Abstract
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common non-Hodgkin lymphoma and is an aggressive malignancy with heterogeneous outcomes. Diverse methods for DLBCL outcomes assessment ranging from clinical to genomic have been developed with variable predictive and prognostic success.
Areas covered
The authors provide an overview of the various methods currently used to estimate prognosis in DLBCL patients. Models incorporating cell of origin, genomic features, sociodemographic factors, treatment effectiveness measures, and machine learning are described.
Expert opinion
The clinical and genetic heterogeneity of DLBCL presents distinct challenges in predicting response to therapy and overall prognosis. Successful integration of predictive and prognostic tools in clinical trials and in a standard clinical workflow for DLBCL will likely require a combination of methods incorporating clinical, sociodemographic, and molecular factors with the aid of machine learning and high-dimensional data analysis.
Keywords: B-cell lymphoma, non-Hodgkin lymphoma, diffuse large B-cell lymphoma, DLBCL, outcomes prediction, prognosis
1. Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common non-Hodgkin lymphoma (NHL) and is fatal without treatment.1 DLBCL is characterized by heterogeneous clinical2 and molecular subgroups3 with disparate outcomes, and the rapid and accurate identification of high-risk groups has important ramifications for prognosis and therapeutic decision-making. However, the clinical and genetic heterogeneity of DLBCL often presents challenges for risk stratification and prognostic modeling. Over the past three decades, methods of outcomes prediction in DLBCL have utilized a diverse array of data sources including clinical factors,4 cell-of-origin (COO) subtypes,5 and genetic subgroups,6-9 and it remains unclear what combination of these or other techniques will ultimately yield optimal prognostic models for integration into clinical trials and practice. Here, we review methods, remaining challenges, and future directions in outcomes prediction for DLBCL.
2. Prognostic modeling based on clinical characteristics
2.1. International Prognostic Index and beyond
Developed more than 25 years ago using stepwise regression, the International Prognostic Index (IPI; Tables 1 and 2)4 provides DLBCL risk assessment according to clinical characteristics (age, stage, serum lactic dehydrogenase level [LDH] level, performance status, and number of extranodal disease sites) and is routinely used in clinical practice. However, survival outcomes have changed markedly with the addition of rituximab to frontline chemotherapy regimens.10 In addition, IPI scores may have insufficient granularity for predicting the course of some DLBCLs, including the more aggressive cases that most warrant the development of individualized treatment strategies.11 To address these issues, two variations of the IPI have been proposed: the revised-IPI,10 which re-groups the original IPI scores into three risk groups, and the National Comprehensive Cancer Network–IPI (NCCN-IPI),12 which assigns incremental scores to increasing levels of age and LDH values and includes specific high-risk extranodal sites. A case series of >1,000 DLBCL patients treated with first-line chemoimmunotherapy investigated the interactions between clinical and genomic prognostic factors and found that the original IPI categories stratified patients, suggesting that the original factors remain prognostic in large populations.9 The inclusion of additional prognostic factors such as albumin serum levels have also been reported to increase prognostic accuracy in some models.13,14 A comparison of the IPI, R-IPI, and NCCN-IPI using individual patient-level data from 7 multicenter trials involving patients with aggressive B-cell lymphoma (n=2561; 86% DLBCL) treated with front-line with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) or variants was performed to determine which clinical scoring systems best discriminated overall survival (OS). This analysis showed that the NCCN-IPI produced the greatest absolute difference in OS estimates between the highest and lowest risk groups at 1, 3, and 5 years, and best discriminated OS (c-index = 0.63), but was only marginally better than the IPI (c-index = 0.62).15
Table 1. Prognostic methods for DLBCL.
Italicized methods incorporate a prognostic model.
| Method | Variables in prognostic method | Comments |
|---|---|---|
| Clinical | ||
| IPI4 | Age, stage, LDH, ECOG PS, no. of extranodal sites | 4 risk groups: low (score 0-1, 4-year OS 82%), low-intermediate (2, 81%), high-intermediate (3, 49%), high (4-5, 59%). |
| R-IPI10 | Age, stage, LDH, ECOG PS, no. of extranodal sites | 3 risk groups: very good (score 0, 4-year OS 94%), good (1-2, 79%), poor (3-5, 55%). |
| NCCN-IPI12 | Age, stage, LDH, ECOG PS, no. of extranodal sites | 4 risk groups: low (score 0-1, 5-year OS 96%), low-intermediate (2-3, 82%), high-intermediate (4-5, 64%), high (6-8, 33%). |
| Biccler et al.16 | Age, stage, LDH, ECOG PS, no. & type of extranodal sites, B symptoms, WBC, ALC, Hb, albumin, sex, tumor diameter | Combined multiple models using a stacking algorithm; lymphomapredictor.org. |
| Howlader et al.17 | Age, stage, sex, race, Hispanic ethnicity, marital status, poverty, initial therapy | 3 risk groups by 10-year OS: low (80%), medium (60%), high (36%). |
| Fitness status19 | Comprehensive Geriatric Assessment | Predictive of OS, ORR, and toxicities in older patients. |
| LMR32 | ALC and monocyte counts | Low LMR associated with inferior OS. |
| Vitamin D37 | Serum 25-hydroxyvitamin D level | Low 25(OH)D associated with inferior EFS and OS. |
| Molecular | ||
| COO subtype56 | Non-GCB- (by IHC) and ABC-DLBCL (by GEP) vs GCB-DLBCL | Non-GCB & ABC subtypes associated with inferior outcomes. |
| OxPhos67 | Gene expression signature | OxPhos subtype associated with insensitivity to BCR signaling inhibition. |
| DHL/THL72 | MYC & BCL2 and/or BCL6 rearrangement | DHL and THL associated with poor outcomes. |
| DHITsig73 | Gene expression signature | 5-year TTP 57% in DHITsig+, 81% in DHITsig− GCB-DLBCL. |
| DEL78 | Expression of MYC and BCL2 by IHC | Coexpression of BCL2 & MYC associated with inferior OS & PFS. |
| NGS | ||
| Shipp et al.97 | Gene expression in tumors | Identifies patients within IPI risk groups with a greater probability of cure or dying of DLBCL. |
| Reddy et al.9 | Gene expression markers (COO, MYC, and BCL2) and 150 genetic driver genes | 3 risk groups (low, medium, and high). |
| Schmitz et al.7 | Genetic abnormalities | MCD and N1 subtypes with inferior outcomes; BN2 and EZB with favorable survival. |
| Chapuy et al.8 | Genetic abnormalities | 5 genetic subsets with outcomes independent of IPI. |
| Arthur et al.6 | Genetic abnormalities | FCGR2B overexpression associated with poor outcomes. |
| Sociodemographic | ||
| Race100 | African American vs. white vs. other | African American race associated with inferior 5-year OS. |
| SES101 | Neighborhood SES by census-block group | Lower SES associated with increased mortality risk. |
| Residence103 | Urban, metro, and rural residence location | Urban and rural residence associated with inferior outcomes. |
| Insurance status102 | Private vs. Medicaid vs. no insurance | Medicaid & no insurance associated with inferior outcomes. |
| DTI104 | Disease-to-treatment interval | Shorter DTI associated with adverse clinical factors & worse outcomes. |
Abbreviations: 25(OH)D, 25-hydroxyvitamin D; ABC, activated B cell-like; ALC, absolute lymphocyte count; BCR, B cell receptor; COO, cell-of-origin; DEL, double-expressor lymphoma; DHITsig, double-hit signature; DHL, double-hit lymphoma; DLBCL, diffuse large B-cell lymphoma; DTI, diagnosis-to-treatment interval; ECOG PS, Eastern Cooperative Oncology Group performance status; EFS, event-free survival; GCB, germinal center B cell-like; GEP, gene expression profiling; IHC, immunohistochemistry; IPI, International Prognostic Index; LDH, lactate dehydrogenase; LMR, lymphocyte/monocyte ratio; NCCN, National Comprehensive Cancer Network; NGS, next-generation sequencing; OS, overall survival; ORR, overall response rate; OxPhos, oxidative phosphorylation; PFS, progression-free survival; R-IPI, revised IPI; SES, socioeconomic status; THL, triple-hit lymphoma; WBC, white blood cell count.
Table 2. International Prognostic Index variants.
| Prognostic index | Risk factors assessed | Risk groups |
|---|---|---|
| IPI4 | Age > 60 years Ann Arbor stage III–IV ECOG PS ≥ 2 Serum LDH level > 1× normal > 1 extranodal site |
0 or 1 risk factor: Low 2: Low intermediate 3: High intermediate 4 or 5: High |
| AA-IPI (age ≤ 60 years)4 | Ann Arbor stage III–IV ECOG PS ≥ 2 Serum LDH level > 1× normal |
0 risk factors: Low 1: Low intermediate 2: High intermediate 3: High |
| R-IPI10 | Age > 60 years Ann Arbor stage III–IV ECOG PS ≥ 2 Serum LDH level > 1× normal > 1 extranodal site |
0 risk factors: Very good 1 or 2: Good 3–5: Poor |
| NCCN-IPI12 | Age, years > 40 to ≤ 60 (+1 to NCCN-IPI score) > 60 to ≤ 75 (+2) > 75 (+3) Ann Arbor stage III–IV (+1) ECOG PS ≥ 2 (+1) Serum LDH level > 1× to ≤ 3× normal (+1) > 3× normal (+2) Extranodal disease (+1) |
Total score 0 or 1: Low 2 or 3: Low intermediate 4 or 5: High intermediate 6–8: High |
Abbreviations: AA-IPI, age-adjusted IPI; ECOG PS, Eastern Cooperative Oncology Group performance status; IPI, International Prognostic Index; LDH, lactate dehydrogenase; NCCN-IPI, National Comprehensive Cancer Network IPI; R-IPI, revised IPI.
Recently, Biccler et al. developed a new prognostic model based on clinical data using a modern machine learning method known as a stacking algorithm.16 As an ensemble learning method, the stacking algorithm aggregates several regression models for obtaining survival curves and therefore eliminates the need for the specification of one prognostic modeling approach. The authors incorporated clinical data from Danish and Swedish nationwide lymphoma registries in defining their algorithm and reported that their stacking-based prognostic model was superior to the NCCN-IPI and IPI models (available as a web-based tool at https://lymphomapredictor.org).
