Abstract
HMCES and yedK were recently identified as sensors of abasic sites in ssDNA. In this study, we present multiple crystal structures captured in the apo-, nonspecific-substrate-binding, specific-substrate-binding, and product-binding states of yedK. In combination with biochemical data, we unveil the molecular basis of AP site sensing in ssDNA by yedK. Our results indicate that yedK has a strong preference for AP site-containing ssDNA over native ssDNA and that the conserved Glu105 residue is important for identifying AP sites in ssDNA. Moreover, our results reveal that a thiazolidine linkage is formed between yedK and AP sites in ssDNA, with the residues that stabilize the thiazolidine linkage important for the formation of DNA-protein crosslinks between yedK and the AP sites. We propose that our findings offer a unique platform to develop yedK and other SRAP domain-containing proteins as tools for detecting abasic sites in vitro and in vivo.
INTRODUCTION
DNA damage from various endogenous and exogenous sources can adversely affect genome stability and cell viability (1–7). Apurinic or apyrimidinic sites (AP or abasic sites) are one of the most common DNA lesions and are generated by spontaneous base loss or base damage caused by ionizing radiation (IR), UV radiation (UV) and alkylating agents such as methyl methanesulfonate (MMS) (8–10). Whilst AP sites in double-stranded DNA (dsDNA) are primarily repaired by the base excision repair (BER) pathway (10,11), their repair mechanism in single-stranded DNA (ssDNA) is only just being uncovered (3,12).
Recently, a human protein known as 5-hydroxymethylcytosine (5hmC) binding ES-cell-specific protein (HMCES) (13) and its bacterial ortholog yedK in Escherichia coli were validated as AP site sensors in ssDNA at stalled replication forks (12). Both HMCES and yedK contain a highly conserved SOS response-associated peptidase (SRAP) domain with a putative catalytic triad (Cys2-His160-Glu105 in yedK) (14). The peptidase activity of the SRAP domain is thought to cause the autoproteolytic cleavage of the first methionine to expose the catalytic residue Cys2 (14,15). The HMCES and yedK SRAP domains have been reported to form a DNA-protein crosslink (DPC) with the sugar moiety of the AP site in ssDNA via the catalytic Cys2 residue. For HMCES, this effectively shields lesions from the action of AP endonucleases and translesion DNA synthesis (TLS) polymerases (16,17), maintaining genome integrity by promoting the error-free repair of AP sites in ssDNA (12). Thus, HMCES-deficient cells exhibit delayed AP site repair, accumulate DNA damage, are hypersensitive to AP site-generating genotoxins, and have increased genetic instability (12). However, the mechanism by which HMCES and yedK SRAP domains sense AP sites in ssDNA and the chemical nature of the covalent linkage between SRAP domains and AP sites remain unknown.
Here, we focused on the E. coli yedK system and solved multiple crystal structures of yedK, as well as binary complexes of yedK with native ssDNA, ssDNA containing a tetrahydrofuran (THF) AP site mimic, and three different ssDNA substrates each containing a natural AP site covalently linked to yedK. Our results indicate that these structures correspond to the apo-, nonspecific-substrate-binding, specific-substrate-binding, and product-binding states of yedK, respectively. Moreover, our results demonstrate that yedK has a strong preference for AP site-containing ssDNA over native ssDNA. Interestingly, the strictly conserved Glu105 residue is found to be important for yedK when discriminating between AP sties and native ssDNA via electrostatic repulsion. A single substitution from Glu105 to alanine causes yedK to lose its preference for AP sites over native ssDNA. Moreover, our study reveals that a thiazolidine linkage is formed between yedK and AP sites in ssDNA and identifies residues important for the formation and stabilization of this thiazolidine linkage between yedK and AP sites. Together with biochemical data, our study reveals the molecular basis for the dynamic process by which the yedK SRAP domain senses AP sites in ssDNA and provides a comprehensive understanding of the process.
MATERIALS AND METHODS
Cloning and protein preparation in bacteria
The full-length E. coli yedK (a.a. 1–222; NCBI reference sequence: YP_025310.1) and Uracil-DNA glycosylases (UDG; a.a. 1–229; NCBI reference sequence: NP_417075.1) genes were isolated from BL21 (DE3)-RIL genomic DNA. The full-length yedK (1–222) was cloned into a modified pET22b plasmid with a C-terminal 3C protease cleavage site followed by a 6xHis-tag, while the yedK (2–222) was cloned into a modified pET28a plasmid with an N-terminal 6xHis-tag followed by a non-canonical TEV protease cleavage site (E-N-L-Y-F-Q-Cys; 18). The UDG (1–229) was cloned into pET22b with a C-terminal 6xHis-tag, and then the UDG Tyr66 was mutated to tryptophan to improve the enzymatic activity (19). All mutations were introduced using a standard PCR procedure and verified by DNA sequencing.
All the protein used in this study were expressed in BL21 (DE3)-RIL cell strain (Stratagene) and cultured in Luria-Bertani (LB) medium with proper antibiotics at 37°C to OD600 ∼1.0–1.2. Protein expression was induced with 0.5 mM isopropyl-β-d-thiogalactoside (IPTG) and cells were further incubated overnight at 20°C. The expressed protein was first purified with Ni-sepharose affinity beads (GE Healthcare). After removing the 6xHis-tag by 3C protease or TEV protease, the protein was further purified on a pre-equilibrated HiLoad Superdex 200 16/600 column (GE Healthcare). The UDG was purified with Ni-sepharose affinity beads followed by Hiload superdex 200 16/600 column. Purified protein samples were flash-frozen in liquid nitrogen and stored in −80°C for further usage.
Crystallization
All protein samples used for crystallization were in the same buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl) and at the same protein concentration of 12 mg/mL. The ssDNA substrates used for co-crystallization were purchased from Sangon Biotech (Shanghai) and dissolved in the same buffer mentioned above. The ssDNA sequences used in this study are listed in Table 1. All crystals were obtained using the sitting-drop vapor-diffusion method at 20°C.
The crystals of apo-yedK were crystallized in 0.2 M dl-malic acid pH 7.0, 20% PEG3350;
To form the ssDNA-bound complex, the yedK protein was incubated with ssDNA at a molar ratio of 1:1.2 on ice for 30 min before setting up crystallization. The crystals of yedK in complex with ssDNA (hereafter denoted as yedK–ssDNA complex) were crystallized in 0.1 M citric acid pH 4.0, 3% PEG 6000;
The crystals of yedK in complex with ssDNA containing a tetrahydrofuran (THF) AP site mimic (20) (yedK–THF complex) were crystallized in 0.02 M magnesium chloride hexahydrate, 0.1 M HEPES pH 7.5, 22% w/v Poly(acrylic acid sodium salt) 5100;
To get the complex of yedK covalently linked to ssDNA containing an AP site (yedK–Xlink complex), the yedK protein, ssDNA containing an uracil base and UDG protein were mixed together at a molar ratio of 100:120:1.2, and incubated for 24 h at 37°C. The final concentration of yedK in the reaction mixture was about 1.2 mg/ml. After incubation, the yedK–Xlink complex was efficiently formed which was checked by SDS-PAGE. Then, the yedK–Xlink complex was concentrated to about 12 mg/mL (estimated by using UV spectrometer at a wavelength of 260 nm) before setting up crystallization. The crystals of yedK–Xlink complex were crystallized in 0.2 M sodium formate, 20% PEG 3350.
