Abstract
Plants monitor changes in day length to coordinate their flowering time with appropriate seasons. In Arabidopsis, the diel and seasonal regulation of CONSTANS (CO) protein stability is crucial for the induction of FLOWERING LOCUS T (FT) gene in long days. FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1) and ZEITLUPE (ZTL) proteins control the shape of CO expression profile antagonistically, although regulation mechanisms remain unknown. In this study, we show that GIGANTEA (GI) protein modulates the stability and nuclear function of FKF1, which is closely related to the stabilization of CO in the afternoon of long days. The abundance of FKF1 protein is decreased by the gi mutation, but increased by GI overexpression throughout the day. Unlike the previous report, the translocation of FKF1 to the nucleus was not prevented by ZTL overexpression. In addition, the FKF1-ZTL complex formation is higher in the nucleus than in the cytosol. GI interacts with ZTL in the nucleus, implicating the attenuation of ZTL activity by the GI binding and, in turn, the sequestration of FKF1 from ZTL in the nucleus. We also found that the CO-ZTL complex presents in the nucleus, and CO protein abundance is largely reduced in the afternoon by ZTL overexpression, indicating that ZTL promotes CO degradation by capturing FKF1 in the nucleus under these conditions. Collectively, our findings suggest that GI plays a pivotal role in CO stability for the precise control of flowering by coordinating balanced functional properties of FKF1 and ZTL.
Keywords: CONSTANS, FKF1, flowering, GIGANTEA, ZEITLUPE
INTRODUCTION
Many plants transit from the vegetative to reproductive phase in the most favorable season to maximize reproductive fitness (Song et al., 2015). The LOV domain blue light photoreceptors, FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1) and ZEITLUPE (ZTL), play important roles in seasonal flowering of Arabidopsis, a facultative long-day plant (Imaizumi et al., 2005; Kim et al., 2005; 2013; Song et al., 2012; 2014; Takase et al., 2011). FKF1 acts as the photoperiod sensor that positively controls the robust FLOWERING LOCUS T (FT) mRNA expression in a day length-dependent manner by forming a multiple feed-forward motif (Song et al., 2012). FT protein is the mobile floral signal, which is synthesized in leaves and translocated to the shoot apical meristem, resulting in acceleration of flowering (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007). In contrast to FKF1, ZTL negatively regulates FT expression (Kim et al., 2005; 2013; Takase et al., 2011).
CONSTANS (CO) gene encodes a transcription factor that directly binds to the promoter region of FT gene and activates its transcription (Hayama et al., 2017; Song et al., 2012; Tiwari et al., 2010). The abundance of CO mRNA shows a diel expression pattern in long days (Suárez-López et al., 2001). The CYCLING DOF FACTORs (CDFs) repress the expression of CO gene in the morning, and this repression is relieved by the redundant function of FKF1 and ZTL proteins in the afternoon, allowing CO gene to be expressed (Fornara et al., 2009; Goralogia et al., 2017; Imaizumi et al., 2005; Sawa et al., 2007). FKF1 and ZTL proteins possess an E3 ubiquitin ligase activity that mediates the degradation of CDF repressors (Fornara et al., 2009; Han et al., 2004; Imaizumi et al., 2005; Ito et al., 2012). The activities of FKF1 and ZTL largely depend on the function of GIGANTEA (GI) protein (Kim et al., 2007; Sawa et al., 2007). Both FKF1 and ZTL interact with GI through their LOV and F-box domains, and these interactions are blue light-dependent (Kim et al., 2007; 2013; Krahmer et al., 2019; Pudasaini et al., 2017; Sawa et al., 2007). The FKF1-GI complex presents in the cytosol and the nucleus, while the ZTL-GI interaction is proposed to occur in the cytosol exclusively (Kim et al., 2007; 2013). The GI binding facilitates FKF1 function to relieve the transcriptional repression of CO whereas inhibits ZTL-mediated degradation of circadian clock components (Kim et al., 2007; Sawa et al., 2007). Additionally, GI regulates ZTL stability positively and reciprocally (Kim et al., 2007; 2013).
In contrast to the functional redundancy in the transcriptional regulation of CO, FKF1 and ZTL proteins depict an antagonistic role in the posttranslational regulation of CO (Song et al., 2014). FKF1 interacts with CO by using the LOV domain that absorbs blue light (Imaizumi et al., 2003; Ito et al., 2012; Song et al., 2012). This interaction is enhanced by blue light and facilitates the stabilization of CO protein in the afternoon, which is closely related to the induction of FT expression under these conditions (Song et al., 2012). In contrast, ZTL function mediates the degradation of CO earlier in the day by binding to it (Song et al., 2014). Like FKF1, GI forms a coherent feedforward loop to control the induction of FT expression (Alon, 2007; Sawa and Kay, 2011; Sawa et al., 2007). However, unlike FKF1, GI negatively influences CO stabilization (Song et al., 2014). The diel profile of CO protein abundance in the gi mutant resembles to that in the ztl mutant (Song et al., 2014). Increased CO abundance in the gi mutant is offset by the fkf1 mutation, suggesting the complicated inter-relationships among FKF1, GI, and ZTL (Song et al., 2014).
Although the roles of FKF1, GI, and ZTL in the photoperiodic flowering are known (Imaizumi et al., 2003; 2005; Kim et al., 2005; Lee et al., 2018; Sawa et al., 2007; Song et al., 2012; 2014; Takase et al., 2011), their relationships between functionalities and biochemical properties associated with the regulation of CO stability still remain underexplored. Given that the protein expression of FKF1, GI, and ZTL coincides in the afternoon (Kim et al., 2007; Sawa et al., 2007), the crucial timing for CO stabilization under the same conditions (Song et al., 2012; 2014), it is critical to understand how the activity of these positive and negative regulators of CO stability is modulated. Here, we demonstrate that GI influences the stability of FKF1 in the cytosol as well as in the nucleus in Arabidopsis. The translocation of FKF1 to the nucleus was not inhibited by ZTL overexpression, and, inconsistent with the previous report (Takase et al., 2011), the tight binding of FKF1 to ZTL was observed in the nucleus. In addition, nuclear GI forms a protein complex with ZTL in Arabidopsis. Our results indicate that GI function is crucial for the balanced operation of ZTL family members involved in shaping daily CO stability.
