Abstract
Sink strength optimizes sucrose import, which is fundamental to support developing seed grains and increase crop yields, including those of rice (Oryza sativa). In this regard, little is known about the function of vacuolar invertase (VIN) in controlling sink strength and thereby seed size. Here, in rice we analyzed mutants of two VINs, OsVIN1 and OsVIN2, to examine their role during seed development. In a phenotypic analysis of the T-DNA insertion mutants, only the OsVIN2 mutant osvin2-1 exhibited reduced seed size and grain weight. Scanning electron microscopy analysis revealed that the small seed grains of osvin2-1 can be attributed to a reduction in spikelet size. A significant decrease in VIN activity and hexose level in the osvin2-1 spikelets interfered with spikelet growth. In addition, significant reduction in starch and increase in sucrose, which are characteristic features of reduced turnover and flux of sucrose due to impaired sink strength, were evident in the pre-storage stage of osvin2-1 developing grains. In situ hybridization analysis found that expression of OsVIN2 was predominant in the endocarp of developing grains. A genetically complemented line with a native genomic clone of OsVIN2 rescued reduced VIN activity and seed size. Two additional mutants, osvin2-2 and osvin2-3 generated by the CRISPR/Cas9 method, exhibited phenotypes similar to those of osvin2-1 in spikelet and seed size, VIN activity, and sugar metabolites. These results clearly demonstrate an important role of OsVIN2 as sink strength modulator that is critical for the maintenance of sucrose flux into developing seed grains.
Keywords: rice, seed, sink strength, sucrose flux, vacuolar invertase
INTRODUCTION
Sucrose is a major transitory reserve form of photoassimilates, the product of photosynthetic carbon fixation, and the main carbohydrate mobilized to sink organs via phloem loading. Its proper production, consumption, and partitioning among cellular organelles and various tissues/organs are essential for feeding metabolic processes and supporting plant growth and development. It also functions in photosynthetic regulation (Cottage et al., 2010; Sheen, 1990), anthocyanin biosynthesis (Shin et al., 2013; Solfanelli et al., 2006) and carbon-nitrogen balancing (Cheng et al., 1992; Hanson et al., 2008) as a signaling molecule.
In particular, sugar partitioning between source and sink organs is fundamental to support developing seed grains and increase crop yields. Optimized source supply and partitioning of sugar to sink organs depend largely on sink strength, which is the ability of a sink to import photoassimilates required for growth, development, and maintenance (Chamont, 1993; Chang et al., 2017). That is, enhanced sink strength in seed grains improves sucrose import and results in increased crop yields. This is supported by research findings that optimization of sucrose flux via proper transport from source to sink is fundamental in controlling crop yield (Braun et al., 2014). For instance, mutation of the tonoplast-localized sucrose transporter (SUT) attenuates symplasmic phloem loading activity, accumulates sucrose in source leaves, and reduces yields in rice (Oryza sativa) and populus (Eom et al., 2011; Payyavula et al., 2011). In wheat, overexpression of a barley SUT by an endosperm-specific promoter increases sucrose import and storage protein synthesis, improving grain quality (Weichert et al., 2010). In rice, enhanced capacity of apoplastic phloem loading improves sucrose flux and sink strength, resulting in increased grain size and substantially increased grain yield (Wang et al., 2015).
Sink strength in plants can be regulated by sucrose cleavage enzymes, invertase (β-fructofuranosidase, EC 3.2.1.26) and sucrose synthase (SuSy, EC 2.4.1.13) that can control the amount of sucrose. SuSy degrades sucrose in the presence of uridine diphosphate (UDP) into UDP-glucose and fructose, and its primary function is the synthesis of sugar polymers such as cellulose (Coleman et al., 2009). Unlike SuSy, invertase is known to regulate plant growth and development as an irreversible hydrolyase of sucrose (Barratt et al., 2009; González et al., 2005). Invertase is classified by its localization, solubility, optimal pH, and isoelectric point (pI) in plants. Its isoforms—cell wall invertase (CIN), neutral invertase (NIN), and vacuolar invertase (VIN)—are localized in the cell wall, cytosol and vacuole, respectively. VIN and CIN cleave sucrose most efficiently in acidic conditions, but a neutral pH is optimal for sucrose hydrolysis with NIN. VIN and NIN are soluble and have an acidic pI, while CIN is insoluble and has a basic pI (Cho et al., 2005; Ji et al., 2005).
Among the three invertases, CIN, which is relatively well characterized, contributes to sink strength, growth and development, and thus plays an important role in grain filling. CIN is expressed in immature seeds and its production plays important roles in the early developmental stages of grain filling in rice, cotton, and maize (Hirose et al., 2002; Li et al., 2013; Wang and Ruan, 2012; Wang et al., 2008). Overexpression of CIN in seeds increases seed grain productivity (Li et al., 2013; Wang et al., 2008), and similar results can be obtained by suppressing expression of CIN inhibitors (Jin et al., 2009; Tang et al., 2017). Unlike the well-established role of CIN in sink strength (Hirose et al., 2002; Jin et al., 2009; Wang and Ruan, 2012), few studies have been conducted on VIN’s contribution. In tomatoes, VIN leads to hexose accumulation during fruit ripening and controls fruit size (Klann et al., 1993; 1996). It is also required for stamen and seed development in cotton (Wang and Ruan, 2016) and may play a role in sucrose storage, phloem unloading, and transport (with its hydrolysis in vacuoles) in carrot and maize (Kim et al., 2000; Sturm et al., 1995).
Rice has two VINs, OsVIN1 and OsVIN2, whose recombinant proteins function as invertases (Ji et al., 2005; 2007). In the present study, we characterized the function of rice VINs with their mutants in detail. On the basis of our results, we propose an important role of OsVIN2 in controlling sink strength and sugar partitioning in rice.
MATERIALS AND METHODS
Plant materials
Mutant and transgenic lines of OsVIN1 and OsVIN2, with a japonica cultivar (cv.) Dongjin serving as wild type (WT), were used in the experiments. Rice plants were grown in an environmental growth chamber at 30°C and 20°C during the day and night, respectively, in a light/dark cycle of 14/10 h or in a paddy field for living modified organisms (LMOs) at Kyung Hee University under natural environmental conditions during summer.
