Abstract
Somatic mutations in acute myeloid leukemia are acquired sequentially and hierarchically. First, pre-leukemic mutations, such as t(8;21) that encodes AML1-ETO, are acquired within the hematopoietic stem cell (HSC) compartment, while signaling pathway mutations, including KRAS activating mutations, are late events acquired during transformation of leukemic progenitor cells and are rarely detectable in HSC. This raises the possibility that signaling pathway mutations are detrimental to clonal expansion of pre-leukemic HSC. To address this hypothesis, we used conditional genetics to introduce Aml1-ETO and K-RasG12D into murine HSC, either individually or in combination. In the absence of activated Ras, Aml1-ETO-expressing HSC conferred a competitive advantage. However, activated K-Ras had a marked detrimental effect on Aml1-ETO-expressing HSC, leading to loss of both phenotypic and functional HSC. Cell cycle analysis revealed a loss of quiescence in HSC co-expressing Aml1-ETO and K-RasG12D, accompanied by an enrichment in E2F and Myc target gene expression and depletion of HSC self-renewal-associated gene expression. These findings provide a mechanistic basis for the observed absence of KRAS signaling mutations in the pre-malignant HSC compartment.
Introduction
Acute myeloid leukemia (AML) is a poor prognosis hematopoietic malignancy caused by the uncontrolled proliferation of differentiation-arrested myeloid cells.1,2 Genome sequencing studies have comprehensively characterized the mutational landscape of AML, identifying many somatically acquired recurrent driver mutations.3 Whist AML is a genetically complex disease, a number of general principles underlie the clonal evolution in AML. Genes mutated in AML can be classified into distinct categories such as chromatin modifiers, transcription factor fusions, and signal transduction genes,3 with most patients showing co-mutation of genes within at least two of these functional groups. Genomic data from sequencing studies, together with mechanistic studies using mouse models,4–6 support the concept that certain classes of mutation frequently co-occur during leukemia development, whereas mutations of the same functional group are often mutually exclusive.7
Acute myeloid leukemia has long been recognized as a hierarchically organized, stem cell-propagated disease.8 However, more recently, analysis of purified hematopoietic stem cells (HSC) and progenitor populations from AML patients have revealed that leukemia-initiating mutations, which include balanced translocations and mutations in epigenetic regulators, are frequently acquired within the HSC compartment as early events in disease evolution, generating so called “pre-leukemic” stem cells.9–12 In particular, the t(8;21) translocation, which generates the fusion protein AML1-ETO (also known as RUNX1-RUNX1T1 and AML1-MTG8) occurs in approximately 7% of adult AML patients.13 Several lines of evidence sug gest that AML1-ETO is acquired in pre-leukemic HSC. First, AML1-ETO mRNA could still be detected in AML patients who had been in clinical remission for up to 150 months.14 Secondly, AML1-ETO remains stable in patients who relapse, while additional mutations were highly dynamic with mutations both gained and lost at relapse.15 Finally, evidence from mouse models support the concept that pre-leukemic mutations confer a competitive advantage to cells within the phenotypic HSC compartment, without causing transformation of downstream progenitor cells.16,17 In particular, Aml1-ETO knock-in mice did not develop leukemia, but Aml1-ETO-expressing cells had an enhanced in vitro replating ability, indicating greater self-renewal capacity.16
In contrast, signaling transduction mutations of genes such as FLT3, KIT or KRAS occur as late events that are detected in the transformed leukemic progenitors but rarely detected in the pre-leukemic HSC compartment.11,12 RAS mutations also frequently co-occur with t(8;21) (NRAS = 12.9%, KRAS = 4.3%).15 In AML patients who achieve remission, RAS mutations are unstable and often lost at subsequent relapse, with gain of a novel signaling transduction mutation (e.g. FLT3-ITD), while the initiating translocation remains. This is consistent with RAS mutations occurring as a late event during leukemic transformation.15 Finally, the KrasG12D/+;Mx1-Cretg/+mouse model develops a fatal myeloproliferative neoplasm (MPN); however, these mice do not develop AML.18,19 Collectively, these studies provide evidence that RAS mutations are secondary events in AML development and are not present within pre-leukemic HSC. Mouse models in which activating signaling pathway mutations were introduced into wild-type (WT) HSC have revealed both cell-intrinsic and cell-extrinsic effects on the HSC compartment, usually resulting in a depletion of HSC.20–24 However, the impact of signaling transduction mutations on pre-leukemic HSC remains unclear. This is of considerable importance for understanding why signaling mutations are absent from the pre-leukemic HSC compartment.
We hypothesized that the absence of signaling mutations in the HSC may reflect a detrimental impact of such mutations on pre-leukemic HSC. To address this question, we used conditional mouse genetics to introduce Aml1-ETO and K-RasG12D separately or in combination, both expressed from their endogenous loci, into WT HSC, to determine the effect of K-Ras activation on a well-defined pre-leukemic HSC population. While Aml1-ETO expression enhanced the long-term repopulating ability of HSC, expression of K-RasG12D in Aml1-ETO-expressing HSC led to loss of quiescence and self-renewal-associated gene expression, and was detrimental to their function. Such functional impairment would limit clonal expansion of pre-malignant HSC co-expressing AML1-ETO and activated RAS, providing a molecular and cellular basis for the observed absence of activating RAS mutations in pre-leukemic HSC.