MATERIALS AND METHODS
Plant materials and growth conditions
All Arabidopsis thaliana plants, wild type, 35S:HA-FKF1 / fkf1 #10 (Sawa et al., 2007), 35S:HA-FKF / fkf1 gi-2 #10 (Sawa et al., 2007), 35S:HA-FKF1 35S:GI-TAP / fkf1 #18 (Sawa et al., 2007), pFKF1:HA-FKF1 / fkf1 #24 (Sawa et al., 2007), pGI:GI-TAP / gi-2 #30 (Sawa et al., 2007), pFKF1:HA-FKF1 35S:Myc-ZTL / fkf1, pZTL:HA-ZTL pGI:GI-TAP / gi-2, pZTL:HA-ZTL pGI:GI-TAP / fkf1 gi-2, pZTL:3FLAG-ZTL pFKF1:HA-FKF1 / fkf1, pZTL:3FLAG-ZTL pFKF1:HA-FKF1 pGI:GI-TAP / fkf1 gi-2, 35S:3HA-CO #7, #10, #22 (Song et al., 2012), 35S:3HA-CO 35S:MYC-ZTL, 35S:3HA-CO / ztl-4 #14 (Song et al., 2014), and 35S:3HA-CO / ztl-4 fkf1-2 lkp2-1, used in this paper are Columbia (Col-0) ecotype.
To generate the pZTL:HA-ZTL pGI:GI-TAP / gi-2 and pZTL:HA-ZTL pGI:GI-TAP / fkf1 gi-2 lines, the 1.5 kb segment of the ZTL promoter region was cloned into pENTR 5′-TOPO (Invitrogen, USA) and sequenced. HA-ZTL full-length cDNA was amplified using the ZTL forward primer that contains the nucleotide sequences encoding a HA epitope tag (5′-CACCATGTACCCATACGATGTTCCTGACTATGCGGCCATGGAGTGGGACAGT GGTTCC-3′, the sequences encoding the HA epitope tag are underlined) and ZTL reverse primer that contains the BamHI restriction enzyme site (5′-GGATCCCTAATGAGGAAGAAAGAAGAAGAAGGAC-3′, ZTL specific sequences are underlined). The amplified HA-ZTL cDNA was cloned into the pENTR/D-TOPO (Invitrogen) vector and sequenced. Both the HA-ZTL cDNA and ZTL promoter were transferred into the R4pGWB501 vector (Nakagawa et al., 2008) using multi-Gateway reactions. The R4pGWB501 plasmid-carrying the pZTL:HA-ZTL construct was introduced into the pGI:GI-TAP / gi-2 #30 and pGI:GI-TAP / fkf1 gi-2 plants (Sawa et al., 2007) by conventional Agrobacterium-mediated transformation method. For the pZTL:3FLAG-ZTL pFKF1:HA-FKF1 / fkf1 and pZTL:3FLAG-ZTL pFKF1:HA-FKF1 pGI:GI-TAP / fkf1 gi-2 plants, we first introduced the nucleotide sequences encoding 3xFLAG epitope tags into the pENTR/D-TOPO vector to generate the pENTR-3xFLAG vector. ZTL full-length cDNA was amplified using a primer set (KpnI-ZTL forward, 5′-GGTACCAAATGGAGTGGGACAGTGGTTC-3′ [ZTL specific sequences are underlined] and BamHI-ZTL reverse described above). The amplified ZTL cDNA was digested by KpnI and BamHI restriction enzymes and then inserted to the KpnI-BamHI sites of the pENTR-3xFLAG vector to make an in-frame fusion with 3xFLAG. After confirming sequences, together with the ZTL promoter, the ZTL cDNA was transferred to the R4pGWB501 binary vector. The R4pGWB501 plasmid-harboring the pZTL:3FLAG-ZTL construct was transformed in the pFKF1:HA-FKF1 / fkf1 #24 and pFKF1:HA-FKF1 pGI:GI-TAP / fkf1 gi-2 plants (Sawa et al., 2007). To produce the 35S:3HA-CO 35S:Myc-ZTL and 35S:3HA-CO / ztl-4 fkf1-2 lkp2-1 lines, the pH7WG2 vector harboring the 3HA-CO overexpression cassette described previously (Song et al., 2012) was transformed into the 35S:Myc-ZTL (Song et al., 2014) and the ztl-4 fkf1-2 lkp2-1 mutant plants (Baudry et al., 2010).
All plants including Nicotiana benthamiana were grown on soil or 0.5× Linsmaier and Skoog (LS) media (Caisson, USA) containing 1% sucrose or 0.5× Murashige and Skoog (MS) medium (Duchefa Biochemie, The Netherlands) containing 1% sucrose for gene and protein analysis and coimmunoprecipitation assays in plant incubators at 22°C under full-spectrum white fluorescent light (F017/950/24”, Octron Osram Sylvania) or full-spectrum white LED light (4,470 K) with a fluence rate of 80–90 μmol m−2 s−1 in long days (16-h light/8-h dark) and 75–115 μmol m−2 s−1 in short days (8-h light/16-h dark).
RNA isolation and gene expression analysis
For gene expression analysis, seedlings were grown on LS agar plates in long days and collected at every 4 h during the daytime from ZT0 on day 10. Ground plant tissues were used for RNA extraction using Higene Total RNA Prep Kit (BioFact, Korea), and 2 μg of total RNA was reverse-transcribed using DiaStar RT kit (SolGent, Korea) to synthesize cDNA. Methods for quantitative real-time polymerase chain reaction (qRT-PCR) and primer information for gene expression analysis of IPP2, CO, and FT were described previously (Song et al., 2012).