Isolation of the OsVIN1 and OsVIN2 mutants
T-DNA insertion lines of OsVIN1 and OsVIN2 knockout mutants osvin1 and osvin2-1 were identified from the SIGNAL Database (http://signal.salk.edu/cgi-bin/RiceGE) (Jeon et al., 2000; Jeong et al., 2006). Homozygous mutants were isolated by genomic DNA polymerase chain reaction (PCR) analysis. The gene-specific primers used for the genotyping were, 5′-GTTAGGCAAGTTGTTGTGATGCTAATAGG-3′ as forward and 5′-GTTGTTGTGTATGTGTATGGCACACTTTT-3′ as reverse for osvin1; and 5′-AGTTCTATGCCTCCAAGACCTTCTACGAT-3′ as forward and 5′-TCGATCGAGATACAAAATTAAAGCAGAGA-3′ as reverse for osvin2-1. The T-DNA-specific primer was 5′-ATCCAGACTGAATGCCCACAGG-3′.
RNA isolation and RT-PCR analysis
Total RNA was extracted from the collected WT and mutant samples using the TRIzol reagent and then reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad, USA). First-strand cDNAs were used in reverse-transcriptase (RT) PCR reactions with gene-specific primers and control primers for the housekeeping gene OsUBQ5 (Jain et al., 2006). The gene-specific primers used were: OsVIN1, 5′-AAGCAAACGATCTAGTTAAGAGAG-3′ and 5′-TTCCCATTACATTAAAAATGATCT-3′; OsVIN2, 5′-GACATCGTCAAGAGGGTCG-3′ and 5′-CCATCCATGATCCATCATCC-3′; and OsUBQ5, 5′-GACTACAACATCCAGAAGGAGTC-3′ and 5′-TCATCTAATAACCAGTTCGATTTC-3′. RT-PCR analyses were repeated at least three times, giving similar results using previously described methods (Cho et al., 2005).
Genetic complementation experiment
To complement the osvin2-1 mutant, a native full-length OsVIN2 genomic DNA of WT was amplified by PCR using the 5′-CACCTTTATGAAAGCTTTTTGAAAGATG-3′ and 5′-GTTATGAAGTTGCAATTTGACTTT-3′ primers, with underlined sequences indicating overhang sequences for TOPO® cloning. The PCR product was then subcloned into the pENTRTM/D-TOPO® vector (Invitrogen, USA) and transferred into a modified NC4300 binary vector carrying the phosphomannose isomerase (PMI) gene as a second selectable marker (Eom et al., 2011) using the Gateway system. The resulting construct was used to transform the osvin2-1 as described by Lucca et al. (2001).
Scanning electron microscopy analysis
The young spikelet samples of WT and mutant were harvested, fixed in a 3.5% glutaraldehyde solution, and dehydrated using a series of ethanol (60% to 100%). The samples were then dried with a critical-point drier (Chemical Free FDCF; Operon, Korea) and coated with platinum (Pt) using a sputter. Scanning electron microscopy (SEM) images were observed using a focused ion-beam scanning transmission electron microscope (Strata 400 STEM; FEI, USA).
Measurement of net photosynthetic activity
Net photosynthetic activity of the most recent fully expanded leaves of eight-week-old rice plants was measured using a portable gas-exchange system (Li-6400; Li-Cor, USA) at 1,200 μmol m−2 s−1 photon flux density. Leaf temperature was 25°C, and the reference CO2 concentration was 400 μmol mol−1.
Determination of soluble sugars and starch
Approximately 50 mg of palea/lemma at an early developmental stage and rice seeds at the pre-storage stage (three to six days after fertilization [DAF]), respectively, were harvested from plants grown in the LMO paddy field. Glucose, fructose, sucrose, and starch were measured in the soluble and insoluble fractions using enzymatic methods after preparing ethanol-water extracts (Lee et al., 2005). The measured metabolite contents were normalized to fresh weights.
Measurement of VIN activity
VIN activity was measured according to Kocal et al. (2008) with slight modifications. Twenty mg of leaves, young palea/lemma, and developing seeds at the pre-storage stage (three to six DAF), respectively, were homogenized with 50 mM of a Tris buffer, pH 6.8, containing 5 mM MgCl2, 5 mM dithiothreitol, 1 mM EDTA, 1 mM EGTA, 15% (v/v) glycerine, and a proteinase inhibitor. The extracts were centrifuged for 10 min at 13,000 rpm at 4°C. The supernatant was desalted by a Sephadex G-25 medium equilibrated in an extraction buffer. After desalting, VIN activities were determined using a method reported by Zrenner et al. (1995) with slight modifications. To remove invertase inhibitor complexes, the desalting samples were incubated at 4°C for 60 min. The samples were mixed with 20 mM of a sodium acetate buffer, pH 4.7, containing 100 mM of sucrose and incubated at 37°C for 90 min. The mixture was neutralized by adding a sodium phosphate solution, pH 7.2, and the reaction was stopped by heat inactivation at 95°C for 5 min. Glucose was measured as according to previously described enzymatic methods (Lee et al., 2005).
In situ hybridization
Spikelet samples from WT at different stages were fixed for 16 h in 5% acetic acid, 50% ethanol, and 3.7% formaldehyde in 4°C water, dehydrated through an ethanol series, embedded in Paraplast Plus, and sectioned into 8 μm slices. The OsVIN2 cDNA fragment generated by PCR was used to prepare antisense and sense probes under the T7 promoter with RNA polymerase using a DIG RNA labeling kit (Roche, Switzerland). The sense and antisense primers used were: OsVIN2 sense, 5′-TAATACGACTCACTATAGGGAGGAGATGGTGAGGCTGATG-3′ and 5′-ACTTGGTCCAGTTGGTGAGG-3′; and OsVIN2 antisense, 5′-AGGAGATGGTGAGGCTGATG-3′ and 5′-TAATACGACTCACTATAGGGACTTGGTCCAGTTGGTGAGG-3′. RNA hybridization and immunological detection of the hybridized probes were performed according to Li et al. (2006), and photographed with an Olympus E600 microscope (Olympus, Japan).
Creation of OsVIN2 mutants using the CRISPR/Cas9 system
To find an effective protospacer adjacent motif and avoid any off-targets, we screened possible target sequences using the CRISPRdirect program (Naito et al., 2015; http://crispr.dbcls.jp/). Designed guide RNA (5′-CTCGCCGTGGCGGTCCTCTC-3′) was cloned into an entry vector, pOs-sgRNA, and then cloned into a destination vector, pH-Ubi-cas9-7, using the Gateway system (Miao et al., 2013). The resulting vector was transformed with Agrobacterium tumefaciens (LBA4404). Cells grown on AB media containing 50 mg L−1 of hygromycin were used for rice transformation according to the Agrobacterium-mediated cocultivation method (Jeon et al., 2000). Transgenic rice plants were regenerated from the transformed calli on selection media containing 50 mg L−1 of hygromycin and 250 mg L−1 of cefotaxime.