In an effort to risk-stratify DLBCL patients in terms of cure rate rather than survival per se, Howlader et al. used the large population-based dataset available through the SEER registry to develop a prognostic model that included several additional non-IPI risk factors such as gender, race, Hispanic ethnicity, marital status, and a population-level measure of poverty.17 Utilizing this model, Howlader et al. estimated 10-year survival and cure rates for high-, medium-, and low-risk groups and calculated mortality risks using standardized mortality ratios for various noncancer causes. The SEER data used in this study did not include several clinical factors known to be associated with poor survival, such as high IPI score. However, IPI data are available for a subset of DLBCL patients in SEER and have been utilized in other prognostic models using SEER data.18
2.2. Fitness status
Determination of fitness status using the Comprehensive Geriatric Assessment (CGA) has demonstrated prognostic and predictive significance among older patients with DLBCL and may inform therapeutic decision making.19 The CGA categorizes patients as fit, unfit, or frail and incorporates a variety of attributes including performance status, functional status, comorbidities, nutritional status, cognitive function, psychological state, social support, and polypharmacy.20 Prospective studies incorporating modified CGA have shown stratification by fitness status to OS, overall response rate (ORR), and toxicities associated with therapy in older DLBCL patients.21-24 CGA was more effective than clinical judgment in highlighting fit patients who would tolerate anthracycline-based treatment with curative intent.23 Additional analysis is needed to identify preferred dose adjustments or alternate regimens for unfit and frail patients. Guidelines for oncologic treatment of elderly patients advise future integration of CGA tools,25,26 though application of the full CGA is impractical in a clinical setting. Simplified CGA models may be applicable in a clinical workflow and require validation in future studies. Incorporation of CGA in clinical trials has been limited,27,28 and additional prospective clinical trials are required prior to wide clinical adoption of CGA. Alternate methods for assessment of fitness status are in development and include the Vulnerable Elders Survey 13 (VES-13), which has demonstrated an association between vulnerable status and adverse outcomes in NHL.29
2.3. Hematologic parameters
Other studies have used hematologic parameters as a marker of prognosis. For example, there is evidence that the ratio of lymphocytes to monocytes in the serum at diagnosis of DLBCL may reflect the immune microenvironment and can serve as a predictor of response to standard chemoimmunotherapy with R-CHOP, independent of IPI score.30-33 While these indicators of tumor milieu may be helpful in predicting disease progression, identifying molecular abnormalities will be vital to analyzing putative drivers of poor-risk tumor microenvironments and developing therapies that target them.34-36
2.4. Vitamin D
Low levels of 25-hydroxyvitamin D [25(OH)D] are associated with inferior outcomes in DLBCL.37 Vitamin D deficiency is thought to impair rituximab-mediated cytotoxicity,38 and in vitro supplementation with vitamin D yields greater rituximab-mediated antibody-dependent cytotoxicity in B-cell lymphoma cells.39 In a prospective study analyzing serum 25(OH)D levels in 983 patients with newly diagnosed NHL, 25(OH)D insufficiency (< 25 ng/mL) demonstrated an association with inferior event-free survival (EFS) and OS in patients with DLBCL.37 A prospective cohort study assessed the impact of normalization of serum 25(OH)D levels in patients with aggressive lymphoma treated with R-CHOP.40 Of the 155 patients included in analysis, 128 patients had a diagnosis of DLBCL, not otherwise specified. Two-thirds of patients were found to be deficient (< 20 ng/mL) in 25(OH)D, and 25(OH)D deficiency was independently associated with inferior EFS. Normalization of 25(OH)D following weekly doses of 25,000 IU of cholecalciferol led to improved EFS in comparison with patients who had ongoing vitamin D deficiency or insufficiency, indicating the importance of vitamin D repletion in patients treated with R-CHOP. Studies are underway evaluating whether vitamin D repletion can impact OS in DLBCL.
3. Molecular prognostic classification
3.1. Cell-of-origin subtype
Gene expression profiling (GEP) has provided significant insight into biologic factors underlying divergent clinical outcomes in DLBCL. Seminal work by Alizadeh et al. identified two molecularly distinct subgroups of DLBCL based on GEP: germinal center B cell-like (GCB)-DLBCL, with an expression profile resembling that of normal germinal center B cells, and activated B cell-like (ABC)-DLBCL, which resembles normal activated B cells.5 Despite identical histologic appearance of the ABC and GCB subtypes, ABC-DLBCL has been shown to have significantly worse outcomes, exhibiting a 5-year OS rate of 35% following anthracycline-based chemotherapy compared to a 60% 5-year OS rate for GCB-DLBCL.41 Subsequent studies have since allowed for a better understanding of the dysregulated molecular pathways that characterize ABC- and GCB-DLBCLs, and clinical trials are underway examining subtype-specific therapeutic targets.42,43 At present, GEP is widely used in bench research, but it has only recently been incorporated into clinical trials and has not yet been adopted in clinical practice due to limitations of cost, turnaround time, and accessibility.
To assess DLBCL COO subtypes in the clinical setting, immunohistochemical (IHC) classification systems have been developed as surrogates for GEP (Table 3). The first and most widely used IHC classification system was developed in 2004 by Hans et al. and uses the differential expression of the proteins CD10, BCL6, and MUM1 to separate GCB and non-GCB phenotypes.44 By combining these IHC stains sequentially, the Hans algorithm identifies GCB and non-GCB DLBCLs with sensitivity of 70% and 87%, with positive predictive values of 84% and 75%, respectively. This classification system demonstrates 86% concordance with GEP, thus misclassifying 14% of patients. Eight other IHC classification systems have subsequently been developed that reported a higher concordance with molecular-based classification of DLBCL, though none have been as readily used in clinical practice as the Hans algorithm, likely due to its ease of use;45-51 these include the Choi algorithm, which adds GCET1 and FOXP1 to the Hans algorithm, and the Tally algorithm, which creates a score using CD10, MUM1, GCET1, FOXP1 and LMNO2. Many of the IHC classification systems have failed to reproduce the prognostic significance of COO subtyping seen with GEP.52
Table 3. Immunohistochemical classifiers for DLBCL.
| IHC classifier | Antibodies in algorithm |
COO subtypes identified |
|---|---|---|
| Choi48 | GCET1 MUM1 CD10 BCL6 FOXP1 |
GCB, non-GCB, ABC |
| Choi modified49 | FOXP1 GCET1 CD10 MUM1 |
GCB, ABC |
| Hans44 | CD10 BCL6 MUM1 |
GCB, non-GCB |
| Hans modified49 | CD10 MUM1 |
GCB, non-GCB |
| Muris45 | BCL2 CD10 MUM1 |
GCB, ABC |
| Natkunam46 | LMO2 | GCB, ABC |
| Nyman47 | MUM1 FOXP1 |
ABC, Other |
| Tally49 | CD10 GCET1 MUM1 FOXP1 |
GCB, ABC |
| Visco-Young50 | CD10 FOXP1 BCL6 |
GCB, non-GCB |
Abbreviations: ABC, activated B cell-like; COO, cell-of-origin; GCB, germinal center B cell-like; IHC, immunohistochemistry.
IHC staining is now considered essential for DLBCL prognostication in the clinical setting; however, it remains rife with challenges, as detailed throughout this section. Such classification systems also struggle to adapt to the evolving understanding of DLBCL biology. Informatics approaches have the potential to facilitate more objective and accurate classification systems. Approaches utilizing computerized image segmentation techniques have already began to show success in generating more reliable prognostic information in follicular lymphoma.53 Computerized image analysis tools may be able to play a similar role in classification of COO in DLBCL. For instance, the survival convolutional neural network combines deep learning with traditional survival models to learn survival-related patterns from histology images.54 Large whole-slide images are generated by digitizing IHC and hematoxylin and eosin (H&E)-stained glass slides, and a web-based viewer is used to manually identify representative regions of interest in the image. High-power fields are sampled from those regions and used to train a neural network to predict patient survival. The network includes convolutional layers that learn visual patterns related to survival and a Cox proportional hazards layer that models time-to-event data, such as OS. Predictions are then compared with actual patient outcomes to adaptively train the network weights that interconnect the layers. Our group has begun utilizing these image analysis tools with machine learning methods to classify COO for DLBCL in a consistent and accurate manner,53 while others have used similar techniques for identifying the DLBCL proliferation index.55 These methods will begin to show more value in the clinical setting as digital pathology tools, which were recently FDA-approved for use in hospitals, gain more widespread traction.
3.2. Clinical integration of IHC and GEP methods for prognostication
Given the prognostic importance of identifying COO subtypes and other DLBCL subgroups with predictive significance, there is clear benefit to incorporating IHC and GEP methods into a clinical workflow. However, historical challenges related to feasibility and reproducibility have prevented clinical adoption of these laboratory techniques. IHC methods have shown poor inter-assay and inter-laboratory concordance in multiple studies. Gutierrez-Garcia et al. compared the Colomo, Hans, Muris, Choi, and Tally IHC algorithms and showed that none of these IHC methods effectively risk-stratified patients by COO subtype or OS.56 Coutinho et al. assessed nine IHC algorithms and found low concordance and poor prognostic performance across all methods.51 While IHC algorithms and materials are readily accessible and available for clinical use, early IHC methods were considered too unreliable for clinical adoption.57
Clinical implementation of GEP technology has also proven difficult. Initial GEP methods were impractical in a clinical setting due to reliance on fresh frozen tissue with microarrays. In addition, the enormity and complexity of data obtained from cancer-related gene expression studies present great challenges in analyses and subsequent applications to a practical clinical environment. Data analysis methodologies that will efficiently aid in extracting relevant biological information in high-dimensional settings are required. In 2010, Williams et al. compared GEP with fresh frozen tissue to GEP using formalin-fixed paraffin-embedded tissue (FFPET) with 98% concordance.58 The successful use of FFPET, which is readily available from routine biopsy, meant that GEP integration in a clinical setting was more feasible than before, and subsequent research has focused on adapting DLBCL molecular COO subtyping to the bedside. The Lymphoma/Leukemia Molecular Profiling Project developed and validated Lymph2Cx, a digital gene expression-based assay that can determine COO subtype using FFPET biopsies with both consistency and accuracy.59 Using the NanoString multiplexed gene expression analysis platform, the Lymph2Cx assay was able to maintain prognostic significance of COO subtypes.60
However, FFPET-based GEP methods have also met challenges that hinder ready adoption in the clinical setting. Barriers to clinical implementation include the requirement of sufficient tissue for GEP beyond the initial diagnostic workup, potential for increased cost, and necessary access to equipment for this specialized technique.57 Despite successful performance of the NanoString platform, cost-effectiveness analysis of the Lymph2Cx assay has not been conducted, and GEP has not yet gained traction in clinical practice. Of note, a recent study on early-stage breast cancer found a substantially higher cost-effectiveness ratio for GEP use in community practice. 61 Given these challenges, the 2016 WHO classification system includes IHC algorithms as acceptable methods for COO categorization62 despite better historical performance of GEP.
In 2019, Robetorye et al. published clinical validation of GEP in a routine clinical setting through implementation of the Lymph2Cx digital GEP assay.63 The authors outline a workflow that proceeds from biopsy to immunophenotyping using the Hans algorithm to flow cytometry and FISH. If sufficient tissue remained at this stage (60+% lymphoma remaining in the specimen), the authors conducted the Lymph2Cx digital GEP assay. They reported analysis of 90 clinical cases and concluded that incorporation of Lymph2Cx in their clinical workflow was accurate, rapid, and reproducible.
