β-Hemoglobinopathies result from mutations in the β-globin (HBB) gene.1 Whereas causative mutations may be corrected by precise gene correction based on homology-directed repair, imprecise disruption of genome elements by non-homologous end joining is inherently more efficient and more suitable for long-term repopulating cells.2 This has already prompted the pursuit of disruption-based reactivation of the HBB paralog γ-globin as a potentially universal genome-editing strategy to treat patients with β-hemoglobinopathies,3 which is as yet unproven in the clinic. The common β-thalassemia allele IVSI-110 (HBBIVS-110(G>A)) has an aberrant splice acceptor site that leads to abnormal splicing.4 Here we investigated the use of a mutation-specific and disruption-based approach to correct HBBIVS-110(G>A). Based on both transcription activator-like effector nucleases (TALEN) and CRISPR/Cas9 RNA-guided HBBIVS-110(G>A)-targeting nucleases we analyzed non-homologous end joining-based indel events at on-and off-target sites, and the efficiency of functional correction in patient-derived CD34+-derived HBBIVS-110(G>A)-homozygous erythroblasts. Both platforms showed significant correction at the RNA, protein and morphological levels, with up to 95% on-target disruption, using a design that minimized δ-globin (HBD) off-target activity. The present study establishes suitable target sequences for effective restoration of normal splicing and validates gene disruption by virus-and DNA-free delivery of nucleases as potential therapy for HBBIVS-110(G>A) thalassemia.
The HBBIVS-110(G>A) mutation resides 19 nucleotides upstream of the normal intron-1 splice acceptor site. We identified one CRISPR/Cas9 and two TALEN-pair target sites compatible with platform-specific sequence constraints, proximity of exon 2, and the need to discern HBB from HBD for therapy by disruption (Figure 1, Online Supplementary Figure S1). Predicted double-stranded break sites were adjacent to the aberrant splice acceptor site for the RNA-guided nuclease (RGN) and upstream for TALEN pairs, TALEN R1/L1 (R1/L1) and TALEN R1/L2 (R1/L2), and had the potential to render the aberrant splice acceptor site non-functional and promote normal splicing. All nucleases, including three alternative TALEN R monomers with specificity-enhancing repeat-variable-diresidue (RVD) substitutions for combination with L1 and L2 monomers (Online Supplementary Figure S2A),5 gave significant disruption for an episomal HBBIVSI-110(G>A)-green fluorescent protein (GFP) reporter construct in HEK 293T cells (Online Supplementary Figure S2B–F), but only R1/L1 and R1/L2 reduced GFP fluorescence almost to background levels. R1/L1, R1/L2 and the RGN were then selected for evaluation in patient-derived HBBIVS-110(G>A)-homozygous CD34+ cells. This would allow the assessment of translatable nuclease delivery, off-targeting, endogenous HBB expression and phenotypic correction after erythroid differentiation in therapeutically relevant cells.
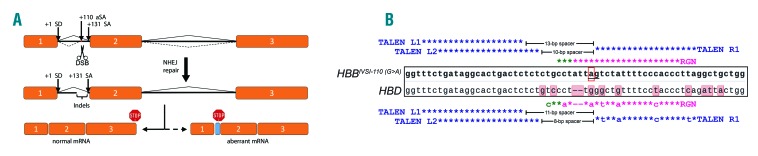
Figure 1.
Principle of non-homologous end joining-based functional correction of HBBIVSI-110(G>A). (A) Diagram illustrating the HBBIVSI-110(G>A) gene structure (exons as orange boxes) with normal (solid lines) and aberrant (hashed lines) splice events, and the effect of disruption on HBB mRNA splicing. (B) Alignment of HBBIVSI-110(G>A) and HBD sequences (central, boxed sequences with shaded letters indicating mismatches for HBD) and corresponding nuclease recognition sites (colored letters and asterisks). Three TALEN monomers (L1, L2 and R1) were employed as two differentially spaced active dimers, R1/L1 and R1/L2, to induce double-stranded breaks upstream of the HBBIVSI-110(G>A) mutation (red box). The CRISPR/Cas9 guide RNA binding sequence encompasses the mutation close to its protospacer-adjacent motif (PAM), creating double-stranded breaks immediately adjacent to the +110 aberrant splice acceptor site (ag). Target-dependent TALEN spacer lengths for HBB and HBD targets are indicated; spacer length below 10 bp impairs TALEN-mediated disruption. asterisks–perfect complementarity; colored letters: mismatches; blue–TALEN target sequence; purple–RGN target sequence; green–PAM sequence. aSA: aberrant splice acceptor site; SA: splice acceptor site; NHEJ: non-homologous end joining; TALEN: transcription activator-like effector nucleases; RGN: RNA-guided nuclease.
