Abstract
Background and Aims
The advent of molecular breeding is advocated to improve the productivity and sustainability of second-generation bioenergy crops. Advanced molecular breeding in bioenergy crops relies on the ability to massively sample the genetic diversity. Genotyping-by-sequencing has become a widely adopted method for cost-effective genotyping. It basically requires no initial investment for design as compared with array-based platforms which have been shown to offer very robust assays. The latter, however, has the drawback of being limited to analyse only the genetic diversity accounted during selection of a set of polymorphisms and design of the assay. In contrast, genotyping-by-sequencing with random sampling of genomic loci via restriction enzymes or random priming has been shown to be fast and convenient but lacks the ability to target specific regions of the genome and to maintain high reproducibility across laboratories.
Methods
Here we present a first adoption of single-primer enrichment technology (SPET) which provides a highly efficient and scalable system to obtain targeted sequence-based large genotyping data sets, bridging the gaps between array-based systems and traditional sequencing-based protocols. To fully explore SPET performance, we conducted a benchmark study in ten Zea mays lines and a large-scale study of a natural black poplar population of 540 individuals with the aim of discovering polymorphisms associated with biomass-related traits.
Key Results
Our results showed the ability of this technology to provide dense genotype information on a customized panel of selected polymorphisms, while yielding hundreds of thousands of untargeted variable sites. This provided an ideal resource for association analysis of natural populations harbouring unexplored allelic diversities and structure such as in black poplar.
Conclusion
The improvement of sequencing throughput and the development of efficient library preparation methods has made it feasible to carry out targeted genotyping-by-sequencing experiments cost-competitively with either random complexity reduction systems or traditional array-based platforms, while maintaining the key advantages of both technologies.
Keywords: SPET, Allegro, genotyping-by-sequencing, targeted genotyping, bioenergy crops, Zea mays, Populus nigra, SNP, GWAS
Introduction
The ability to sample genetic polymorphisms in an efficient manner is the basis for the discovery of genotype–phenotype associations or for the assessment of population structure and patterns of adaptation. The availability of a large number of genetic markers has been the basis for the wide application of novel breeding technologies. Marker-assisted selection (MAS) and genomics selection (GS) have been widely used to improve yield and sustainability of several species of agronomic importance. The use of novel breeding technologies holds an even greater potential in many bioenergy crops that do not have a history of breeding and selection (Allwright and Taylor, 2016), and that could greatly benefit from the availability of efficient high-throughput genotyping approaches. While some efforts towards large-scale genotyping have been made in poplar, they have been limited to a very small number of candidate genes (Marroni et al., 2011), and do not meet the need for sampling large portions of the genome.
Since the late 1990s, high-density DNA arrays have become a promising tool to assess genetic variability at a massive scale (Wang et al., 1998). After their first use in human genetics, they have been widely adopted for non-human research, including plant and animal breeding (Deschamps et al., 2012). While high-density DNA arrays have been instrumental in the advancement of genetics and genomics, they suffer from ascertainment bias, a phenomenon that has been extensively observed in humans (Albrechtsen et al., 2010) and maize (Ganal et al., 2011), but is probably present in most DNA arrays. As the first next-generation sequencing (NGS) platforms appeared on the market, genotyping by short read sequencing was soon adopted to detect polymorphisms in a reduced representation of the target genome. One of the first implementations was CRoPS (van Orsouw et al., 2007) which substantially extended the amplified fragment length polymorphism (AFLP) preparation to 454 (Roche) pyrosequencing technology. However, the throughput was limited and not suitable for a very large cohort of samples. The widespread genotyping-by-sequencing (GBS) by Elshire and colleagues (2011) is a simplified version of complexity reduction with only one restriction enzyme. It was one of the first implementations of GBS on the Illumina Genome Analyzer platform and allowed genotyping of hundreds of samples per run. Modification of the original concept came in the following years. RAD-Seq (Baird et al., 2008) relies on both mechanical shearing and enzymatic restriction to improve coverage uniformity across loci. Moreover, the staggered paired-ends extended the sequencing space, thus increasing the likelihood of finding polymorphic sites and facilitating contig reconstruction in species lacking a reference genome. ddRAD (Peterson et al., 2012) instead, similarly to the AFLP method, relies on two restriction enzymes and provides more control on the abundance and distribution of loci; a feature that found further improvement in the targeted GBS (tGBS) protocol by Ott et al. (2017) with the reduction carried by extra selective bases during amplification cycles. In RESTseq, a second restriction enzyme is used to cleave adaptor-ligated fragments, thus reducing the fraction of the sampled genome by the depletion of functional fragments (Stolle and Moritz, 2013). Other methods such as NextRAD (Russello et al., 2015) directly leverage an amplification step to reduce the number of genomic loci to be considered using tagmentation-cleaved fragments as a template.