Protein preparation, immunoblot analysis, and protein quantification
To analyze protein expression of CO, FKF1, GI, and ZTL, 10-day-old plants, except the 35S:3HA-CO 35S:Myc-ZTL line, grown on LS or MS agar plates under long days were harvested at each time point indicated. The 35S:3HA-CO 35S:Myc-ZTL seedlings were grown in short days and harvested at ZT0, 0.5, 4, 8, 12, 16, and 20 on day 14. Protocols for extracting cytosolic proteins, isolation of nuclei, and immunoblot experiments were previously described (Song et al., 2014). Protein amounts were imaged and quantified with ChemiDoc Touch Imaging System and Image Lab software (Bio-Rad, USA).
Coimmunoprecipitation experiments
Procedures for separation of the cytosolic and nuclei-enriched fractions and coimmunoprecipitation assays were previously described in Song et al. (2014). We performed coimmunoprecipitation experiments with slight modifications. Briefly, Arabidopsis and tobacco tissues were extracted in coimmunoprecipitation buffer (50 mM Na-phosphate [pH 7.4], 100 mM NaCl, 10% [vol/vol] glycerol, 5 mM EDTA, 1 mM DTT, 50 μM MG-132, 1 mM NaF, 2 mM NaVO3 and protease inhibitor tablet [Roche, Switzerland]). After separating by centrifugation, the supernatant was kept at 4°C to use as the cytosolic fraction. Then the pellet was resuspended in the same coimmunoprecipitation buffer and sonicated five times for 15 s with pulse-on 3 s and pulse-off 2 s on ice. After centrifuging for 5 min, the supernatant was used as the nuclei-enriched fraction.
RESULTS
GI is involved in the stabilization of FKF1
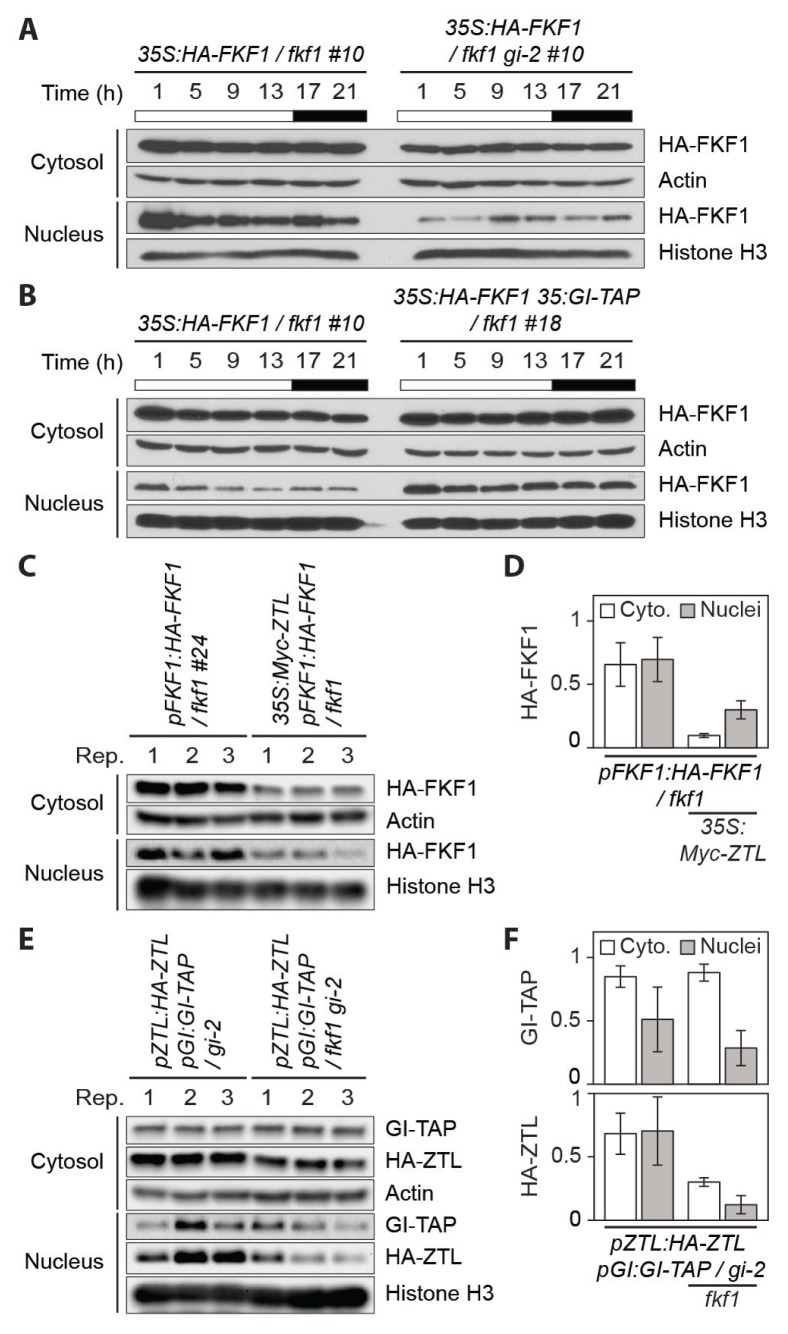
The binding of ZTL to GI contributes to increased cytosolic retention of GI in Arabidopsis, thereby preventing the nuclear roles of GI (Kim et al., 2013). The reciprocal regulation of GI and ZTL stability is critical to their roles in flowering and robust circadian oscillations (Kim et al., 2007; 2013). Moreover, it is proposed that ZTL recruits FKF1 to the cytoplasmic speckles through a physical interaction, which, in turn, inhibits FKF1-mediated removal of floral repressors in the nucleus (Takase et al., 2011). This indicates that balanced nucleocytosolic partitioning of FKF1, GI, and ZTL proteins controlled by inter-relationships among them is essential for the proper timing of CO stabilization (Song et al., 2014). We investigated the effect of GI on FKF1 stability changes to elucidate the regulatory mechanisms that determine balanced spatial activities of the proteins. FKF1 overexpression lines in wild type, the gi mutant, and the 35S:GI-TAP overexpression plants, in which tandem affinity purification (TAP)-tagged GI was expressed by the Cauliflower Mosaic Virus 35S promoter, were used. The levels of FKF1 protein were reduced in the gi mutant throughout the day in long days, and this reduction is more obvious in the nucleus (Fig. 1A). On the contrary, the FKF1 abundance was greatly increased in the nuclear fraction of Arabidopsis transgenic plants overexpressing functional hemagglutinin (HA)-tagged FKF1 and GI-TAP in the fkf1 deletion mutant (Fig. 1B), indicating that GI controls FKF1 stability. We next generated the pFKF1:HA-FKF1 35S:Myc-ZTL / fkf1 plants that the expression of HA-FKF1 and Myc-ZTL was controlled by the FKF1 promoter and the 35S promoter, respectively, to test how ZTL influences the nucleocytoplasmic partitioning of FKF1. In contrast to the previous report (Takase et al., 2011), the translocation of FKF1 to the nucleus was not hindered by ZTL overexpression, at least under our experimental conditions (Figs. 1C and 1D). We also explored whether FKF1 is involved in the stability and spatial partitioning of GI and ZTL. To study this, the pZTL:HA-ZTL pGI:GI-TAP / gi-2 and pZTL:HA-ZTL pGI:GI-TAP / fkf1 gi-2 transgenic plants were generated. The fkf1 mutation seems to affect the stability and localization of GI and ZTL proteins but not strongly (Figs. 1E and 1F), indicating that FKF1 activity may not be a limiting factor for functions of GI and ZTL.