Given the prognostic and potentially predictive significance of COO subtype, IHC and GEP methods have been increasingly implemented in randomized controlled trials for DLBCL. The PHOENIX trial assessing R-CHOP ± the BTK inhibitor ibrutinib in non-GCB patients (ClinicalTrials.gov identifier ) utilized a Hans-based IHC kit to distinguish non-GCB study participants from GCB patients. Development of such IHC kits may standardize IHC methods and minimize discordance across laboratories. The REMoDL-B trial comparing R-CHOP ± bortezomib in ABC-DLBCLs (ClinicalTrials.gov identifier ) utilized a GEP assay designed by Barrans et al.64 to assign COO subtype. In the ECOG/ACRIN E1412 (ClinicalTrials.gov identifier ) and ROBUST (ClinicalTrials.gov identifier ) trials examining R-CHOP ± lenalidomide, the NanoString platform categorized patients according to COO. Results of IHC and GEP application in these and other trials may guide future clinical workflows for rapid identification of COO subtypes and assignment of patients to subtype-specific therapeutic approaches.
3.3. Oxidative phosphorylation subgroup
Revisiting prior gene expression-based analyses beyond COO subtypes may provide prognostic and predictive value. The oxidative phosphorylation (OxPhos)-DLBCL subgroup has been shown to exhibit a gene expression profile that is independent of COO classification and may inform therapeutic decision making.65-67 OxPhos-DLBCLs do not respond to B cell receptor (BCR) signaling inhibition68 but may respond to disruption of fatty acid oxidation pathways, glutathione synthesis, and PPAR-γ.66 GEP-based identification of this subgroup may yield important clinical and research insight regarding appropriate therapies. Of particular note, the glycylcycline antibiotic tigecycline, which has demonstrated antitumor effects in other cancers69 and has been shown to have therapeutic synergy with venetoclax in lymphomas with rearrangements of MYC and BCL2,70 exhibited toxicity in OxPhos-DLBCLs at doses known to be tolerable in humans.67 This approach for classifying DLBCL has rarely been used in research or clinical settings, but could be more widely adopted if GEP characterization of OxPhos-DLBCLs can identify patients who are responsive to pharmacologic interference of fatty acid oxidation, glutathione synthesis, or mitochondrial translation and who display insensitivity to disruption of BCR signaling.
3.4. Rearrangements of MYC and BCL2 and/or BCL6
So-called “double-hit” lymphomas (DHL), which exhibit dual rearrangement of MYC and either BCL2 or BCL6, and “triple-hit” lymphomas (THL), which possess simultaneous rearrangement of all three genes, are associated with poor outcomes after standard frontline chemoimmunotherapy and have historically required fluorescence in situ hybridization (FISH) for detection.71,72 Ennishi et al. have described a gene expression-based methodology derived from RNAseq analysis of double- and triple-hit high-grade B-cell lymphomas with BCL2 rearrangements (HGBL-DH/TH-BCL2) that identifies a GCB subgroup with distinct biological and clinical characteristics.73 In this study, 27% of GCB DLBCLs demonstrated a HGBL-DH/TH-BCL2 gene expression profile according to a 104-gene double hit signature (“DHITsig”). Intriguingly, only half of this GCB subgroup exhibited MYC and BCL2 rearrangements. DHITsig positivity was associated with poor outcomes among GCB-DLBCLs according to time to progression, disease-specific survival, and OS. Notably, the DHITsig-negative GCB subgroup exhibited a 5-year disease-specific survival rate of 90%, suggesting that R-CHOP is sufficient therapy for these patients following DHITsig stratification. Incorporation of the DHITsig gene expression-based assay in clinical practice and future clinical trials may allow for identification of twice as many patients with the HGBL-DH/TH-BCL2 gene expression signature as would be identified by FISH analysis of MYC and BCL2 rearrangements alone. In addition, clinical implementation might distinguish patients in the DHITsig-negative GCB subgroup who could remain on R-CHOP without therapy escalation.
3.5. Double-expressor lymphoma
Independent of COO classification or IPI score, positive IHC staining for BCL2 and MYC have been shown to be strong predictors of inferior outcomes.74-77 DLBCLs with high expression of both BCL2 and MYC, referred to as double-expressor lymphomas (DELs), have been reported to portend a more aggressive disease course with a risk of death elevated as high as nine times that of those with low BLC2 and MYC expression.78,79 However, pathologist scoring of these two important IHC stains has been inconsistent. Interpretation of the BCL2 IHC stain varies significantly with an agreement rate of just 47% (κ = 0.23).80 Furthermore, ideal cut-off for a positive BCL2 IHC stain has not yet been universally agreed upon. A recent study has attempted a new scoring system for BCL2 hoping to improve consistency.81 Meanwhile, studies looking at inter-rater reliability of pathologists on scoring whole slides for MYC positivity yielded almost 40% discordant cases.82 Accurate prognostication of DLBCL will require more consistency of scoring the MYC and BCL2 IHC stains that define double-expressor lymphomas.
4. Next-generation sequencing
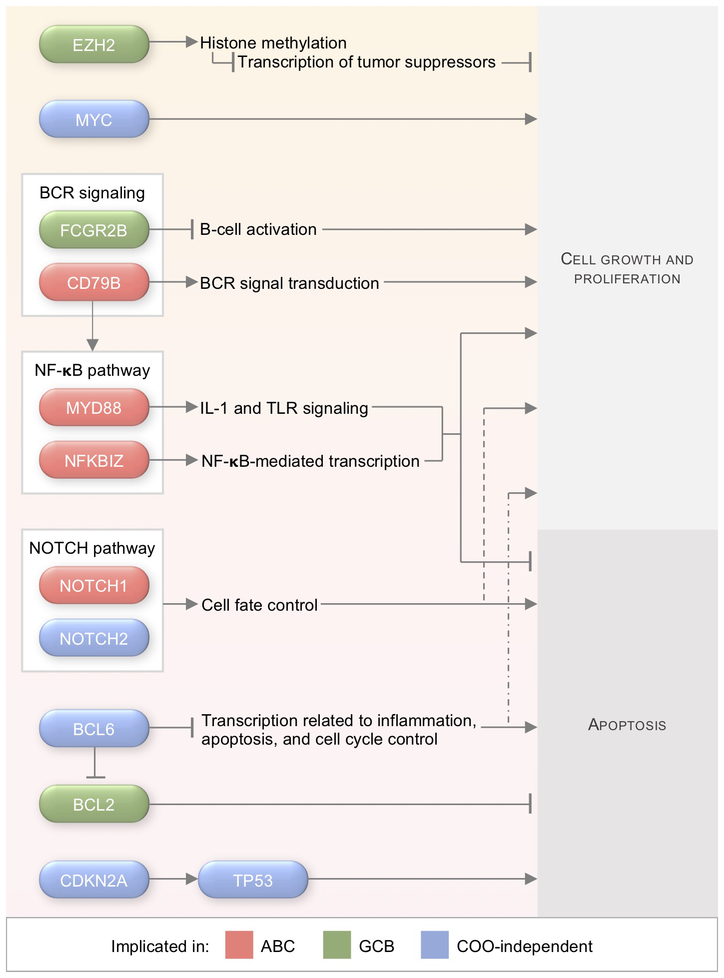
Next-generation sequencing has allowed clinicians and researchers to identify driving genetic aberrations and to determine which pathways and rearrangements demonstrate preferential distribution among DLBCL subtypes (Figure 1). For example, ABC-DLBCLs preferentially express somatic mutations in CD79A/B and MYD88 that result in constitutive BCR signaling and canonical NF-κB activation, while GCB-DLBCLs have much higher rates of mutated EZH2, resulting in suppressed apoptosis.83 Identifying and validating potential predictive biomarkers will be an important component of future personalized medicine strategies for DLBCL. Molecular hallmarks based on genetic aberrations have increasingly been accepted as nascent biomarkers for selecting precision medicine therapies.84-87
Figure 1. Select genetic alterations in DLBCL and associations with COO subtype.
Abbreviations: ABC, activated B cell-like; BCR, B-cell receptor; COO, cell-of-origin; DLBCL, diffuse large B-cell lymphoma; GCB, germinal center B cell-like; IL-1, interleukin-1; NF-κB, nuclear factor κB; TLR, toll-like receptor.
Progress in identifying putative mutations in DLBCL has been rapid. Four recent high-impact papers have each analyzed a large sample of DLBCLs to characterize the genomic and exomic landscape of the disease6-9 (Table 4). Reddy et al. performed whole-exome and transcriptome sequencing of 1,001 DLBCL patients and then established a prognostic model with the 150 driver mutations identified. Their genomic risk model was highly predictive, especially in regard to long-term mortality outcomes, which was lacking from the clinical IPI model. This prognostic model is discussed in greater detail later in the current review. Similarly, Schmitz et al. performed whole-exome and transcriptome sequencing alongside array-based DNA copy-number analysis as well as targeted amplicon resequencing of 574 DLBCL samples to define novel genomic subtypes beyond the standard COO classification scheme. The Schmitz et al. algorithm was based on co-occurring genetic aberrations and defined four subtypes: MCD (exhibiting mutations in MYD88 and CD79B), BN2 (exhibiting BCL6 fusions and NOTCH2 mutations), N1 (exhibiting NOTCH1 mutations), and EZB (exhibiting EZH2 mutations and BCL2 translocations). Chapuy et al. analyzed a cohort of 304 primary DLBCLs to identify 98 driver mutations that defined 5 novel DLBCL subsets: low-risk ABC-DLBCLs of extrafollicular/marginal zone origin, two phenotypically distinct subsets of GCB-DLBCL, and one COO subtype-independent group with biallelic inactivation of TP53 and CDKN2A loss. The most recent of these DLBCL genomics studies by Arthur et al. analyzed 338 de novo DLBCL cases and identified novel cis- regulatory sites, implicated recurrent mutations in the 3′ UTR of NFKBIZ as a NF-κB pathway activator in ABC-DLBCLs, and associated over-expression of FCGR2B with poor outcomes, particularly in GCB-DLBCL. New efforts are underway to use the results of these studies to identify potential genome-directed therapies as part of a precision medicine approach.88
Table 4. Next-generation sequencing studies in DLBCL.
| Study | Methods | n |
|---|---|---|
| Reddy et al.9 | Transcriptome and whole-exome sequencing; functional genomic analysis using CRISPR screen | 1,001 newly diagnosed DLBCL patients treated with rituximab-containing therapies |
| Schmitz et al.7 | Transcriptome and exome sequencing; DNA copy-number analysis; targeted amplicon resequencing | 574 DLBCL biopsy samples (transcriptome and exome sequencing); 374 genes (amplicon resequencing) |
| Chapuy et al.8 | Whole-exome sequencing; consensus clustering | 304 primary DLBCLs |
| Arthur et al.6 | Integrative analysis of whole genomes, exomes, and transcriptomes | 491 DLBCL tumor/normal pairs not previously exposed to rituximab (whole genome sequencing) compared with whole-exome-sequencing data from > 1,000 DLBCL cases |
Abbreviations: CRISPR, clustered regularly interspaced short palindromic repeats; DLBCL, diffuse large B-cell lymphoma.