Methods
Animals
All mouse lines were maintained on a C57Bl/6J genetic background. Conditional knock-in mice expressing Aml1-ETO (Aml1ETO/+)16 and K-Ras (KrasG12D/+),25 either individually or combined (Aml1ETO/+;KrasG12D/+), were crossed to the Mx1-Cre mouse line.26 All mice were bred and maintained in accordance with UK Home Office regulations. Experiments were conducted following approval by the University of Oxford Animal Welfare and Ethical Review Body (project license n. 30/3103).
Competitive transplantation
Competitive transplants were performed as previously described.27 See Online Supplementary Appendix for further details.
Serial transplantations were performed by co-transplanting 1.25×105 CD45.2 fetal liver (FL) cells with 5x106 CD45.1 WT bone marrow (BM) competitor cells into lethally irradiated recipients (2x500rads). Bulk secondary and tertiary transplants were performed by transplanting 3x106 BM cells from primary and secondary recipients respectively into lethally irradiated recipients at eight weeks post-poly(I:C) for secondary transplants or 12 weeks post-transplantation for tertiary transplants. Tertiary transplanted mice were analyzed 12 weeks post-transplantation.
Flow cytometry and fluorescence-activated cell sorting
Details of antibodies and viability dyes are shown in Online Supplementary Table S1. All antibodies were used at pre-determined optimal concentrations. Hematopoietic stem and progenitor cells were analyzed as previously described.28 Cell acquisition and analysis were performed on a BD LSRFortessa (BD Biosciences, San Jose, CA, USA) using BD FACSDiva™ software (BD Biosciences). Cell sorting was performed on a BD FACSAriaII cell sorter (BD Biosciences). Cells used in cell sorting experiments were c-Kit-enriched (MACS Miltenyi Biotec, Bergisch Gladbach, Germany) and were stained with specific antibodies following initial Fc-block incubation. Gates were set using a combination of fluorescence minus one controls and populations known to be negative for the antigen.
For HSC cell cycle staining, BM cells were c-Kit-enriched and stained following initial Fc-block incubation. Stained cells were then fixed and permeabilized using BD cytofix/cytoperm fixation and permeabilization solution (BD Biosciences). Cells were stained with Ki-67 PE (BD Biosciences) overnight. Cells were then stained with 4′,6-diamidino-2-phenylindole (DAPI) (0.5 mg/mL) (ThermoFisher Scientific, Waltham, MA, USA) for one hour before analysis.
In vitro serial replating assay
Serial replating was performed as previously described.5 Briefly, 100 CD45.2 Lineage−Sca1+cKit+ (LSK) BM cells were sorted from mice transplanted with 2.5x105 FL cells and 1x106 CD45.1 WT BM competitor cells eight weeks post-poly(I:C). Cells were seeded into 1 mL of methylcellulose medium (Methocult, M3434, STEM-CELL Technologies, Vancouver, BC, Canada) and incubated in 37°C, in 5% CO2, with ≥95% humidity. Colonies (≥30 cells) were counted after eight days. Cells were re-suspended and re-plated at 1x104 cells per 1 mL of methylcellulose medium. Cells were then counted and re-plated after 6-7days.
RNA-sequencing
Fifty cells per biological replicate were sorted into 4 mL of lysis buffer containing; 0.2% Triton X-100 (Sigma-Aldrich, St Louis, MO, US), 2.5 mM OligodT (Biomers, Ulm, Germany), 2.5 mM dNTPs (ThermoFisher Scientific), RNase Inhibitor 20 U (Takara Bio USA Inc., Mountain View, CA, USA) and ERCC spike-in 1:4x106 (ThermoFisher Scientific). For details on cDNA synthesis, library preparation and data analysis see Online Supplementary Appendix.
Statistical analysis
Unless otherwise indicated, statistical significance of differences between samples was determined using an analysis of variance (ANOVA) for multiple comparisons following Tukey’s multiple comparisons test.
Results
Aml1-ETO ameliorates key features of the myeloproliferative neoplasm phenotype caused by K-RasG12D
To study the effect of combining Aml1-ETO with mutant K-Ras, whilst avoiding the previously described spontaneous recombination in primary KrasG12D/+;Mx1-Cretg/+ mice,19,20 we generated E14.5 FL cells with the necessary genotypes: Aml1ETO/+;Mx1-Cretg/+ (AM genotype); KrasG12D/+;Mx1-Cretg/+ (KM genotype); Aml1ETO/+;KrasG12D/+;Mx1-Cretg/+ (AKM genotype); and Mx1-Cretg/+ controls (CON genotype); all CD45.2 allotype. FL cells were used as these have been previously shown to have either no or minor spontaneous recombination using Mx1-Cre.29 To create a scenario where competitive advantage and disadvantage of mutant HSC could be observed, mice were generated where the hematopoietic system was partially repopulated by experimental cells and par tially by WT competitor cells (CD45.1 allotype). This was done by competitively transplanting 2.5x105 FL cells (CD45.2) and 1x106 WT BM competitor cells (CD45.1) into lethally irradiated CD45.1 recipients, as previously described.27 Recombination was induced four weeks post-transplantation by subcutaneous injection of poly(I:C), with all groups of mice treated, including controls. Long-term monitoring of primary transplanted mice was not possible as both KM and AKM developed a T-cell leukemia (data not shown), as previously reported for KM.20 Therefore, the hematopoietic phenotype was analyzed eight weeks post-poly(I:C) before T-cell leukemia development occurred (Figure 1A).