Fig. 1. GI regulates FKF1 protein abundance in long days.
(A and B) Immunoblot assays for daily expression profiles of FKF1 protein in the cytosol and the nucleus. The 35S:HA-FKF1 #10, 35S:HA-FKF1 / fkf1 gi-2 #10, and 35S:HA-FKF1 35S:GI-TAP / fkf1 #18 plants were grown for 10 days in long days. Effects of the gi mutation (A) and the GI overexpression (B) on FKF1 stability and translocation. Similar results were observed from two biological replicates. White and black bars represent the light and dark conditions, respectively. (C) FKF1 proteins in the cytosolic and nuclei fractions of the pFKF1:HA-FKF1 / fkf1 #24 and pFKF1:HA-FKF1 35S:Myc-ZTL / fkf1 plants grown for 10 days in long days and collected in the afternoon (ZT13). Rep., replication. (D) Quantification for the amounts of HA-FKF1 protein in the cytosol (abbreviated as Cyto.) and the nucleus with or without Myc-ZTL overproduction. (E) The effect of the fkf1 mutation on changes in the stability of GI and ZTL proteins between the cytosol and the nucleus. The pZTL:HA-ZTL pGI:GI-TAP / gi-2 and pZTL:HA-ZTL pGI:GI-TAP / fkf1 gi-2 plants grown in long days were harvested at ZT13 on day 10. (F) Quantification for the amounts of GI-TAP and HA-ZTL in the cytosol and the nucleus with or without the fkf1 mutation. Numbers in (C) and (E) represent each protein extract from three biological replicates. (D) and (F) Quantification data was calculated from the results of (C) and (E), respectively. Actin and Histone H3 antibodies were used for loading controls.
Interactions among FKF1, GI, and ZTL occur in the nucleus
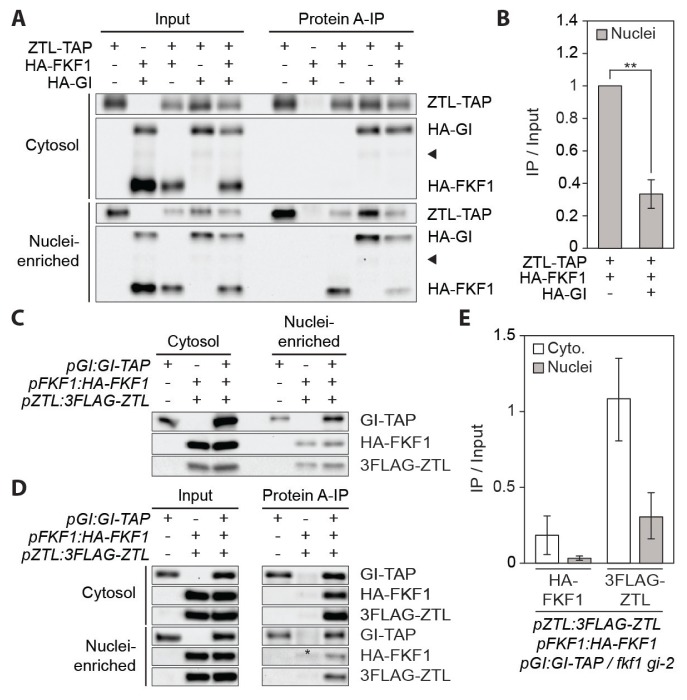
As the FKF1 translocation is not inhibited by ZTL function (Figs. 1C and 1D), we attempted to validate the interactions among FKF1, GI, and ZTL in planta using a N. benthamiana transient expression system. HA-FKF1 was coimmunoprecipitated with ZTL-TAP from the cytosolic and nuclei-enriched fractions. Interestingly, coimmunoprecipitated HA-FKF1 protein was mainly observed in the nuclei-enriched fraction (Fig. 2A), indicating that the FKF1-ZTL complex formation preferentially occurs in the nucleus. The GI-ZTL interaction existed in both the cytosol and the nucleus (Fig. 2A). The nuclear specific interaction between FKF1 and ZTL was weakened by the presence of GI protein (Figs. 2A and 2B), indicating that GI facilitates the release of FKF1 from ZTL in the nucleus. Since we only observed the ZTL-FKF1 interaction in the nuclei-enriched fraction, this result also indicates that a small portion of ZTL, which interacts with FKF1, exists in the nucleus. This ZTL nuclear localization could be controlled by FKF1, which possesses the nuclear localization sequence (NLS) (Figs. 1E and 1F). Together, these data suggest that GI may be the key player modulating the functions of FKF1 and ZTL in photoperiodic flowering.
Fig. 2. GI inhibits the FKF1-ZTL interaction in the nucleus.