The covalent complex of yedK with polyA ssDNA containing an AP site (yedK–polyA complex) was crystallized in 0.05 M calcium chloride dihydrate, 0.1 M Bis–Tris pH 6.5, 30% PEG monomethyl ether 550.
The covalent complex of yedK with a random ssDNA containing an AP site (yedK–Xlink2 complex) was crystallized in 0.2 M ammonium formate, 20% PEG 3350.
Table 1.
DNA used for crystallization and assays.
| ID | Length (nt) | Sequence | Experiments |
|---|---|---|---|
| ssDNA | 10 | 5′-CGGTCGATTC-3′ | Crystallization, crosslinking assays, and ITC |
| ssDNA_THF | 10 | 5′-CGGT(THF)GATTC-3′ | Crystallization and ITC |
| ssDNA_dU | 10 | 5′-CGGT(dU)GATTC-3′ | Crystallization and crosslinking assays |
| ssDNA_polyA | 11 | 5′-AAAAA(dU)AAAAA-3′ | Crystallization |
| ssDNA2_dU | 10 | 5′-GATTC(dU)GTCG-3′ | Crystallization |
| 3′ junction | 31 + 15 | 5′-ATGACTCTTCTGGTC(THF)GGATGGTAGTTAAGT-3′ | ITC |
| 5′-ACTTAACTACCATCC-3′ | |||
| 5′ junction | 31 + 15 | 5′-ATGACTCTTCTGGTC(THF)GGATGGTAGTTAAGT-3′ | ITC |
| 5′-GACCAGAAGAGTCAT-3′ |
All crystals were soaked in cryoprotectants made from the mother liquors supplemented with 15–30% glycerol before flash freezing in liquid nitrogen.
Data collection and structure determination
Diffraction data for all crystals were collected at a wavelength of 0.979 Å at beamline 19U (BL19U1) at Shanghai Synchrotron Radiation Facility (SSRF), China (21). X-ray data sets were subsequently processed with the program HKL3000 (22). The structure for apo-yedK was solved by molecular replacement in PHASER (23) with the structure of selenium-labelled E. coli yedK (PDB 2ICU; this entry was deposited by structural genomics in year 2006, which should be acknowledged.) as a search model and manually refined and built using Coot (24). The other structures were determined by molecular replacement in PHASER with our structure of apo-yedK as the search model. The final structures of apo-yedK and binary complexes of yedK–ssDNA, yedK–THF, yedK–Xlink, yedK–polyA and yedK–Xlink2 were refined to 2.10, 1.60, 1.65, 1.22, 1.70 and 1.58 Å, respectively, by PHENIX (25). Table 2 summarizes the statistics for data collection and structure refinement. All structure figures were prepared using PyMOL (The PyMOL Molecular Graphics System, Schrödinger).
Table 2.
Data collection and refinement statistics
| Apo-yedK (6KBU) | yedK–ssDNA (6KBS) | yedK–THF (6KBZ) | yedK–Xlink (6KBX) | yedK–polyA (6KCQ) | yedK–Xlink2 (6KIJ) | |
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space group | P1211 | C121 | P1211 | P1211 | P43 | P1211 |
| Cell dimensions | ||||||
| a, b, c (Å) | 46.7, 67.3, 74.9 | 91.9, 50.3, 54.5 | 40.5, 111.4, 104.1 | 47.1, 44.1, 54.7 | 70.7, 70.7, 44.4 | 47.4, 44.1, 55.1 |
| α, β, γ (°) | 90.0, 98.8, 90.0 | 90.0, 99.1, 90.0 | 90.0, 96.1, 90.0 | 90.0, 101.7, 90.0 | 90.0, 90.0, 90.0 | 90.0, 102.3, 90.0 |
| Resolution (Å) | 40–2.10 (2.18–2.10)a | 40–1.60 (1.66–1.60) | 40–1.65 (1.71–1.65) | 50–1.22 (1.26–1.22) | 40–1.70 (1.76–1.70) | 30–1.58 (1.64–1.58) |
| R pim (%) | 7.2 (23.9) | 2.5 (26.2) | 3.6 (37.1) | 3.8 (29.9) | 2.7 (22.0) | 5.0(30.0) |
| I/σI | 9.0 (2.5) | 20.6 (1.8) | 19.4 (1.5) | 18.5 (2.0) | 23 (2.5) | 16.5 (2.0) |
| Completeness (%) | 97.4 (88.7) | 99.4 (99.6) | 99.0 (98.4) | 98.8 (93.6) | 99.1 (94.3) | 99.8 (92.8) |
| Redundancy | 5.2 (4.1) | 6.6 (5.9) | 6.1 (4.8) | 6.5 (4.6) | 8.4 (3.7) | 6.7 (5.8) |
| No. of reflections (total/unique) | 136 928/26 234 | 214 311/32 253 | 660 240/108 392 | 416 830/64 468 | 202 281/24 080 | 204 929/30 514 |
| Refinement | ||||||
| R work/ R free (%) | 16.7/24.0 | 17.3/20.1 | 16.0/19.6 | 15.5/16.5 | 16.0/20.4 | 15.2/18.3 |
| No. atoms | ||||||
| Protein | 3364 | 1839 | 7111 | 1794 | 1757 | 1806 |
| DNA | 185 | 708 | 96 | 116 | 70 | |
| Glycerol | 12 | 6 | ||||
| Magnesium | 4 | |||||
| Water | 270 | 237 | 1086 | 302 | 218 | 323 |
| B-factors | ||||||
| Protein | 29.3 | 26.8 | 18.2 | 18.2 | 16.8 | 16.0 |
| DNA | 33.4 | 20.1 | 28.7 | 33.9 | 24.8 | |
| Glycerol | 47.3 | 29.4 | ||||
| Magnesium | 35.2 | |||||
| Water | 32.6 | 35.4 | 29.1 | 31.9 | 25.6 | 30.9 |
| R.m.s deviations | ||||||
| Bond lengths (Å) | 0.007 | 0.007 | 0.006 | 0.005 | 0.007 | 0.006 |
| Bond angles (°) | 0.85 | 0.85 | 0.83 | 0.92 | 0.89 | 0.90 |
aValues in parentheses are for the highest-resolution shell. One crystal was used for each data set.
Isothermal titration calorimetry (ITC)
The wild-type yedK protein and its mutants were buffer-exchanged to 20 mM Tris–HCl pH 7.5, 150 mM NaCl. The ssDNA and ssDNA_THF were dissolved in the same buffer. ITC experiments were performed at 25°C on a MicroCal PEAQ-ITC (Malvern Panalytical Ltd) using 25 injections of 1.6 μl. A 50 μl of 300 μM solution of the wild-type yedK protein or its mutants were loaded in the syringe and a 250 μl of 30 μM solution of ssDNA or ssDNA_THF was loaded into the cell. The measured heat changes of the binding reactions were integrated and processed using the standard ‘one set of sites’ model implemented in the Origin software package (OriginLab) to determine the binding stoichiometry, N, and the equilibrium dissociation constant, Kd.
