M., Glibowski P., Targoński Z. (2014). A new insight into the physiological role of bile salt hydrolase among intestinal bacteria from the genus Bifidobacterium. PLoS One 9, e114379.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandler O. (1983). Carbohydrate metabolism in lactic acid bacteria. Antonie Van Leeuwenhoek 49, 209–224. [DOI] [PubMed] [Google Scholar]
- Klaassen C. D., Aleksunes L. M. (2010). Xenobiotic, bile acid, and cholesterol transporters: Function and regulation. Pharmacol. Rev. 62, 1–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer S. A., Goodwin B., Willson T. M. (2002). The nuclear pregnane X receptor: A key regulator of xenobiotic metabolism. Endocr. Rev. 23, 687–702. [DOI] [PubMed] [Google Scholar]
- Kliewer S. A., Moore J. T., Wade L., Staudinger J. L., Watson M. A., Jones S. A., McKee D. D., Oliver B. B., Willson T. M., Zetterstrom R. H., et al. (1998). An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92, 73–82. [DOI] [PubMed] [Google Scholar]
- Koppel N., Bisanz J. E., Pandelia M. E., Turnbaugh P. J., Balskus E. P. (2018). Discovery and characterization of a prevalent human gut bacterial enzyme sufficient for the inactivation of a family of plant toxins. Elife 7, e33953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppel N., Maini Rekdal V., Balskus E. P. (2017). Chemical transformation of xenobiotics by the human gut microbiota. Science 356, 1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier J. C., Million M., Hugon P., Armougom F., Raoult D. (2012). Human gut microbiota: Repertoire and variations. Front. Cell. Infect. Microbiol. 2, 136.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahtela J. T., Arranto A. J., Sotaniemi E. A. (1985). Enzyme inducers improve insulin sensitivity in non-insulin-dependent diabetic subjects. Diabetes 34, 911–916. [DOI] [PubMed] [Google Scholar]
- Langille M. G., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J. A., Clemente J. C., Burkepile D. E., Vega Thurber R. L., Knight R., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S., Matamoros S., Cani P. D., Neyrinck A. M., Jamar F., Starkel P., Windey K., Tremaroli V., Backhed F., Verbeke K., et al. (2014). Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc. Natl. Acad. Sci. U.S.A. 111, E4485–E4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire G., Mnif W., Pascussi J. M., Pillon A., Rabenoelina F., Fenet H., Gomez E., Casellas C., Nicolas J. C., Cavailles V., et al. (2006). Identification of new human pregnane X receptor ligands among pesticides using a stable reporter cell system. Toxicol. Sci. 91, 501–509. [DOI] [PubMed] [Google Scholar]
- Li C. Y., Cheng S. L., Bammler T. K., Cui J. Y. (2016). Editor's highlight: Neonatal activation of the xenobiotic-sensors PXR and CAR results in acute and persistent down-regulation of PPARalpha-signaling in mouse liver. Toxicol. Sci. 153, 282–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Chiang J. Y. (2013). Nuclear receptors in bile acid metabolism. Drug Metab. Rev. 45, 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Chiang J. Y. (2014). Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 66, 948–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickteig A. J., Csanaky I. L., Pratt-Hyatt M., Klaassen C. D. (2016). Activation of constitutive androstane receptor (CAR) in mice results in maintained biliary excretion of bile acids despite a marked decrease of bile acids in liver. Toxicol. Sci. 151, 403–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbaum J., Rund D. G., Butler V. P. Jr, Tse-Eng D., Saha J. R. (1981). Inactivation of digoxin by the gut flora: Reversal by antibiotic therapy. N. Engl. J. Med. 305, 789–794. [DOI] [PubMed] [Google Scholar]
- Little M. S., Pellock S. J., Walton W. G., Tripathy A., Redinbo M. R. (2018). Structural basis for the regulation of beta-glucuronidase expression by human gut Enterobacteriaceae. Proc. Natl. Acad. Sci. U.S.A. 115, E152–E161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin A., Bok C. M., Aronsson L., Bjorkholm B., Gustafsson J. A., Pott S., Arulampalam V., Hibberd M., Rafter J., Pettersson S. (2008). Gut flora, Toll-like receptors and nuclear receptors: A tripartite communication that tunes innate immunity in large intestine. Cell. Microbiol. 10, 1093–1103. [DOI] [PubMed] [Google Scholar]
- Makki K., Deehan E. C., Walter J., Backhed F. (2018). The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 23, 705–715. [DOI] [PubMed] [Google Scholar]