(A) In planta interactions among FKF1, GI, and ZTL proteins in the cytosolic and nuclei-enriched fractions. Proteins were transiently expressed in N. benthamiana for 3 days. FKF1 and GI proteins in the ZTL immune complex were probed with anti-HA and anti-protein A antibodies, respectively. Arrow heads indicate non-specific bands. (B) Relative amounts of the FKF1-ZTL complex with or without GI as shown in (A) were quantified. Bar graphs represent amounts of coimmunoprecipitated HA-FKF1, calculated by (HA-FKF1IP/ZTL-TAPIP)/(FKF1Input/ZTL-TAPInput). **P < 0.01 (one-tailed t-test). (C–E) The GI-FKF1 and GI-ZTL interactions in Arabidopsis. Ten-day-old pGI:GI-TAP / gi-2 #30, pZTL:3FLAG-ZTL pFKF1:HA-FKF1 / fkf1, and pZTL:3FLAG-ZTL pFKF1:HA-FKF1 pGI:GI-TAP / fkf1 gi-2 plants grown in long days were harvested at ZT13. Proteins were extracted and fractionated for coimmunoprecipitation. (C) Input proteins were loaded to compare relative protein levels between the cytosolic and nuclei-enriched fractions. (D) HA-FKF1 and 3FLAG-ZTL proteins in the GI-TAP immune complex. Long exposed immunoblot images for the nuclei-enriched fractions were used to visualize similar amounts of input proteins for comparing relative amounts of protein complexes between the cytosol and the nucleus. An asterisk denotes a non-specific band. (E) Quantification of the interactions of GI with FKF1 and ZTL in the cytosolic and nuclei-enriched fractions. Means ± SEM were calculated from three biological replicates.
FKF1 and ZTL bind to GI at similar times in the day (Kim et al., 2007; Sawa et al., 2007). As the GI binding is critical to the functional activities of FKF1 and ZTL, we further analyzed the spatial distribution and relative abundance of protein complexes in transgenic Arabidopsis plants that HA-FKF1, GI-TAP, and 3xFLAG-tagged ZTL (3FLAG-ZTL) were expressed by the FKF1, GI, and ZTL promoters, respectively. The pGI:GI-TAP / gi-2 #30, pZTL:3FLAG-ZTL pFKF1:HA-FKF1 / fkf1, and pZTL:3FLAG-ZTL pFKF1:HA-FKF1 pGI:GI-TAP / fkf1 gi-2 plants were grown in long days and harvested at zeitgeber time ZT13 (i.e., 13 hours after light onset on a given day), when all three proteins are highly expressed. The levels of all three proteins in the cytosol were higher than those in the nucleus (Fig. 2C). HA-FKF1 and 3LAG-ZTL proteins were coimmunoprecipitated with GI-TAP protein in the cytosol as well as the nucleus, although the cytosolic interactions were much higher than the nuclear interactions (Figs. 2D and 2E). In addition, the amount of coimmunoprecipitated 3FLAG-ZTL protein was greater than HA-FKF1 protein in both the cytosol and the nucleus (Figs. 2D and 2E), indicating that GI favorably interacts with ZTL in Arabidopsis. These results could account for the reduction in the nuclear interaction between FKF1 and ZTL in the presence of GI (Fig. 2A).
Day length-dependent regulation of CO protein by FKF1, GI, and ZTL
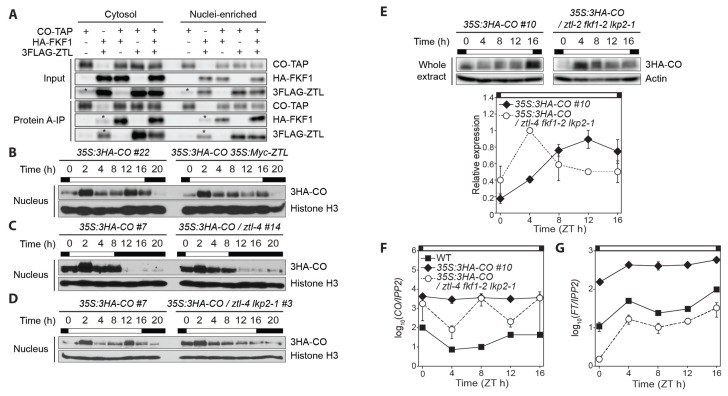
CO is the main transcriptional activator of FT gene (Samach et al., 2000). During long days, CO protein expression fluctuates throughout the day (Song et al., 2012; 2018; Valverde et al., 2004). Since FKF1 and ZTL directly bind to CO and modulate its daily abundance in the nucleus (Song et al., 2012; 2014), we therefore examined their relationship to the CO binding. CO-TAP, HA-FKF1, and 3FLAG-ZTL were coexpressed as appropriate combinations in N. benthamiana leaves. Both HA-FKF1 and 3FLAG-ZTL proteins interacted with CO in the nuclei-enriched fraction (Fig. 3A). In addition, the CO-ZTL complex formation was not affected in the nucleus when FKF1 was expressed together, and vice versa. The amount of CO-ZTL complex was reduced in the cytosol but not in the nucleus by the presence of FKF1 (Fig. 3A), indicating that FKF1 competes with ZTL for binding to CO in the cytosol, although the biological relevance needs to be addressed.
Fig. 3. GI and the ZTL group members regulate the daytime CO protein stability.
(A) Interactions among CO, FKF1, and ZTL in the cytosolic and nuclei-enriched fractions. N. benthamiana leaves constantly expressing CO-TAP, HA-FKF1, and 3FLAG-ZTL proteins were collected 3 days after infiltration. Asterisks denote non-specific bands. (B–E) Immunoblot assays for diel expression profiles of 3HA-CO protein in Arabidopsis transgenic plants. Similar expression patterns were observed in two biological replicates except (E). Nuclear 3HA-CO protein signals were detected by anti-HA antibody, and histone H3 protein was used for an internal control in the nucleus. (B) Long day-grown 35S:3HA-CO #22 and 35S:3HA-CO 35S:Myc-ZTL plants were harvested on day 10 for nuclei isolation. (C) The 35S:3HA-CO #7 and 35:3HA-CO / ztl-4 #14 transgenic plants were grown in short days for 14 days. (D) The 35S:3HA-CO #7 and 35S:3HA-CO / ztl-4 lkp2-1 #3 plants were collected at the time points indicated in long days. (E) The daytime expression of 3HA-CO protein in long days was compared between two CO overexpression lines in wild type and the ztl-4 fkf1-2 lkp2-1 mutant background. Immunoblot images (the upper panel) and quantified 3HA-CO protein abundance (the lower panel) in whole protein extract are shown. Actin protein was used as a loading control for normalization. A direct comparison between two CO transgenic plants is not valid. Means ± SEM were calculated from three independent replicates. (F and G) CO (F) and FT (G) mRNA levels in 10-day-old wild type, 35S:3HA-CO #10, and 35S:3HA-CO / ztl-4 #14 seedlings grown in long days. The mRNA levels were measured by quantitative real-time polymerase chain reaction and normalized against IPP2 gene. Daytime gene expression profiles are represented in a logarithmic scale. Means of three biological trials ± SEM.