Crosslinking assays
Crosslinking assays for wild-type yedK and mutants were performed in the reaction buffer of 20mM Tris pH 8.0, 50 mM NaCl and followed a procedure as previously reported (12). The yedK protein, ssDNA_dU (5′-CGGT(dU)GATTC-3′) and UDG protein were mixed at a molar ratio of 1:1.2:0.06. The native ssDNA was mixed with yedK at the same molar ratio but without UDG as a control. The total reaction volume was 50 μl and the final concentration of the yedK protein was 5 μM. Then, the reaction mixtures were incubated at 37°C for 12 h. Finally, the reaction products were added with equivalent volume of 2× SDS loading buffer and resolved by 12% SDS-PAGE.
RESULTS
Overall structure of the yedK–ssDNA complex
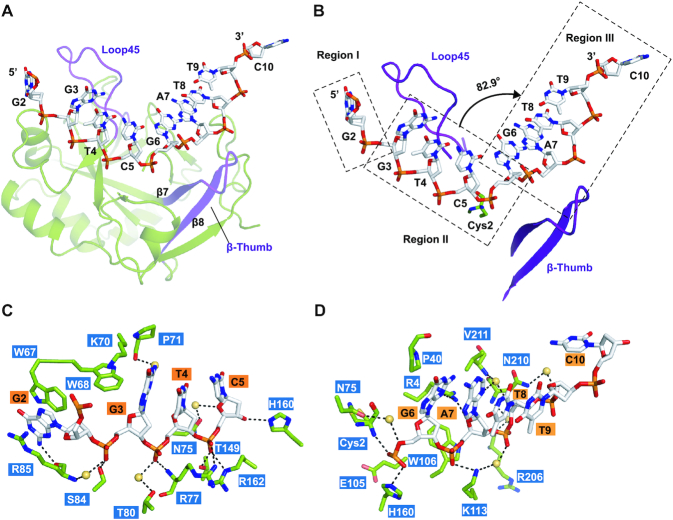
In order to elucidate how yedK and HMCES achieve a strong preference for ssDNA over dsDNA (12), we sought to obtain structural information for the yedK protein and different ssDNA substrates (Table 1). We successfully obtained a structure of yedK in complex with a 10nt-ssDNA substrate (ssDNA, 5′-CGGTCGATTC-3′) refined to 1.60 Å (Table 2), consisting of one yedK and one ssDNA molecules in an asymmetric unit. In this complex structure, we were able to model all yedK residues and nine ssDNA nucleotides (G2 to C10; stereoview in Figure 1A). The structure of yedK contains twelve β-strands, with the β3, β4, β5, β6, β9, β10, and β11 strands forming a β-barrel core flanked by four α-helices (α1, α2, α3 and α4) and the β1, β2, β7, β8 and β12 strands constituting a separate, relatively flat β-sheet structure stabilized by a 310-helix (η1 helix; Figure 1A and topology diagram in Figure 1B). The β-sheet is almost perpendicular to the β6 strand of the β-barrel, positioning the N-terminus and catalytic Cys2 residue of yedK at the center of a shallow, positively charged groove (Figure 1C). The ssDNA strand lies exactly in the positively charged groove of yedK and has major interactions with its ribose-phosphate backbone.
Figure 1.
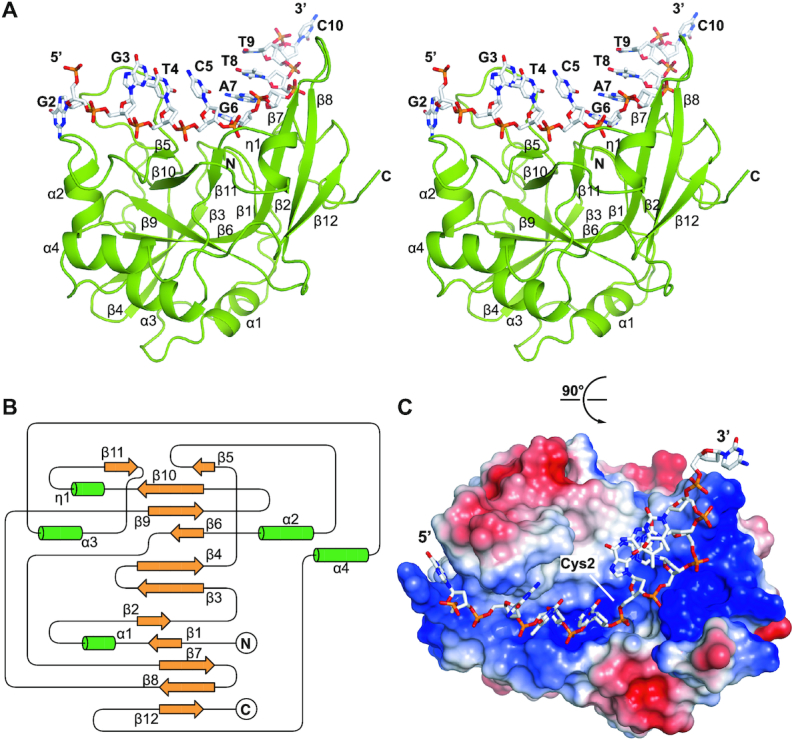
The overall structure of yedK complexed with ssDNA. (A) Stereoview of the yedK–ssDNA complex structure. The yedK protein is shown in green and the ssDNA is shown in white with its atoms colored by element in PyMOL. (B) Topological diagram of the overall fold of the yedK protein. The β-strands and α-helices are shown in yellow and green, respectively. (C) Electrostatic surface representation of yedK and stick representation of the ssDNA. The protein surface is colored according to surface potential, with red and blue indicating negative and positive charges, respectively. The ssDNA is colored as in panel A.
Loop45 and β-thumb kink the ssDNA strand
Detailed analysis of the interactions in the yedK–ssDNA complex structure identified two structural elements that may determine ssDNA conformation. The first is ‘Loop45’, a long loop between β4 and β5, and the other is ‘β-thumb’, composed of the β7 and β8 strands (Figure 2A). Loop45 and β-thumb contact the ssDNA strand via various stacking interactions and act as obstacles to guide the ssDNA strand through the positively charged groove by kinking it at two points: one between the G2 and G3 nucleotides, the other between C5 and G6, respectively (Figure 2B). Based on these observations, the ssDNA strand is divided into three regions, with a ∼83° bend between regions II and III. The kink between C5 and G6 is on top of the catalytic Cys2 residue (Figure 2B), suggesting that the kink may facilitate the positioning of the AP site towards the yedK active site. Interestingly, both Loop45 and β-thumb are partially disordered in the apo-yedK structure, as revealed by superimposing the yedK–ssDNA and apo-yedK structures, further suggesting that ssDNA binding stabilizes the two structural elements (Table 2 and Supplementary Figure S1A).
Figure 2.
Intermolecular interactions between yedK and ssDNA. (A) Two structural elements are important for the conformation of the ssDNA strand. The Loop45 and β-thumb elements are shown in purple. (B) ssDNA strand conformation can be divided into three regions, as indicated by rectangles. Loop45 and β-thumb are shown in purple. The torsion angle between the C5 and G6 nucleotides is indicated by the black arrow. The catalytic Cys2 residue is shown with two conformers. (C, D) Detailed interactions between yedK and ssDNA. Residues and ssDNA are shown as green and white sticks, respectively. Water molecules are shown as yellow spheres.