RESULTS
Characterization of OsVIN mutants
OsVIN1 is expressed in various vegetative and reproductive organs, while the OsVIN2 transcript is expressed in specific organs, including panicles, during heading and flowering stages of growth (Ishimaru, 2005; Ji et al., 2005; 2007). A digital expression pattern generated with an Affymetrix chip-based gene expression profile (http://ricexpro.dna.affrc.go.jp/) revealed a high level of expression of OsVIN2 in developing ovaries, while expression of OsVIN1 was relatively low (OsVIN1: http://ricexpro.dna.affrc.go.jp/GGEP/graph-view.php?featurenum=5172, OsVIN2: http://ricexpro.dna.affrc.go.jp/GGEP/graph-view.php?featurenum=4559). These observations suggest a role for OsVIN2 in panicle and seed development.
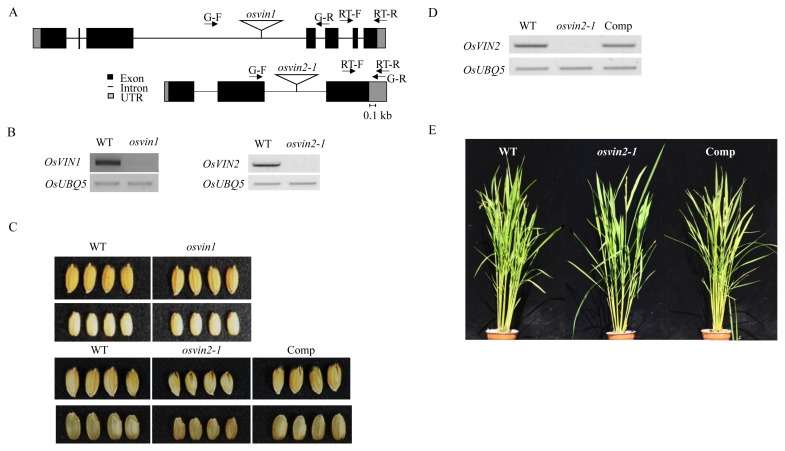
To elucidate the function of OsVIN1 and OsVIN2 during growth and development, we isolated the T-DNA insertion mutants osvin1 (serial number PFG_4A-50469) and osvin2-1 (PFG_2D-30640) from our T-DNA mutant pool (Jeon et al., 2000). Homozygous mutants isolated from a segregating progeny population by genomic DNA PCR analysis confirmed that the T-DNA was inserted into the third and second intron in osvin1 and osvin2-1, respectively (Fig. 1A). Transcripts of osvin1 and osvin2 were not detectable, indicating that they are null mutant alleles (Fig. 1B). Compared with segregating WT plants, osvin2-1 showed smaller seed grains, while osvin1 exhibited normal vegetative and reproductive growth (Fig. 1C). osvin2-1 showed decreased spikelet hull (palea/lemma) and grain size (Fig. 1C). The average weight of 1,000 grains was reduced by approximately 25% in osvin2-1 (Table 1). osvin2-1 did not show any remarkable growth defects before heading as it grew normally during vegetative growth (Fig. 1E).
Fig. 1. T-DNA mutant analysis of OsVIN1 and OsVIN2.
(A) Schematic representation of T-DNA insertions on OsVIN1 and OsVIN2 genomic structures in osvin1 and osvin2-1 mutants. G-F, genotyping forward primer; G-R, genotyping reverse primer; RT-F, RT-PCR forward primer; RT-R, RT-PCR reverse primer; UTR, untranslated region. (B) RT-PCR analysis of T-DNA insertion mutants. OsUBQ5 is a PCR control. (C) Representative seed grain phenotype. Only osvin2-1 showed small seed grain compared to WT. (D) RT-PCR analysis of osvin2-1 and complementation (Comp) lines. (E) Representative mature plants of osvin2-1 mutant.
Table 1.
Grain weight, length, and width of WT, osvin2-1 heterozygote and homozygote, and Comp lines
| Plant genotype | 1,000-Grain weight (g) | Grain length (mm) | Grain width (mm) |
|---|---|---|---|
| OsVIN2/OsVIN2 | 21.54 ± 0.90 | 5.23 ± 0.19 | 2.91 ± 0.09 |
| osvin2-1/osvin2-1 | 14.58 ± 0.79* | 4.52 ± 0.20* | 2.52 ± 0.08* |
| OsVIN2/osvin2-1 | 20.87 ± 0.65 | 5.16 ± 0.18 | 2.88 ± 0.11 |
| Comp | 21.08 ± 0.78 | 5.20 ± 0.19 | 2.88 ± 0.14 |
Each data point represents the mean ± SD from five different plants (*P < 0.05, Student’s t-test).
To determine if the small grain was controlled by either maternally or filially defective function of OsVIN2 during seed grain development, we measured seed grain size and weight of the progeny of self-fertilized WT (OsVIN2/OsVIN2), heterozygous (OsVIN2/osvin2-1) and homozygous (osvin2-1/osvin2-1) plants (Table 1). Only homozygous mutant bore small seed grains and exhibited decreased grain weight compared with WT and heterozygote plants, neither of which produced small grains. This suggests that the small grains of osvin2-1 were the result of a maternally defective OsVIN2 function.
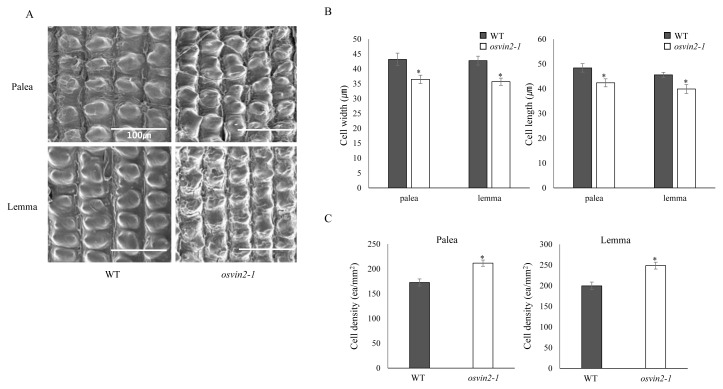
Changes in size and density of spikelet hull cells are known to affect the size and shape of seed grains (Peng et al., 2017). We therefore examined spikelet hulls of WT and osvin2-1 plants using SEM (Fig. 2). The average width and length of cells in osvin2-1 were significantly decreased compared with those of WT (Figs. 2A and 2B) plants. Consistently, the average cell number per unit area of outer epidermal surface of palea/lemma in osvin2-1 was reversely substantially increased than WT (Figs. 2A and 2C). This suggests that a decrease in the cell size of spikelet hulls contributed to the reduction of spikelet size and thereby grain seed in osvin2-1.