All the GBS techniques mentioned above share the principle of being an open, ascertainment bias-free system, unlike array-based methods, with no dependence on an original catalogue of polymorphisms. However, while all these methods provide elegant techniques to control the stringency and to reduce genomic loci under analysis, none of them is capable of efficiently targeting specific regions of a genome. Single genes, gene families, promoters and enhancers, gene clusters and non-coding genes are the genomic fractions that probably contain polymorphisms that are causative of or tightly associated with phenotypic variability. Therefore, sampling the genetic variability in a random fashion, mostly outside of these regions, is prone to impair the detection of valuable markers, especially in very complex and repetitive genomes or when the reproductive/crossing model generates a rapid linkage decay. This issue was easy to circumvent on an array platform at the cost of a strong ascertainment bias and poor re-usability when changing the genetic pool under analysis.
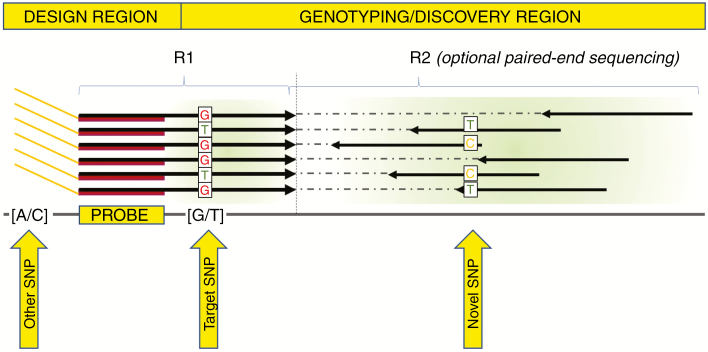
Here we present the first benchmark and large-scale adoption of a novel GBS concept, where a targeted approach, with high reproducibility and cost efficiency, is coupled with the sampling of unexplored genetic variability at each round of analysis. This system, which is commercialized under the name of Allegro (NuGEN Technologies, San Carlos, CA, USA), utilizes a single primer extension reaction to perform multiplex enrichment of a set of thousands to tens of thousands of target loci. The final library construct allows for stacked sequencing of readouts from such primers, thus maximizing the coverage use efficiency and enabling the targeting of known polymorphisms. In state-of-art sequencing platforms such as NextSeq500 or NovaSeq6000, a single-end sequencing of 150 bp can provide up to 110 bp of template sequence. This allows the discovery of novel polymorphisms, along with the target polymorphism, which can be simultaneously genotyped at the same depth of coverage (Fig. 1). Paired ends, which are on a randomly fragmented edge, can be optionally sequenced in order to augment the probability of finding novel polymorphisms. Moreover, a locus-specific complexity reduction method can provide unprecedented reproducibility across experiments and laboratories, with complete alignment across data sets. In other methods, such as restriction-associated methods, steps in the protocol are inherently drivers of variability in the representation of enriched loci: PCR cycles, gel-based selection and bead-based purifications can influence the population of DNA fragments retained for the sequencing.
Fig. 1.
Allegro sequencing layout. A design region is identified based on the read length and distance from the target SNP site. Annealing sites are designed avoiding other known polymorphic sites to prevent amplification bias. The sequencing template of each first-in-pair read is composed of 40 bp representing the probe itself, while from the 41st base onwards the genomic template is sequenced to genotype both target and novel SNP sites. All first-in-pair reads are stacked with the same 5′ mapping co-ordinates of a given locus. Second-in-pair reads (optionally sequenced), originating from mechanical/enzymatic fragmentation, are scattered in a 200–300 bp region in the 3′ direction of the first read, further contributing to the discovery of new polymorphisms. For the ‘Ovation® Target Enrichment’ system, a different layout is depicted in Supplementary Data Fig. S1.
Here we use Allegro to genotype two different data sets with the aim of evaluating the performance and the power of this new genotyping technology. First, we conducted a benchmark experiment on maize inbred and hybrid lines for which polymorphism data were already generated by means of the Illumina MaizeSNP50 (Ganal et al., 2011; Dell’Acqua et al., 2015) with the aim of assessing the level of accuracy and reproducibility at varying depths of coverage. Afterwards, we conducted a massive experiment to genotype hundreds of thousands of polymorphic markers in a collection of 540 Populus nigra samples to be used in a genome-wide association study (GWAS) for biomass-related traits.
MATERIALS AND METHODS
Study samples
The benchmark study sample is composed of five Zea mays inbred lines (F7, H99, HP301, Mo17 and W153R) and five F1 crosses (A632 × B73, B73 × B96, B73 × F7, B73 × Mo17 and W153 × HP301). To evaluate single-primer enrichment technology (SPET) reproducibility, each sample was analysed in two replicates. Replicates of the inbred lines were generated starting from the same DNA, while replicates of the F1 crosses were generated using DNA extracted from two different plants. Three inbred lines (F7, Mo17 and HP301) and the five crosses have already been genotyped using the Illumina MaizeSNP50 array (Ganal et al., 2011; Dell’Acqua et al., 2015). The extensive study was conducted on 540 Populus nigra plants that have been collected during a large association study in the framework of the WATBIO project. Geographical origin of poplar samples is summarized in Supplementary Data Table S1.