Taken together, the unique configuration adopted by the ssDNA strand upon binding yedK might prevent an integrated B-form dsDNA binding, consistent with the strong preference of yedK for ssDNA over dsDNA (12).
Interactions between yedK and ssDNA
The ssDNA substrate is bound to yedK via extensive interactions: the ribose-phosphate backbone of the ssDNA is bound to the positively-charged groove of yedK via numerous hydrogen bonds and salt-bridges. Although most nucleotide bases point to the outside of the groove, they do contribute to interactions with some hydrophobic yedK residues (Figure 2C and D).
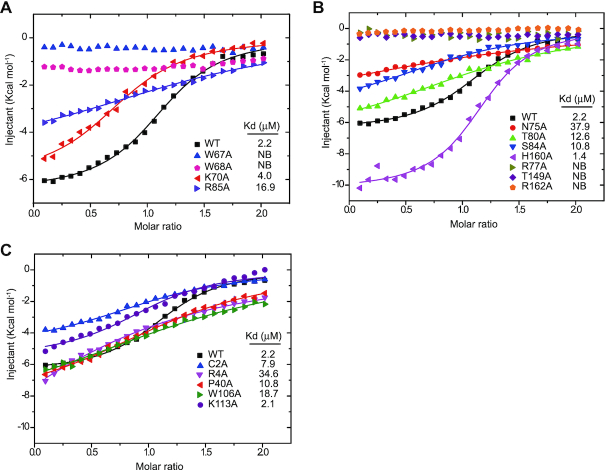
In region I, the G2 base is sandwiched by the side chains of yedK Trp67 in Loop45 and Arg85 by π-π stacking, with the N2 atom of G2 interacting with the Arg85 main-chain carbonyl via a hydrogen bond (Figure 2C). Moreover, the Trp68 and Lys70 side chains in Loop45 are wedged into the ssDNA strand, adding strong hydrophobic interactions to the base stacking interactions from G3 to C5 in region II and potentially stabilizing the kink between G2 and C3. Consistently, the Loop45 Trp67 and Trp68 residues are conserved across species (Supplementary Figure S2A). Mutagenesis studies by ITC experiments showed that, compared with wild-type yedK (Kd of 2.2 μM), the yedK W67A and W68A mutants disrupted ssDNA binding, R85A dramatically reduced binding (Kd of 16.9 μM), and K70A slightly reduced binding (Kd of 4.0 μM), highlighting the importance of Loop45 in ssDNA recognition (Figure 3A and Table 3).
Figure 3.
Biochemical characterization of the interactions between yedK and ssDNA. (A–C) Each interacting residue shown in Figure 2C and D were mutated to alanine and their interactions with ssDNA were accessed using ITC experiments. NB, no binding.
Table 3.
Thermodynamic parameters for wild-type yedK and mutants binding different DNA substrates from ITC experiments
| Protein | DNA | N | ΔH (kcal/mol) | ΔS (cal/mol/K) | K d (μM) |
|---|---|---|---|---|---|
| WT | ssDNA | 1.14 | –6.46 | 4.37 | 2.2 |
| C2A | ssDNA | 1.08 | –4.95 | 6.75 | 7.9 |
| R4A | ssDNA | 0.93 | –15.97 | –33.2 | 34.6 |
| P40G | ssDNA | 1.26 | –8.60 | –6.10 | 10.8 |
| W67A | ssDNA | – | – | – | NB |
| W68A | ssDNA | – | – | – | NB |
| K70A | ssDNA | 0.82 | –5.93 | 4.81 | 4.0 |
| N75A | ssDNA | 1.36 | –5.95 | 0.28 | 37.9 |
| R77A | ssDNA | – | – | – | NB |
| T80A | ssDNA | 1.24 | –7.05 | –1.22 | 12.6 |
| S84A | ssDNA | 0.93 | –5.42 | 4.54 | 10.8 |
| R85A | ssDNA | 1.50 | –4.97 | 5.17 | 16.9 |
| E105A | ssDNA | 1.26 | –9.55 | –0.43 | 0.12 |
| W106A | ssDNA | 1.48 | –9.30 | –9.53 | 18.7 |
| K113A | ssDNA | 1.14 | –6.46 | 4.37 | 2.1 |
| T149A | ssDNA | – | – | – | NB |
| H160A | ssDNA | 1.17 | –10.27 | –7.68 | 1.4 |
| R162A | ssDNA | – | – | – | NB |
| WT | ssDNA_THF | 1.19 | –14.0 | –15.9 | 0.17 |
| E105A | ssDNA_THF | 1.34 | –13.60 | –13.0 | 0.07 |
| H160A | ssDNA_THF | 1.11 | –16.16 | –22.0 | 0.09 |
| WT | 5′ junction | – | – | – | NB |
| WT | 3′ junction | 0.95 | –6.12 | 10.3 | 0.18 |
NB: No binding. Please note that all raw data from the ITC experiments are shown in Supplementary Figure S3A-W.
In region II, the G3 phosphate group forms direct and water-mediated hydrogen bonds with the Ser84 side chain and the Arg85 main-chain amino group, respectively, whilst the N2 atom of G3 is hydrogen bonded to the Pro71 main-chain carbonyl via water (Figure 2C). The T4 phosphate group is also hydrogen bonded to the Arg77 main-chain amino group and the Thr80 side chain via water. For the interactions between the C5 nucleotide and yedK, the O4’ atom of the sugar moiety is bound to the Asn75 side chain via water whilst its O3’ atom forms a hydrogen bond with the His160 side chain. Remarkably, the C5 phosphate group forms a strong hydrogen bond with the Thr149 side chain and establishes two prominent salt-bridges with the Arg77 and Arg162 side chains. Interestingly, most of the residues involved in interactions, including Asn75, Arg77, Thr149, His160 and Arg162, are strictly conserved (Supplementary Figure S2A). ITC showed that the yedK R77A, T149A, and R162A mutants disrupted ssDNA interactions, the N75A, T80A and S84A mutants reduced interactions (Kd of 37.9, 12.6 and 10.8 μM, respectively), and the H160A mutant marginally increased interactions (Kd of 1.4 μM) (Figure 3B and Table 3).
In region III, the bases between G6 and T9 stack together, with G6 forming further stacking interactions with the Pro40 side chain and its N3 atom being hydrogen bonded to the Arg4 side chain (Figure 2D). The G6 phosphate group establishes direct hydrogen bonds with the Cys2 amino group and the His160 side chain and forms a water-mediated hydrogen bond with the Asn75 side chain. Meanwhile the phosphate group is ∼5.5 Å away from the Glu105 side chain carboxyl group, which may produce electrostatic repulsion contributing to yedK substrate selectivity (see below). The G6 sugar moiety forms hydrophobic interactions with Trp106 in β-thumb to further stabilize the ssDNA conformation in region III. Moreover, the O4’ atom of the A7 sugar moiety contacts the Arg4 side chain, whilst the N3 atom of A7 and the O4’ atom of the T8 sugar moiety contact the Val211 main-chain amino group via water. The A7, T8 and T9 phosphate groups form direct or water-mediated interactions with Lys113 in β-thumb, Arg206, and Asn210, whilst C10 has no obvious interaction with yedK. Consistently, the interacting residues Cys2, Arg4, Pro40, Asn75, Trp106 and His160 are strictly conserved and Lys113 is conserved in residue type (Supplementary Figure S2A). ITC revealed that the yedK C2A, R4A, P40G and W106A mutants had reduced ssDNA binding (Kd of 7.9, 34.6, 10.8 and 18.7 μM, respectively) but the K113A mutant had no effect on binding (Kd of 2.1 μM) (Figure 3C and Table 3). It is noteworthy that the catalytic Cys2 residue has two conformers, both of which have its thiol group buried in a hydrophobic pocket (Supplementary Figure S1B).