We nucleofected expanded primary CD34+ cells,6 either with pre-assembled RNP complexes for the RGN, or with in vitro synthesized mRNA for TALEN pairs and for the GFP transfection control, reaching transfer efficiencies greater than 90% (98.4±1.0% GFP+ cells) and a viability of approximately 95% (Online Supplementary Figure S3). Initial experiments investigated potential HBD off-target activity, reported as problematic elsewhere.7 The number and position of RGN mismatches with HBD (Figure 1B) would predictably prevent any HBD cleavage,8 restricting these analyses to L1/R1 and R1/L2 (Online Supplementary Figure S4). Measurement of HBB and HBD disruption by a T7E1 assay confirmed high (70–80%) on-target activity by both TALEN, but also substantial (≈18%) HBD disruption by R1/L1. By contrast, R1/L2 HBD disruption was negligible and comparable to controls, attributable to the suboptimal 8-bp R1/L2 spacer for HBD (Figure 1B). For three additional samples, R1/L1 gave on average 52.0±7.5% HBBIVS-110(G>A) on-target and 25.8±6.2% HBD off-target disruption. Concurrent cleavage and corresponding ≈7.4-kb deletion would create a chimeric HBD/HBB gene, which we confirmed by specific, fusion-spanning polymerase chain reaction and sequencing (Online Supplementary Figure S4C, D). The detected HBD/HBB fusion gene comprised the therapeutically immaterial promoter and the 5’ region of HBD up to its intron-1 off-target site, and the corresponding 3’ region of HBB. Accordingly, reversed phase high performance liquid chromatography (RP-HPLC) analysis for R1/L1 showed a reduction of the HBD/HBA ratio to 0.068 (approximately-53% compared with control levels: 0.16±0.04), concurrent with an increase in combined γ-globin chains HBG/HBA (0.39, +32.7%) and with a marginal decrease in HBB/HBA levels (0.37,-2.2%) (Online Supplementary Figure S4E,F). These findings reflected inadvertent HBD targeting superimposed on normal competition of HBB-like globin expression9 and led to exclusion of R1/L1 from further analyses.
We then performed deep sequencing analysis of off-target activity at HBD and at the additional respective top ten predicted off-target sites (Online Supplementary Table S1)10,11 for R1/L2-and RGN-treated cells (Figure 2). At HBBIVSI-110(G>A) on-target disruption efficiencies of 68.2% and 88.3%, respectively, R1/L2 gave significant detection only in HBD (Figure 2A), albeit at a marginal frequency (0.19% vs. 0.11% for control background, P=0.001), while the RGN gave significant detection at the top three predicted off-target sites (Figure 2B). RGN off-target sites CAS_OFF1 (54.1% vs. 0.08% for the untransfected control, P<0.0001), CAS_OFF2 (0.4% vs. 0.35%, P<0.05) and CAS_OFF3 (0.07% vs. 0.04%, P<0.05) were all identified as intronic.12 CAS_OFF1 lies on chromosome 13, within the 331-kb intron 3 of RNF219_AS1 (Online Supplementary Figure S5A), which encodes a long non-coding RNA with mainly cerebral expression and without disease association (Online Supplementary Figure S5B).13 Off-target site CAS_OFF2 lies in intron 2 of DGKK and is mainly expressed in the brain and pituitary gland,13 and CAS_OFF3, in intron 13 of CDC42BPB, is expressed ubiquitously, and weakly in whole blood.13
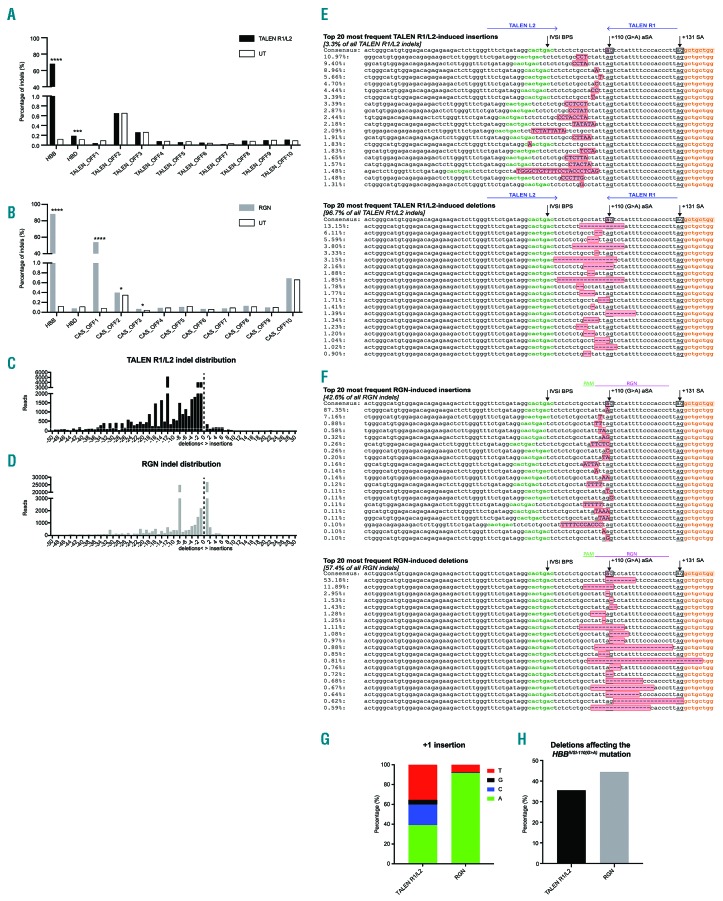
Figure 2.