Fig. 2. SEM observation of the outer surface of WT and osvin2-1 spikelet hulls.
(A) SEM analysis of the outer surface of palea/lemma of WT and osvin2-1. (B) Average width and length of cells. (C) Average number of cells per mm2 (density) on the outer surface of palea/lemma. Each data point represents the mean ± SD from five different plants (*P < 0.05, Student’s t-test).
Genetic complementation of the OsVIN2 mutant
To confirm if the small seed phenotype was due to impaired function of OsVIN2, the entire WT OsVIN2 genomic DNA regulated by its native promoter and terminator was introduced into osvin2-1 by PMI selection system (Eom et al., 2011). Among transgenic plants produced, we selected the osvin2-1-complementation (Comp) line that showed higher expression of OsVIN2 for further analysis (Fig. 1D). In the Comp line with normal plant growth, seed grain size and 1,000-grain weight were both rescued to levels comparable to those of the WT (Figs. 1C–1E and Table 1) samples. Thus, the genetic complementation experiment confirmed that mutation in OsVIN2 caused small seed grain in osvin2-1.
Photosynthesis rate of the OsVIN2 mutant
To determine if the small seed grain phenotype in osvin2-1 was related to reduced photosynthetic activity, we assessed the net photosynthesis rates in osvin2-1, Comp, and WT by measuring CO2 consumption at 45 min intervals from zeitgeber time 1 (ZT 1) to midday in the leaves of eight-week old plants (Table 2). The osvin2-1 mutant did not show a significant change in net photosynthetic activity compared with WT and Comp plants, indicating that the small size of seed grain is not due to changes in photosynthetic activity in osvin2-1.
Table 2.
Net photosynthetic activity of eight-week-old WT, osvin2-1, and Comp lines
| Photosynthetic rate (μmol m−2 s−1), mean ± SD | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| 1:00 | 1:45 | 2:30 | 3:15 | 4:00 | 4:45 | 5:30 ZT (time) | |
| Wild type | 8.26 ± 0.71 | 21.50 ± 0.49 | 26.05 ± 1.36 | 26.65 ± 1.12 | 27.10 ± 1.08 | 17.44 ± 1.91 | > 10 |
| osvin2-1 | 8.81 ± 0.64 | 22.70 ± 1.06 | 26.80 ± 1.24 | 27.20 ± 1.30 | 27.60 ± 1.97 | 16.45 ± 1.84 | > 10 |
| Comp | 8.35 ± 0.59 | 19.95 ± 2.12 | 27.00 ± 1.56 | 27.10 ± 1.84 | 26.80 ± 1.82 | 18.95 ± 1.41 | > 10 |
The reference CO2 concentration was 400 μmol mol−1, and the photosynthetic photon flux density (PPFD) was 1,200 (μmol m−2 s−1).
VIN activity of the OsVIN2 mutant
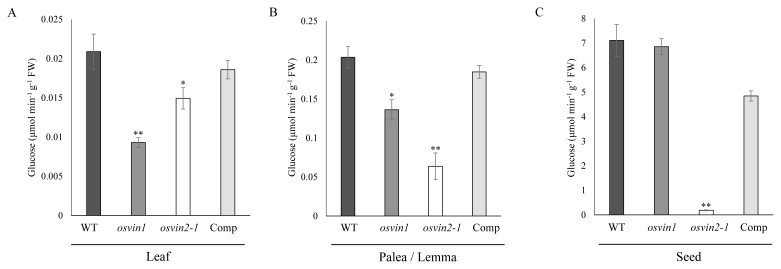
Altered sugar flux results in changes in seed grain size in various plants (Jin et al., 2009; Li et al., 2013; Wang et al., 2015). In osvin2-1, the small seed grain was determined maternally specifically at the seed development stage (Fig. 2 and Table 1). Among the maternal factors that can influence seed size during seed development are spikelet hull size and seed coat, including pericarp (epicarp, mesocarp, and endocarp) and testa (integument and nucellar tissue) (Li and Li, 2015; Wu et al., 2016a). In osvin mutants, Comp, and WT plants, we measured VIN activity from their palea/lemma and developing seed at the pre-storage stage of three to six DAF, as well as from the leaves of six-week-old plants.
Analysis of leaf VIN activity showed osvin2-1 retained 70% to 80% of VIN activity and slightly decreased VIN activity compared with WT and Comp plants, whereas osvin1 showed greatly reduced VIN activity, indicating that VIN1 is responsible for the majority of VIN activity in rice leaves (Fig. 3A). The absolute activity of VIN in leaves was relatively low, suggesting it is unlikely that VIN activity contributes to leaf growth as much as to seed development.
Fig. 3. Analysis of VIN activity in WT, osvin2-1 and Comp lines.
(A) Six-week-old rice leaves. (B) Palea/lemma. (C) Three to six DAF (pre-storage stage) developing seeds. Each data point represents the mean ± SD from five different plants (*P < 0.05, **P < 0.01, Student’s t-test).
In contrast, in palea/lemma, osvin2-1 decreased VIN activity to less than 40%, while osvin1 retained 70% of VIN activity compared with WT and Comp plants (Fig. 3B). In developing seeds, osvin2-1 decreased VIN activity significantly, to less than 5% of the seeds of the WT, Comp, and osvin1 lines, which retained high VIN activity (Fig. 3C). In WT plants, absolute VIN activity in spikelet hulls and developing seed grains was about 10-fold and 340-fold higher, respectively than that in leaves, suggesting a more prominent role of VIN during reproductive growth involving seed development. This result supports that OsVIN2 activity is critical for normal seed grain development.