Probe design for single-primer enrichment technology
For the Z. mays study, single nucleotide polymorphisms (SNPs) have been selected in order to cover the majority of the positions included in the Maize Infinium Chip. After successful conversion from V3 to V4 assembly co-ordinates (http://ensembl.gramene.org/), a list of 55 198 target sites was obtained. With the aim of avoiding probe 3′ ends overlapping with other known variants, we generated a Z. mays genome-wide catalogue of polymorphisms by performing SNP detection on 40 different Z. mays lines for which short reads were available on NCBI’s Short Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra/). The complete list of analysed lines with the respective SRA accession is reported in Supplementary Data Table S2. SNP calling was performed using the software package GATK version 3.3-0 (McKenna et al., 2010). The GATK utilities ‘RealignerTargetCreator’ and ‘IndelRealigner’ were used to define the intervals in the proximity of indels and to perform the local realignment of reads spanning small indels, respectively. The variant discovery tool ‘UnifiedGenotyper’ was used in order to call SNPs in each Z. mays line (DePristo et al., 2011). Nucleotide differences called by GATK were classified as SNPs if (1) the position had a coverage ranging between 0.5 and 1.5 times the modal coverage of the sample; (2) the Phred-scaled quality score was >50; and (3) the variant allele had a frequency of at least 0.25. In total 15 083 641 polymorphic positions have been detected and provided to Nugen along with target sites in order to optimize probe design with its proprietary algorithm. Another prerequisite for probe design was to avoid their 3′ end (15 bp) overlapping with other known variants. It was also requested to design, where possible, two probes targeting the same site from both sides. A final set of 71 201 probes passed the design pipeline, targeting 46 358 unique sites, with 24 843 sites being enriched from both strands.
For black poplar, the selection was based on sites identified in 51 black poplar accessions (Faivre-Rampant et al., 2016) which have been filtered on minor allele frequency (MAF >0.15) and prioritized on genomic features to optimize the GWAS experiment (Taylor G., Allwright M.; unpubl. res.). All available SNPs were annotated using SnpEff (v4.1a) based on P. trichocarpa V3 gene models, and a prioritization scheme was applied as follows: (1) based on gene length quartiles, 1–4 SNPs sites were requested for each gene; (2) each SNP had to be at least 500 bp from any other selected SNP; (3) for the first selected SNP of each gene, the order of prioritization (the first category available is taken) was (a) non_synonymous_coding, (b) 5′ untranslated region (UTR), (c) synonymous_coding, (d) intron, 3′UTR, (e) 1500 bp upstream and (f) 1000 bp downstream; and (4) for the second to fourth SNP in gene selection, the order was instead (a) 5′UTR, (b) 1500 bp upstream, (c) non_synonymous_coding, (d) synonymous_coding, (e) intron, 3′UTR and (f) 1000 bp downstream. A final set of 98 134 probes was successfully designed and synthesized from a selection set of 112 724 SNPs, each probe targeting a different SNP site. In both experiments, the maximum distance between an SNP and its probe 3′ end was constrained based on the available read length excluding probe and linker readthrough. In this article, we describe two distinct experiments, carried out with the same technology but different configurations. The Z. mays experiment was carried with the genotyping-centric ‘Allegro targeted genotyping’ kit, which focuses on maximized yields of informative reads with first-in-pair reads (Fig. 1). For black poplar, the ‘Ovation® Target Enrichment’, which is based on the same SPET technology but with an inverted sequencing layout, was used. In fact, for Z. mays, a single-end sequencing was sufficient to genotype target SNPs, while for black poplar paired-end sequencing was necessary as read stacks on target sites are accessible by means of second-in-pair reads (Supplementary Data Fig. S1). The technology adopted in this study is also described in the latest issued patent US10036012.
DNA extraction, library preparation and sequencing
Zea mays leaves were ground in liquid nitrogen, and high molecular weight genomic DNA was extracted from nuclei as previously described (Zhang et al., 1995). The protocol was improved with the addition of PVP40 both in the wash (5 %) and in the lysis (2 %) buffers. Libraries were prepared using the ‘Allegro Targeted Genotyping’ protocol from NuGEN Technologies, using 150 ng of DNA as input and following the manufacturer’s instructions. Libraries were quantified through quantitative PCR using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA).
Populus nigra libraries were prepared starting from genomic DNA in the range of 200–400 ng, following the standard protocol for the ‘Ovation® Target Enrichment’ (NuGEN Technologies), with minor modification during sonication (four cycles of 15 min/90 min in 100 μL of TE solution, Bioruptor – Diagenode) and fragment purification steps (0.7 vols of Ampure XP beads). Fragment libraries were quantified with the GloMax® Explorer System (Promega) after adaptor ligation and pooled as 8-plex, based on quantification ranks. Real-time PCR with Illumina P5 and P7 primers (37 °C for 10 min, 95 °C for 3 min, 35 cycles of 95 °C for 30 s, 62°C for 15 s and 72°C for 20 s) was used to determine the necessary number of amplification cycles (15 or 16, depending on pools) by the detection of the inflection point in the log-transformed amplification curve as described in the ‘Ovation® Target Enrichment’ manual.