FKF1 stabilizes CO whereas ZTL destabilizes it (Song et al., 2012; 2014). In long days, the protein abundance of FKF1 and ZTL peaks in the afternoon, when the CO protein level is also high. Because the FKF1-ZTL heterodimerization mainly exists in the nucleus (Fig. 2A), we hypothesized that ZTL restricts the FKF1-dependent CO stabilization in the afternoon by capturing FKF1 in the nucleus. In such a case, a reduction in CO protein levels is expected owing to the ZTL overproduction during the afternoon in long days. To validate this hypothesis, we constitutively expressed 3HA-CO cDNA in wild type and the 35S:Myc-ZTL transgenic line. As expected, the levels of nuclear CO in the 35S:3HA-CO 35S:Myc-ZTL plants were decreased dramatically and specifically in the afternoon of long days compared to those in the 35S:3HA-CO #22 plants (Fig. 3B), resembling CO protein profile in the fkf1 mutant (Song et al., 2012). We next investigated if the effect of ZTL on the stability changes in CO is long day-specific. In short days, CO protein abundance in the ztl mutant was similar to that in the wild type (Fig. 3C), indicating that ZTL regulates CO stability in a day length-dependent manner, like FKF1 (Song et al., 2012).
All the three ZTL family members including FKF1 and LOV KELCH PROTEIN 2 (LKP2) interact with CO in yeast and in planta (Fukamatsu et al., 2005; Song et al., 2012; 2014). As LKP2 synergistically delays flowering with ZTL and is proposed to inhibit FKF1 function by the direct interaction (Takase et al., 2011), we further examined the changes in CO stability using CO overexpression line in the ztl-4 lkp2-1 double mutant background. The daily profile of CO protein in the ztl-4 lkp2-1 mutant grown in long days was similar to that in the ztl mutant, suggesting a marginal function of LKP2 on CO stability (Fig. 3D). The ZTL/LKP2/FKF1 proteins bind to GI (Kim et al., 2007; Krahmer et al., 2019; Sawa et al., 2007; Song et al., 2014). Moreover, FKF1 promotes the timing of flowering whereas LKP2 and ZTL delay it (Kim et al., 2005; Imaizumi et al., 2003; Takase et al., 2011). We therefore investigated a combined effect of these positive and negative regulators on CO stabilization and the induction of FT expression, which determines flowering in long days. 3HA-CO protein was constitutively expressed in the ztl-4 fkf1-2 lkp2-1 triple mutant, and the amount of CO protein in the mutant was increased in the morning (Fig. 3E), resembling the gi or the ztl mutants in which the amount of GI protein is low (Kim et al., 2007; Song et al., 2014). In addition, the levels of FT mRNA in the 35S:3HA-CO / ztl-4 fkf1-2 lkp2-1 line were lower than those in wild type plants, although the amount of CO mRNA was higher than the plants (Figs. 3F and 3G). These data suggest that not only the modulation of the FKF1-ZTL dimerization by GI but also GI-independent pathways control the timing stabilization of CO and FT expression (Jung et al., 2007; Lee et al., 2017; Sawa and Kay, 2011).
DISCUSSION
In order to understand day length-dependent flowering regulation, we aimed to determine functional relationships among the ZTL/LKP2/FKF1 and GI proteins that positively and negatively contribute to CO stability, which is closely related to the induction of florigen gene, FT (Song et al., 2012; 2014; Valverde et al., 2004). We have previously shown that the gi mutation causes increased and decreased CO protein abundance in the morning and the afternoon in long days, respectively (Song et al., 2014). This complicated phenotype was similar to the ztl mutant (Song et al., 2014). Given that GI promotes flowering but ZTL inhibits it (Jung et al., 2007; Sawa et al., 2007; Takase et al., 2011), it would be worth investigating molecular mechanisms that finely adjust stoichiometric relationships among the ZTL family proteins and GI protein. Our current results suggest that GI interrupts the heterodimerization between FKF1 and ZTL proteins in the nucleus by simultaneously interacting with the proteins (Figs. 2A and 2B). The GI binding seems to be the process activating FKF1 and inactivating ZTL functions (Kim et al., 2007; Sawa et al., 2007; Song et al., 2014). Previously, ZTL has been proposed to capture FKF1 and GI in the cytosol and prevent their translocation to the nucleus, which is important for the expression of CO and FT genes (Kim et al., 2013; Takase et al., 2011). However, the FKF1-ZTL interaction mainly occurs in the nucleus of tobacco leaf tissues, as observed under our conditions (Figs. 2A and 2B). In addition, the intracellular localization of FKF1 was not prevented by ZTL overexpression in the Arabidopsis transgenic line (Figs. 1C and 1D). The discrepancy related to the intracellular distribution of FKF1 may be due to the use of Arabidopsis whole plants versus Arabidopsis protoplasts isolated from suspension-cultured cells. As FKF1 and ZTL proteins are blue light photoreceptors, perception of light is crucial for their functions in many physiological events including daylength-dependent flowering (Ito et al., 2012). Living plants use light as sources of blue wavelengths and photoperiods. In contrast, protoplast-based transient expression assays are often performed in darkness, which is the same experimental condition as previously employed by Takase et al. (2011).