Sugar and starch metabolites of OsVIN2 mutant
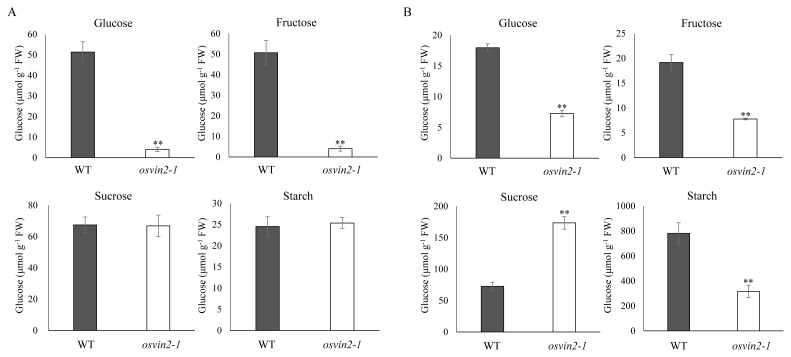
To determine if decreased VIN activity affected sugar metabolism, we measured steady-state levels of primary sugar metabolites and starch in palea/lemma and developing seeds in WT and osvin2-1 plants (Fig. 4). The samples were harvested at the end of day of 14 h light/10 h dark growth conditions when photosynthetic activity is complete. osvin2-1 showed significantly reduced levels of hexoses, glucose, and fructose in both palea/lemma (Fig. 4A) and seed grains (Fig. 4B). In addition, osvin2-1 reduced starch significantly and accumulated sucrose in developing seed grains (Fig. 4B). This suggests that OsVIN2 functions predominantly in starch synthesis via sucrose hydrolysis at an early stage of seed grain development. However, there were no differences in the amount of sucrose and starch in palea/lemma of osvin2-1 compared with WT (Fig. 4A). It is therefore likely that the reduced size of osvin2-1 hulls was due to low levels of hexoses, while the transitory reserves of sucrose and starch were compensated by remaining VIN activity and other sucrose cleavage enzymes, such as CINs, NINs, and SuSys.
Fig. 4. Soluble sugar and starch contents in WT and osvin2-1 mutant.
(A) Palea/lemma. (B) Three to six DAF developing seeds. Each data point represents the mean ± SD from five different plants (**P < 0.01, Student’s t-test).
In situ RNA hybridization of OsVIN2 in developing seeds
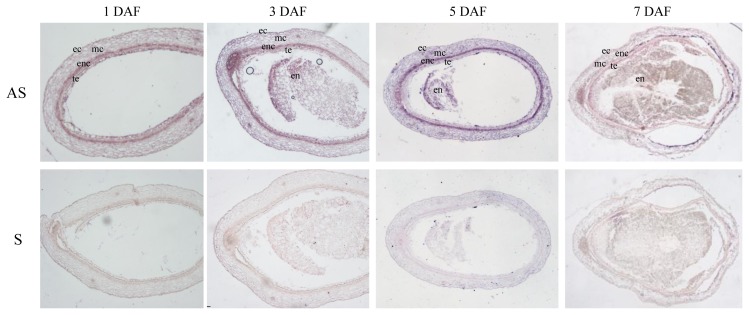
OsVIN2 is believed to function in sucrose degradation and subsequent starch synthesis in developing seeds. To further examine detailed expression of OsVIN2, we performed in situ RNA hybridization experiments in developing seeds (Fig. 5). Specific expression of OsVIN2 was detected in the endocarp of one to seven DAF developing seed grains (Fig. 5, top). The sense control experiment showed no clear signals (Fig. 5, bottom).
Fig. 5. RNA in situ hybridization of 1, 3, 5, and 7 DAF developing rice seeds with OsVIN2 antisense and sense probes.
AS, antisense; S, sense; ec, epicarp; mc, mesocarp; enc, endocarp; te, testa; en, endosperm.
Generation of additional OsVIN2 mutants by CRISPR/Cas9
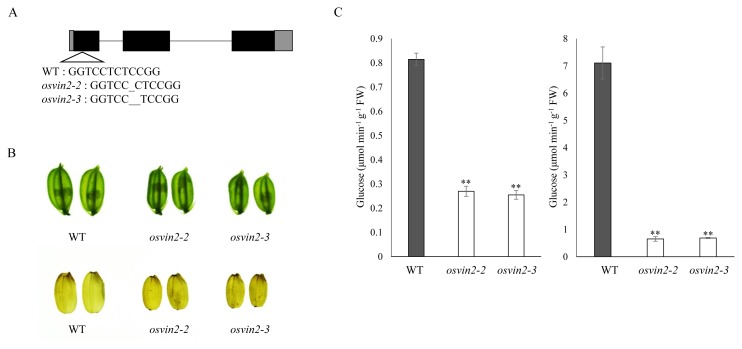
To further confirm the role of OsVIN2 in seed development, we employed the CRISPR/Cas9 method to produce additional allelic mutants of OsVIN2. The 160th nucleotide of the first exon was selected as the start of the guide RNA target. Among 38 independent transgenic lines, we found two homozygous mutants, osvin2-2 and osvin2-3, with mutations at the target site (Fig. 6A).
Fig. 6. Characterization of osvin2-2 and osvin2-3 mutants created by CRISPR/cas9.
(A) Schematic depiction of mutation positions in osvin2-2 and osvin2-3. (B) Spikelet (top) and brown rice (bottom) phenotype of osvin2-2 and osvin2-3. (C) VIN activity of osvin2-2 and osvin2-3 in developing hulls (left) and seed grains (right). Each data point represents the mean ± SD from five different plants (**P < 0.01, Student’s t-test).
One- and two-nucleotide deletions were found in osvin2-2 and osvin2-3, respectively, which eventually induce premature termination codons. As expected, the size of spikelet hulls and seed grains was reduced in osvin2-2 and osvin2-3, as was the case with osvin2-1 (Fig. 6B). VIN activities measured in osvin2-2 and osvin2-3 were consistently and significantly reduced in both developing hulls (Fig. 6C, left) and seed grains (Fig. 6C, right). Levels of VIN activity in osvin2-2 and osvin2-3 were reduced to 33% and 10%, respectively, in palea/lemma and developing seeds, compared with WT.
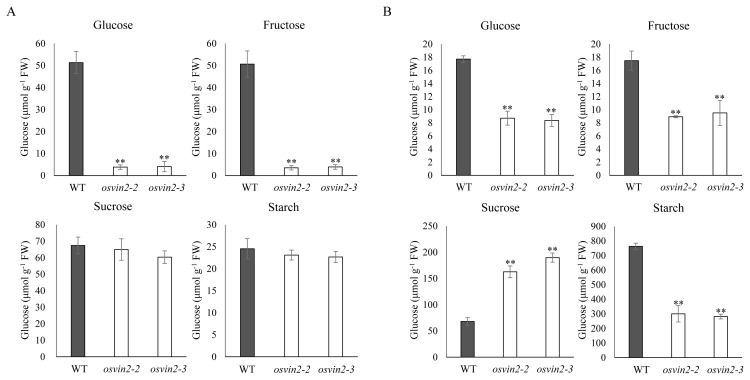
We measured sugar metabolites such as sucrose, glucose, fructose, and starch in developing hulls and seed grains of WT, osvin2-2 and osvin2-3. As with the results of T-DNA insertion mutant, osvin2-1 (Fig. 4), low levels of hexoses were measured in osvin2-2 and osvin2-3 samples, and accumulated sucrose with reduced starch was found in developing seeds but no changes were noted in palea/lemma compared with WT (Fig. 7). From these results it is clear that OsVIN2 plays a critical role in developing spikelet hulls and seed grains.