Taken together, our results reveal the molecular basis of the interactions between yedK and ssDNA and highlight their sequence-independent binding properties.
Recognition of ssDNA THF AP sites by yedK
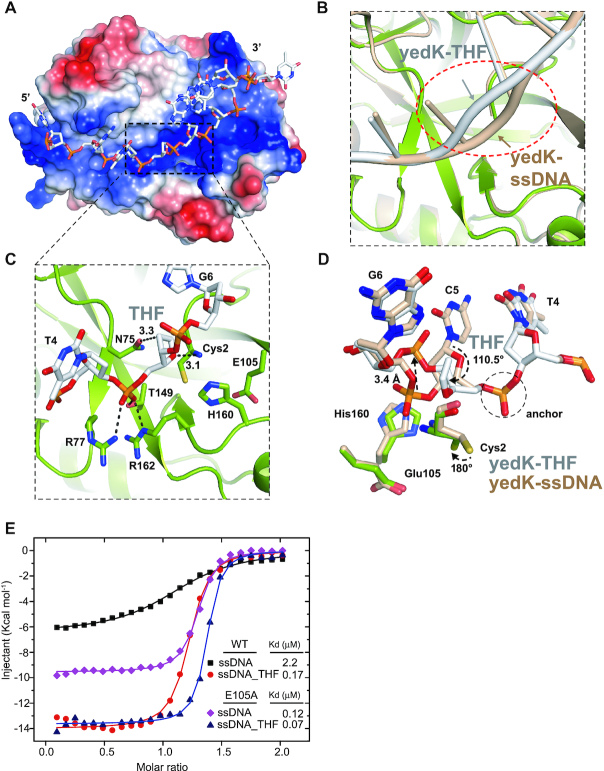
To further elucidate the process by which yedK senses AP sties in ssDNA, we determined the structure of yedK in complex with the same ssDNA containing a tetrahydrofuran AP site mimic (ssDNA_THF, 5′-CGGT(THF)GATTC-3′). The yedK–THF complex structure was refined to 1.65 Å, with a high enough resolution to resolve the molecular interactions of the THF AP site (Figure 4A and Supplementary Figure S4A; Table 2). The yedK protein itself in the yedK–THF complex shows little structural conformational change compared to that in the yedK–ssDNA structure, with a small r.m.s. deviation (rmsd) of ∼0.26 Å (Supplementary Figure S4B). In contrast, the ssDNA strand has significant conformational changes around the THF AP site (Figure 4B). The conformation of the THF AP site phosphate group is similar to that of C5 in the yedK–ssDNA structure, with both stabilized by the same prominent interactions with Arg77, Thr149 and Arg162 in yedK (Figures 2C and 4C). Thus, this phosphate group appears to be an ‘anchor’ that helps stabilize the conformation of the G2, G3 and T4 nucleotides 5′ to the anchor (Figure 4D and Supplementary Figure S4B).
Figure 4.
Structure and interactions between yedK and ssDNA containing a THF AP site. (A) Surface representation of the yedK–THF complex colored as in Figure 1C. The region around the THF AP site and catalytic site is indicated by a black dashed box. (B) ssDNA conformational changes between the yedK–THF and yedK–ssDNA structures. ssDNA strands are indicated by arrows. The structure of yedK–ssDNA is shown in wheat. yedK and ssDNA in the yedK–THF complex structure are shown in green and white, respectively. (C) The binding environment of the THF AP site. Contacts between the THF sugar moiety and the catalytic Cys2 residue of yedK are highlighted. ssDNA is shown in white and the environmental residues are shown in green. (D) Comparisons of the conformational changes between the ssDNA containing a THF AP site and native ssDNA. The phosphate group anchor is circled by a black dashed line, and the conformational changes are indicated by black arrows. The Cys2, Glu105 and His160 residues are also highlighted for comparison. (E) ITC results for yedK WT and E105A with the native ssDNA and ssDNA containing a THF AP site.
Remarkably, the sugar moiety of the THF AP site rotates by ∼110.5° compared to the yedK–ssDNA structure and moves towards the catalytic Cys2 residue. Meanwhile the Cys2 thiol group previously buried in a hydrophobic environment flips 180° to contact the THF sugar moiety (Figure 4D). These conformational changes position the THF sugar moiety in the catalytic site, where it forms extensive van der Waals contacts with the catalytic Cys2 residue (Figure 4C). In particular, the THF sugar moiety C1’ and O4’ atoms are within 3.6 and 3.1 Å of the amino group and 4.8 and 3.7 Å of the thiol group of Cys2, respectively, a configuration that may facilitate the catalytic reaction. Moreover, the G6 phosphate group moves 3.4 Å out of the catalytic site and loses contact with Cys2 and His160 to avoid clashing with the THF sugar moiety in the catalytic site; meanwhile the phosphate group is far away from the Glu105 side chain carboxyl (Figure 4D). Interestingly, the Asn75 side chain carbonyl group is 3.3 Å away from the C1’ atom of the THF sugar moiety (Figure 4C). The native AP site has a hydroxyl group on the C1’ atom which may be bound to the Asn75 side chain via a strong hydrogen bond and would play a role in the catalytic reaction. Collectively, the THF sugar moiety gains multiple contacts with Cys2 and Asn75 in yedK and the G6 phosphate group releases its potential electrostatic repulsion with Glu105, thus may explain how yedK discriminates between AP sties and native ssDNA.
Besides the conformational changes around the THF AP site, the C10 nucleotide replaces T9 to stack with T8, squeezing T9 out of the stacking sequence (Supplementary Figure S4B). This conformational change may have been due to crystal packing.
Structural analyses reveal that the THF AP site may be a specific yedK substrate and shed light on the process by which yedK senses AP sites. We propose that the yedK–THF and yedK–ssDNA structures correspond to the specific-substrate-binding and nonspecific-substrate-binding states, respectively.
YedK Glu105 plays a key role in sensing abasic sites in ssDNA
The structural analyses reveal two prominent processes: (i) the THF sugar moiety of the AP site gains contacts with the catalytic Cys2 residue; and (ii) the phosphate group 3′ to the AP site releases electrostatic repulsion with Glu105 of yedK (Figure 4D). We used ITC experiments to validate these interactions. Wild-type yedK had a much stronger preference for ssDNA containing a THF AP site (Kd of 0.17 μM) over native ssDNA (Kd of 2.2 μM; Figure 4E and Table 3). This in vitro observation supports that the AP site in ssDNA is the natural substrate of yedK (12). Moreover, a single substitution from Glu105 to alanine, designed to release electrostatic repulsion, increased the binding affinity of yedK for native ssDNA (Kd of 0.12 μM) and further increased its binding affinity for ssDNA containing a THF AP site (Kd of 0.07 μM; Figure 4E and Table 3), supporting the idea that the electrostatic repulsion between Glu105 and the ssDNA phosphate group helps yedK discriminate between AP sties and native ssDNA.