Light activating FKF1 facilitates CO stabilization in the afternoon by the physical interaction with CO and by the formation of a protein complex with CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), resulting in the inhibition of COP1 dimerization that targets CO for proteasome-dependent degradation (Lee et al., 2017; Song et al., 2012), albeit the involvement of GI in these FKF1 roles is still unknown. In contrast, ZTL binds to CO and mediates its degradation (Fig. 3B) (Song et al., 2014). The mode of action for the morning stabilization of CO in the ztl mutant needs to be explored in future (Song et al., 2014). As mentioned earlier, CO abundance in the ztl mutant is low in the afternoon. This might be explained by the large reduction of GI in the mutant at the same time, leading to decreases in FKF1 stability and activity (Fig. 1A) (Sawa et al., 2007). Therefore, the attenuation of functional FKF1 in the ztl mutant, and the gi mutant, may decrease the amount of protein complexes with CO and COP1, which are important for CO stabilization in the afternoon of long days (Lee et al., 2017; Song et al., 2012).
ZTL overexpression increases the cytosolic retention of GI (Kim et al., 2013). Our results show that GI preferentially binds to ZTL as compared to FKF1, and a substantial amount of GI-ZTL complex exists in the nucleus, although the majority of the complex is present in the cytosol (Figs. 2D and 2E). The overexpression of ZTL likely leads to a change in the ratio of GI-ZTL complex between the cytosol and the nucleus. With the depletion of functional FKF1 in the gi mutant, the similarity in CO daily profile between the ZTL overexpression line and the fkf1 mutant, which CO abundance is dramatically reduced in the afternoon of long days, strongly supports the notion that the binding of GI to ZTL in the nucleus plays the key role for the timing stabilization of CO by sequestering FKF1 from ZTL and by activating FKF1 in a day length-dependent manner (Figs. 1A and 3B) (Sawa et al., 2007; Song et al., 2012; 2014).
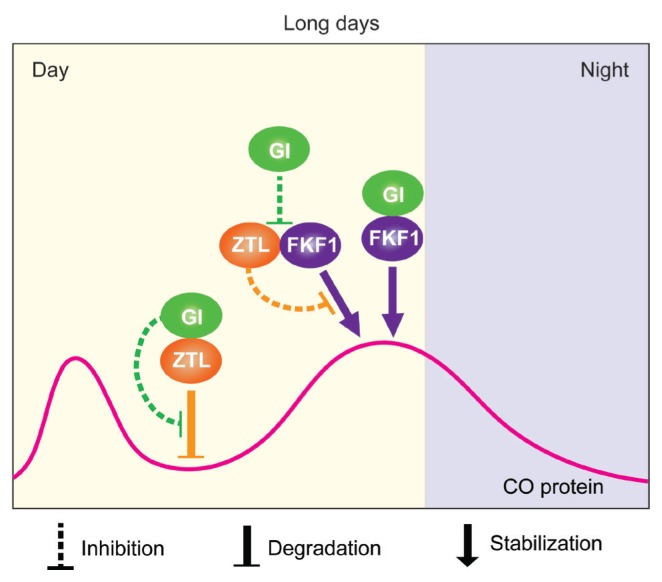
Despite the distinct roles of the ZTL/LKP2/FKF1 family members and GI in the regulation of seasonal flowering and circadian clock, their diverged functions lead to complicated redundancy and antagonistic relationships (Kim et al., 2007; Lee et al., 2018; Sawa et al., 2007; Song et al., 2012; 2014; Takase et al., 2011). Here we report the evidence that GI conveys timing information for CO stabilization in the afternoon of long days by altering the ZTL-FKF1 complex in the nucleus (Fig. 4). This new regulatory mechanism provides an important clue to understand the functional complexity. Together with possible implications of GI function for the regulation of developmental age- and temperature-dependent flowering (Balasubramanian et al., 2006; Jung et al., 2007; Mishra and Panigrahi, 2015; Sawa and Kay, 2011; Song et al., 2013), the roles of GI in photoperiodic flowering comprise sophisticated mechanisms that enable plants to represent adaptive plastic adjustment for reproductive success.
Fig. 4. A model for the regulation of CO protein expression by relationships among FKF1, GI, and ZTL in long days.
The temporal expression profile of CO in the nucleus under long-day conditions, which shows bimodal peaks in the early morning and late afternoon. ZTL mediates the degradation of CO in the morning by directly binding to it. Once FKF1 and GI proteins are expressed in the afternoon, the proteins form an active protein complex. GI preferentially interacts with ZTL and inactivates its function, leading to sequestration of CO from ZTL. The function of FKF1 stabilizes CO in this condition through forming a protein complex with it. In addition, ZTL can interact with FKF1 and inhibit the FKF1-mediated CO stabilization, resulting in destabilization of CO. The preferential binding of GI to ZTL also interferes the complex formation between FKF1 and ZTL. All together, these multilayered and sophisticated regulatory mechanisms allow CO protein to be highly accumulated at the late afternoon of long days.
ACKNOWLEDGMENTS
This project was supported by the Next-Generation BioGreen 21 Program (SSAC, project No. PJ013386 to T.I. and Y.H.S., Rural Development Administration, Republic of Korea), the Ajou University Research Fund (to S.P. and Y.H.S.), NIH grant (GM079712 to T.I.), NSF grants (IOS-1656076 to T.I.), NRF grant (NRF-2018R1D1A1A09083990 to Y.H.S.), and the Nuclear R&D Program of the Ministry of Science and ICT (MSIT, Republic of Korea) (to S.L., S.S.L., and Y.H.S.).