Sequencing was performed at IGA Technology Services (IGATech, Udine, Italy) facilities. The maize library pool showed an average size of 738 bp and it was sequenced using either a HiSeq2000 or a NextSeq500 platform (Illumina, San Diego, CA, USA) in single-end mode (150 bp). Poplar libraries, with an average library size ranging from 700 to 800 bp depending on pools (16-plex), were sequenced on the HiSeq2500 platform in paired-ends mode, with 130 bp reads. BCL files from the instruments were processed using the manufacturer’s pipeline software to generate FASTQ sequence files. For black poplar libraries, a third read containing the six random bases, generally known as the unique molecular identifier (UMI), which are present after the eight bases of the index, was produced. Zea mays FASTQ files are available at the SRA database under the accession number SRP157896 while P. nigra sequences will soon released.
Short read mapping and SNP detection for SPET benchmark
Adaptor sequences and low-quality 3′ ends were removed from short reads using cutadapt (Martin, 2011) and ERNE-FILTER (http://erne.sourceforge.net), respectively, with default parameters. After trimming, reads longer than 50 bp were aligned to the Z. mays v4 reference genome using the short read aligner BWA-MEM (Li and Durbin, 2009) with default parameters. The mean individual coverage at target sites was calculated using the utility ‘multiBamCov’ included in bedtools v2.26.0 (Quinlan and Hall, 2010). SNP calling was performed on uniquely aligned reads using the software package GATK version 3.8 (McKenna et al., 2010). The GATK utilities ‘RealignerTargetCreator’ and ‘IndelRealigner’ were used to define the intervals in the proximity of indels and to perform the local realignment of reads spanning small indels, respectively. The variant discovery tool ‘UnifiedGenotyper’ was used to call SNPs in each sample. Low-quality variant calls were excluded using GATK ‘VariantFiltration’ (filterExpression: ‘QD < 2.0 || MQ < 40.0 || MQRankSum < –12.5’) and ‘SelectVariants’.
SPET benchmark data analysis
The SPET performance was evaluated using genotypes called with an array-based technology as gold standard. Since maize array has been developed using the Z. mays v3 reference genome, the co-ordinates of the array-based genotypes have been translated to the newer version of the genome (B73 RefGen_v4). To select a high confidence list of loci to work with, a sub-set of positions genotyped with the array technology was selected in order to avoid: (1) positions located in the unanchored part of the genome; (2) positions for which the B73 genotype from the SNP chip was not in agreement with the reference genome (B73) sequence; and (3) positions in which array-based genotypes called in the inbred lines and in the corresponding F1 crosses showed Mendelian inconsistency (i.e. hybrids showing an inconsistent genotype from the expected parental inheritance). Residual heterozygosity (RH) in advanced maize inbred progenies is a well-known phenomenon (Eichten et al., 2011; Liu et al., 2018). Thus, whole-genome resequencing data of five inbred lines (F7, H99, HP301, Mo17 and W153R) were employed to estimate RH at a genome-wide level in 100 kb contiguous windows. To avoid possible biases in the called genotypes due to RH, genotypes belonging to windows in which the percentage of heterozygous sites was higher than 20 % in the corresponding sample or in one of the two parental lines were not considered. The accuracy of SPET genotyping was measured as the percentage of sites in which genotypes called with array-based technology and SPET were in accordance. To evaluate the effect of the sequencing coverage, accuracy was measured on sub-sets of positions fulfilling an increasing coverage threshold. Towards this aim, the coverage threshold (c) was progressively increased from 1 to 100 and accuracy was measured in all positions with a coverage included in the interval c ± (0.25 × c). Reproducibility was calculated as the fraction of genotype calls in accordance between the two replicates of each sample. For reproducibility, the effect of coverage was tested by progressively increasing the coverage threshold (c) from 1 to 100 and evaluating only positions with a coverage included in the interval c ± (0.5 × c) in both replicates.
Short read mapping and SNP detection for massive genotyping: case study
Cutadapt was used to remove adaptor leftover by searching alignment matches to either a standard Illumina adaptor or the linker sequence residing between each probe and the remaining Illumina-like adaptor (first 15 bp sequenced by ‘second-in-pair’ reads). Low-quality ends were removed using ERNE-FILTER, as described before in the benchmark analysis. Alignment of paired reads on the reference genome of P. trichocarpa was performed with the BWA MEM algorithm (0.7.10). A custom script was used to remove PCR duplicates by means of the 5′ position of ‘first-in-pair’ reads and their corresponding random six bases sequenced after the index. SNPs were called using GATK UnifiedGenotyper (v. 3.3.0) after local realignment around indels (as described above); all SNP calls residing in the region of primer hybridization were removed: this step was taken in order to avoid false SNP calls due to the presence of errors or mismatches in the primer sequence since it is part of the readout. SNP sites were retained up to a maximum distance of 1000 bp from each probe 3′ end. Genotype calls were further refined on the basis of (1) a minimum coverage of six reads to call homozygous samples; (2) a minor allele count >0.20 to call heterozygous genotypes; and (3) a minimum quality of 500 for target SNPs or 1000 for off-target SNPs. Principal component analysis (PCA) was performed using ‘smartpca’ from the software package EIGENSOFT v6.0.1 (Patterson et al., 2006; Price et al., 2006).