Collectively, the structural and ITC analyses identify Glu105 as the key residue by which yedK senses abasic sites in ssDNA.
Formation of a thiazolidine linkage between yedK and AP sites in ssDNA
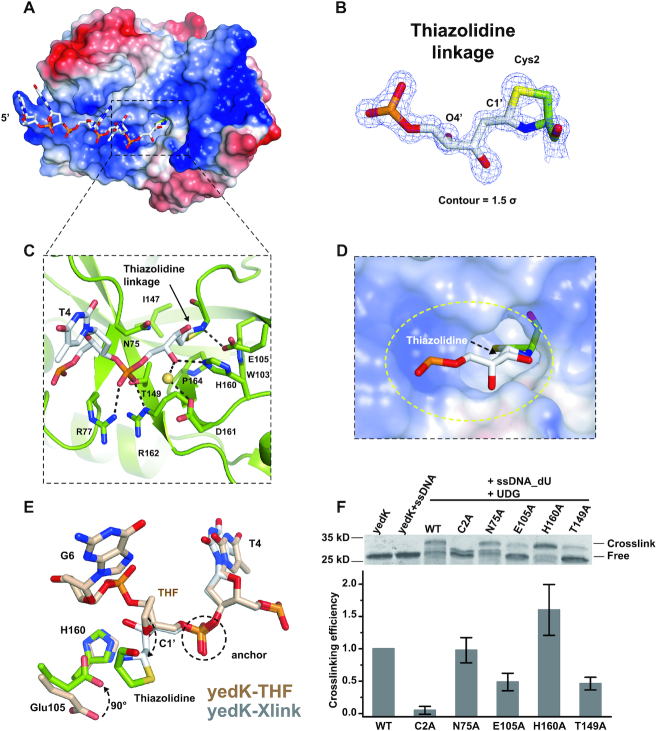
Although both yedK and HMCES have been shown to form covalent linkages with AP sites in ssDNA (12), the chemical nature of the covalent linkage remains unknown; therefore, we decided to explore this further. Firstly, yedK, ssDNA containing an uracil base (ssDNA_dU: 5′-CGGT(dU)GATTC-3′), and UDG were incubated together to produce a covalently-linked yedK–Xlink complex, according to a previously established protocol (12). We successfully obtained well-diffracted yedK–Xlink complex crystals and determined its structure at 1.22 Å (Table 2). The 5′ ssDNA fragment including the C1 to T4 nucleotides and the crosslinked AP site could be modeled due to high quality electron density; however, the remaining 3′ nucleotides exhibited very weak electron density and could not be modeled (Figure 5A).
Figure 5.
DPC formation between yedK and the AP site in ssDNA. (A) Surface representation of the yedK–Xlink structure. The binding pocket for the thiazolidine linkage is indicated with a black dashed box. (B) Chemical nature and high-quality density map of the thiazolidine linkage formed between the AP site and the catalytic Cys2 residue. The 2|Fo|–|Fc| σ-weighted map is contoured at 1.5σ. (C) Detailed interactions between yedK and the thiazolidine linkage. YedK residues that participate in binding with the covalent linkage are shown as green sticks. (D) Enlarged view of the surface representation of the hydrophobic pocket accommodating the thiazolidine linkage. The pocket is circled by a yellow dashed line. (E) Conformational comparison between ssDNA around the thiazolidine linkage of the yedK–Xlink structure and the THF AP site of the yedK–THF structure. ssDNA with the THF AP site is shown in wheat and ssDNA with the thiazolidine linkage is shown in white. The Glu105 and His160 yedK residues are also highlighted for comparison. (F) Crosslinking efficiency of yedK WT and mutants with ssDNA containing an AP site. The upper panel shows a representative SDS-PAGE gel. The lower panel shows the quantification results of crosslinking bands in the SDS-PAGE gels from 6 independent experiments. The original and uncropped gels are shown in Supplementary Figure S6A-F.
The high-resolution structure reveals that the AP site C1’ atom is covalently linked to both the amino and thiol groups of the catalytic Cys2 residue, forming a thiazolidine linkage (Figure 5B). Interestingly, the thiazolidine ring is inserted into a hydrophobic pocket surrounded by Trp103, Ile147, and Pro164, with the Thr149 side chain also contributing hydrophobic contacts to the thiazolidine ring (Figure 5C and D). The amine group of the thiazolidine ring (previously the Cys2 amino group) forms a strong hydrogen bond (distance of 2.7 Å) with the Glu105 side chain carboxyl and the AP site O4’ atom forms direct and water-mediated hydrogen bonds with the His160 and Asp161 side chains, respectively (Figure 5C).
Superimposing the yedK–Xlink and yedK–THF structures reveals that the protein undergoes a minor conformational change (rmsd = 0.35 Å; Supplementary Figure S5A). The phosphate anchor 5′ to the AP site has a similar conformation; however, the AP site sugar moiety in the yedK–Xlink structure has opened to form a thiazolidine linkage with the catalytic Cys2 residue, whilst the Glu105 side chain carboxyl has flipped 90° to interact with the thiazolidine linkage (Figure 5E).
We mutated several strictly conserved yedK residues, including Cys2, Glu105, His160, Asn75, and Thr149 to test their effect on thiazolidine linkage formation between yedK and the AP site. The C2A mutant reduced crosslinking efficiency to background levels, E105A and T149A reduced efficiency to 49 and 46%, respectively, N75A slightly influenced efficiency, and H160A increased efficiency to 160% (Figure 5F). This increase may be partially explained by the fact that H160A AP site binding was stronger than that of wild-type yedK (Kds: 0.09 versus 0.17 μM; Table 3 and Supplementary Figure S3S and U). These results confirm that conserved yedK residues, including Cys2, Glu105, and Thr149, are important for thiazolidine linkage formation.
Collectively, our very high-resolution structure of yedK–Xlink (product-binding or post-reaction state) reveals that the covalent bond between yedK and the AP site in ssDNA is a thiazolidine linkage. Our biochemical data further suggests that some of the conserved residues stabilizing the thiazolidine linkage are important for DPC formation between yedK and the AP sites in ssDNA.
Conservative recognition mode of the thiazolidine linkage
To confirm that the recognition mode of the thiazolidine linkage in yedK is conservative and not dependent on the ssDNA sequence, we produced two more covalent structures of yedK with different ssDNA substrates, following the procedures described above. One structure was generated using an 11nt-ssDNA with a polyA sequence (ssDNA_polyA: 5′-AAAAA(dU)AAAAA-3′) and the other using a 10nt-ssDNA with a random sequence (ssDNA2_dU: 5′-GATTC(dU)GTCG-3′), referred to hereafter as yedK–polyA and yedK–Xlink2, respectively (Table 2). Structural comparisons reveal that their overall structures were essentially the same as yedK–Xlink, with small rmsds of 0.18 and 0.06 Å, respectively (Supplementary Figure S5B and C). It is noted that two more nucleotides 3′ to the AP site could be modeled in the yedK–polyA structure with poor density.