Characterization of R1/L2-and RGN-edited HBBIVSI-110(G>A)-homozygous hematopoietic stem and progenitor cells by targeted deep sequencing. (A) and (B) Genome modifications (indels) as the percentage of all reads in HBB, HBD and each of the top-ten in silico-predicted off-target sites, for untransfected control (UT) (white bars), R1/L2 [in (A); black bars] and RGN [in (B); gray bars]. Elevated bar height compared to UT indicates nuclease-specific indels. Significant comparisons for z-score analysis: *P<0.05, ***P<0.001, ****P<0.0001. (C) and (D) Distribution of R1/L2 (C) and RGN (D) HBB indels based on the type (insertion and deletions) and size, omitting combined editing events of insertions and deletions. (E) and (F) Alignments and percentages in R1/L2-(E) and RGN-(F) modified cells of the top 20 most frequent insertion (top) or deletion (bottom) events at the HBBIVSI-110(G>A) target site, showing intron 1 (unshaded), the intron-1 branch point site (IVSI BPS; green), exon 2 (orange), the HBBIVSI-110(G>A) mutation (pink in consensus sequence, only), non-homologous end joining-induced indels (pink with red outline), aberrant splice acceptor sites [+110(G>A) aSA] and splice acceptor sites (+131 SA) as underlined boxed sequences in the consensus sequence. Events combining insertions (upper-case letters) and deletions are not shown. Binding sites for TALEN (E) and RGN genomic RNA and protospacer-adjacent motif (PAM) sequence (F) are indicated above each consensus sequence. The frequencies shown are frequencies within each class of indels. (G) Percentage of different +1 nucleotide insertions at the HBBIVSI-110(G>A) target site in R1/L2-and RGN-edited hematopoietic stem and progenitor cells. (H) Percentage of deletions removing the HBBIVSI-110(G>A) mutation in TALEN R1/L2-(black) and RGN-(gray) modified cells. TALEN: transcription activator-like effector nuclease; RGN: . RNA-guided nuclease
Analysis of on-target activity additionally employed Human Splicing Finder (HSF) for prediction of changes in the aberrant splice acceptor site consensus motif and splice-related binding sites.14 R1/L2 and the RGN produced distinctive patterns of on-target indels (Figure 2C, D), similar to patterns detected for three additional samples by TIDE-based analysis (Online Supplementary Figure S6) and as exemplified by the 20 most frequent events detected for each nuclease (Figure 2E, F). For R1/L2, the majority of indels (96.7%) were deletions of various lengths ≥1 bp (Figure 2C, E). For the RGN, the indel pattern was more balanced between insertions (42.7%) and deletions (57.3%) (Figure 2D, F). The commonest RGN-induced event was a 1-bp insertion immediately upstream of the mutation (40.4% of all events), with clear preference for adenine (91.8%) and thymidine (7.6%) (Figure 2G). As shown in Figure 2H, 44.5% of RGN-mediated events and only 35.5% of TALEN R1/L2-mediated events abolished the aberrant splice acceptor site. Importantly, indel events above 5% relative frequency invariably preserved the splice acceptor site core motif while weakening aberrant splice acceptor site-related splice motifs overall (Online Supplementary Figure S7), thus favoring functional correction even if the primary mutation is preserved. For both nucleases, few deletions extended into the splice acceptor site or beyond, suggesting a good safety profile for on-target activity and further predicting a high level of functional correction.