RESULTS
SPET genotyping benchmark
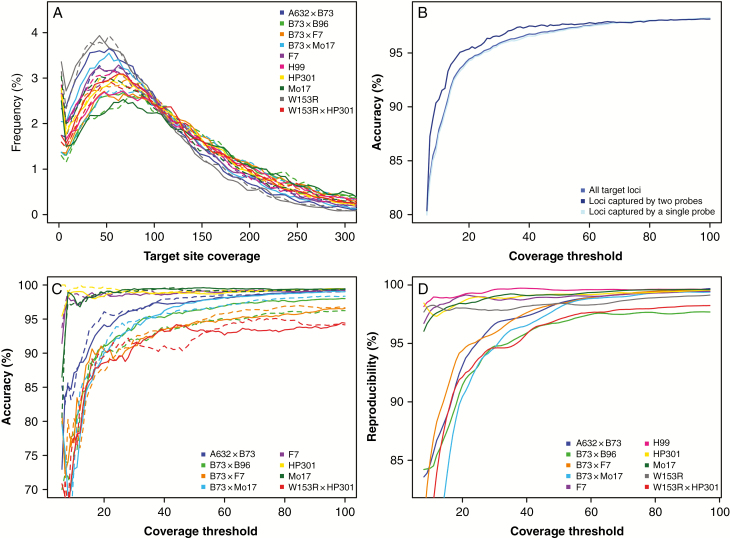
For Zea mays, SPET reads were generated in excess as compared with a real-world use in order to evaluate the performance at varying levels of coverage. The average was 25 million reads per sample, ranging from 20 to 30 million (Supplementary Data Table S3). The alignment rate on the whole genome was stable between 88 and 91 %. On average, 24.3 % of the 46 358 target sites were covered by less than three reads. Considering only the 24 843 sites targeted by two probes, the percentage of uncovered sites decreased to 5.3% (Supplementary Data Table S4). Since the probe set used during the experiment also included accessory probes for the generation of other data outside the scope of this work, metrics on effective enrichment efficiency cannot be provided for Z. mays, and we refer to the black poplar data as bona fide results. We analysed the performance of SPET genotyping on 27 236 sites for which we have the genotypes called by both SPET and array-based technology. Of them, 12 762 (46.9%) were targeted by a single probe, while the remainder were targeted by two distinct probes, one located upstream and the other located downstream of the target site. The accuracy of SPET genotyping was evaluated on three inbred lines (F7, HP301 and Mo17) and five F1 crosses (A632 × B73, B73 × B96, B73 × F7, B73 × Mo17 and W153 × HP301) for which genotypes called using the array technology were already available. The distribution of the coverage obtained at target sites was very similar between all samples, with most of the sites having a coverage >50× (Fig. 2A). The accuracy showed a marked improvement by progressively increasing the coverage threshold, and reached a plateau at about 50×, with an accuracy of 97.2 % (Fig. 2B). However, by lowering the SNP detection coverage threshold to a broadly used depth of 30×, the accuracy is only slightly reduced (95.9 %). Accuracy measured only on loci targeted by two different probes was higher (96.5 % by applying a coverage threshold of 30×). We analysed the accuracy separately for each sample (Fig. 2C) and we notice that the accuracy obtained for the three inbred lines (F7, HP301 and Mo17) was higher with respect to that obtained in crosses. For example, at a coverage threshold of 30×, in inbred lines the mean accuracy was 99.0 % while in F1 crosses it was 93.3 %. In particular, the worst performing F1 cross was W153R × HP301 for which we obtained an accuracy of 91% at a coverage threshold of 30×. We observed that loci with discordant genotype calls between the two technologies were often clustered and probably resulted from the high levels of RH of the line W153R (Dell’Acqua et al., 2015). We measured the reproducibility of the SPET analysis by comparing, for each sample, the genotype calls obtained for the two replicates (Fig. 2D). By increasing the coverage threshold, the reproducibility progressively increased and, similarly to the accuracy, reached a plateau at about 50×. By requiring a minimum coverage of 30× we obtained a reproducibility ranging from 94.2 % of cross B73 × Mo17 to 99.3 % of sample H99. As observed for accuracy, in inbred lines we obtained a higher degree of reproducibility in comparison with the F1 crosses.
Fig. 2.
SPET genotyping benchmark results. (A) Target site coverage distribution obtained for each genotyped Z. mays sample; solid and dashed lines represent the two replicates of each sample. (B) Accuracy of SPET genotyping measured by increasing the coverage threshold considering all sites, sites targeted by a single probe and sites targeted by two distinct probes. (C) Accuracy of SPET genotyping at different coverage thresholds obtained in each sample; solid and dashed lines represent the two replicates of each sample. (D) Reproducibility of SPET genotyping measured at different coverage thresholds.
Massive genotyping case study
Different library pools were prepared separately and sequenced on different HiSeq2500 runs. Despite the abundance of samples deriving from different geographical areas, sampling time and extractions, the final sequencing yields turned out to be highly homogeneous, with an average of 9.7 million reads per sample, and with 508 samples (94 %) within the 5–16 million range (Supplementary Data Fig. S2). The enrichment efficiency (i.e. the fraction of reads aligned on target loci) was very similar between the different samples (Supplementary Data Fig. S3), with an average of 88.4 %, while the average rate of PCR duplicates was 24.4 %.