Fig. 7. Soluble sugar and starch contents in osvin2-2 and osvin2-3 mutants.
(A) Palea/lemma of WT, osvin2-2 and osvin2-3. (B) Three to six DAF developing seeds of WT, osvin2-2 and osvin2-3. Each data point represents the mean ± SD from five different plants (**P < 0.01, Student’s t-test).
DISCUSSION
In this study, we discovered a novel function of OsVIN2 involving in reproductive growth, in particular seed grain development, in rice. Among two rice VIN isoforms, only a loss of function of OsVIN2 displayed a visible phenotype of small spikelet hulls and seed grains (Figs. 1 and 2, Table 1). OsVIN2 contributed consistently to a large portion of total VIN activities in spikelet hulls and seed grains (Figs. 3 and 6). Importantly, osvin2 mutants grew normally but with a reproductive stage-specific defect of impaired growth and development of spikelet hulls and seed size and grain weight. This observation is consistent with the finding that the net photosynthesis rate was not changed in osvin2 mutant compared with control, WT, and Comp lines (Table 2). It is therefore evident that OsVIN2 function is necessary for normal growth and development of reproductive organs.
In cereal crops including rice, a decrease in seed size is influenced mainly by maternal factors of parental genotype, along with environmental factors (Li and Li, 2015; 2016). We found that only the homozygous mutants of OsVIN2 produced consistently small seed grains (Table 2). The cell size of osvin2 spikelet hulls appeared to be clearly reduced compared with WT in SEM analysis, which causes small spikelet hulls (Fig. 2). Thus, the small seed phenotype in osvin2 homozygous mutant is supported by the notion that seed size in rice is determined by the space within spikelet hulls, which limits how large seed grains can grow (Shomura et al., 2008; Song et al., 2007).
In the analysis of sugars and starch, we observed a remarkable decrease in hexose levels in the spikelets of osvin2 mutants compared with WT (Figs. 4 and 7). High hexose levels mediated by VIN activity are reportedly involved in cell expansion (Andersen et al., 2002; Roitsch and González, 2004; Ruan et al., 2010; Sergeeva et al., 2006; Wang and Ruan, 2012). A high intracellular concentration of hexose induces high osmotic activity, which allows water to enter the cell, leading to cell expansion. It is likely that reduced levels of hexose in osvin2 mutants can affect cell expansion of spikelets. It is also well known that glucose is associated with auxin, an important plant hormone that regulates cell growth and expansion (Wang and Ruan, 2013). Glucose can increase a number of indole-3-acetic acid precursors, metabolites, and conjugates in auxin synthesis and is also involved in auxin distribution and signal transduction (Mishra et al., 2009; Sairanen et al., 2012). Thus, these findings suggest that the reduced level of glucose may affect cell size in osvin2 spikelet hulls.
Growth and development of sink organs depend on sucrose import from source organs. Thus, controlling sink strength to keep sucrose importing from source organs is an important regulatory mechanism to optimize sugar partitioning and thereby support growth and development of reproductive organs. Changes in the ability of sugar partitioning can therefore dramatically alter productivity. For instance, mutation of the rice vacuolar sucrose transporter OsSUT2 accumulated sucrose within leaf vacuoles, limited availability of sucrose toward phloem loading, and resulted in growth retardation and yield reduction (Eom et al., 2011). An increase of sink strength enhanced apoplastic phloem loading of sucrose, increased seed grain size, and substantially improved grain yield in transgenic rice plants expressing the phloem-specific Arabidopsis sucrose transporter (AtSUC2) (Wang et al., 2015). At the pre-storage stage of developing seeds, increased sucrose and decreased starch amounts were observed in osvin2 mutants compared with WT (Figs. 4 and 7). Together with reduced levels of hexose, this suggests that flux and turnover of sucrose to starch is not properly maintained in osvin2 mutants, which limits growth and development of seed grains.
OsVIN2 expression was predominant in the endocarp at one to seven DAF in in situ hybridization (Fig. 5). Starch granules are accumulated in the seed coat at six DAF and disappeared after ten DAF (Wu et al., 2016b). Starch granules are also observed in the seed coat in barley and wheat (Radchuk et al., 2009; Xiong et al., 2013). Thus, the inner pericarp-specific expression of OsVIN2 indirectly suggests that osvin2 mutation interferes with starch synthesis due to reduced flux and turnover of sucrose in developing seed coats and thereby limits endosperm growth and development.
In conclusion, the present study demonstrates that in addition to CIN, VIN activity contributes to control of sink strength and sucrose flux into seed grains in rice. The OsVIN2 gene under the control of appropriate spatiotemporal-specific promoters, can be utilized to improve sink strength and yield potential in rice.
ACKNOWLEDGMENTS
This work was supported by grants from the Next Generation BioGreen 21 Program of the Rural Development Administration of Korea (PJ013172012018) and from the Mid-Career Researcher Program of the National Research Foundation of Korea (NRF2017R1A2B4009687).