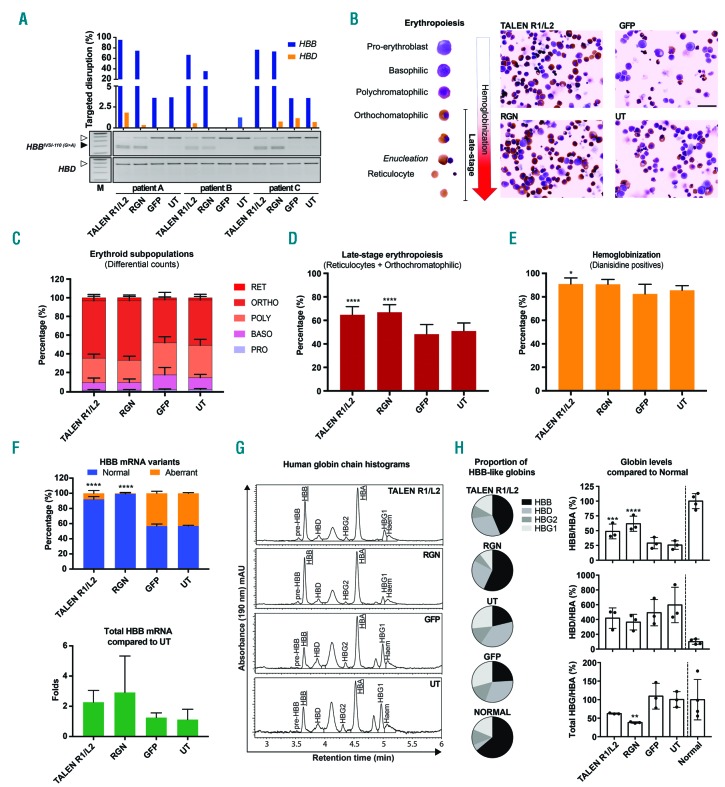
We then assessed functional correction of HBBIVSI-110(G>A) homozygous primary cells by R1/L2 and by the RGN in additional samples (n=3), at high HBB-targeted disruption efficiency (R1/L2: 66.6–95.4% and RGN: 35.6–74.6%) according to T7E1 assays and at marginal or undetectable HBD off-targeting (Figure 3A). Complementary analyses by TIDE confirmed high disruption efficiency and additionally revealed consistent indel patterns across multiple experiments, showing that 86% of R1/L2 events were deletions and that 84.1% of RGN events were almost equally split between an 8-bp deletion and 1-bp insertions (Online Supplementary Figure S6). We analyzed treatment-related functional correction based on key disease parameters of HBBIVSI-110(G>A) thalassemia, specifically erythropoiesis and hemoglobinization by differential microscopic scoring, HBB mRNA splicing by real-time quantitative PCR and expression of individual globin species by RP-HPLC.4,15 Microscopy consistently showed morphology indicative of more advanced erythroid differentiation after R1/L2 and RGN treatment (Figure 3B), which based on stalling of thalassemic progenitors at the polychromatophilic stage of erythropoiesis is a diagnostic gold standard for disease correction, at moderate sample requirements.4 Treatment-blinded scoring of thousands of cells from each culture (R1/L2: 4,579; RGN: 2,287; GFP: 4,230; untransfected control: 4,570) showed significant correction of late-stage erythroid differentiation for R1/L2 (64.8±6.9%, P<0.0001) and for the RGN (67.0±6.4%, P<0.0001) compared with controls (51.0±6.9) (Figure 3C, D). Likewise, hemoglobinization was increased with the RGN (90.7±4.1%, P=0.0688) and significantly increased with R1/L2 (91.0±5.1%, P=0.0272) compared with controls (85.6±3.3%) (Figure 3C–E), although this represents a less sensitive indicator of functional correction for β-thalassemias with residual β-globin expression.4 RNA analysis of variant ratios revealed significantly corrected HBB pre-mRNA splicing in bulk populations, from a control ratio of 56.8±0.9% to 92.0±3.7% (P<0.0001) for R1/L2 and to 99.3±1.3% (P<0.0001) for RGN treatment (Figure 3F, top), the latter with a real-time quantitative PCR readout for aberrant mRNA close to the limit of detection. As an additional measurement, total HBB mRNA indicated variably increased expression by a factor of 2.19±1.39 for R1/L2 and of 2.36±2.16 for RGN treatment (Figure 3F, bottom). Protein analysis by RP-HPLC showed that both, R1/L2 and RGN, restored HBB expression significantly and to therapeutic levels (Figure 3G, H). In the absence of detectable HBB disruption (Figure 3A), competition by increased HBB levels reduced HBD/HBA by 30% for R1/L2 (P=0.2677) and by 38.8% for the RGN (P=0.1342) (Figure 3H), reduced HBG/HBA by 37.8% for R1/L2 (P=0.0689) and significantly reduced HBG/HBA by 61.9% for the RGN (P=0.0086) treatment. Whereas GFP and untransfected controls only reached 29.0±9.5% and 25.6 ±7.3%, respectively, of normal HBB/HBA levels, R1/L2-treated cells achieved on average 48.8±12.5% (P=0.0009) and the RGN 61.8±12.8% (P=0.0001), with 76.5% peak levels for RGN-edited bulk populations.