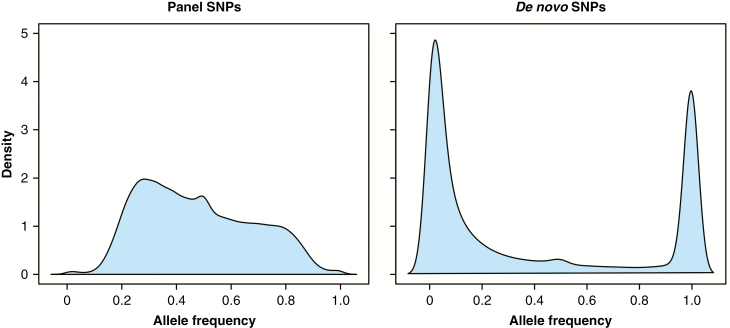
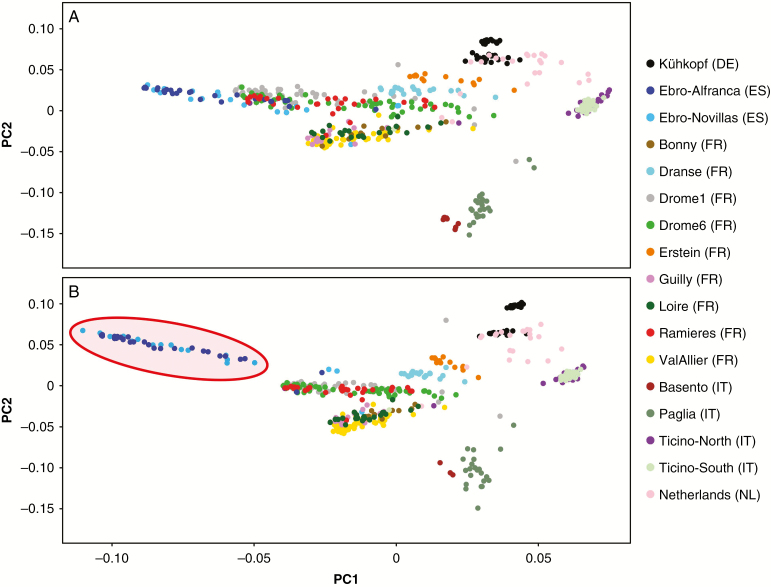
Among the target sites, 66 922 SNPs were called following the standard filtering criteria (see the Materials and Methods). Furthermore, 453 170 polymorphic sites were provided from non-target positions. Adopting more stringent filtering criteria, i.e. requiring a minimum coverage of eight reads to call homozygous sites and a minimum ratio of called individuals of 75 %, the numbers of SNPs were reduced to 51 943 and 203 478, respectively. In general, samples belonging to groups Dranse and Loire showed a lower performance as a measure of coverage distribution despite comparable sequencing yields (Table 1). The analysis of coverages at target sites showed that a fraction (approx. 10 %) of probes provided zero coverage, suggesting a failure in a functional annealing/extension to the region, caused by either variability in the assayed samples compared with the reference genome (indeed we used P. trichocarpa to analyse P. nigra samples) or, even with lower probability, a sequence error in the reference sequence. Excluding these two poorly performing groups, the rate of target sites was between 73 and 79 %, providing a final coverage of at least six non-duplicated reads. We analysed allele frequencies in the whole population stratifying for those coming from selected sites (panel) or from de novo discovered sites (Fig. 3). Not surprisingly, we found that the target site allele frequencies in the population were somewhat bounded to the applied selection criteria, which included a minimum allele frequency of 0.15 in the panel of 51 samples used for the discovery (Pinosio S., unpubl. res.). In contrast, de novo genotyped sites showed a great abundance of very low and very high allele frequencies (excluding AF = 0 and AF = 1 to avoid species-specific calls since P. trichocarpa was used as a reference). To further investigate the value of such a secondary set of de novo genotyped SNPs, we carried out two parallel PCAs, one with with panel sites (Fig. 4A) and the other with de novo sites (Fig. 4B). The analysis carried out with de novo SNPs showed an augmented performance in population clustering, especially for those groups that were not highly represented in the selection sample panel. These refer to Spanish genotypes, which were not well represented in the selection panel as it was mostly based on genotypes sampled in France, Germany, Italy and The Netherlands.
Table 1.