Footnotes
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Andersen M.N., Asch F., Wu Y., Jensen C.R., Naested H., Mogensen V.O., Koch K.E. Soluble invertase expression is an early target of drought stress during the critical, abortion-sensitive phase of young ovary development in maize. Plant Physiol. 2002;130:591–604. doi: 10.1104/pp.005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt D.H.P., Derbyshire P., Findlay K., Pike M., Wellner N., Lunn J., Feil R., Simpson C., Maule A.J., Smith A.M. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc Natl Acad Sci U S A. 2009;106:13124–13129. doi: 10.1073/pnas.0900689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun D.M., Wang L., Ruan Y.L. Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J Exp Bot. 2014;65:1713–1735. doi: 10.1093/jxb/ert416. [DOI] [PubMed] [Google Scholar]
- Chamont S. Sink strength: the key for plant yield modeling. Plant Cell Environ. 1993;16:1033–1034. doi: 10.1111/j.1365-3040.1996.tb02056.x. [DOI] [Google Scholar]
- Chang T.G., Zhu X.G., Raines C. Source-sink interaction: a century old concept under the light of modern molecular systems biology. J Exp Bot. 2017;68:4417–4431. doi: 10.1093/jxb/erx002. [DOI] [PubMed] [Google Scholar]
- Cheng C.L., Acedo G.N., Cristinsin M., Conkling M.A. Sucrose mimics the light induction of Arabidopsis nitrate reductase gene transcription. Proc Natl Acad Sci U S A. 1992;89:1861–1864. doi: 10.1073/pnas.89.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J.I., Lee S.K., Ko S., Kim H.K., Jun S.H., Lee Y.H., Bhoo S.H., Lee K.W., An G., Hahn T.R., et al. Molecular cloning and expression analysis of the cell-wall invertase gene family in rice (Oryza sativa L.) Plant Cell Rep. 2005;24:225–236. doi: 10.1007/s00299-004-0910-z. [DOI] [PubMed] [Google Scholar]
- Coleman H.D., Yan J., Mansfield S.D. Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc Natl Acad Sci U S A. 2009;106:13118–13123. doi: 10.1073/pnas.0900188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottage A., Mott E.K., Kempster J.A., Gray J.C. The Arabidopsis plastid-signalling mutant gun1 (genomes uncoupled1) shows altered sensitivity to sucrose and abscisic acid and alterations in early seedling development. J Exp Bot. 2010;61:3773–3786. doi: 10.1093/jxb/erq186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom J.S., Cho J.I., Reinders A., Lee S.W., Yoo Y., Tuan P.Q., Choi S.B., Bang G., Park Y.I., Cho M.H., et al. Impaired function of the tonoplast-localized sucrose transporter in rice, OsSUT2, limits the transport of vacuolar reserve sucrose and affects plant growth. Plant Physiol. 2011;157:109–119. doi: 10.1104/pp.111.176982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González M.C., Roitsch T., Cejudo F.J. Circadian and developmental regulation of vacuolar invertase expression in petioles of sugar beet plants. Planta. 2005;222:386–395. doi: 10.1007/s00425-005-1542-4. [DOI] [PubMed] [Google Scholar]
- Hanson J., Hanssen M., Wiese A., Hendriks M.M.W.B., Smeekens S. The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J. 2008;53:935–949. doi: 10.1111/j.1365-313X.2007.03385.x. [DOI] [PubMed] [Google Scholar]
- Hirose T., Takano M., Terao T. Cell wall invertase in developing rice caryopsis: molecular cloning of OsCIN1 and analysis of its expression in relation to its role in grain filling. Plant Cell Physiol. 2002;43:452–459. doi: 10.1093/pcp/pcf055. [DOI] [PubMed] [Google Scholar]
- Ishimaru T. Expression patterns of genes encoding carbohydrate-metabolizing enzymes and their relationship to grain filling in rice (Oryza sativa L.): comparison of caryopses located at different positions in a panicle. Plant Cell Physiol. 2005;46:620–628. doi: 10.1093/pcp/pci066. [DOI] [PubMed] [Google Scholar]
- Jain M., Nijhawan A., Tyagi A.K., Khurana J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun. 2006;345:646–651. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- Jeon J.S., Lee S., Jung K.H., Jun S.H., Jeong D.H., Lee J., Kim C., Jang S., Yang K., Nam J., et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000;22:561–570. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- Jeong D.H., An S., Park S., Kang H.G., Park G.G., Kim S.R., Sim J., Kim Y.O., Kim M.K., Kim S.R., et al. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 2006;45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- Ji X., Van den Ende W., Schroeven L., Clerens S., Geuten K., Cheng S., Bennett J. The rice genome encodes two vacuolar invertases with fructan exohydrolase activity but lacks the related fructan biosynthesis genes of the Pooideae. New Phytol. 2007;173:50–62. doi: 10.1111/j.1469-8137.2006.01896.x. [DOI] [PubMed] [Google Scholar]
- Ji X., Van den Ende W., Van Laere A., Cheng S., Bennett J. Structure, evolution, and expression of the two invertase gene families of rice. J Mol Evol. 2005;60:615–634. doi: 10.1007/s00239-004-0242-1. [DOI] [PubMed] [Google Scholar]
- Jin Y., Ni D.A., Ruan Y.L. Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. Plant Cell. 2009;21:2072–2089. doi: 10.1105/tpc.108.063719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Mahé A., Brangeon J., Prioul J.L. A maize vacuolar invertase, IVR2, is induced by water stress. Organ/tissue specificity and diurnal modulation of expression. Plant Physiol. 2000;124:71–84. doi: 10.1104/pp.124.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann E.M., Chetelat R.T., Bennett A.B. Expression of acid invertase gene controls sugar composition in tomato (Lycopersicon) fruit. Plant Physiol. 1993;103:863–870. doi: 10.1104/pp.103.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann E.M., Hall B., Bennett A.B. Antisense acid invertase (TIV1) gene alters soluble sugar composition and size in transgenic tomato fruit. Plant Physiol. 1996;112:1321–1330. doi: 10.1104/pp.112.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocal N., Sonnewald U., Sonnewald S. Cell wall-bound invertase limits sucrose export and is involved in symptom development and inhibition of photosynthesis during compatible interaction between tomato and Xanthomonas campestris pv vesicatoria. Plant Physiol. 2008;148:1523–1536. doi: 10.1104/pp.108.127977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.W., Lee D.S., Bhoo S.H., Jeon J.S., Lee Y.H., Hahn T.R. Transgenic Arabidopsis plants expressing Escherichia coli pyrophosphatase display both altered carbon partitioning in their source leaves and reduced photosynthetic activity. Plant Cell Rep. 2005;24:374–382. doi: 10.1007/s00299-005-0951-y. [DOI] [PubMed] [Google Scholar]
- Li B., Liu H., Zhang Y., Kang T., Zhang L., Tong J., Xiao L., Zhang H. Constitutive expression of cell wall invertase genes increases grain yield and starch content in maize. Plant Biotechnol J. 2013;11:1080–1091. doi: 10.1111/pbi.12102. [DOI] [PubMed] [Google Scholar]