Figure 3.
Non-homologous end joining-based correction of HBBIVSI-110(G>A)-homozygous hematopoietic stem and progenitor cells using R1/L2 and RGN. (A) T7E1-based assessment of targeted disruption of HBBIVSI-110(G>A) (blue bars) and HBD (orange bars) in patient-derived hematopoietic stem and progenitor cells (HSPC) for treatment with R1/L2, RNA-guided nucleases (RGN), green fluorescent protein (GFP) and untransfected control (UT). (B) Left. Stages of erythropoiesis detected during in vitro erythroid differentiation of HSPC with progressing hemoglobinization. Right. Images of treated, differentiated (day 3), cytocentrifuged and stained HSPC from patient A. Size marker for right panel: 10 μm. (C) Average percentages of erythroid subpopulations of HSPC across patients A–C as illustrated in (B). PRO: pro-erythroblast; BASO: basophilic; POLY: polychromatophilic; ORTHO: orthochomatophilic; RET: reticulocytes. (D) Average percentages of cells in late-stage erythropoiesis (orthochromatophilic and reticulocytes) extracted from (C) and compared to the UT by standard unmatched one-way analysis of variance (ANOVA) and the Dunnett multiple comparison test. Significant comparisons: R1/L2 and RGN ****P<0.0001. (E) Average percentages of hemoglobinized (o-dianisidine-positive) HSPC across patients A–C as illustrated in (B), compared with UT by an unmatched Kruskal-Wallis test and the Dunn multiple comparison analysis. Significant comparison: *P=0.0272. (F) Splice correction shown at the transcript level as the mean (± standard deviation) proportion of normal and aberrant HBB mRNA of total HBB mRNA (top) and as total HBB mRNA expression compared to UT (bottom). Statistical comparison to UT was performed by matched one-way ANOVA and the Dunnett multiple comparison test. Significant comparisons: ****P<0.0001. (G) Representative reversed phase high performance liquid chromatography-based detection of human globin chains in patient-derived HSPC cultures on day 7 of induced erythroid differentiation after the treatments indicated. (H) Quantification of mean HBB-like/HBA globin chain ratios as determined in (G) across experiments (n=3), shown as fractions of total HBB-like globin chains (left) and as HBB-like/HBA globin chain ratios given relative to normal controls (n=4). Statistical comparison with UT of treated HSPC across patients A–C was performed by matched one-way ANOVA and the Dunnett multiple comparison test. Significant comparisons **P=0.0086; *** P=0.0009; RGN **** P=0.0001.