As shown above, the thiazolidine linkage is inserted into a hydrophobic pocket lined by Trp103, Ile147, Thr149 and Pro164 of yedK and further stabilized by the Glu105 and His160 side chains via hydrogen bonds. Local structural comparisons of different covalent structures around the AP sites reveal that the thiazolidine linkage positions and their yedK interacting residues are superimposed quite well (Figure 6A and B).
Figure 6.
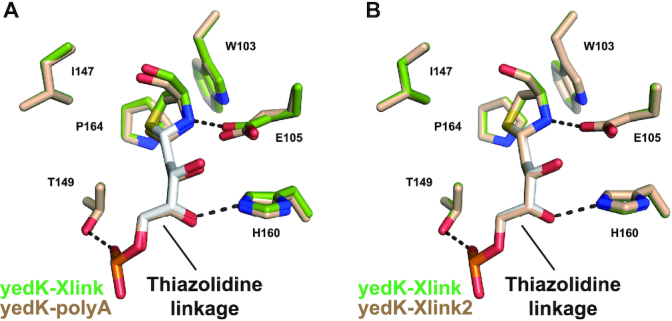
Thiazolidine linkage recognition is conservative. (A) Local structural comparison of the yedK–Xlink and yedK–polyA structures focusing on the AP site. (B) Local structural comparison of the yedK–Xlink and yedK–Xlink2 structures focusing on the AP site.
These results suggest that the recognition mode of the thiazolidine linkage by yedK is conservative and indicate that that yedK has no ssDNA sequence preference, only preferring the AP site.
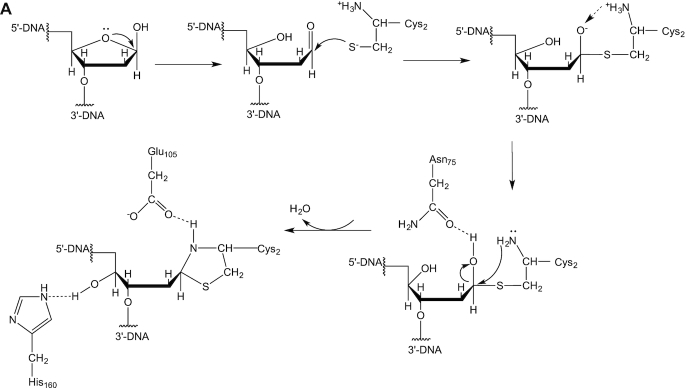
Reaction scheme
Based on our results, we propose a potential reaction scheme for DPC formation between yedK and AP sites in ssDNA (Figure 7A; 26,27). The thiol group of the catalytic Cys2 residue of yedK is easily deprotonated by its α-amino group to form a thiolate anion that is poised for catalysis. The AP site sugar moiety is in equilibrium between its cyclic furanose and open-chain aldehyde forms (20). The Cys2 thiolate anion attacks the AP site C1’ position to form a covalent bond. The Cys2 α-amino group proton then transfers to the oxygen which is stabilized by the Asn75 side chain. The O4’ rotates to a new position and interacts with the His160 side chain. The Cys2 α-amino group then attacks the C1’ position and transfers a proton to the hydroxyl group to release a water molecule, meanwhile forming a covalent bond with the C1’ atom. Thus, a thiazolidine linkage is formed between the AP site C1’ atom and the Cys2 amino and thiol groups. The Glu105 side chain carboxyl stabilizes the thiazolidine linkage via a strong hydrogen bond with its amine group.
Figure 7.
Probable reactions for DPC formation between yedK and AP sites in ssDNA.
DISCUSSION
Cells typically detect DNA lesions using highly specific sensors that detect distinct types of damage with high affinity (5). Recently, human HMCES has been reported to crosslink AP sites in ssDNA and form covalent DPC products alongside its orthologous protein yedK in E. coli (12). Here we illustrate the molecular mechanism by which yedK acts as an AP site sensor for ssDNA using multiple sets of complex structures. Our structures capture different stages of the AP site sensing process in ssDNA by yedK. First, yedK scans and loosely binds native ssDNA in a nonspecific-substrate-binding state. YedK has a much stronger preference for AP site-containing ssDNA than native ssDNA, which is conferred by the conserved Glu105 via electrostatic repulsion. Once an AP site is detected, it is positioned in the catalytic site, releasing the electrostatic repulsion between Glu105 and the ssDNA and thus forming a tight interaction between yedK and the AP site; this corresponds to a specific-substrate-binding state. The cyclic sugar ring of the bound AP site opens to form an open-chain aldehyde to start the reaction with the catalytic Cys2 residue in the intermediate state. Finally, the thiazolidine linkage is readily formed, penetrates the hydrophobic pocket formed by Trp103, Ile147, Thr149 and Pro164, and is stabilized by the strictly conserved Glu105 and His160 side chains in the product-binding state. Our study indentifies that Glu105 is not only the key residue by which yedK senses abasic sites in ssDNA via electrostatic repulsion, but is also involved in the recognition/stabilization of the thiazolidine linkage, consistent with the observation that yedK E105A reduces crosslinking efficiency to the AP site in ssDNA.
Since most of the residues involved in the recognition of the THF AP site and thiazolidine linkage are strictly conserved among bacteria, yeast, plants, and animals (Supplementary Figure S2A), the mechanism deduced from our studies can be extended to other SRAP domain-containing proteins and human HMCES. The stable chemical nature of the thiazolidine linkage (28) correlates well with the way HMCES effectively shields ssDNA AP sites from AP endonucleases and translesion DNA synthesis (TLS) polymerases, thus protecting genome integrity by promoting error-free AP site repair (12).
YedK has no sequence preference and is a specific sensor that can discriminate between AP sites and native ssDNA (Figure 4E); therefore, it could be developed as a tool to detect AP sites in vitro and in vivo. Such a tool could also be used to detect and enrich specific DNA or RNA base modifications as long as they can be converted into abasic sites by chemical agents (29,30).