- Martin G., Kolida S., Marchesi J. R., Want E., Sidaway J. E., Swann J. R. (2018). In vitro modeling of bile acid processing by the human fecal microbiota. Front. Microbiol. 9, 1153.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J., Fang S., Bae Y., Kemper J. K. (2006). Functional inhibitory cross-talk between constitutive androstane receptor and hepatic nuclear factor-4 in hepatic lipid/glucose metabolism is mediated by competition for binding to the DR1 motif and to the common coactivators, GRIP-1 and PGC-1alpha. J. Biol. Chem. 281, 14537–14546. [DOI] [PubMed] [Google Scholar]
- Mistry R. H., Verkade H. J., Tietge U. J. (2017). Reverse cholesterol transport is increased in germ-free mice-brief report. Arterioscler. Thromb. Vasc. Biol. 37, 419–422. [DOI] [PubMed] [Google Scholar]
- Moore J. T., Moore L. B., Maglich J. M., Kliewer S. A. (2003). Functional and structural comparison of PXR and CAR. Biochim. Biophys. Acta 1619, 235–238. [DOI] [PubMed] [Google Scholar]
- Moore L. B., Parks D. J., Jones S. A., Bledsoe R. K., Consler T. G., Stimmel J. B., Goodwin B., Liddle C., Blanchard S. G., Willson T. M., et al. (2000). Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J. Biol. Chem. 275, 15122–15127. [DOI] [PubMed] [Google Scholar]
- Nies A. T., Keppler D. (2007). The apical conjugate efflux pump ABCC2 (MRP2). Pflugers Arch. 453, 643–659. [DOI] [PubMed] [Google Scholar]
- Niu B., Coslo D. M., Bataille A. R., Albert I., Pugh B. F., Omiecinski C. J. (2018). In vivo genome-wide binding interactions of mouse and human constitutive androstane receptors reveal novel gene targets. Nucleic Acids Res. 46, 8385–8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Q., Li P., Hao S., Zhang Y., Kim S. W., Li H., Ma X., Gao S., He L., Wu W., et al. (2015). Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci. Rep. 5, 9938.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacyniak E. K., Cheng X., Cunningham M. L., Crofton K., Klaassen C. D., Guo G. L. (2007). The flame retardants, polybrominated diphenyl ethers, are pregnane X receptor activators. Toxicol. Sci. 97, 94–102. [DOI] [PubMed] [Google Scholar]
- Petrick J. S., Klaassen C. D. (2007). Importance of hepatic induction of constitutive androstane receptor and other transcription factors that regulate xenobiotic metabolism and transport. Drug Metab. Dispos. 35, 1806–1815. [DOI] [PubMed] [Google Scholar]
- Pollet R. M., D'Agostino E. H., Walton W. G., Xu Y., Little M. S., Biernat K. A., Pellock S. J., Patterson L. M., Creekmore B. C., Isenberg H. N., et al. (2017). An Atlas of beta-glucuronidases in the human intestinal microbiome. Structure 25, 967–977 e965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M., Biagi E., Heilig H. G., Kajander K., Kekkonen R. A., Tims S., de Vos W. M. (2011). Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 141, 1792–1801. [DOI] [PubMed] [Google Scholar]
- Repa J. J., Berge K. E., Pomajzl C., Richardson J. A., Hobbs H., Mangelsdorf D. J. (2002). Regulation of ATP-binding cassette sterol transporters ABCG5 and ABCG8 by the liver X receptors alpha and beta. J. Biol. Chem. 277, 18793–18800. [DOI] [PubMed] [Google Scholar]
- Ridlon J. M., Harris S. C., Bhowmik S., Kang D. J., Hylemon P. B. (2016). Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7, 22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon J. M., Kang D. J., Hylemon P. B. (2006). Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47, 241–259. [DOI] [PubMed] [Google Scholar]
- Riottot M., Sacquet E. (1985). Increase in the ileal absorption rate of sodium taurocholate in germ-free or conventional rats given an amylomaize-starch diet. Br. J. Nutr. 53, 307–310. [DOI] [PubMed] [Google Scholar]
- Rius M., Hummel-Eisenbeiss J., Hofmann A. F., Keppler D. (2006). Substrate specificity of human ABCC4 (MRP4)-mediated cotransport of bile acids and reduced glutathione. Am. J. Physiol. Gastrointest. Liver Physiol. 290, G640–G649. [DOI] [PubMed] [Google Scholar]
- Russell J. Q., Klaassen C. D. (1972). Species variation in the biliary excretion of ouabain. J. Pharmacol. Exp. Ther. 183, 513–519. [PubMed] [Google Scholar]
- Saha J. R., Butler V. P. Jr, Neu H. C., Lindenbaum J. (1983). Digoxin-inactivating bacteria: Identification in human gut flora. Science 220, 325–327. [DOI] [PubMed] [Google Scholar]
- Saini S. P., Sonoda J., Xu L., Toma D., Uppal H., Mu Y., Ren S., Moore D. D., Evans R. M., Xie W. (2004). A novel constitutive androstane receptor-mediated and CYP3A-independent pathway of bile acid detoxification. Mol. Pharmacol. 65, 292–300. [DOI] [PubMed] [Google Scholar]