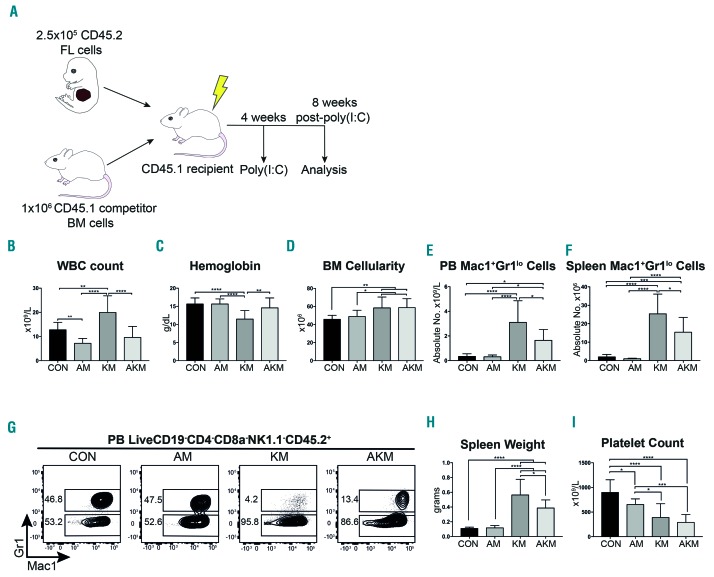
Figure 1.
Aml1-ETO ameliorates the key features of the myeloproliferative neoplasm phenotype caused by K-RasG12D. (A) Schematic representation of in vivo competitive transplant experiment. (B-I) Analysis of recipients of Mx1-Cretg/+ controls; CON (n=13), Aml1ETO/+;Mx1-Cretg/+; AM (n=14), KrasG12D/+;Mx1-Cretg/+; KM (n=12) and Aml1ETO/+;KrasG12D/+;Mx1-Cretg/+; AKM fetal liver (FL) (n=14) for (B) peripheral blood (PB) white blood cell (WBC) count. (C) PB hemoglobin levels. (D) Bone marrow (BM) cellularity per tibia and femur. (E and F) CD45.2 Mac1+Gr1lo myeloid cells as absolute number in the PB (E) and spleen (F). (G) Representative FACS plots of Mac1+Gr1+ and Mac1+Gr1lo myeloid cells as a percentage of LiveCD19−CD4−CD8a−NK1.1−CD45.2+ cells across all experiments in the PB. (H) Spleen weight. (I) Platelet count. Results were generated in three independent experiments. The results were analyzed using multiple comparison ANOVA and are presented as mean±Standard Deviation. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
AM-transplanted recipients displayed mild leukopenia compared to CON-transplanted mice (Figure 1B), due to a decrease in B and T cells in the peripheral blood (PB) (Online Supplementary Figure S1A and B), whereas no difference in hemoglobin levels, BM cellularity, Mac1+Gr1lo myeloid cells in the PB or spleen, or spleen weight was observed (Figure 1C-H). However, AM showed a 27% decrease in platelet count compared to CON (Figure 1I). These results suggest Aml1-ETO affects B and T cell, as well as platelet development, in line with known functions of AML1 (also known as RUNX1).30,31
In line with previous reports,18,19 expression of K-RasG12D (KM genotype) caused a myeloproliferative phenotype, consisting of leukocytosis, anemia, increased BM cellularity, increase in the Mac1+Gr1lo myeloid cells in the PB and spleen, splenomegaly, and thrombocytopenia (Figure 1B-I and Online Supplementary Table S2).
Strikingly, co-expression of Aml1-ETO and K-RasG12D did not result in a more aggressive disease, but ameliorated key features of the phenotype associated with K-RasG12D expression, including restoration of white blood cell (WBC) count and hemoglobin levels (Figure 1B and C), reduction in the Mac1+Gr1lo myeloid cells in the PB and spleen, and reduced spleen weight (Figures 1E-H). Platelet count was, however, further reduced compared to CON (Figure 1I) and mice still showed an increase in BM cellularity (Figure 1D).