Whilst preparing our manuscript for submission, two related studies have been published online (31,32), both of which reveal the thiazolidine linkage in the DPC of HMCES and yedK, respectively, and elucidate the interactions of HMCES and yedK with 3′ junction DNA. Consistently, our ITC results showed that yedK can bind AP sites immediately adjacent to the 3′ ssDNA-dsDNA junction as efficiently as the AP site in ssDNA (Kd of 0.18 and 0.17 μM, respectively) but not AP sites immediately adjacent to the 5′ ssDNA-dsDNA junction (Table 3 and Supplementary Figure S3V and W). Interestingly, four ssDNA nucleotides (G6-A7-T8-T9) 3′ to the AP site in the yedK–ssDNA structure are found to form a dsDNA-like structure with G6’-A7’-T8’-T9’ in the crystallographic symmetry-related ssDNA’ molecule (Supplementary Figure S7A and B), and a similar dsDNA-like structure are also found in the yedK–THF structure (Supplementary Figure S7C and D), supporting the model that yedK and HMCES can recognize a DNA duplex structure 3′ to the AP site as previously proposed (31,32). Notably, we showed that the yedK H160A mutant increased crosslinking efficiency to AP sites in ssDNA, whereas Thompson et al. found that it reduced crosslinking efficiency (31). The inconsistency between these two studies might be due to the different buffers and ssDNA substrates used for crosslinking assays.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structures have been deposited in the RCSB PDB (www.rcsb.org) with the following accession numbers: apo-yedK (6KBU), yedK–ssDNA (6KBS), yedK–THF (6KBZ), yedK–Xlink (6KBX), yedK–polyA (6KCQ) and yedK–Xlink2 (6KIJ). Other data are available upon reasonable request.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to gratefully thank the staffs from beamlines BL19U, BL18U and BL17U of Shanghai Synchrotron Radiation Facility (Shanghai, China) for assistance during data collection and prof. Xin-Yuan Liu at SUSTech for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2018YFC1004500]; Shenzhen Government ‘Peacock Plan’ [Y01226136 to H.H.]; Thousand Young Talents Program [to H.H.]; Chinese National Natural Science Foundation [31800619 to H.B.]. Funding for open access charge: the Thousand Young Talents Program.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001; 411:366–374. [DOI] [PubMed] [Google Scholar]
- 2. Friedberg E.C., Elledge S.J., Lehmann A., Lindahl T., Muzi-Falconi M.. DNA repair, mutagenesis, and other responses to DNA damage. 2013; 1 editionCold Spring Harbor Laboratory Press. [Google Scholar]
- 3. Cortez D. Replication-coupled DNA repair. Mol. Cell. 2019; 74:866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stingele J., Jentsch S.. DNA-protein crosslink repair. Nat. Rev. Mol. Cell Biol. 2015; 16:455–460. [DOI] [PubMed] [Google Scholar]
- 5. Stingele J., Bellelli R., Boulton S.J.. Mechanisms of DNA-protein crosslink repair. Nat. Rev. Mol. Cell Biol. 2017; 18:563–573. [DOI] [PubMed] [Google Scholar]
- 6. Fielden J., Ruggiano A., Popovic M., Ramadan K.. DNA protein crosslink proteolysis repair: from yeast to premature ageing and cancer in humans. DNA Repair (Amst.). 2018; 71:198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ide H., Nakano T., Salem A.M.H., Shoulkamy M.I.. DNA-protein cross-links: Formidable challenges to maintaining genome integrity. DNA Repair (Amst.). 2018; 71:190–197. [DOI] [PubMed] [Google Scholar]
- 8. Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993; 362:709–715. [DOI] [PubMed] [Google Scholar]
- 9. Dianov G. Repair of abasic sites in DNA. Mutat. Res. 2003; 531:157–163. [DOI] [PubMed] [Google Scholar]
- 10. Friedberg E.C., Walker G.C., Siede W., Wood R.D.. DNA Repair and Mutagenesis. 2005; American Society for Microbiology Press. [Google Scholar]
- 11. Krokan H.E., Bjoras M.. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013; 5:a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mohni K.N., Wessel S.R., Zhao R., Wojciechowski A.C., Luzwick J.W., Layden H., Eichman B.F., Thompson P.S., Mehta K.P.M., Cortez D.. HMCES maintains genome Integrity by shielding abasic sites in single-strand DNA. Cell. 2019; 176:144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spruijt C.G., Gnerlich F., Smits A.H., Pfaffeneder T., Jansen P.W., Bauer C., Munzel M., Wagner M., Muller M., Khan F. et al.. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013; 152:1146–1159. [DOI] [PubMed] [Google Scholar]
- 14. Aravind L., Anand S., Iyer L.M.. Novel autoproteolytic and DNA-damage sensing components in the bacterial SOS response and oxidized methylcytosine-induced eukaryotic DNA demethylation systems. Biol. Direct. 2013; 8:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kweon S.M., Zhu B., Chen Y., Aravind L., Xu S.Y., Feldman D.E.. Erasure of Tet-oxidized 5-methylcytosine by a SRAP nuclease. Cell Rep. 2017; 21:482–494. [DOI] [PubMed] [Google Scholar]
- 16. Sale J.E. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013; 5:a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schaaper R.M., Kunkel T.A., Loeb L.A.. Infidelity of DNA synthesis associated with bypass of apurinic sites. Proc. Natl. Acad. Sci. U.S.A. 1983; 80:487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kapust R.B., Tozser J., Copeland T.D., Waugh D.S.. The P1' specificity of tobacco etch virus protease. Biochem. Biophys. Res. Commun. 2002; 294:949–955. [DOI] [PubMed] [Google Scholar]
- 19. Acharya N., Talawar R.K., Saikrishnan K., Vijayan M., Varshney U.. Substitutions at tyrosine 66 of Escherichia coli uracil DNA glycosylase lead to characterization of an efficient enzyme that is recalcitrant to product inhibition. Nucleic Acids Res. 2003; 31:7216–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilson D.M. 3rd, Barsky D.. The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat. Res. 2001; 485:283–307. [DOI] [PubMed] [Google Scholar]
- 21. Wang Q.-S., Zhang K.-H., Cui Y., Wang Z.-J., Pan Q.-Y., Liu K., Sun B., Zhou H., Li M.-J., Xu Q. et al.. Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nucl. Sci. Tech. 2018; 29:68. [Google Scholar]
- 22. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M.. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D, Biol. Crystallogr. 2006; 62:859–866. [DOI] [PubMed] [Google Scholar]
- 23. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D, Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 25. Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C.. PHENIX: building new software for automated crystallographic structure determination. Acta crystallogr. D, Biol. Crystallogr. 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 26. Ratner S., Clarke H.T.. The action of formaldehyde upon cysteine. J. Am. Chem. Soc. 1937; 59:200–206. [Google Scholar]
- 27. Mackenzie C.G., Harris J.. N-formylcysteine synthesis in mitochondria from formaldehyde and L-cysteine via thiazolidinecarboxylic acid. J. Biol. Chem. 1957; 227:393–406. [PubMed] [Google Scholar]
- 28. Bermejo-Velasco D., Nawale G.N., Oommen O.P., Hilborn J., Varghese O.P.. Thiazolidine chemistry revisited: a fast, efficient and stable click-type reaction at physiological pH. Chem. Commun. 2018; 54:12507–12510. [DOI] [PubMed] [Google Scholar]
- 29. Hofer A., Liu Z.J., Balasubramanian S.. Detection, structure and function of modified DNA bases. J. Am. Chem. Soc. 2019; 141:6420–6429. [DOI] [PubMed] [Google Scholar]
- 30. Zhang L.S., Liu C., Ma H., Dai Q., Sun H.L., Luo G., Zhang Z., Zhang L., Hu L., Dong X. et al.. Transcriptome-wide mapping of internal N(7)-methylguanosine methylome in mammalian mRNA. Mol. Cell. 2019; 74:1304–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thompson P.S., Amidon K.M., Mohni K.N., Cortez D., Eichman B.F.. Protection of abasic sites during DNA replication by a stable thiazolidine protein-DNA cross-link. Nat. Struct. Mol. Biol. 2019; 26:613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Halabelian L., Ravichandran M., Li Y., Zeng H., Rao A., Aravind L., Arrowsmith C.H.. Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition. Nat. Struct. Mol. Biol. 2019; 26:607–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures have been deposited in the RCSB PDB (www.rcsb.org) with the following accession numbers: apo-yedK (6KBU), yedK–ssDNA (6KBS), yedK–THF (6KBZ), yedK–Xlink (6KBX), yedK–polyA (6KCQ) and yedK–Xlink2 (6KIJ). Other data are available upon reasonable request.