4.1. Translating high-dimensional genomic data into clinical predictions
Although the aforementioned prognostic models such as the IPI score have demonstrated the utility of combining clinical and population-based data with statistical methods for prognostic prediction, they are imperfect in the identification of high-risk DLBCL patients, and they fail to capture the intrinsic molecular heterogeneity of DLBCL. With the rapid advent of genomic technologies, efforts have shifted to include gene expression and mutational profiles in prognostic modeling for more accurate discrimination between high- and low-risk DLBCL.
Utilization of DNA microarray and, more recently, high-throughput sequencing technologies have spawned large amounts of gene expression data. Integration of gene expression data with clinical, histological, imaging, demographic, and epidemiological information could provide insights for improving cancer diagnosis and prognosis. However, the enormity and complexity of data obtained from cancer-related gene expression studies present great challenges in making accurate predictions of clinical outcomes. Machine learning methods are designed to organize, process, and discover actionable knowledge in high-dimensional settings. As such, several different types of machine learning methods have been adapted to achieve three fundamental predictive tasks in cancer research: 1) prediction of cancer susceptibility (risk assessment); 2) prediction of cancer recurrence; and 3) prediction of cancer survival outcomes.89,90 An important challenge in translating high-dimensional data into accurate predictions for clinical decision-making is to identify informative features (e.g., clinical risk factors and genes) that contribute most to the prediction. Firstly, a more compact model will be more useful and interpretable in predicting outcomes for future patients. Secondly, selecting informative features is critical to avoiding overfitting and improving the accuracy and speed of prediction systems. Lastly, informative features allow investigators to understand the underlying cancer mechanisms that generated the data.
To overcome the difficulty of constructing accurate predictive models with high-dimensional data, there are two dimensionality-reduction techniques that are often used: feature selection, and feature extraction. Feature selection in genomic microarray analysis has been extensively studied in the last two decades for the prediction of cancer survivability, susceptibility and subtypes.91-94 Methods of feature selection vary in terms of performance and computational load and include filtering based on statistically defined relevance scores, stepwise selection, Lasso regression, and others. In contrast to feature selection, feature extraction creates new features as combinations of others to reduce the dimensionality of the selected features. Commonly used approaches for feature extraction include clustering methods and principal component analysis (PCA). As illustrated in Alizadeh et al., hierarchical clustering can be used to find meaningful clusters of features without the knowledge of clinical outcome, and these clusters can later be used in conjunction with the outcome to build a predictive model.5 An important drawback of both clustering and PCA for dimensionality reduction is that new features are created in an unsupervised manner: there is no guarantee that the new features will be predictive of the clinical outcome. To overcome this, some have proposed supervised principal component analysis, which selects the principal components based on the clinical outcome. This method has been used to predict patient survival using microarray data.95,96
In the DLBCL domain, several high-impact studies have combined genomics and machine learning for prediction of outcomes. Shipp et al.97 combined gene expression data of 58 DLBCL patients from oligonucleotide microarrays with classification analysis to predict a dichotomous clinical outcome (cured versus fatal/refractory disease). Feature selection methods were used to determine the predictive genes. However, this analysis was limited by the low number of genes and lacked some of the clinical data elements described above that are known to be associated with survival. In a more recent study, Reddy et al. performed an integrative analysis of whole-exome sequencing and transcriptome sequencing in a cohort of 1,001 DLBCL patients to comprehensively define the landscape of the disease.9 A supervised learning approach including an embedded feature selection method was used to develop a predictive model for survival that included clinical information, 150 genetic driver genes, and gene expression markers (COO, MYC, and BCL2). Although these studies demonstrate the utility of combining genomic data and machine learning for DLBCL risk assessment, future efforts can expand upon this approach by incorporating additional classification methods (i.e., other than linear models), comprehensively assessing all possible combinations of features and classification methods to identify the best possible model, and using alternative feature selection algorithms capable of capturing the molecular heterogeneity of DLBCL. Recent advances in prognostic modeling for DLBCL include the Continuous Individualized Risk Index (CIRI), which dynamically integrates personalized risk factors at pretreatment, treatment, and end-of-treatment phases.98 CIRI showed improved outcomes prediction in comparison with other models and may be applicable to cancers beyond DLBCL.
With major advances in the fields of epidemiology, genomics, and clinical research, large amounts of heterogeneous data have become available in various healthcare organizations. Therefore, there is a profound need for unified machine learning-based platforms incorporating vast amounts of mixed data types (e.g., imaging, histological, clinical, and genomic). The neural networks-based approaches, broadly described as deep learning, have been successfully implemented in areas such as image recognition, natural language processing, and robotics. Due to its ability to effectively leverage large data sets, the application of deep learning for precision genomic medicine is rapidly developing and has shown promise for the prediction of clinical outcomes with genomics.54,99 Future efforts should also aim to integrate robust machine learning-based platforms into clinical use to improve the risk stratification of DLBCL patients in a manner that can eventually translate to more effective and personalized treatment strategies.
5. Sociodemographic disparities in DLBCL presentation and outcomes
5.1. Race
Population-based epidemiology studies in the U.S. have demonstrated that DLBCL in African Americans displays different characteristics compared to DLBCL in white patients. The National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) Program data have shown that African American patients presented at 10 years younger, exhibited more advanced stage, and had inferior 5-year survival relative to individuals of the white population.100 Historically, these differences have been attributed to confounding social, environmental, and behavioral factors, but biologic factors may also play a role in racially disparate outcomes.
5.2. Socioeconomic status
According to a retrospective cohort analysis of 33,032 patients diagnosed with DLBCL in California from 1988 to 2009, DLBCL patients living in neighborhoods with lower socioeconomic status (SES) had increased risks of mortality compared to patients in higher SES neighborhoods.101 After adjusting for insurance status, the association of neighborhood SES with mortality risk was attenuated but remained statistically significant, suggesting an impact of SES beyond barriers related to being uninsured or underinsured. These findings are corroborated by a retrospective study using the National Cancer Database (NCDB), which demonstrated that uninsured patients (hazard ratio [HR]1.39, p<0.05) and Medicaid-insured patients (HR 1.48, p<0.05) had lower survival than patients with private insurance after adjusting for sociodemographic factors such as age, sex, race, ZIP code area, and level of education.102 These findings highlight the association of SES and mortality risk in DLBCL and suggest that barriers to optimal outcomes include but are not limited to inadequate insurance coverage among disadvantaged patients.
5.3. Place of residence
Moreover, place of residence affects the survival of lymphoma patients. Ritter et al. analyzed outcomes of 83,108 DLBCL patients in the NCDB who were classified as rural (county population less than 2,500), urban (county population more than 2,500 but outside metropolitan areas), or metro (at least 50,000 urbanized population in county).103 Rural and urban DLBCL patients were more likely than metro populations to have lower SES, Medicaid insurance, advanced stage at diagnosis, and more comorbidities. Rural and urban populations exhibited inferior 5-year OS compared to metro patients, although risk was attenuated by SES, insurance status, and treatment facility type. Neighborhood SES may affect health outcomes directly or indirectly through mechanisms such as availability and accessibility of healthcare, healthy foods, recreational facilities, environmental pollution, health literacy, and social support. It is therefore important to address neighborhood SES when developing strategies for rural/urban DLBCL patients.
5.4. Diagnosis-to-treatment interval
Recently, Maurer and colleagues analyzed 986 DLBCL patients included in the Molecular Epidemiology Resource (MER) database at the Mayo Clinic and the University of Iowa from 2002 to 2013. 104 They found that shorter diagnosis-to-treatment interval (DTI) was associated with adverse clinical factors and worse outcomes. The prognostic effect of DTI independent of IPI may indicate high-risk disease features that are not entirely captured by standard prognostic assessments such as age, disease stage, or performance status. This study is limited by its observational nature and lack of information regarding patient comorbidity and reason for treatment delay, and requires further validation. However, these findings may have important implications for clinicians and researchers designing and interpreting clinical trials in DLBCL.
5.5. Association between genomic alterations and ancestry
Extensive prior work has explored SES contributions to DLBCL disparities. However, our knowledge of the extent to which genetic mechanisms may contribute to the observed disparities has been limited. Analysis of genetic contributions to demographic disparities in DLBCL is challenging in part because the majority of genomic characterization studies primarily incorporate patients with European ancestry. This limited racial diversity precludes the detection of genomic patterns that are unique in underrepresented African American patients. Recently, Lee et al. utilized genetically-determined African ancestry rather than self-reported race to examine differences in mutational profiles of 150 DLBCL driver genes.105 Distinct prevalence and patterns of genomic alterations occurring in African Americans suggest involvement of different oncogenic genes and pathways in African American populations. These divergent patterns may constitute possible mechanisms that contribute to racial differences in disease incidence, patterns of presentation, and survival.
5.6. Merging sociodemographic and molecular factors in DLBCL outcomes research
With the identification of putative DLBCL driver mutations and pathways through whole-genome and -exome sequencing, novel molecular factors and sociodemographic factors should now be incorporated together in DLBCL prediction models. In an ongoing effort to define the interplay between clinical, epidemiologic, host genetic, tumor, and treatment factors in determining patient outcomes in lymphoma, the Lymphoma Epidemiology of Outcomes (LEO) Cohort Study was established in 2016 (; https://leocohort.org/). The LEO cohort is currently accruing at eight medical centers across the U.S., with a goal enrollment of 13,900 patients. This comprehensive prospective study catalogs clinical factors (e.g., body mass index and co-morbid diseases), epidemiologic factors (including race, lifestyle, and exposures), pathology (tumor bank and peripheral blood sample), and treatment data. Notably, quality-of-life scores as well as NHL molecular subtype are included—factors not available in SEER or any other large, prospective U.S. NHL cohort. With >9,000 NHL patients already enrolled, this unique study will enable examination of the interactions among a broad array of clinical and molecular factors and their impact on outcomes in DLBCL.
6. Measures of treatment effectiveness in DLBCL outcomes prediction
6.1. Interim PET/CT
While pre-treatment risk factors will continue to play an important role in prognostication, metrics associated with treatment response can provide valuable, dynamic information for predicting DLBCL outcomes. The current Lugano criteria for DLBCL staging rely on the use of [18F]-fluorodeoxyglucose (FDG)-positron emission tomography (PET)/computed tomography (CT) obtained prior to treatment. Assessment of pretreatment total metabolic tumor volume using [18F]-FDG PET/CT has been shown to predict OS in DLBCL patients,106 and baseline tumoral metabolic heterogeneity on PET/CT may be prognostic in B-cell lymphomas.107 The utility of PET/CT in restaging patients after completion of therapy is well established.108 Likewise, it is generally accepted that the use of PET/CT for routine surveillance after treatment does not provide clinical benefit and is currently not recommended.109,110 The use of interim PET/CT (i.e., at some time between cycles 2 and 4 of frontline therapy) is an attractive approach given the possibility to ‘adapt’ the treatment based on ongoing responses, with the goal of achieving improved response. Indeed, the use of this strategy has produced improved outcomes in Hodgkin lymphoma111-113 and is now considered standard of care in that disease.
