Abstract
Summary
We introduce YeastSpotter, a web application for the segmentation of yeast microscopy images into single cells. YeastSpotter is user-friendly and generalizable, reducing the computational expertise required for this critical preprocessing step in many image analysis pipelines.
Availability and implementation
YeastSpotter is available at http://yeastspotter.csb.utoronto.ca/. Code is available at https://github.com/alexxijielu/yeast_segmentation.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The accurate segmentation of a microscopy image into single cells is an important preprocessing step for many image analysis pipelines (Meijering, 2012). As a model organism, the budding yeast Saccharomyces cerevisiae is often used in imaging experiments, some of which can generate tens of thousands of images (Dubreuil et al., 2018; Koh et al., 2015; Riffle and Davis, 2010; Weill et al., 2018). To analyze these images, a range of segmentation options have emerged, often tailored to specific datasets. Some integrate assumptions specialized to screens, such as the presence of fluorescent markers (Handfield et al., 2013), edge patterns (Dimopoulos et al., 2014; Wang et al., 2018), or assumptions specific to microfluidics experiments (Bakker et al., 2018). Others require the laborious specification of many manual parameters (Carpenter et al., 2006); indeed, most methods for brightfield images require extensive parameter tuning for optimal performance (Versari et al., 2017).
For a cell biologist, the wide choice and complexity of segmentation methods may lead them to manual quantification if the effort required for automation appears disproportionate to the scale of their experiments. We envisioned a tool that could produce reasonable segmentations for most images with minimal effort. Toward this goal, we designed YeastSpotter (yeastspotter.csb.utoronto.ca), a web application that generalizes to images from different microscopes and imaging modalities, without the need to specify any parameters: the user simply submits their images and obtains a segmentation. Despite its simple use, we obtain comparable performance to specialized state-of-the-art methods on benchmarks for segmentation of both fluorescent and brightfield images.
2 Materials and methods
Our underlying segmentation method is based upon transferring publicly available convolutional neural networks from the 2018 Kaggle Data Science Bowl competition. In this competition, contestants trained models to segment images of mostly human nuclei, using image set BBBC038v1 from the Broad Bioimage Benchmark Collection (Ljosa et al., 2013) as training data. Despite not being trained on yeast cells, we found that these models transferred well without fine-tuning. We used a pre-trained mask-RCNN model (He et al., 2017) by the third-place winner, the Deep Retina team, which we chose due to its simplicity and easily extensible code. To make this model more accessible to the community, we implemented YeastSpotter as a web application to run images through this model.
To use YeastSpotter, the user simply uploads their image, which redirects them to a page that tracks the progress of their request and produces segmentation results once ready. A preview image on the result page shows the outlines of the segmentation overlaid on the original input. The user can then download the segmentation, which is stored as an integer-signed tiff file (pixels with a value of 0 correspond to the background, while pixels belonging to each unique cell are each assigned a different integer value). On the website, we provide instructions for loading these fines into ImageJ and scripts to read them in Python, Matlab and R.
YeastSpotter is intended for low-throughput use and only accepts a single image per request. For batch segmentation, we also provide user-friendly Python code (www.github.com/alexxijielu/yeast_segmentation).
3 Results
To understand the accuracy and run-time of the segmentations produced by our method, we used a set of 4305 ellipses manually drawn around yeast cells in fluorescent micrographs (Handfield et al., 2013). We compared segmentations from YeastSpotter to previously reported results for segmentation software specially designed for this dataset by Handfield et al. (Handfield et al., 2013), and for segmentations obtained through CellProfiler (Carpenter et al., 2006) in Table 1, using parameters previously optimized by Chong et al. (Chong et al., 2015). These results suggest that our method segments fluorescent micrographs of yeast cells more accurately than established methods, with no manual tuning of parameters.
Table 1.
Benchmark results on fluorescent yeast micrographs
| Method | Ellipses matched | Mean | Standard deviation | Correlation | Run time |
|---|---|---|---|---|---|
| YeastSpotter | 97.5% | 1.58 | 0.99 | 0.969 | 1172 |
| Handfield et al. (2013) | 92.3% | 1.41 | 1.21 | 0.928 | 13 851 |
| CellProfiler | 89.0% | 2.23 | 1.80 | 0.876 | 231 |
We report the percent of manual ellipses with a matched single-cell segmentation within ten pixels, the mean and standard deviation of distance (in pixels) between the centers of the manual ellipse and segmentation, the correlation between their areas and the time (in seconds) to process the evaluation image set (68 images).
To test the generalization capacity, we evaluated our segmentations on detecting cell centers in brightfield images from the Yeast Image Toolkit benchmark (Versari et al., 2017, Supplementary Fig. S1). We achieved comparable performance to most tools, even though they have been extensively optimized for brightfield images (Versari et al., 2017), while YeastSpotter was not. We note that YeastSpotter does not achieve state-of-the-art performance, so expert users may still want to optimize tools for their images.