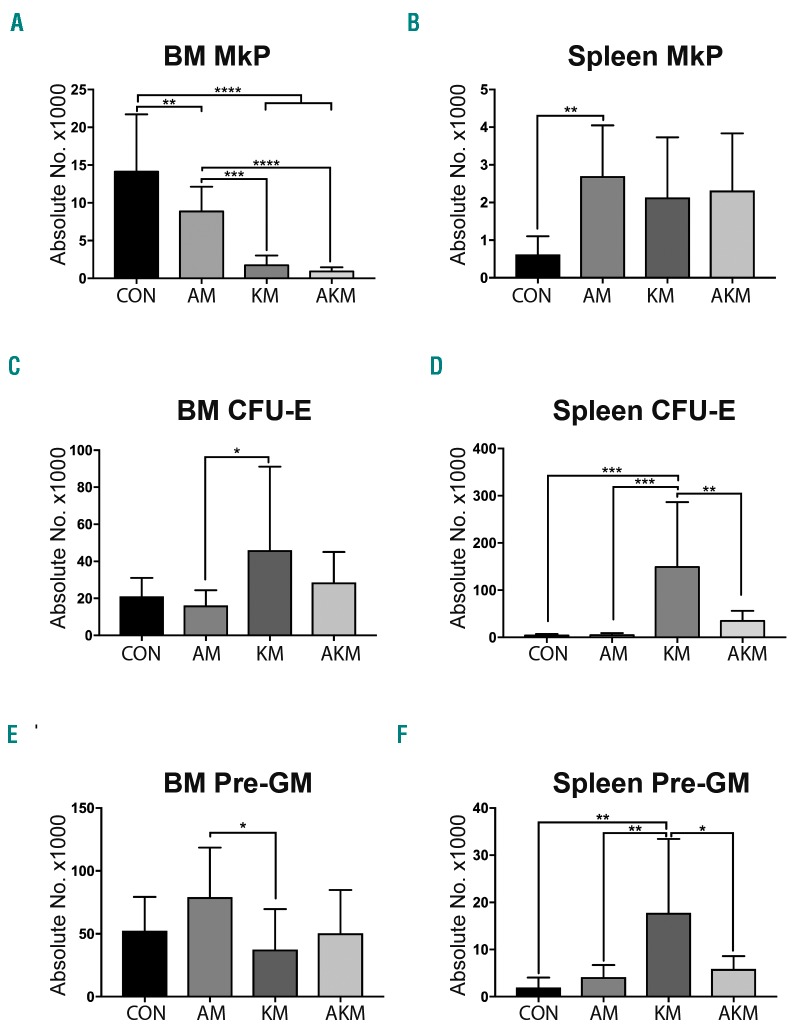
Myeloerythroid progenitors were analyzed by flow cytometry (Figure 2 and Online Supplementary Figure S2). We found both Aml1-ETO (2-fold, P=0.0093) and K-RasG12D (8-fold, P<0.0001) expression decreased the number of megakaryocyte progenitors (MkP), and when co-expressed resulted in a further reduction (15-fold, P<0.0001) in the BM compared to CON (Figure 2A). This paralleled the decrease in platelet counts found in the PB (Figure 1I). Aml1-ETO also increased in the number of MkP in the spleen compared to CON (2-fold, P=0.005) (Figure 2B). BM colony forming unit erythrocytes (CFU-E) were not affected in any genotypes compared to controls (Figure 2C). However, K-RasG12D caused an increase in CFU-E in the spleen compared to CON (36-fold increase, P=0.0004) (Figure 2D), indicating stress erythropoiesis, which was reversed in AKM mice. BM pre-granulocyte/macrophage progenitors (pre-GM) were unperturbed in all three genotypes compared to CON (Figure 2E). However, K-RasG12D expression caused an increase in pre-GM in the spleen compared to CON (9-fold increase, P<0.0001) (Figure 2F). The increase in pre-GM was reversed when K-RasG12D was co-expressed with Aml1-ETO (3-fold decrease, P<0.0001) (Figure 2F). There was no difference in BM lymphoid-primed multi-potent progenitor (LMPP) across all genotypes; however, there was an increase in the number of spleen LMPP in KM mice compared to CON, likely reflecting increased mobilization associated with myeloproliferation. The increase in LMPP was reversed in AKM mice (Online Supplementary Figure S3). Together, these data demonstrate that when Aml1-ETO is co-expressed with K-RasG12D the myeloproliferative phenotype caused by K-RasG12D is ameliorated rather than enhanced.
Figure 2.