Footnotes
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S., Sureshkumar S., Lempe J., Weigel D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2006;2:e106. doi: 10.1371/journal.pgen.0020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A., Ito S., Song Y.H., Strait A.A., Kiba T., Lu S., Henriques R., Pruneda-Paz J.L., Chua N.H., Tobin E.M., et al. F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell. 2010;22:606–622. doi: 10.1105/tpc.109.072843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Fornara F., Panigrahi K.C., Gissot L., Sauerbrunn N., Rühl M., Jarillo J.A., Coupland G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009;17:75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Fukamatsu Y., Mitsui S., Yasuhara M., Tokioka Y., Ihara N., Fujita S., Kiyosue T. Identification of LOV KELCH PROTEIN2 (LKP2)-interacting factors that can recruit LKP2 to nuclear bodies. Plant Cell Physiol. 2005;46:1340–1349. doi: 10.1093/pcp/pci144. [DOI] [PubMed] [Google Scholar]
- Goralogia G.S., Liu T.K., Zhao L., Panipinto P.M., Groover E.D., Bains Y.S., Imaizumi T. CYCLING DOF FACTOR 1 represses transcription through the TOPLESS co-repressor to control photoperiodic flowering in Arabidopsis. Plant J. 2017;92:244–262. doi: 10.1111/tpj.13649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Mason M., Risseeuw E.P., Crosby W.L., Somers D.E. Formation of an SCF(ZTL) complex is required for proper regulation of circadian timing. Plant J. 2004;40:291–301. doi: 10.1111/j.1365-313X.2004.02207.x. [DOI] [PubMed] [Google Scholar]
- Hayama R., Sarid-Krebs L., Richter R., Fernandez V., Jang S., Coupland G. PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length. EMBO J. 2017;36:904–918. doi: 10.15252/embj.201693907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T., Schultz T.F., Harmon F.G., Ho L.A., Kay S.A. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Tran H.G., Swartz T.E., Briggs W.R., Kay S.A. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature. 2003;426:302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- Ito S., Song Y.H., Imaizumi T. LOV domain-containing F-Box proteins: light-dependent protein degradation modules in Arabidopsis. Mol Plant. 2012;5:47–56. doi: 10.1093/mp/sss013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger K.E., Wigge P.A. FT protein acts as a long-range signal in Arabidopsis. Curr Biol. 2007;17:1050–1054. doi: 10.1016/j.cub.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Jung J.H., Seo Y.H., Seo P.J., Reyes J.L., Yun J., Chua N.H., Park C.M. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell. 2007;19:2736–2748. doi: 10.1105/tpc.107.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Geng R., Gallenstein R.A., Somers D.E. The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development. 2013;140:4060–4069. doi: 10.1242/dev.096651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.Y., Fujiwara S., Suh S.S., Kim J., Kim Y., Han L., David K., Putterill J., Nam H.G., Somers D.E. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449:356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- Kim W.Y., Hicks K.A., Somers D.E. Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiol. 2005;139:1557–1569. doi: 10.1104/pp.105.067173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer J., Goralogia G.S., Kubota A., Zardilis A., Johnson R.S., Song Y.H., MacCoss M.J., Le Bihan T., Halliday K.J., Imaizumi T., et al. Time-resolved interaction proteomics of the GIGANTEA protein under diurnal cycles in Arabidopsis. FEBS Lett. 2019;593:319–338. doi: 10.1002/1873-3468.13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.D., Kim M.R., Kang M.Y., Cha J.Y., Han S.H., Nawkar G.M., Sakuraba Y., Lee S.Y., Imaizumi T., McClung C.R., et al. The F-box protein FKF1 inhibits dimerization of COP1 in the control of photoperiodic flowering. Nat Commun. 2017;8:2259. doi: 10.1038/s41467-017-02476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.M., Feke A., Li M.W., Adamchek C., Webb K., Pruneda-Paz J., Bennett E.J., Kay S.A., Gendron J.M. Decoys untangle complicated redundancy and reveal targets of circadian clock F-Box proteins. Plant Physiol. 2018;177:1170–1186. doi: 10.1104/pp.18.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Küttner F., Schmid M. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol. 2007;17:1055–1060. doi: 10.1016/j.cub.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Mishra P., Panigrahi K.C. GIGANTEA: an emerging story. Front Plant Sci. 2015;6:8. doi: 10.3389/fpls.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Nakamura S., Tanaka K., Kawamukai M., Suzuki T., Nakamura K., Kimura T., Ishiguro S. Development of R4 gateway binary vectors (R4pGWB) enabling high-throughput promoter swapping for plant research. Biosci Biotechnol Biochem. 2008;72:624–629. doi: 10.1271/bbb.70678. [DOI] [PubMed] [Google Scholar]
- Pudasaini A., Shim J.S., Song Y.H., Shi H., Kiba T., Somers D.E., Imaizumi T., Zoltowski B.D. Kinetics of the LOV domain of ZEITLUPE determine its circadian function in Arabidopsis. Elife. 2017;6:e21646. doi: 10.7554/eLife.21646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288:1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- Sawa M., Kay S.A. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2011;108:11698–11703. doi: 10.1073/pnas.1106771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M., Nusinow D.A., Kay S.A., Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Estrada D.A., Johnson R.S., Kim S.K., Lee S.Y., MacCoss M.J., Imaizumi T. Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of Constans stability in Arabidopsis photoperiodic flowering. Proc Natl Acad Sci U S A. 2014;111:17672–17677. doi: 10.1073/pnas.1415375111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Ito S., Imaizumi T. Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 2013;18:575–583. doi: 10.1016/j.tplants.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Kubota A., Kwon M.S., Covington M.F., Lee N., Taagen E.R., Laboy Cintron D., Hwang D.Y., Akiyama R., Hodge S.K., et al. Molecular basis of flowering under natural long-day conditions in Arabidopsis. Nat Plants. 2018;4:824–835. doi: 10.1038/s41477-018-0253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Shim J.S., Kinmonth-Schultz H.A., Imaizumi T. Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol. 2015;66:441–464. doi: 10.1146/annurev-arplant-043014-115555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Smith R.W., To B.J., Millar A.J., Imaizumi T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science. 2012;336:1045–1049. doi: 10.1126/science.1219644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- Takase T., Nishiyama Y., Tanihigashi H., Ogura Y., Miyazaki Y., Yamada Y., Kiyosue T. LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under non-inductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1. Plant J. 2011;67:608–621. doi: 10.1111/j.1365-313X.2011.04618.x. [DOI] [PubMed] [Google Scholar]
- Tiwari S.B., Shen Y., Chang H.C., Hou Y., Harris A., Ma S.F., McPartland M., Hymus G.J., Adam L., Marion C., et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010;187:57–66. doi: 10.1111/j.1469-8137.2010.03251.x. [DOI] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]