Several studies have been performed to assess the prognostic and predictive value of an interim PET/CT in the management of DLBCL with varying outcomes,114-117 which may stem from differences in factors such as patient population, sample size, tumor heterogeneity, number of treatment cycles prior to re-imaging, and approach to subsequent therapy for interim PET-positive patients. A recent retrospective study from our group examining DLBCL patients treated with R-CHOP as first-line therapy showed that interim PET with full resolution of metabolic tumor was highly correlated with achieving complete remission of DLBCL by the end of treatment, as well as with improved survival.116 However, the degree of decrease from pre-treatment PET to interim PET did not relate to outcomes. Studies are underway to test novel compounds117,118 or the addition of laboratory measurements119 that could improve upon traditional FDG-PET/CT. Further studies addressing the use of interim imaging assessments in the context of tumor genetics may also help identify the patient population who may benefit most from a PET-adapted approach.
6.2. Cell-free DNA
The presence of extracellular nucleic acids in humans has been known for decades and has been detected in multiple types of bodily fluids.120 Most of these DNA fragments are short, although some can be >30 kb long, and are present at very low concentrations (in the ng/mL range) due to a short half-life in the circulation.120 In certain illnesses such as cancer, however, the amount of plasma cell-free DNA increases. This increase in cell-free DNA originates mostly from the tumor and is thus termed circulating tumor DNA (ctDNA). Despite the knowledge of the existence of cell-free DNA, it was not until recently that improvements in DNA sequencing technologies enabled accurate detection and sequencing of such small and infrequent fragments, allowing for the detection of mutations and epigenetic changes in a variety of applications.121-124
The use of ctDNA for tumor diagnosis, monitoring, and detection of relevant mutations is an attractive approach as it is minimally invasive, allows for serial sampling, and can be sensitive enough to detect subclinical disease. Armand and colleagues first showed that ctDNA can be detected in DLBCL patients with newly diagnosed disease, and became undetectable in patients after treatment, positing ctDNA as a useful biomarker for assessing treatment response.125 The use of ctDNA as a tool for treatment response assessment and post-remission surveillance was also illustrated in a recent restrospective analysis by Roschewski et al. They found that detection of VDJ segments of tumor immunoglobulin genes after 2 treatment cycles correlated with disease progression by 5 years, with presence of ctDNA detected a median of 3.5 months before clinical evidence of disease in individuals undergoing surveillance after therapy.126 Similarly, Kurtz and colleagues found prospectively that detection of molecular disease in the plasma preceded PET/CT detection of relapsed disease in DLBCL patients.127
The use of ctDNA also allows for the assessment of the entirety of a given tumor’s genetics, providing information about tumor heterogeneity and clonal evolution. This was recently illustrated by Rossi et al., who were able to detect many mutations in the plasma that were undetectable in the tissue biopsy, presumably due to spatial tumor heterogeneity. They also detected new mutations in ctDNA in treatment-resistant patients, potentially reflecting the mechanisms that confer resistance to treatment.128 In other applications, ctDNA can be used for genotyping, allowing for identification of COO, and to distinguish patterns of clonal evolution distinguishing transformed lymphomas from their indolent counterpart.129,130 In the era of precision medicine, the possibility to identify targetable mutations and monitor treatment response through minimally invasive methods could prove very useful in the design of clinical trials and identification of patients who would benefit most from targeted approaches. However, additional studies are needed to determine whether the routine use of ctDNA for surveillance is cost-effective and improves clinical outcomes.
7. Conclusions
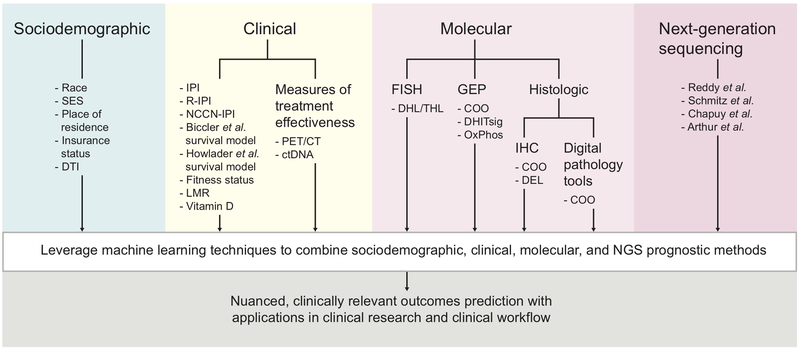
The clinical and genetic heterogeneity of DLBCL presents significant challenges for accurate outcomes prediction. Several non-overlapping DLBCL subgroup classifications including cell-of-origin subtype, double- and triple-hit status, and, more recently, genetic clusters, exhibit independent predictive and prognostic significance. Furthermore, technological advances are steadily ushering in various classification methods from research laboratories to DLBCL clinical trials and daily clinical workflow. In particular, recent advances in DLBCL genomics may have far-reaching ramifications for prognostic modeling and therapeutic decision-making in the molecular subtyping era. Ongoing hurdles for successful integration of subtyping methods into clinical trials and standard practice include demonstration of cost-effectiveness, inter-method and inter-laboratory concordance, and standard protocols that enable consistency across hospitals and clinics. Beyond molecular and histologic subtypes, sociodemographic and clinical decision-making factors such as racial and ethnic disparities, socioeconomic status, and diagnosis-to-treatment interval are known to play important roles in DLBCL outcomes. These factors should thus be considered in estimating patient prognosis and in designing future DLBCL clinical trials. Interim PET/CT and ctDNA present additional attractive options for predicting outcomes in DLBCL by allowing for longitudinal measures of therapeutic efficacy, but more research is needed prior to widespread adoption of these technologies for the development of dynamic, adaptive treatment strategies. Prognostic models in DLBCL are diverse and mirror the clinical and genetic heterogeneity of the disease through incorporation of clinical, histologic, and genetic factors. Nuanced, accurate outcomes prediction in DLBCL will likely leverage advances in machine learning techniques to combine clinical, sociodemographic, tumor microenvironment, and genetic factors in comprehensive but easy-to-use prognostic models (Figure 2).
Figure 2. Summary schematic for optimized integration of prognostic methods in DLBCL.
Abbreviations: COO, cell of origin; CT, computed tomography; ctDNA, circulating tumor DNA; DEL, double-expressor lymphoma; DHITsig, double-hit signature; DHL, double-hit lymphoma; DLBCL, diffuse large B-cell lymphoma; DTI, diagnosis-to-treatment interval; FISH, fluorescence in situ hybridization; GEP, gene expression profiling; IHC, immunohistochemistry; IPI, International Prognostic Index; LMR, lymphocyte/monocyte ratio; NCCN, National Comprehensive Cancer Network; NGS, next-generation sequencing; OxPhos, oxidative phosphorylation; PET, positron emission tomography; R-IPI, revised IPI; SES, socioeconomic status; THL, triple-hit lymphoma.
8. Expert opinion
The introduction of next-generation sequencing has facilitated discovery and characterization of oncologic genomic landscapes at a rapid pace and at relatively low costs.131 Genomic sequencing may elucidate mechanisms of lymphomagenesis by associating genetic aberrations with dysregulated molecular pathways and thus has the potential to generate novel therapeutic targets. Incorporation of next-generation sequencing into prognostic modeling and therapeutic decision-making in the management of DLBCL patients may allow definition of more precise disease subsets based on actionable mutation groups, which could be useful for reviving subtype-based therapies. Large genomic studies will be instrumental to define rational biomarkers for DLBCL therapy selection. These genomic data can emerge from various sources, including FFPE blocks at diagnosis and ctDNA at diagnosis, between treatment cycles, and at the end of treatment. Determining the optimal means for collecting and utilizing tumor genomic information should be addressed in future clinical trials.
After identifying predictive biomarkers and potential targets for precision medicine in DLBCL, there remain external considerations for the rigorous design of precision medicine clinical trials. Novel approaches to define cross-talk between molecular pathways via bioinformatics and computational methods may identify relevant genes or groups of genes that act as biomarkers for therapeutic response to targeted agents.132-135 In addition, future trials that incorporate DLBCL genomics will benefit from standardized sequencing methods and informatics pipelines across multi-center collaborations to optimize the reproducibility of findings.134 Importantly, DLBCL clinical trials using genomic biomarkers will require quick turnaround of DLBCL genomic analysis given the aggressive nature of the disease and the frequent need for urgent treatment.
Bridging clinical trial results that leverage DLBCL genomics with clinical standards of care will require collaborative relationships between diverse groups of professionals, including clinicians and bioinformaticians.136 Clinical integration of next-generation sequencing technologies will necessitate a systematic method for balancing clinical and biological prognostic factors when determining treatment strategies by subtype or patient.137 Furthermore, conversion of this “big data” into a streamlined, clinically relevant report that is integrated into the electronic medical record and clinical workflow will be essential in facilitating clinicians’ use of genomic data during medical decision-making.138 This becomes particularly relevant for patients in rapid need of therapy, since current research indicates that patients with the shortest DTI have adverse outcomes, and thus may have the greatest need for novel molecularly-targeted therapeutic approaches. Moreover, ensuring rapid and affordable access to genomic sequencing will be necessary for clinicians to confirm whether patients express biomarkers linked to specific treatment strategies in a cost-effective and efficient manner.139
Mounting evidence from studies examining disparities in DLBCL suggests that sociodemographic factors including race, insurance status, and rural status also play a significant role in lymphoma-related survival. Further examination of practice patterns and health outcomes in treatment of DLBCL may inform public policy to improve access to care for poor-risk populations. Situations where access to care is a key determinant of outcome may be further exacerbated by the advances in molecular technologies described above if these are inaccessible by certain patient groups. In addition to individual-level and neighborhood-level SES, analysis of the impact of living conditions and environmental exposures are needed to guide public health policy and preventive programs. At present, little is known about the community infrastructures (e.g., transportation, sick pay) necessary to reduce barriers to care for patients with DLBCL. Ultimately, a more thorough understanding of the interaction between biological, clinical, and socioeconomic factors that lead to inferior patient outcomes will help identify the most effective strategies to eliminate disparities and improve survival in DLBCL. In the next 5 years, we expect that advances in sequencing technology, robust population-level capture of multi-level clinical and sociodemographic factors, and informatics accessibility of these data sources will allow for widespread, real-time incorporation of complex genomic and patient-specific data into prognostic models, leading to targeted treatment algorithms used by lymphoma clinicians and patients for decision-making.
Article highlights:
The clinical and genetic heterogeneity of diffuse large B-cell lymphoma presents significant challenges for accurate outcomes prediction.
Several non-overlapping DLBCL subgroup classifications, including cell-of-origin subtype, “double-hit” rearrangements of MYC, BCL2, and/or BCL6, and newly defined genetic clusters, exhibit independent predictive and prognostic significance.
Technological advances in high-throughput sequencing are steadily ushering in various classification methods from research laboratories to DLBCL clinical trials and daily clinical workflow.