Aml1-ETO reverses some of the myeloerythroid progenitor cell phenotypes caused by K-RasG12D. (A, C, E) Absolute number of CD45.2 megakaryocyte progenitor (MkP) (A), CFU-E (C) and Pre-GM (E) in the bone marrow (BM) from recipients of CON (n=12), AM (n=14), KM (n=12) and AKM FL (n=14). Results were generated in three independent experiments. (B, D, F) Absolute numbers of CD45.2 MkP (B), colony forming unit-erythrocyte (CFU-E) (D) and pre-granulocyte-monocyte (Pre-GM). (F) in the spleen from recipients of CON (n=8), AM (n=10), KM (n=8) and AKM FL (n=10). Results were generated in two independent experiments. The results were analyzed using multiple comparison ANOVA and are presented as the mean±Standard Deviation. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
K-RasG12D reverses the hematopoietic stem cell expansion associated with Aml1-ETO
As the co-expression of Aml1-ETO and K-RasG12D was insufficient to induce acute leukemic transformation, this provided an ideal model to study the impact of these mutations on pre-leukemic HSC. Here, expression of the mutations from their endogenous loci (rather than through viral transduction) is crucial in order to ensure faithful expression level of the mutations within the hematopoietic hierarchy, including the HSC compartment. We reasoned that the myeloproliferative phenotype may have been ameliorated when Aml1-ETO and K-RasG12D were co-expressed due to loss of disease propagating HSC. We therefore analyzed LSKCD150+Flt3− phenotypic HSC eight weeks post-poly(I:C). The SLAM marker CD48 was not used as it was previously reported that CD48 expression is dysregulated in Aml1 deficient HSC.32
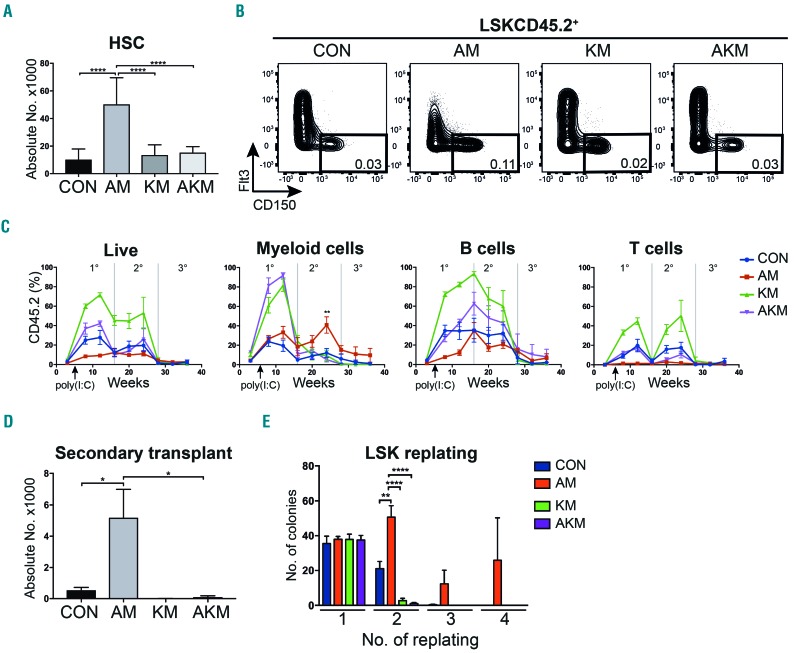
We observed an expansion of HSC expressing Aml1-ETO compared to CON (5-fold increase, P<0.0001) (Figure 3A and B). There was no significant difference in HSC number expressing K-RasG12D compared to CON. However, when K-RasG12D was co-expressed with Aml1-ETO the HSC expansion caused by Aml1-ETO was reversed (3-fold decrease, P<0.0001) (Figure 3A and B).
Figure 3.
Hematopoietic stem cell (HSC) expansion caused by Aml1-ETO is reversed by K-RasG12D. (A) Absolute number of CD45.2 HSC in the bone marrow (BM) from recipients of CON (n=11), AM (n=14), KM (n=13) and AKM fetal liver (FL) cells (n=14). Results were generated in three independent experiments; (B) Representative FACS plots showing gating used to quantify HSC as a percentage of the BM mononuclear cells across all experiments. (C) Percentage reconstitution of total CD45.2 cells, CD45.2 myeloid (LiveCD19−CD4−CD8a−NK1.1−), CD45.2 B cells (LiveNK1.1-Mac1-CD19+) and CD45.2 T cell (LiveNK1.1−Mac1−CD4+CD8a+) compartments in primary, secondary and tertiary transplantations. (D) Absolute number of CD45.2 HSC in secondary recipients of CON (n=9 recipient mice in 2 independent experiments), AM (n=10 recipient mice in 3 independent experiments), KM (n=4 recipient mice in 2 independent experiments), and AKM FL cells (n=7 recipient mice in 2 independent experiments). (E) Replating efficiency of CD45.2 LSK BM cells. Average number of colonies is shown for 5-6 biological replicates per genotype in two independent experiments. The results were analyzed using multiple comparison ANOVA. The results are presented as the mean±Standard Error of Mean. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
To determine if K-RasG12D is detrimental to the function of Aml1-ETO-expressing HSC we performed serial transplantations. In secondary recipients, Aml1-ETO-expressing cells showed an increase in myeloid reconstitution over time and a 10-fold increase in phenotypic HSC number compared to CON (Figure 3C and D), indicating a competitive advantage, possibly due to an enhanced self-renewal capacity. In contrast, secondary transplant with K-RasG12D-expressing cells, or in combination with Aml1-ETO, showed markedly decreased myeloid reconstitution and HSC number (Figure 3C and D). The lack of engraftment following bulk BM transplantation from AKM mice also supports the concept that AKM progenitor cells (which would be included in bulk BM transplants) do not acquire aberrant self-renewal capacity. Furthermore, Aml1-ETO-expressing LSK cells showed increased replating potential in vitro (Figure 3E), in keeping with previous reports.16 In contrast, the additional expression of K-RasG12D abrogated this enhanced replating potential (Figure 3E). Collectively, these results support the concept that Aml1-ETO expression is associated with increased self-renewal of HSC. But in the additional presence of K-RasG12D, HSC are at a competitive disadvantage, consistent with functional impairment of AKM HSC.