Coverage metrics of all target sites for the most abundant sample groups
| Group | Median | Mean | s.d. | Non-zero coverage | Minimum six reads |
|---|---|---|---|---|---|
| Drome1 | 15 | 23.18 | 29.70 | 89 % | 73 % |
| Drome6 | 22 | 31.34 | 37.05 | 90 % | 79 % |
| ValAllier | 26 | 36.71 | 41.78 | 90 % | 79 % |
| Dranse | 7 | 14.01 | 23.74 | 79 % | 56 % |
| Guilly | 15 | 23.09 | 29.56 | 89 % | 73 % |
| Kühkopf | 21 | 31.82 | 38.82 | 88 % | 76 % |
| Ticino-North | 23 | 34.73 | 41.72 | 90 % | 78 % |
| The Netherlands | 17 | 24.79 | 31.94 | 89 % | 75 % |
| Paglia | 22 | 32.20 | 37.94 | 90 % | 78 % |
| Ramieres | 19 | 28.59 | 36.49 | 89 % | 76 % |
| Ticino-South | 21 | 33.57 | 42.90 | 89 % | 76 % |
| Loire | 9 | 24.18 | 45.20 | 83 % | 60 % |
| Ebro-Alfranca | 18 | 27.91 | 35.10 | 85 % | 72 % |
| Ebro-Novillas | 15 | 27.69 | 37.85 | 86 % | 69 % |
Data are given after data filtering, including removal of duplicated reads.
Fig. 3.
Distribution of allele frequencies. Polymorphic sites of the target panel (on the left) and de novo genotyped sites (on the right). De novo sites show a prominent sampling of rare alleles.
Fig. 4.
Principal component analysis (PCA) as calculated using target sites (A) or de novo sites (B). Samples collected in Spain are highlighted in red. Samples with an undefined origin (see Supplementary Data Table S1) are not reported.
Discussion
We have assessed the capability of SPET as a tool for a new paradigm of GBS experiments. Unlike other methods, this technique allows for the execution of cost-effective genotyping experiments while providing full control on target sites. This is mainly achieved by the combination of (1) a high efficiency enrichment system to target a pre-defined number of loci; (2) the convenient stack of reads at the same mapping co-ordinate of a given locus; (3) the scalability to tens of thousands of probes in a single reaction; and (4) the removal of PCR duplicates even in the absence of variation of the mapping co-ordinates for a given locus. This is the first reported large cohort GBS protocol that merges the economy of stringent complexity reduction as provided by restriction enzyme-based methods such as GBS, RADSeq and ddRAD with the ability for on-target analysis. This opens the door to a new era of genotyping, with experimental designs that offer complete reproducibility across laboratories and operators, while avoiding ascertainment bias. In fact, this sequencing-based genotyping shares with random complexity reduction systems the ability to sample new genetic diversity and, with arrays, the ability to target specific genomic sites. Allele frequency distribution among de novo sites in poplar data reflected the distribution predicted by population genetics theory (Nielsen, 2005; Marth et al., 2011), suggesting that they are free from the ascertainment bias often observed in SNP chip data (Albrechtsen et al., 2010). In addition, we observed that diversity estimates are significantly more aligned to geographical origin when using de novo genotyped SNPs in 540 black poplars. The method already showed a certain uniformity of coverage across probes; however, the possibility to re-synthesize the probe pools without further investment allows the empiric optimization of the latter, toward the most uniform coverage and thus cost-effectiveness. In the benchmark study, the accuracy of SPET (using previous data on SNP genotyping arrays as a gold standard) was high but did not reach 100 %. Considering that the two technologies were applied on DNA extracted from different plant material and that RH in advanced maize inbred progenies is a well-known phenomenon (Eichten et al., 2011; Liu et al., 2018), we are not expecting a 100 % concordance between the genotypes called by the SNP array and those called by our SPET experiment. In fact, the presence of RH in the inbred lines is expected to generate genotypic differences between the two data sets. Differences are also expected when comparing the genotypes obtained in the two replicates of the F1 crosses, for which we used DNA extracted from different plants. In fact, in F1 crosses we steadily obtained lower levels of both accuracy and reproducibility if compared with inbred lines (Fig. 2C, D). In addition, a fraction of the discordant genotype calls between SPET and array-based technology is attributable to real differences between the different plants used in the two studies; thus, we suggest that the reported accuracy is an underestimate of the real accuracy of the SPET genotyping.
In our experiments we showed that an average real coverage of 30× to 50× was sufficient to obtain high-quality genotype calls. This means that some 5 million reads are sufficient per sample using a panel of 100 000 probes, which means a single S1 flowcell of the Illumina NovaSeq platform can run 260 sample in a single run, while an S4 flowcell can load >1000 samples. Paired-end sequencing, which is not required, can be used as a complementary data source when it is the intention to yield as many additional variant sites and haplotypes as possible. Overall, the technology demonstrated very promising performance and suitability to be a valid replacement of random complexity reduction methods and array platforms to overcome their respective limitations while maintaining a low cost per sample and complete scalability.