In the future, nuanced prediction of clinically relevant outcomes for patients with DLBCL will likely leverage advances in machine learning techniques to combine clinical, sociodemographic, tumor microenvironment, and genetic factors in comprehensive, multilevel prognostic models that are easily used by patients and providers.
Acknowledgments
Funding
This work was supported in part by the National Center for Advancing Translational Sciences of the National Institutes of Health under RA Harkin’s Award Numbers UL1TR002378 and TL1TR002382; the Winship Cancer Institute of Emory University Nell W. and William S. Elkin Fellowship to A Chang; A Chang’s National Institutes of Health National Cancer Institute(NCI) grant T32CA160040; and CR Flowers’ NCI Award K24CA208132.
Footnotes
Declaration of interests
CR Flowers has served as a consultant for: Abbvie, AstraZeneca, Bayer, BeiGene, Celgene (unpaid), Denovo Biopharma, Genentech/Roche (unpaid), Gilead, OptumRx, Karyopharm, MEI Pharmaceuticals, Pharmacyclics/ Janssen, Spectrum. CR Flower has also received research funding from: Abbvie, Acerta, BeiGene, Celgene, Gilead, Genentech/Roche, Janssen Pharmaceutical, Millennium/Takeda, Pharmacyclics, TG Therapeutics, Burroughs Wellcome Fund, Eastern Cooperative Oncology Group, National Cancer Institute, and the V Foundation. JL Koff has received research funding from the American Association for Cancer Research and the Lymphoma Research Foundation. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or conflict with the subject matter or materials discussed in this manuscript apart from those disclosed.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as either of interest (*) or of considerable interest (**) to readers.
- 1.Teras LR et al. 2016 US lymphoid malignancy statistics by World Health Organization subtypes. CA: a cancer journal for clinicians, doi: 10.3322/caac.21357 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Flowers CR, Sinha R & Vose JM Improving outcomes for patients with diffuse large B-cell lymphoma. CA: a cancer journal for clinicians 60, 393–408, doi: 10.3322/caac.20087 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Pasqualucci L & Dalla-Favera R Genetics of diffuse large B-cell lymphoma. Blood 131, 2307–2319, doi: 10.1182/blood-2017-11-764332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Non-Hodgkin's Lymphoma Prognostic Factors, P. A predictive model for aggressive non-Hodgkin's lymphoma. New England Journal of Medicine 329, 987–994, doi: 10.1056/NEJM199309303291402 (1993). [DOI] [PubMed] [Google Scholar]
- **5.Alizadeh AA et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511, doi: 10.1038/35000501 (2000).Seminal work defining cell-of-origin subtypes of DLBCL by gene expression profiling.
- 6.Arthur SE et al. Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nature communications 9, 4001, doi: 10.1038/s41467-018-06354-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- *7.Schmitz R et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N Engl J Med 378, 1396–1407, doi: 10.1056/NEJMoa1801445 (2018).Important next-generation sequencing work that defined novel subtypes of DLBCL beyond cell-of-origin.
- *8.Chapuy B et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nature medicine 24, 679–690, doi: 10.1038/s41591-018-0016-8 (2018).Important next-generation sequencing work that defined novel subtypes of DLBCL beyond cell-of-origin.
- **9.Reddy A et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 171, 481–494.e415, doi: 10.1016/j.cell.2017.09.027 (2017).Important next-generation sequencing work that combined sequencing, clinical, and histologic data to generate a novel prognostic algorithm for DLBCL.
- 10.Sehn LH et al. The revised International Prognostic Index (R-IPI) is a better predictor of outcome than the standard IPI for patients with diffuse large B-cell lymphoma treated with R-CHOP. Blood 109, 1857–1861, doi: 10.1182/blood-2006-08-038257 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Vaidya R & Witzig TE Prognostic factors for diffuse large B-cell lymphoma in the R(X)CHOP era. Annals of oncology : official journal of the European Society for Medical Oncology 25, 2124–2133, doi: 10.1093/annonc/mdu109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Z et al. An enhanced International Prognostic Index (NCCN-IPI) for patients with diffuse large B-cell lymphoma treated in the rituximab era. Blood 123, 837–842, doi: 10.1182/blood-2013-09-524108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gang AO et al. A clinically based prognostic index for diffuse large B-cell lymphoma with a cut-off at 70 years of age significantly improves prognostic stratification: population-based analysis from the Danish Lymphoma Registry. Leukemia & lymphoma 56, 2556–2562, doi: 10.3109/10428194.2015.1010078 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi T et al. The Kyoto Prognostic Index for patients with diffuse large B-cell lymphoma in the rituximab era. Blood Cancer Journal 6, e383–e383, doi: 10.1038/bcj.2015.111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruppert AS et al. Comparison of clinical scoring systems in aggressive B-cell lymphomas (BCL): An individual patient-level analysis across international trials (SEAL). Journal of Clinical Oncology 37, 7544–7544, doi: 10.1200/JCO.2019.37.15_suppl.7544 (2019). [DOI] [Google Scholar]
- 16.Biccler JL et al. Optimizing Outcome Prediction in Diffuse Large B-Cell Lymphoma by Use of Machine Learning and Nationwide Lymphoma Registries: A Nordic Lymphoma Group Study. JCO Clinical Cancer Informatics, 1–13, doi: 10.1200/CCI.18.00025 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Howlader N et al. Cancer-specific mortality, cure fraction, and noncancer causes of death among diffuse large B-cell lymphoma patients in the immunochemotherapy era: Population-Based Outcomes in DLBCL. Cancer 123, 3326–3334, doi: 10.1002/cncr.30739 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Chen Q et al. Population-specific prognostic models are needed to stratify outcomes for African-Americans with diffuse large B-cell lymphoma. Leukemia & lymphoma 57, 842–851, doi: 10.3109/10428194.2015.1083098 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin RJ, Behera M, Diefenbach CS & Flowers CR Role of anthracycline and comprehensive geriatric assessment for elderly patients with diffuse large B-cell lymphoma. Blood 130, 2180–2185, doi: 10.1182/blood-2017-05-736975 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrison VA et al. Approach to therapy of diffuse large B-cell lymphoma in the elderly: the International Society of Geriatric Oncology (SIOG) expert position commentary. Annals of oncology : official journal of the European Society for Medical Oncology 26, 1058–1068, doi: 10.1093/annonc/mdv018 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Spina M et al. Modulated chemotherapy according to modified comprehensive geriatric assessment in 100 consecutive elderly patients with diffuse large B-cell lymphoma. The oncologist 17, 838–846, doi: 10.1634/theoncologist.2011-0417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olivieri A et al. Tailored therapy in an unselected population of 91 elderly patients with DLBCL prospectively evaluated using a simplified CGA. The oncologist 17, 663–672, doi: 10.1634/theoncologist.2011-0355 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tucci A et al. A comprehensive geriatric assessment is more effective than clinical judgment to identify elderly diffuse large cell lymphoma patients who benefit from aggressive therapy. Cancer 115, 4547–4553, doi: 10.1002/cncr.24490 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Tucci A et al. Comprehensive geriatric assessment is an essential tool to support treatment decisions in elderly patients with diffuse large B-cell lymphoma: a prospective multicenter evaluation in 173 patients by the Lymphoma Italian Foundation (FIL). Leukemia & lymphoma 56, 921–926, doi: 10.3109/10428194.2014.953142 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Mohile SG et al. Practical Assessment and Management of Vulnerabilities in Older Patients Receiving Chemotherapy: ASCO Guideline for Geriatric Oncology. Journal of Clinical Oncology 36, 2326–2347, doi: 10.1200/jco.2018.78.8687 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrison VA et al. Diffuse large B-cell lymphoma in the elderly: impact of prognosis, comorbidities, geriatric assessment, and supportive care on clinical practice. An International Society of Geriatric Oncology (SIOG) expert position paper. Journal of geriatric oncology 6, 141–152, doi: 10.1016/j.jgo.2014.11.004 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Merli F et al. Cyclophosphamide, doxorubicin, vincristine, prednisone and rituximab versus epirubicin, cyclophosphamide, vinblastine, prednisone and rituximab for the initial treatment of elderly “fit” patients with diffuse large B-cell lymphoma: results from the ANZINTER3 trial of the Intergruppo Italiano Linfomi. Leukemia & lymphoma 53, 581–588, doi: 10.3109/10428194.2011.621565 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Merli F et al. Outcome of frail elderly patients with diffuse large B-cell lymphoma prospectively identified by Comprehensive Geriatric Assessment: results from a study of the Fondazione Italiana Linfomi. Leukemia & lymphoma 55, 38–43, doi: 10.3109/10428194.2013.788176 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Fama A et al. Prevalence and clinical correlates of vulnerable status using the Vulnerable Elders Survey 13 (VES-13) in newly diagnosed adult non-Hodgkin lymphoma (NHL) patients: A LEO cross-sectional analysis. Journal of Clinical Oncology 36, 10042–10042, doi: 10.1200/JCO.2018.36.15_suppl.10042 (2018). [DOI] [Google Scholar]
- 30.Wang J et al. Lymphocyte-to-monocyte ratio is associated with prognosis of diffuse large B-cell lymphoma: correlation with CD163 positive M2 type tumor-associated macrophages, not PD-1 positive tumor-infiltrating lymphocytes. Oncotarget 8, 5414–5425, doi: 10.18632/oncotarget.14289 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilcox RA et al. The absolute monocyte and lymphocyte prognostic score predicts survival and identifies high-risk patients in diffuse large-B-cell lymphoma. Leukemia 25, 1502–1509, doi: 10.1038/leu.2011.112 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Xia WK et al. Prognostic significance of lymphocyte-to-monocyte ratio in diffuse large B-cell lymphoma: a systematic review and meta-analysis. FEBS Open Bio 6, 558–565, doi: 10.1002/2211-5463.12066 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porrata LF et al. Absolute monocyte/lymphocyte count prognostic score is independent of immunohistochemically determined cell of origin in predicting survival in diffuse large B-cell lymphoma. Leukemia & lymphoma 53, 2159–2165, doi: 10.3109/10428194.2012.690605 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Danish M, Sissung T, Price DK & Figg WD Genetic stability of tumor microenvironment. Cancer Biol Ther 7, 331–332 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Thorsson V et al. The Immune Landscape of Cancer. Immunity 48, 812–830 e814, doi: 10.1016/j.immuni.2018.03.023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wellenstein MD & de Visser KE Cancer-Cell-Intrinsic Mechanisms Shaping the Tumor Immune Landscape. Immunity 48, 399–416, doi: 10.1016/j.immuni.2018.03.004 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Drake MT et al. Vitamin D insufficiency and prognosis in non-Hodgkin's lymphoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 28, 4191–4198, doi: 10.1200/jco.2010.28.6674 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bittenbring JT et al. Vitamin D deficiency impairs rituximab-mediated cellular cytotoxicity and outcome of patients with diffuse large B-cell lymphoma treated with but not without rituximab. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 32, 3242–3248, doi: 10.1200/jco.2013.53.4537 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Bruns H et al. Vitamin D-dependent induction of cathelicidin in human macrophages results in cytotoxicity against high-grade B cell lymphoma. Sci Transl Med 7, 282ra247, doi: 10.1126/scitranslmed.aaa3230 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Hohaus S et al. Vitamin D deficiency and supplementation in patients with aggressive B-cell lymphomas treated with immunochemotherapy. Cancer medicine 7, 270–281, doi: 10.1002/cam4.1166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenwald A et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. The New England journal of medicine 346, 1937–1947 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Lenz G et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med 359, 2313–2323, doi: 10.1056/NEJMoa0802885 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rutherford SC & Leonard JP DLBCL Cell of Origin: What Role Should It Play in Care Today? Oncology (Williston Park, N.Y.) 32, 445–449 (2018). [PubMed] [Google Scholar]
- 44.Hans CP et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 103, 275–282, doi: 10.1182/blood-2003-05-1545 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Muris JJ et al. Immunohistochemical profiling based on Bcl-2, CD10 and MUM1 expression improves risk stratification in patients with primary nodal diffuse large B cell lymphoma. The Journal of pathology 208, 714–723, doi: 10.1002/path.1924 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Natkunam Y et al. LMO2 protein expression predicts survival in patients with diffuse large B-cell lymphoma treated with anthracycline-based chemotherapy with and without rituximab. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 26, 447–454, doi: 10.1200/jco.2007.13.0690 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Nyman H, Jerkeman M, Karjalainen-Lindsberg ML, Banham AH & Leppa S Prognostic impact of activated B-cell focused classification in diffuse large B-cell lymphoma patients treated with R-CHOP. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 22, 1094–1101, doi: 10.1038/modpathol.2009.73 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Choi WW et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clinical cancer research : an official journal of the American Association for Cancer Research 15, 5494–5502, doi: 10.1158/1078-0432.Ccr-09-0113 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyer PN et al. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 29, 200–207, doi: 10.1200/jco.2010.30.0368 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Visco C et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia 26, 2103–2113, doi: 10.1038/leu.2012.83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coutinho R et al. Poor concordance among nine immunohistochemistry classifiers of cell-of-origin for diffuse large B-cell lymphoma: implications for therapeutic strategies. Clinical cancer research : an official journal of the American Association for Cancer Research 19, 6686–6695, doi: 10.1158/1078-0432.Ccr-13-1482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Read JA et al. Evaluating cell-of-origin subtype methods for predicting diffuse large B-cell lymphoma survival: a meta-analysis of gene expression profiling and immunohistochemistry algorithms. Clinical lymphoma, myeloma & leukemia 14, 460–467.e462, doi: 10.1016/j.clml.2014.05.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- *53.Jordan J et al. Informatics Approaches to Address New Challenges in the Classification of Lymphoid Malignancies. JCO Clinical Cancer Informatics, 1–9, doi: 10.1200/cci.17.00039 (2018).Recent review of how informatics approaches can aid in classifying lymphomas.