K-RasG12D expression induces loss of quiescence in Aml1-ETO-expressing hematopoietic stem cells
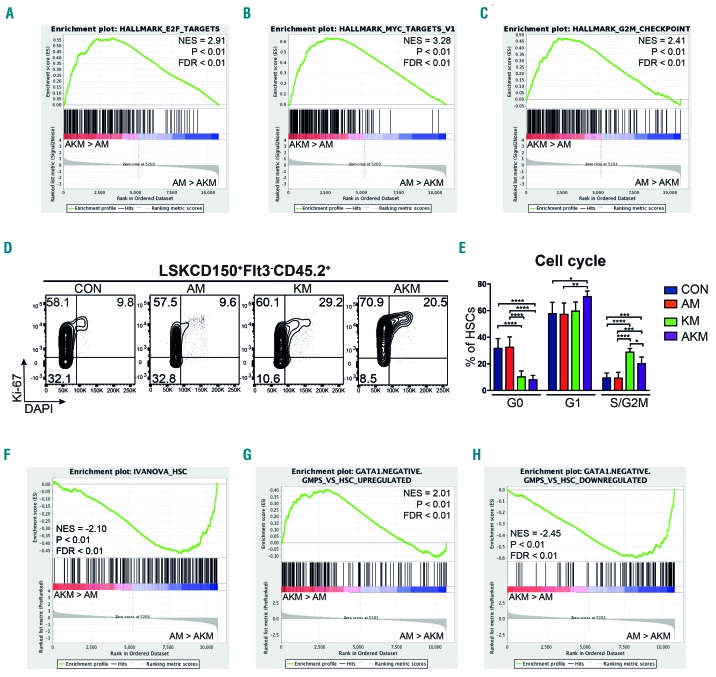
To gain molecular insight into the functional impairment of HSC expressing K-RasG12D, we carried out RNA sequencing of AM, KM, AKM and CON CD45.2 LSKCD150+Flt3− cells from competitively transplanted recipients eight weeks post-poly(I:C) (n=5 replicates per genotype). Gene set enrichment analysis (GSEA) revealed a marked enrichment in E2F, Myc, and G2M checkpoint associated gene expression in AKM compared to AM, likely indicating higher levels of cell cycle activity. In keeping with the observed functional impairment of HSC (Figure 4A-C). Cell cycle activation was confirmed by flow cytometry, showing a 4-fold decrease in quiescent (G0) AKM, compared to AM, HSC (Figure 4D and E). This was accompanied by loss of HSC self-renewal-associated gene expression and an acquisition of gene expression associated with granulocyte-macrophage progenitors (GMP) that lack Gata1 expression that give rise to neutrophils and monocytes (Figure 4F-H).33 This is consistent with phenotypic HSC from AKM mice showing transcriptional signatures of myeloid progenitor cells rather than HSC.
Figure 4.
Hematopoietic stem cells (HSC) co-expressing Aml1-ETO and K-RasG12D are characterized by loss of quiescence and HSC-associated gene expression. (A-C) Bulk CD45.2 LSKCD150+Flt3− cells were subjected to RNA sequencing (5-6 biological replicates per genotype in two independent experiments). Gene set enrichment analysis (GSEA) of AKM versus AM HSC for E2F targets (A), Myc targets (B), and genes associated with G2M checkpoint (C). (D) Representative FACS plots showing cell cycle analysis of CD45.2 LSKCD150+Flt3− phenotypic HSC from the bone marrow (BM) of recipients of CON (n=6 recipient mice in 2 independent experiments), AM (n=9 recipient mice in 3 independent experiments), KM (n=4 recipient mice in 2 independent experiments), and AKM FL (n=6 recipient mice in 3 independent experiments). (E) Percentage of BM CD45.2 LSKCD150+Flt3− cells at each cell cycle stage. The results were analyzed using multiple comparison ANOVA. The results are presented as mean±Standard Deviation. *P<0.05; **P<0.01; ***P<0.001. (F-H) GSEA analysis of AKM versus AM HSC for HSC gene signature (F), genes up-regulated in granulocyte-monocyte progenitor (GMP) that lack Gata1 expression compared to HSC (G), and genes down-regulated in GMP that lack Gata1 expression compared to HSC (H). NES: normalized enrichment score; FDR: false discovery rate.