- Sayin S. I., Wahlstrom A., Felin J., Jantti S., Marschall H. U., Bamberg K., Angelin B., Hyotylainen T., Oresic M., Backhed F. (2013). Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235. [DOI] [PubMed] [Google Scholar]
- Schroeder B. O., Backhed F. (2016). Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 22, 1079–1089. [DOI] [PubMed] [Google Scholar]
- Selwyn F. P., Cheng S. L., Bammler T. K., Prasad B., Vrana M., Klaassen C., Cui J. Y. (2015). Developmental regulation of drug-processing genes in livers of germ-free mice. Toxicol. Sci. 147, 84–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn F. P., Csanaky I. L., Zhang Y., Klaassen C. D. (2015). Importance of large intestine in regulating bile acids and glucagon-like peptide-1 in germ-free mice. Drug Metab. Dispos. 43, 1544–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn F. P., Cui J. Y., Klaassen C. D. (2015). RNA-seq quantification of hepatic drug processing genes in germ-free mice. Drug Metab. Dispos. 43, 1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotaniemi E. A., Karvonen I. (1989). Glucose tolerance and insulin response to glucose load before and after enzyme inducing therapy in subjects with glucose intolerance and patients with NIDDM having hyperinsulinemia or relative insulin deficiency. Diabetes Res. 11, 131–139. [PubMed] [Google Scholar]
- Spanogiannopoulos P., Bess E. N., Carmody R. N., Turnbaugh P. J. (2016). The microbial pharmacists within us: A metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 14, 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger J. L., Goodwin B., Jones S. A., Hawkins-Brown D., MacKenzie K. I., LaTour A., Liu Y., Klaassen C. D., Brown K. K., Reinhard J., et al. (2001). The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl. Acad. Sci. U.S.A. 98, 3369–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman C. A., Liddle C., Coulter S. A., Sonoda J., Alvarez J. G., Moore D. D., Evans R. M., Downes M. (2005). Nuclear receptors constitutive androstane receptor and pregnane X receptor ameliorate cholestatic liver injury. Proc. Natl. Acad. Sci. U.S.A. 102, 2063–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueyoshi T., Li L., Wang H., Moore R., Kodavanti P. R., Lehmler H. J., Negishi M., Birnbaum L. S. (2014). Flame retardant BDE-47 effectively activates nuclear receptor CAR in human primary hepatocytes. Toxicol. Sci. 137, 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Cui W., Woody S. K., Staudinger J. L. (2015). Pregnane X receptor modulates the inflammatory response in primary cultures of hepatocytes. Drug Metab. Dispos. 43, 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swales K., Negishi M. (2004). CAR, driving into the future. Mol. Endocrinol. 18, 1589–1598. [DOI] [PubMed] [Google Scholar]
- Tailford L. E., Crost E. H., Kavanaugh D., Juge N. (2015). Mucin glycan foraging in the human gut microbiome. Front. Genet. 6, 81.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Fukami T., Masuo Y., Brocker C. N., Xie C., Krausz K. W., Wolf C. R., Henderson C. J., Gonzalez F. J. (2016). Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J. Lipid Res. 57, 2130–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S., Piquette-Miller M. (2007). Hepatoprotective role of PXR activation and MRP3 in cholic acid-induced cholestasis. Br. J. Pharmacol. 151, 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakare R., Alamoudi J. A., Gautam N., Rodrigues A. D., Alnouti Y. (2018). Species differences in bile acids I. Plasma and urine bile acid composition. J. Appl. Toxicol. 38, 1323–1335. [DOI] [PubMed] [Google Scholar]
- Thompson T. N., Klaassen C. D. (1995). The effects of hepatic microsomal enzyme inducers on the pharmacokinetics of ouabain after portal and systemic administration to rats. J. Pharm. Pharmacol. 47, 1041–1047. [DOI] [PubMed] [Google Scholar]
- Toda T., Ohi K., Kudo T., Yoshida T., Ikarashi N., Ito K., Sugiyama K. (2009a). Antibiotics suppress Cyp3a in the mouse liver by reducing lithocholic acid-producing intestinal flora. Yakugaku Zasshi 129, 601–608. [DOI] [PubMed] [Google Scholar]
- Toda T., Ohi K., Kudo T., Yoshida T., Ikarashi N., Ito K., Sugiyama K. (2009b). Ciprofloxacin suppresses Cyp3a in mouse liver by reducing lithocholic acid-producing intestinal flora. Drug Metab. Pharmacokinet. 24, 201–208. [DOI] [PubMed] [Google Scholar]
- Toda T., Saito N., Ikarashi N., Ito K., Yamamoto M., Ishige A., Watanabe K., Sugiyama K. (2009). Intestinal flora induces the expression of Cyp3a in the mouse liver. Xenobiotica 39, 323–334. [DOI] [PubMed] [Google Scholar]