In summary, assessments for HBBIVSI-110(G>A) at DNA, RNA, protein and morphological levels indicate disruption of aberrant regulatory elements by TALEN and RGN as a highly efficient gene therapy approach for suitable mutations, at a high level of biosafety in particular for the TALEN R1/L2 pair analyzed here. Further discussion and details of the methods are provided in the Online Supplementary Material.
Acknowledgments
We are indebted to our blood donors, without whom this work would not have been possible, and to Aglaia Athanasiadou for encouraging investigation of mutation-specific gene therapy. Experimentation on human samples was approved by the Cyprus National Bioethics Committee (Applications ΕΕΒΚ/ΕΠ/2012/02 and ΕΕΒΚ/ΕΠ/2013/23) and based on written informed consent from all study subjects.
Footnotes
Funding: this work was co-funded by the Republic of Cyprus through the Research Promotion Foundation through grant agreement ΥΓΕΙΑ/ΒΙΟΣ/0311(ΒΕ)/20 and core funding of The Cyprus Institute of Neurology and Genetics (P. Patsali, P. Papasavva, CCL, CS, SC, MS, MA, CWL, MK), by the TELETHON of Cyprus (CCL), by Eurobank Cyprus Ltd (P. Papasavva, Scholarship 33173114), by the European Union’s Seventh Framework Program for Research, Technological Development and Demonstration under grant agreement n. 306201 (THALAMOSS; P. Patsali, CS, CWL, MK) and by the German Federal Ministry of Education and Research (BMBF-01EO0803; CM, TIC and TC). The present study was awarded the George Stamatoyiannopoulos Prize 2018 of the Hellenic Society of Gene Therapy and Regenerative Medicine.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Kountouris P, Lederer CW, Fanis P, Feleki X, Old J, Kleanthous M. IthaGenes: an interactive database for haemoglobin variations and epidemiology. PLoS One. 2014;9(7):e103020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu M, Rehman S, Tang X, et al. Methodologies for improving HDR Efficiency. Front Genet. 2019;9:691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang K-H, Smith SE, Sullivan T, et al. Long-term engraftment and fetal globin induction upon BCL11A gene editing in bone-marrow-derived CD34 + hematopoietic stem and progenitor cells. Mol Ther Methods Clin Dev. 2017;4:137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patsali P, Papasavva P, Stephanou C, et al. Short-hairpin RNA against aberrant HBBIVSI-110(G>A) mRNA restores β-globin levels in a novel cell model and acts as mono-and combination therapy for β-thalassemia in primary hematopoietic stem cells. Haematologica. 2018;103(9):e419–e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller JC, Zhang L, Xia DF, et al. Improved specificity of TALE-based genome editing using an expanded RVD repertoire. Nat Methods. 2015;12(5):465–471. [DOI] [PubMed] [Google Scholar]
- 6.Stephanou C, Papasavva P, Zachariou M, et al. Suitability of small diagnostic peripheral-blood samples for cell-therapy studies. Cytotherapy. 2017;19(2):311–326. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z, Chen Y, Sun X, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6(5):363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cencic R, Miura H, Malina A, et al. Protospacer adjacent motif (PAM)-distal sequences engage CRISPR Cas9 DNA target cleavage. PLoS One. 2014;9(10):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thein SL. Molecular basis of β thalassemia and potential therapeutic targets. Blood Cells Mol Dis. 2018;70:54–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fine EJ, Cradick TJ, Zhao CL, Lin Y, Bao G. An online bioinformatics tool predicts zinc finger and TALE nuclease off-target cleavage. Nucleic Acids Res. 2014;42(6):e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu PD, Scott DA, Weinstein JA, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zerbino DR, Achuthan P, Akanni W, et al. Ensembl 2018. Nucleic Acids Res. 2018;46(D1):D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguet F, Brown AA, Castel SE, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desmet F-O, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loucari CC, Patsali P, van Dijk TB, et al. Rapid and sensitive assessment of globin chains for gene and cell therapy of hemoglobinopathies. Hum Gene Ther Methods. 2018;29(1):60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]