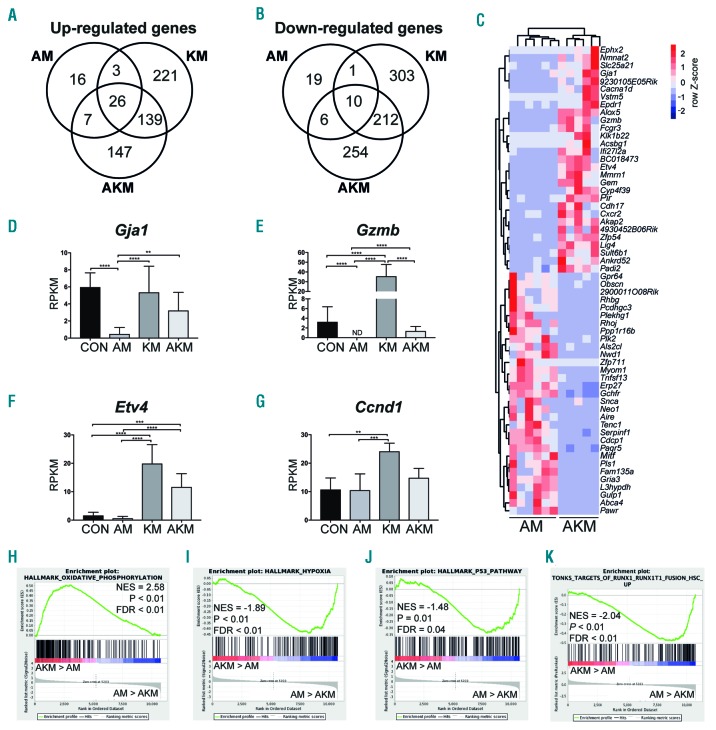
To identify genes that may be involved in the loss of quiescence and HSC function, we performed differential gene expression analysis. Aml1-ETO expression caused an up-regulation of 52 genes and down-regulation of 36 genes in phenotypic HSC compared to CON (Figure 5A C). Among the down-regulated genes were Gja1 and Gzmb (Figure 5D and E). K-RasG12D caused more extensive disruption of gene expression, with up-regulation of 389 genes and down-regulation of 526 genes compared to CON (Figure 5A and B). Among the up-regulated genes were MAPK pathway genes such as Etv4 and Ccnd1 (Figure 5F and G). Genes that were down-regulated by KRAS activation were down-regulated in KM versus CON and AKM versus CON HSC (Online Supplementary Figure S4A and B), in keeping with activation of this signaling pathway by activated K-RasG12D in both Aml1-ETO positive and negative cells.34
Figure 5.
RNA sequencing reveals distinct molecular signatures of hematopoietic stem cells (HSC) co-expressing Aml1-ETO and K-RasG12D. (A and B) Venn-diagram of significantly up-regulated (A) and down-regulated genes (B) in HSC identified by RNA sequencing. (C) Heatmap depicting the read per kilobase of transcript per million (RPKM) values of the top 30 significantly up-regulated and down-regulated genes in AKM HSC versus AM [false discovery rate (FDR) < 0.05]. (D-G) RPKM of selected genes identified from RNA sequencing, Gja1 (D), Gzmb (E), Etv4 (F), and Ccnd1 (G). RPKM and FDR were generated using edgeR package. The results are presented as mean±Standard Deviation. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. (H-K) Gene set enrichment analysis (GSEA) of AKM versus AM HSC for oxidative phosphorylation (H), hypoxia (I), p53 pathway (J), and genes up-regulated in human HSC transduced with AML1-ETO (K). NES: normalized enrichment score.
Hematopoietic stem cells co-expressing Aml1-ETO and K-RasG12D showed up-regulation of 319 genes and down-regulation of 482 genes compared to CON (Figure 5A and B). Many of the up- and down-regulated genes in AKM HSC overlapped with KM HSC (Figure 5A and B), indicating that KRAS confers some of the same transcriptional changes in both Aml1-ETO positive and negative HSC. This was confirmed in a principal component analysis (PCA) and hierarchical clustering of the HSC from all four genotypes which demonstrated that AKM HSC clustered closely with KM HSC indicating that K-RasG12D is driving the separation in gene expression in AKM HSC from CON and AM HSC (Online Supplementary Figure S4C-F). However, the majority of these genes were only dysregulated in the presence of both mutations (Figure 5A and B), indicating that the two mutations together collaborated to induce a distinct pattern of gene expression that only partially overlapped with K-RasG12D or Aml1-ETO regulated genes (Figure 5A-C). Gja1 and Gzmb were up-regulated in HSC co-expressing Aml1-ETO and K-RasG12D when compared to Aml1-ETO (Figure 5C-E).
Gene set enrichment analysis showed an enrichment of oxidative phosphorylation and loss of genes associated with hypoxia in AKM HSC compared to AM (Figure 5H and I and Online Supplementary Table S3). Interestingly, GzmB causes reactive oxygen species (ROS) production,35,36 which can lead to apoptosis and cell death of HSC, suggesting GzmB expression may lead to increased levels of ROS and apoptosis in AKM HSC. Genes associated with the p53 pathway were also down-regulated in AKM HSC compared to AM (Figure 5J). Loss of Gja1 has been shown to increase p53 levels;37 therefore, increased expression of Gja1 in AKM HSC may lead to loss of p53. Analysis of biological GO terms showed a number of metabolic processes up-regulated in AKM versus AM HSC (Online Supplementary Table S4) indicating metabolic dysregulation could also be involved in loss of quiescence in AKM HSC.38
Finally, in order to determine relevance of the genes/pathways described for human AML1-ETO, we studied genes that are up-regulated in human HSC transduced with AML1-ETO.39 There was an enrichment of these genes in AM HSC compared to AKM, suggest a correlation between human and mouse AML1-ETO target genes (Figure 5K).
Collectively, these results demonstrate that K-RasG12D is detrimental to HSC harboring Aml1-ETO, causing a loss of functional HSC, associated with down-regulation of HSC-associated gene expression and loss of quiescence.
Discussion
We have here tested the hypothesis that the observed absence of mutations in signaling pathway genes, such as KRAS, in pre-leukemic HSC from AML patients11,12 is due to such mutations being detrimental not only to normal HSC, but also to pre-leukemic HSC. While Aml1-ETO improved the repopulating capacity of HSC, K-RasG12D had a markedly detrimental effect on Aml1-ETO-expressing pre-leukemic HSC, leading to their eventual depletion, likely due to loss of quiescence and HSC-associated gene expression. The loss of disease-propagating HSC also likely underlies the amelioration of the K-RasG12D-induced myeloproliferative phenotype when the mutations were combined.