- Tzameli I., Pissios P., Schuetz E. G., Moore D. D. (2000). The xenobiotic compound 1,4-bis[2-(3, 5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol. Cell. Biol. 20, 2951–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A., Hamadeh H. K., Webb H. K., Yamamoto Y., Sueyoshi T., Afshari C. A., Lehmann J. M., Negishi M. (2002). Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol. Pharmacol. 61, 1–6. [DOI] [PubMed] [Google Scholar]
- Venkatesh M., Mukherjee S., Wang H., Li H., Sun K., Benechet A. P., Qiu Z., Maher L., Redinbo M. R., Phillips R. S., et al. (2014). Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 41, 296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlstrom A., Al-Dury S., Stahlman M., Backhed F., Marschall H. U. (2017). Cyp3a11 is not essential for the formation of murine bile acids. Biochem. Biophys. Rep. 10, 70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace K., Cowie D. E., Konstantinou D. K., Hill S. J., Tjelle T. E., Axon A., Koruth M., White S. A., Carlsen H., Mann D. A., et al. (2010). The PXR is a drug target for chronic inflammatory liver disease. J. Steroid Biochem. Mol. Biol. 120, 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace B. D., Roberts A. B., Pollet R. M., Ingle J. D., Biernat K. A., Pellock S. J., Venkatesh M. K., Guthrie L., O’Neal S. K., Robinson S. J., et al. (2015). Structure and inhibition of microbiome beta-glucuronidases essential to the alleviation of cancer drug toxicity. Chem. Biol. 22, 1238–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Gong J., Wang W., Long Y., Fu X., Fu Y., Qian W., Hou X. (2014). Are there any different effects of Bifidobacterium, Lactobacillus and Streptococcus on intestinal sensation, barrier function and intestinal immunity in PI-IBS mouse model? PLoS One 9, e90153.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P., Zhang J., Egan-Hafley M., Liang S., Moore D. D. (2000). The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature 407, 920–923. [DOI] [PubMed] [Google Scholar]
- Wistuba W., Gnewuch C., Liebisch G., Schmitz G., Langmann T. (2007). Lithocholic acid induction of the FGF19 promoter in intestinal cells is mediated by PXR. World J. Gastroenterol. 13, 4230–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Zhai Y., Wang J. (2018). The role of PPAR and its cross-talk with CAR and LXR in obesity and atherosclerosis. Int. J. Mol. Sci. 19, e1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y., Kakizaki S., Horiguchi N., Sohara N., Sato K., Takagi H., Mori M., Negishi M. (2007). The role of the nuclear receptor constitutive androstane receptor in the pathogenesis of non-alcoholic steatohepatitis. Gut 56, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari K., Yoda N., Toriyabe T., Yamazoe Y. (2010). Constitutive androstane receptor transcriptionally activates human CYP1A1 and CYP1A2 genes through a common regulatory element in the 5′-flanking region. Biochem. Pharmacol. 79, 261–269. [DOI] [PubMed] [Google Scholar]
- Yu L., Hammer R. E., Li-Hawkins J., Von Bergmann K., Lutjohann D., Cohen J. C., Hobbs H. H. (2002). Disruption of Abcg5 and Abcg8 in mice reveals their crucial role in biliary cholesterol secretion. Proc. Natl. Acad. Sci. U.S.A. 99, 16237–16242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ze X., Duncan S. H., Louis P., Flint H. J. (2012). Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 6, 1535–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelcer N., Saeki T., Bot I., Kuil A., Borst P. (2003). Transport of bile acids in multidrug-resistance-protein 3-overexpressing cells co-transfected with the ileal Na+-dependent bile-acid transporter. Biochem. J. 369, 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Csanaky I. L., Selwyn F. P., Lehman-McKeeman L. D., Klaassen C. D. (2013). Organic anion-transporting polypeptide 1a4 (Oatp1a4) is important for secondary bile acid metabolism. Biochem. Pharmacol. 86, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Klaassen C. D. (2010). Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J. Lipid Res. 51, 3230–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lickteig A. J., Csanaky I. L., Klaassen C. D. (2017). Editor's highlight: Clofibrate decreases bile acids in livers of male mice by increasing biliary bile acid excretion in a PPARalpha-dependent manner. Toxicol. Sci. 160, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lickteig A. J., Csanaky I. L., Klaassen C. D. (2018). Activation of PPARalpha decreases bile acids in livers of female mice while maintaining bile flow and biliary bile acid excretion. Toxicol. Appl. Pharmacol. 338, 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.