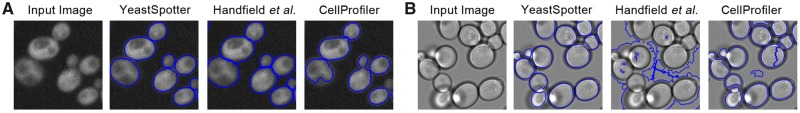
We next qualitatively examined segmentation results on these (Fig. 1) and other image modalities (Supplementary Fig. S2 shows differential interference contrast (DIC) and phase contrast). On fluorescent images (Fig. 1A), CellProfiler (with parameters used for Table 1) under-segments bud cells, grouping the pixels of bud cells with mother cells and does not accurately detect the boundaries of cells with dim vacuoles. Accurately segmenting bud cells is critical for understanding yeast biology, as it permits for the study of the cell-cycle (Handfield et al., 2013). YeastSpotter and the method of Handfield et al. more reliably separate bud cells from mother cells.
Fig. 1.
Qualitative segmentation results for various segmentation algorithms. We show results for fluorescent (A) and brightfield (B) images. In the left-most panels, we show the original input image. In the other panels, we show outlines of the segmentation result from each segmentation method (as labeled) overlaid on the original image in blue
However, as the method of Handfield et al. is engineered for fluorescent images, it fails to generalize to the segmentation of brightfield images (Fig. 1B). The CellProfiler segmentation optimized for fluorescent micrographs is more robust, but still produces many errors, identifying parts of the background as cells and over-segmenting some cells. YeastSpotter performs well on both fluorescent and brightfield images; there are some errors with overlapping or out-of-focus cells in the brightfield images, but most cells are segmented well.
4 Conclusion
Here, we introduced a user-friendly and generalizable web application for the segmentation of yeast microscopy images. We produced high-quality segmentations for both fluorescent and brightfield images using the same model and parameters. These results suggest that YeastSpotter is highly general, as opposed to most previous methods, which have been developed to segment images of a particular type.
YeastSpotter may not outperform carefully optimized methods tailored to specific problems. However, for users without the time or expertise to fine-tune or compare specialized methods, our method offers excellent off-the-shelf performance.
Supplementary Material
Acknowledgements
We thank Yunchen Gong for help in setting up the website, Robert Strome for providing sample images and Purnima Kompella for helpful discussions.
Funding
This work was supported by the National Science and Engineering Research Council, Canada Research Chairs, and the Canadian Foundation for Innovation.
Conflict of Interest: none declared.
References
- Bakker E. et al. (2018) Morphologically constrained and data informed cell segmentation of budding yeast. Bioinformatics, 34, 88–96. [DOI] [PubMed] [Google Scholar]
- Carpenter A.E. et al. (2006) CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol., 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong Y.T. et al. (2015) Yeast proteome dynamics from single cell imaging and automated analysis. Cell, 161, 1413–1424. [DOI] [PubMed] [Google Scholar]
- Dimopoulos S. et al. (2014) Accurate cell segmentation in microscopy images using membrane patterns. Bioinformatics, 30, 2644–2651. [DOI] [PubMed] [Google Scholar]
- Dubreuil B. et al. (2018) YeastRGB: comparing the abundance and localization of yeast proteins across cells and libraries. Nucleic Acids Res., 47, D1245–D1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handfield L.-F. et al. (2013) Unsupervised clustering of subcellular protein expression patterns in high-throughput microscopy images reveals protein complexes and functional relationships between proteins. PLoS Comput. Biol., 9, e1003085.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K. et al. (2017) Mask R-CNN. In: Proceedings of the IEEE international conference on computer vision, pp. 2961–2969. Venice, Italy.
- Koh J.L.Y. et al. (2015) CYCLoPs: a comprehensive database constructed from automated analysis of protein abundance and subcellular localization patterns in Saccharomyces cerevisiae. G3 (Bethesda), 5, 1223–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljosa V. et al. (2013) Comparison of methods for image-based profiling of cellular morphological responses to small-molecule treatment. J. Biomol. Screen, 18, 1321–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E. (2012) Cell segmentation: 50 Years down the road [life sciences]. IEEE Signal Process Mag., 29, 140–145. [Google Scholar]
- Riffle M., Davis T.N. (2010) The yeast resource center public image repository: a large database of fluorescence microscopy images. BMC Bioinformatics, 11, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versari C. et al. (2017) Long-term tracking of budding yeast cells in brightfield microscopy: cellStar and the evaluation platform. J. R. Soc. Interface, 14, 20160705.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. (2018) Segmentation of yeast cell’s bright-field image with an edge-tracing algorithm. J. Biomed. Opt., 23, 1. [DOI] [PubMed] [Google Scholar]
- Weill U. et al. (2018) Genome-wide SWAp-Tag yeast libraries for proteome exploration. Nat. Methods, 15, 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.