- Li N., Li Y. Maternal control of seed size in plants. J Exp Bot. 2015;66:1087–1097. doi: 10.1093/jxb/eru549. [DOI] [PubMed] [Google Scholar]
- Li N., Li Y. Signaling pathways of seed size control in plants. Curr Opin Plant Biol. 2016;33:23–32. doi: 10.1016/j.pbi.2016.05.008. [DOI] [PubMed] [Google Scholar]
- Li N., Zhang D.S., Liu H.S., Yin C.S., Li X., Liang W., Yuan Z., Xu B., Chu H.W., Wang J., et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell. 2006;18:2999–3014. doi: 10.1105/tpc.106.044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucca P., Ye X., Potrykus I. Effective selection and regeneration of transgenic rice plants with mannose as selective agent. Mol Breed. 2001;7:43–49. doi: 10.1023/A:1009661014167. [DOI] [Google Scholar]
- Miao J., Guo D., Zhang J., Huang Q., Qin G., Zhang X., Wan J., Gu H., Qu L.J. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013;23:1233–1236. doi: 10.1038/cr.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra B.S., Singh M., Aggrawal P., Laxmi A. Glucose and auxin signaling interaction in controlling Arabidopsis thaliana seedlings root growth and development. PLoS One. 2009;4:e4502. doi: 10.1371/journal.pone.0004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y., Hino K., Bono H., Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payyavula R.S., Tay K.H.C., Tsai C.J., Harding S.A. The sucrose transporter family in Populus: the importance of a tonoplast PtaSUT4 to biomass and carbon partitioning. Plant J. 2011;65:757–770. doi: 10.1111/j.1365-313X.2010.04463.x. [DOI] [PubMed] [Google Scholar]
- Peng P., Liu L., Fang J., Zhao J., Yuan S., Li X. The rice TRIANGULAR HULL1 protein acts as a transcriptional repressor in regulating lateral development of spikelet. Sci Rep. 2017;7:13712. doi: 10.1038/s41598-017-14146-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk V.V., Borisjuk L., Sreenivasulu N., Merx K., Mock H.P., Rolletschek H., Wobus U., Weschke W. Spatiotemporal profiling of starch biosynthesis and degradation in the developing barley grain. Plant Physiol. 2009;150:190–204. doi: 10.1104/pp.108.133520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch T., González M.C. Function and regulation of plant invertases: sweet sensations. Trends Plant Sci. 2004;9:606–613. doi: 10.1016/j.tplants.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Ruan Y.L., Jin Y., Yang Y.J., Li G.J., Boyer J.S. Sugar input, metabolism, and signaling mediated by invertase: roles in development, yield potential, and response to drought and heat. Mol Plant. 2010;3:942–955. doi: 10.1093/mp/ssq044. [DOI] [PubMed] [Google Scholar]
- Sairanen I., Novák O., Pěnčík A., Ikeda Y., Jones B., Sandberg G., Ljung K. Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell. 2012;24:4907–4916. doi: 10.1105/tpc.112.104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva L.I., Keurentjes J.J.B., Bentsink L., Vonk J., van der Plas L.H.W., Koornneef M., Vreugdenhil D. Vacuolar invertase regulates elongation of Arabidopsis thaliana roots as revealed by QTL and mutant analysis. Proc Natl Acad Sci U S A. 2006;103:2994–2999. doi: 10.1073/pnas.0511015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Metabolic repression of transcription in higher plants. Plant Cell. 1990;2:1027–1038. doi: 10.1105/tpc.2.10.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D.H., Choi M., Kim K., Bang G., Cho M., Choi S.B., Choi G., Park Y.I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013;587:1543–1547. doi: 10.1016/j.febslet.2013.03.037. [DOI] [PubMed] [Google Scholar]
- Shomura A., Izawa T., Ebana K., Ebitani T., Kanegae H., Konishi S., Yano M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 2008;40:1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- Solfanelli C., Poggi A., Loreti E., Alpi A., Perata P. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 2006;140:637–646. doi: 10.1104/pp.105.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X.J., Huang W., Shi M., Zhu M.Z., Lin H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- Sturm A.Šebková VLorenz K., Hardegger M., Lienhard S., Unger C. Development- and organ-specific expression of the genes for sucrose synthase and three isoenzymes of acid β-fructofuranosidase in carrot. Planta. 1995;195:601–610. doi: 10.1007/BF00195721. [DOI] [Google Scholar]
- Tang X., Su T., Han M., Wei L., Wang W., Yu Z., Xue Y., Wei H., Du Y., Greiner S., et al. Suppression of extracellular invertase inhibitor gene expression improves seed weight in soybean (Glycine max) J Exp Bot. 2017;68:469–482. doi: 10.1093/jxb/erw425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Wang J., Zhu X., Hao W., Wang L., Li Q., Zhang L., He W., Lu B., Lin H., et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet. 2008;40:1370–1374. doi: 10.1038/ng.220. [DOI] [PubMed] [Google Scholar]
- Wang L., Lu Q., Wen X., Lu C. Enhanced sucrose loading improves rice yield by increasing grain size. Plant Physiol. 2015;169:2848–2862. doi: 10.1104/pp.15.01170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Ruan Y.L. New insights into roles of cell wall invertase in early seed development revealed by comprehensive spatial and temporal expression patterns of GhCWIN1 in cotton. Plant Physiol. 2012;160:777–787. doi: 10.1104/pp.112.203893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Ruan Y.L. Regulation of cell division and expansion by sugar and auxin signaling. Front Plant Sci. 2013;4:163. doi: 10.3389/fpls.2013.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Ruan Y.L. Critical roles of vacuolar invertase in floral organ development and male and female fertilities are revealed through characterization of GhVIN1-RNAi cotton plants. Plant Physiol. 2016;171:405–423. doi: 10.1104/pp.16.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert N., Saalbach I., Weichert H., Kohl S., Erban A., Kopka J., Hause B., Varshney A., Sreenivasulu N., Strickert M., et al. Increasing sucrose uptake capacity of wheat grains stimulates storage protein synthesis. Plant Physiol. 2010;152:698–710. doi: 10.1104/pp.109.150854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Liu J., Li D., Liu C.M. Rice caryopsis development I: dynamic changes in different cell layers. J Integr Plant Biol. 2016a;58:772–785. doi: 10.1111/jipb.12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Liu J., Li D., Liu C.M. Rice caryopsis development II: dynamic changes in the endosperm. J Integr Plant Biol. 2016b;58:786–798. doi: 10.1111/jipb.12488. [DOI] [PubMed] [Google Scholar]
- Xiong F., Yu X.R., Zhou L., Wang F., Xiong A.S. Structural and physiological characterization during wheat pericarp development. Plant Cell Rep. 2013;32:1309–1320. doi: 10.1007/s00299-013-1445-y. [DOI] [PubMed] [Google Scholar]
- Zrenner R., Salanoubat M., Willmitzer L., Sonnewald U. Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.) Plant J. 1995;7:97–107. doi: 10.1046/j.1365-313X.1995.07010097.x. [DOI] [PubMed] [Google Scholar]