- 54.Mobadersany P et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proceedings of the National Academy of Sciences 115, E2970–E2979, doi: 10.1073/pnas.1717139115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chabot-Richards DS, Martin DR, Myers OB, Czuchlewski DR & Hunt KE Quantitative image analysis in the assessment of diffuse large B-cell lymphoma. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 24, 1598–1605, doi: 10.1038/modpathol.2011.123 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gutierrez-Garcia G et al. Gene-expression profiling and not immunophenotypic algorithms predicts prognosis in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Blood 117, 4836–4843, doi: 10.1182/blood-2010-12-322362 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Scott DW Cell-of-Origin in Diffuse Large B-Cell Lymphoma: Are the Assays Ready for the Clinic? American Society of Clinical Oncology educational book. American Society of Clinical Oncology. Annual Meeting, e458–466, doi: 10.14694/EdBook_AM.2015.35.e458 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Williams PM et al. A novel method of amplification of FFPET-derived RNA enables accurate disease classification with microarrays. J Mol Diagn 12, 680–686, doi: 10.2353/jmoldx.2010.090164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scott DW et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood 123, 1214–1217, doi: 10.1182/blood-2013-11-536433 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scott DW et al. Prognostic Significance of Diffuse Large B-Cell Lymphoma Cell of Origin Determined by Digital Gene Expression in Formalin-Fixed Paraffin-Embedded Tissue Biopsies. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 33, 2848–2856, doi: 10.1200/JCO.2014.60.2383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chandler Y et al. Cost Effectiveness of Gene Expression Profile Testing in Community Practice. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 36, 554–562, doi: 10.1200/jco.2017.74.5034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swerdlow SH et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 127, 2375–2390, doi: 10.1182/blood-2016-01-643569 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robetorye R et al. Incorporation of digital gene expression profiling for cell-of-origin determination (Lymph2Cx testing) into the routine work-up of diffuse large B cell lymphoma. Vol. 12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barrans SL et al. Whole genome expression profiling based on paraffin embedded tissue can be used to classify diffuse large B-cell lymphoma and predict clinical outcome. British journal of haematology 159, 441–453, doi: 10.1111/bjh.12045 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Monti S et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood 105, 1851–1861, doi: 10.1182/blood-2004-07-2947 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Caro P et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer cell 22, 547–560, doi: 10.1016/j.ccr.2012.08.014 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Norberg E et al. Differential contribution of the mitochondrial translation pathway to the survival of diffuse large B-cell lymphoma subsets. Cell death and differentiation 24, 251–262, doi: 10.1038/cdd.2016.116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen L et al. SYK-dependent tonic B-cell receptor signaling is a rational treatment target in diffuse large B-cell lymphoma. Blood 111, 2230–2237, doi: 10.1182/blood-2007-07-100115 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu Z, Yan Y, Li Z, Qian L & Gong Z The Antibiotic Drug Tigecycline: A Focus on its Promising Anticancer Properties. Frontiers in pharmacology 7, 473, doi: 10.3389/fphar.2016.00473 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ravà M et al. Therapeutic synergy between tigecycline and venetoclax in a preclinical model of MYC/BCL2 double-hit B cell lymphoma. Science Translational Medicine 10, eaan8723, doi: 10.1126/scitranslmed.aan8723 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Munoz-Marmol AM et al. MYC status determination in aggressive B-cell lymphoma: the impact of FISH probe selection. Histopathology 63, 418–424, doi: 10.1111/his.12178 (2013). [DOI] [PubMed] [Google Scholar]
- 72.Swerdlow SH Diagnosis of 'double hit' diffuse large B-cell lymphoma and B-cell lymphoma, unclassifiable, with features intermediate between DLBCL and Burkitt lymphoma: when and how, FISH versus IHC. Hematology. American Society of Hematology. Education Program 2014, 90–99, doi: 10.1182/asheducation-2014.1.90 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Ennishi D et al. Double-Hit Gene Expression Signature Defines a Distinct Subgroup of Germinal Center B-Cell-Like Diffuse Large B-Cell Lymphoma. Journal of Clinical Oncology 37, 190–201, doi: 10.1200/jco.18.01583 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barrans S et al. Rearrangement of MYC is associated with poor prognosis in patients with diffuse large B-cell lymphoma treated in the era of rituximab. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 28, 3360–3365, doi: 10.1200/jco.2009.26.3947 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Savage KJ et al. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood 114, 3533–3537, doi: 10.1182/blood-2009-05-220095 (2009). [DOI] [PubMed] [Google Scholar]
- 76.Iqbal J et al. BCL2 expression is a prognostic marker for the activated B-cell-like type of diffuse large B-cell lymphoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 24, 961–968, doi: 10.1200/jco.2005.03.4264 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Iqbal J et al. BCL2 predicts survival in germinal center B-cell-like diffuse large B-cell lymphoma treated with CHOP-like therapy and rituximab. Clinical cancer research : an official journal of the American Association for Cancer Research 17, 7785–7795, doi: 10.1158/1078-0432.Ccr-11-0267 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Johnson NA et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 30, 3452–3459, doi: 10.1200/jco.2011.41.0985 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Green TM et al. Immunohistochemical double-hit score is a strong predictor of outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 30, 3460–3467, doi: 10.1200/jco.2011.41.4342 (2012). [DOI] [PubMed] [Google Scholar]
- 80.de Jong D et al. Immunohistochemical prognostic markers in diffuse large B-cell lymphoma: validation of tissue microarray as a prerequisite for broad clinical applications--a study from the Lunenburg Lymphoma Biomarker Consortium. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 25, 805–812, doi: 10.1200/jco.2006.09.4490 (2007). [DOI] [PubMed] [Google Scholar]
- 81.Tsuyama N et al. BCL2 expression in DLBCL: reappraisal of immunohistochemistry with new criteria for therapeutic biomarker evaluation. Blood 130, 489–500, doi: 10.1182/blood-2016-12-759621 (2017). [DOI] [PubMed] [Google Scholar]
- 82.Mahmoud AZ et al. Scoring of MYC protein expression in diffuse large B-cell lymphomas: concordance rate among hematopathologists. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 28, 545–551, doi: 10.1038/modpathol.2014.140 (2015). [DOI] [PubMed] [Google Scholar]
- 83.Pasqualucci L The genetic basis of diffuse large B-cell lymphoma. Curr Opin Hematol 20, 336–344, doi: 10.1097/MOH.0b013e3283623d7f (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fang B, Mehran RJ, Heymach JV & Swisher SG Predictive biomarkers in precision medicine and drug development against lung cancer. Chin J Cancer 34, 295–309, doi: 10.1186/s40880-015-0028-4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kato S, Subbiah V & Kurzrock R Counterpoint: Successes in the Pursuit of Precision Medicine: Biomarkers Take Credit. J Natl Compr Canc Netw 15, 863–866, doi: 10.6004/jnccn.2017.0127 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ribrag V, Soria J, Michot J, Schmitt A, Postel-Vinay S, Bijou F, Thomson B, Keilhack H, Blakemore SJ, Reyderman L, Kumar P, Fine G, McDonald A, Ho PT , Italiano A Phase 1 Study of Tazemetostat (EPZ-6438), an Inhibitor of Enhancer of Zeste-Homolog 2 (EZH2): Preliminary Safety and Activity in Relapsed or Refractory Non-Hodgkin Lymphoma (NHL) Patients. Blood 126, 473 (2015). [Google Scholar]
- 87.Zenner HP Individual Biomarkers Using Molecular Personalized Medicine Approaches. ORL J Otorhinolaryngol Relat Spec 79, 7–13, doi: 10.1159/000455811 (2017). [DOI] [PubMed] [Google Scholar]
- 88.Patel SP, Harkins RA, Lee MJ & Flowers CR Using Informatics Tools to Characterize Precision Medicine Treatments for Diffuse Large B-Cell Lymphoma (DLBCL). Blood 132, doi: 10.1182/blood-2018-99-119917 (2018). [DOI] [Google Scholar]
- 89.Cruz JA & Wishart DS Applications of Machine Learning in Cancer Prediction and Prognosis. Cancer Inform 2, 117693510600200–117693510600219, doi: 10.1177/117693510600200030 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV & Fotiadis DI Machine learning applications in cancer prognosis and prediction. CSBJ 13, 8–17, doi: 10.1016/j.csbj.2014.11.005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guyon I & Elisseeff A An introduction to variable and feature selection. JMLR 3, 1157–1182 (2003). [Google Scholar]
- 92.Hira ZM & Gillies DF A review of feature selection and feature extraction methods applied on microarray data. Advances in Bioinformatics 2015, 1–13, doi: 10.1155/2015/198363 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saeys Y, Inza I & Larranaga P A review of feature selection techniques in bioinformatics. Bioinformatics (Oxford, England) 23, 2507–2517, doi: 10.1093/bioinformatics/btm344 (2007). [DOI] [PubMed] [Google Scholar]
- 94.Witten DM & Tibshirani R Survival analysis with high-dimensional covariates. Stat Methods Med Res 19, 29–51, doi: 10.1177/0962280209105024 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bair E & Tibshirani R Semi-supervised methods to predict patient survival from gene expression data. PLoS biology 2, e108 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bair E, Hastie T, Paul D & Tibshirani R Prediction by supervised principal components. Journal of the American Statistical Association 101, 119–137 (2006). [Google Scholar]