Molecular breeding promises greatly to increase yield of bioenergy crops (Allwright and Taylor, 2016). This is especially true for crops for which substantial genomic resources are available. The availability of a reference sequence for poplar (Tuskan et al., 2006) also enabled the development of valuable tools such as genotyping arrays (Geraldes et al., 2013). Here, we present a novel genotyping approach which has costs comparable with genotyping arrays but allows for greater flexibility and de novo polymorphism discovery, and which is virtually free from ascertainment bias. The choice of genotyped loci will drive the population of analysed polymorphisms, which will reflect the genomic context in terms of diversity and selective pressure. However, at the population level, the diversity within each locus is sampled without bias since a given number of bases are sequenced without relying on any a priori genotype information. This allows the system to provide any new polymorphism occurring in every locus. Our results suggest that SPET genotyping is a very effective approach for genotyping a large number of loci, and we suggest that it might be a valuable tool for large-scale genotyping studies in virtually all species. This will greatly facilitate molecular breeding approaches for bioenergy crops.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table 1: geographic origin of black poplar samples. Table 2: SRA run accessions for the 40 Zea mays lines used to build the catalogue of variation. Table 3: sequencing and alignment statistics. Table 4: coverage statistics. Figure 1: ‘Ovation® Target Enrichment’ sequencing configuration. Figure 2: distribution of sequencing yields in the P. nigra data set as paired ends. Figure 3: distribution of the percentage of reads aligned on target regions for the most abundant sample groups.
FUNDING
The poplar project was funded by the WATBIO project: Development of improved perennial non-food biomass and bioproduct crops for water stressed environments is an international research project funded by the European Union’s Seventh Framework Programme under the grant agreement FP7-311929.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for Nugen’s extensive support in optimizing this genotyping approach. Thanks to Elisabetta Frascaroli – University of Bologna (DipSA) – for proving seeds of hybrid maize lines, and to Matteo Dell’Acqua for sharing useful information.
LITERATURE CITED
- Albrechtsen A, Nielsen FC, Nielsen R. 2010. Ascertainment biases in SNP chips affect measures of population divergence. Molecular Biology and Evolution 27: 2534–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allwright MR, Taylor G. 2016. Molecular breeding for improved second generation bioenergy crops. Trends in Plant Science 21: 43–54. [DOI] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, et al. 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3: e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Acqua M, Gatti DM, Pea G, et al. 2015. Genetic properties of the MAGIC maize population: a new platform for high definition QTL mapping in Zea mays. Genome Biology 16: 167. doi: 10.1186/s13059-015-0716-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps S, Llaca V, May GD. 2012. Genotyping-by-sequencing in plants. Biology 1: 460–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten SR, Foerster JM, de Leon N, et al. 2011. B73-Mo17 near-isogenic lines demonstrate dispersed structural variation in maize. Plant Physiology 156: 1679–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, et al. 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6: e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre-Rampant P, Zaina G, Jorge V, et al. 2016. New resources for genetic studies in Populus nigra: genome wide SNP discovery and development of a 12k Infinium array. Molecular Ecology Resources 16: 1023–1036. [DOI] [PubMed] [Google Scholar]
- Ganal MW, Durstewitz G, Polley A, et al. 2011. A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS One 6: e28334. doi: 10.1371/journal.pone.0028334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A, Difazio SP, Slavov GT, et al. 2013. A 34K SNP genotyping array for Populus trichocarpa: design, application to the study of natural populations and transferability to other Populus species. Molecular Ecology Resources 13: 306–323. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics (Oxford, England) 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Liu J, Li W, et al. 2018. Intraspecific variation of residual heterozygosity and its utility for quantitative genetic studies in maize. BMC Plant Biology 18: 66. doi: 10.1186/s12870-018-1287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroni F, Pinosio S, Di Centa E, et al. 2011. Large scale detection of rare variants via pooled multiplexed next generation sequencing: towards next generation Ecotilling. The Plant Journal 67: 736–745. [DOI] [PubMed] [Google Scholar]
- Marth GT, Yu F, Indap AR, et al. 2011. The functional spectrum of low-frequency coding variation. Genome Biology 12: R84. doi: 10.1186/gb-2011-12-9-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal 17: 10–12. [Google Scholar]
- McKenna A, Hanna M, Banks E, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. 2005. Molecular signatures of natural selection. Annual Review of Genetics 39: 197–218. [DOI] [PubMed] [Google Scholar]
- van Orsouw NJ, Hogers RCJ, Janssen A, et al. 2007. Complexity reduction of polymorphic sequences (CRoPS™): a novel approach for large-scale polymorphism discovery in complex genomes. PLoS One 2: e1172. doi: 10.1371/journal.pone.0001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott A, Liu S, Schnable JC, Yeh C-T ‘Eddy’, Wang K-S, Schnable PS. 2017. tGBS® genotyping-by-sequencing enables reliable genotyping of heterozygous loci. Nucleic Acids Research 45: e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. 2006. Population structure and eigenanalysis. PLoS Genetics 2: e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. 2012. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One 7: e37135. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics 38: 904–909. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russello MA, Waterhouse MD, Etter PD, Johnson EA. 2015. From promise to practice: pairing non-invasive sampling with genomics in conservation. PeerJ 3: e1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolle E, Moritz RFA. 2013. RESTseq – efficient benchtop population genomics with RESTriction Fragment SEQuencing. PLoS One 8: e63960. doi: 10.1371/journal.pone.0063960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604. [DOI] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, et al. 1998. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280: 1077–1082. [DOI] [PubMed] [Google Scholar]
- Zhang HB, Zhao X, Ding X, Paterson AH, Wing RA. 1995. Preparation of megabase-size DNA from plant nuclei. The Plant Journal 7: 175–184. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.