- 97.Shipp MA et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nature medicine 8, 68–74, doi: 10.1038/nm0102-68 (2002). [DOI] [PubMed] [Google Scholar]
- 98.Kurtz DM et al. Dynamic Risk Profiling Using Serial Tumor Biomarkers for Personalized Outcome Prediction. Cell 178, 699–713.e619, doi: 10.1016/j.cell.2019.06.011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yousefi S et al. Predicting clinical outcomes from large scale cancer genomic profiles with deep survival models. Scientific reports 7, doi: 10.1038/s41598-017-11817-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shenoy PJ et al. Racial differences in the presentation and outcomes of diffuse large B-cell lymphoma in the United States. Cancer 117, 2530–2540, doi: 10.1002/cncr.25765 (2011). [DOI] [PubMed] [Google Scholar]
- 101.Tao L, Foran JM, Clarke CA, Gomez SL & Keegan TH Socioeconomic disparities in mortality after diffuse large B-cell lymphoma in the modern treatment era. Blood 123, 3553–3562, doi: 10.1182/blood-2013-07-517110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Han X et al. Insurance status is related to diffuse large B-cell lymphoma survival. Cancer 120, 1220–1227, doi: 10.1002/cncr.28549 (2014). [DOI] [PubMed] [Google Scholar]
- 103.Ritter AJ, Goldstein JS, Ayers AA & Flowers CR Rural and urban patients with diffuse large B-cell and follicular lymphoma experience reduced overall survival: a National Cancer DataBase study. Leukemia & lymphoma, 1–12, doi: 10.1080/10428194.2018.1546855 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maurer MJ et al. Diagnosis-to-Treatment Interval Is an Important Clinical Factor in Newly Diagnosed Diffuse Large B-Cell Lymphoma and Has Implication for Bias in Clinical Trials. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 36, 1603–1610, doi: 10.1200/jco.2017.76.5198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee MJ et al. Genome-Defined African Ancestry is Associated with Distinct Mutations and Survival in Diffuse Large B-Cell Lymphoma (2019). [DOI] [PMC free article] [PubMed]
- 106.Sasanelli M et al. Pretherapy metabolic tumour volume is an independent predictor of outcome in patients with diffuse large B-cell lymphoma. European journal of nuclear medicine and molecular imaging 41, 2017–2022, doi: 10.1007/s00259-014-2822-7 (2014). [DOI] [PubMed] [Google Scholar]
- 107.Ceriani L et al. Metabolic heterogeneity on baseline 18FDG-PET/CT scan is a predictor of outcome in primary mediastinal B-cell lymphoma. Blood 132, 179–186, doi: 10.1182/blood-2018-01-826958 (2018). [DOI] [PubMed] [Google Scholar]
- 108.Cheson BD Role of functional imaging in the management of lymphoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 29, 1844–1854, doi: 10.1200/jco.2010.32.5225 (2011). [DOI] [PubMed] [Google Scholar]
- 109.Cohen JB, Kurtz DM, Staton AD & Flowers CR Next-generation surveillance strategies for patients with lymphoma. Future oncology (London, England) 11, 1977–1991, doi: 10.2217/fon.15.92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cohen JB, Behera M, Thompson CA & Flowers CR Evaluating surveillance imaging for diffuse large B-cell lymphoma and Hodgkin lymphoma. Blood 129, 561–564, doi: 10.1182/blood-2016-08-685073 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Radford J et al. Results of a trial of PET-directed therapy for early-stage Hodgkin's lymphoma. N Engl J Med 372, 1598–1607, doi: 10.1056/NEJMoa1408648 (2015). [DOI] [PubMed] [Google Scholar]
- 112.Press OW et al. US Intergroup Trial of Response-Adapted Therapy for Stage III to IV Hodgkin Lymphoma Using Early Interim Fluorodeoxyglucose-Positron Emission Tomography Imaging: Southwest Oncology Group S0816. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 34, 2020–2027, doi: 10.1200/jco.2015.63.1119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Johnson P et al. Adapted Treatment Guided by Interim PET-CT Scan in Advanced Hodgkin’s Lymphoma. New England Journal of Medicine 374, 2419–2429, doi: 10.1056/NEJMoa1510093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Adams HJ & Kwee TC Prognostic value of interim FDG-PET in R-CHOP-treated diffuse large B-cell lymphoma: Systematic review and meta-analysis. Critical reviews in oncology/hematology 106, 55–63, doi: 10.1016/j.critrevonc.2016.07.003 (2016). [DOI] [PubMed] [Google Scholar]
- 115.Burggraaff CN et al. Predictive value of interim positron emission tomography in diffuse large B-cell lymphoma: a systematic review and meta-analysis. European journal of nuclear medicine and molecular imaging 46, 65–79, doi: 10.1007/s00259-018-4103-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Islam P, Goldstein J & Flowers CR PET-derived tumor metrics predict DLBCL response and progression-free survival. Leukemia & lymphoma, 1–7, doi: 10.1080/10428194.2018.1562181 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang R et al. Prognostic value of interim fluorodeoxyglucose and fluorothymidine PET/CT in diffuse large B-cell lymphoma. The British journal of radiology 91, 20180240, doi: 10.1259/bjr.20180240 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ahn SY et al. Prognostic Significance of Interim 11C-Methionine PET/CT in Primary Central Nervous System Lymphoma. Clinical nuclear medicine 43, e259–e264, doi: 10.1097/rlu.0000000000002154 (2018). [DOI] [PubMed] [Google Scholar]
- 119.Marcheselli R et al. The prognostic role of end of treatment FDG-PET-CT in patients with diffuse large B cell lymphoma can be improved by considering it with absolute monocyte count at diagnosis. Leukemia & lymphoma, 1–7, doi: 10.1080/10428194.2018.1564049 (2019). [DOI] [PubMed] [Google Scholar]
- 120.Fleischhacker M & Schmidt B Circulating nucleic acids (CNAs) and cancer--a survey. Biochimica et biophysica acta 1775, 181–232, doi: 10.1016/j.bbcan.2006.10.001 (2007). [DOI] [PubMed] [Google Scholar]
- 121.Snyder MW, Kircher M, Hill AJ, Daza RM & Shendure J Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell 164, 57–68, doi: 10.1016/j.cell.2015.11.050 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lo YM et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med 2, 61ra91, doi: 10.1126/scitranslmed.3001720 (2010). [DOI] [PubMed] [Google Scholar]
- 123.Phallen J et al. Direct detection of early-stage cancers using circulating tumor DNA. Science Translational Medicine 9, eaan2415, doi: 10.1126/scitranslmed.aan2415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.De Rubis G, Rajeev Krishnan S & Bebawy M Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends in pharmacological sciences 40, 172–186, doi: 10.1016/j.tips.2019.01.006 (2019). [DOI] [PubMed] [Google Scholar]
- 125.Armand P et al. Detection of circulating tumour DNA in patients with aggressive B-cell non-Hodgkin lymphoma. British journal of haematology 163, 123–126, doi: 10.1111/bjh.12439 (2013). [DOI] [PubMed] [Google Scholar]
- 126.Roschewski M et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. The Lancet. Oncology 16, 541–549, doi: 10.1016/s1470-2045(15)70106-3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kurtz DM et al. Noninvasive monitoring of diffuse large B-cell lymphoma by immunoglobulin high-throughput sequencing. Blood 125, 3679–3687, doi: 10.1182/blood-2015-03-635169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rossi D et al. Diffuse large B-cell lymphoma genotyping on the liquid biopsy. Blood 129, 1947–1957, doi: 10.1182/blood-2016-05-719641 (2017). [DOI] [PubMed] [Google Scholar]
- 129.Scherer F et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med 8, 364ra155, doi: 10.1126/scitranslmed.aai8545 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sun P et al. Mutation Profiling of Malignant Lymphoma by Next-Generation Sequencing of Circulating Cell-Free DNA. Journal of Cancer 10, 323–331, doi: 10.7150/jca.27615 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Siu LL, Conley BA, Boerner S & LoRusso PM Next-Generation Sequencing to Guide Clinical Trials. Clinical cancer research : an official journal of the American Association for Cancer Research 21, 4536–4544, doi: 10.1158/1078-0432.CCR-14-3215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Johnson PWM in Plenary Session 1: Are We Ready for Lymphoma MATCH Trials? https://webcast.aacr.org/s/2018lym/PL2001;jsessionid=2019A2014C2005B2787ED1034B2060F428371A423957B (American Association for Cancer Research, 2018). [Google Scholar]
- 133.Staudt LM in Plenary Session 1: Are We Ready for Lymphoma MATCH Trials? https://webcast.aacr.org/s/2018lym/PL2001;jsessionid=2019A2014C2005B2787ED1034B2060F428371A423957B (American Association for Cancer Research, 2018). [Google Scholar]
- 134.Vermaat JS et al. Precision medicine in diffuse large B-cell lymphoma: hitting the target. Haematologica 100, 989–993, doi: 10.3324/haematol.2015.128371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Younes A in Plenary Session 1: Are We Ready for Lymphoma MATCH Trials? https://webcast.aacr.org/s/2018lym/PL2001;jsessionid=2019A2014C2005B2787ED1034B2060F428371A423957B (American Association for Cancer Research, 2018). [Google Scholar]
- 136.Bowdin S et al. Recommendations for the integration of genomics into clinical practice. Genetics in medicine : official journal of the American College of Medical Genetics 18, 1075–1084, doi: 10.1038/gim.2016.17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.He J et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 127, 3004–3014, doi: 10.1182/blood-2015-08-664649 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Warner JL, Jain SK & Levy MA Integrating cancer genomic data into electronic health records. Genome Med 8, 113, doi: 10.1186/s13073-016-0371-3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Amin AD et al. Diffuse large B-cell lymphoma: can genomics improve treatment options for a curable cancer? Cold Spring Harb Mol Case Stud 3, a001719, doi: 10.1101/mcs.a001719 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]