Signaling mutations are thought to have a negative cell-intrinsic impact on HSC as enhanced proliferation tends to reduce competitiveness and self-renewal potential. Previous studies have shown N-RasG12D increases cell division and reduces the self-renewal in a subset of HSC. However, this negative impact is counteracted as N-RasG12D also increases the self-renewal potential and reduces division in another subset of HSC. This bimodal effect allows NRasG12D-expressing HSC to outcompete WT HSC, in contrast to our observations with K-RasG12D.23 Recently, signaling mutations have also been shown to have a negative cell-extrinsic impact on HSC by disrupting HSC-supporting BM stromal cells and increasing inflammation-associated gene expression.24 Sabnis et al. have shown that K-RasG12D expression cell-intrinsically drives HSC into cycle and reduces HSC frequency; however, long-term fitness of K-RasG12D-expressing HSC was not analyzed due to lethality caused by K-RasG12D-induced myeloproliferation.20
Experimental approaches investigating collaboration between Aml1-ETO and activated Ras have mainly used retroviral expression of oncogenes, showing that such mutations collaborate to cause an acute leukemia.40,41 However, retroviral transduction can lead to expression at non-physiological levels as well as ectopic expression within the cellular hierarchy.4 This method, therefore, may not accurately address the effect of the mutant proteins on HSC function or the ability of mutations expressed at relevant levels to induce transformation of progenitor cells. Consistent with this, in our model system, knock-in of Am1-ETO and K-RasG12D was insufficient to cause transformation of myeloid progenitors. Other approaches using conditional knock-in mutations or gene knock-out to study the collaboration between pre-leukemic mutations, such as the bi-allelic Cebpa mutations, Tet2 and Dnmt3a knock-out, in combination with the signaling mutation Flt3-ITD have resulted in leukemic transformation.4–6 However, Flt3 is not expressed at detectable levels in repopulating mouse HSC.42 Therefore, such modeling cannot measure the intrinsic effect of a signaling mutation on pre-leukemic HSC, whereas cell-extrinsic effects of FLT3-ITD would be likely to play a role.24 Indeed, in order to study pre- leukemic HSC, it is preferable that the model used retains a relatively unperturbed hematopoietic hierarchy without overt leukemia or other malignancy. In addition, the use of “knock-in” models of oncogenes that are expressed from their own promoter at their original loci is important to retain faithful expression patterns within the hematopoietic hierarchy. Both these conditions are fulfilled by the Aml1ETO/+;KrasG12D/+;Mx1-Cretg/+ model used here.
The results from this study differs to other models that demonstrate that Ras mutations collaborate with pre-leukemic mutations to develop an AML, such as Dnmt3a−/−;KrasG12D/+ and CbfbMYH11/+;NrasG12D/+, where both models develop a more aggressive disease when combined.43,44 Both models lead to transformation of myeloid progenitors, in contrast to the lack of transformation seen in AKM mice, even after serial transplantation. Both Dnmt3a−/−;KrasG12D/+ and CbfbMYH11/+;NrasG12D/+ models do, however, result in a loss of LT-HSC which is consistent with our results. The lack of transformation of myeloid progenitors in AKM mice gave us a unique opportunity to study pre-leukemic stem cells functionally in the absence of progenitor cell transformation as seen in the other models.
RNA sequencing revealed genes that may underlie the observed HSC phenotype of AM and AKM mice. Gzmb is a serine protease that has recently been reported to be important in HSC function.35 Knock-out of Gzmb confers enhanced self-renewal to HSC in a cell-intrinsic manner. Gzmb deficient mice also had a better survival rate after administering 5-FU.35 Gja1 encodes gap junction channel protein connexin 43 found on HSC. Gja1 deficient HSC were shown to be more quiescent after 5-FU treatment. Gja1 deficient HSC also developed an accumulation of ROS.37 ROS levels in HSC have been shown to play an important role in hematopoietic reconstitution;45 however, oxidative phosphorylation gene expression was not enriched in AM HSC versus CON suggesting down-regulation of Gja1 in AM HSC did not lead to an increase in ROS. GJA1 has lower expression on CD34+ BM cells from AML patients with AML1-ETO compared to WT CD34+ BM cells.46 Down-regulation of GZMB and Gja1 have also both been identified in human and murine leukemic stem cells, respectively.47–49 Together, down-regulation of GZMB and GJA1 may contribute to the AML1-ETO-associated competitive advantage of HSC that we observed, and could potentially be important for pre-leukemic HSC persistence after chemotherapy. Importantly, expression of both Gzmb and Gja1 were increased in the presence of K-RasG12D, indicating that re-expression of these genes may have contributed to the loss of HSC function and self-renewal. However, as we identified disruption of multiple genes and pathways in AKM HSC, it seems likely that the underlying mechanistic basis for loss of functional Aml1-ETO expressing HSC associated with K-RasG12D is complex, and is unlikely to be attributable to one single target gene and more likely to involve an interplay of many genes and pathways.
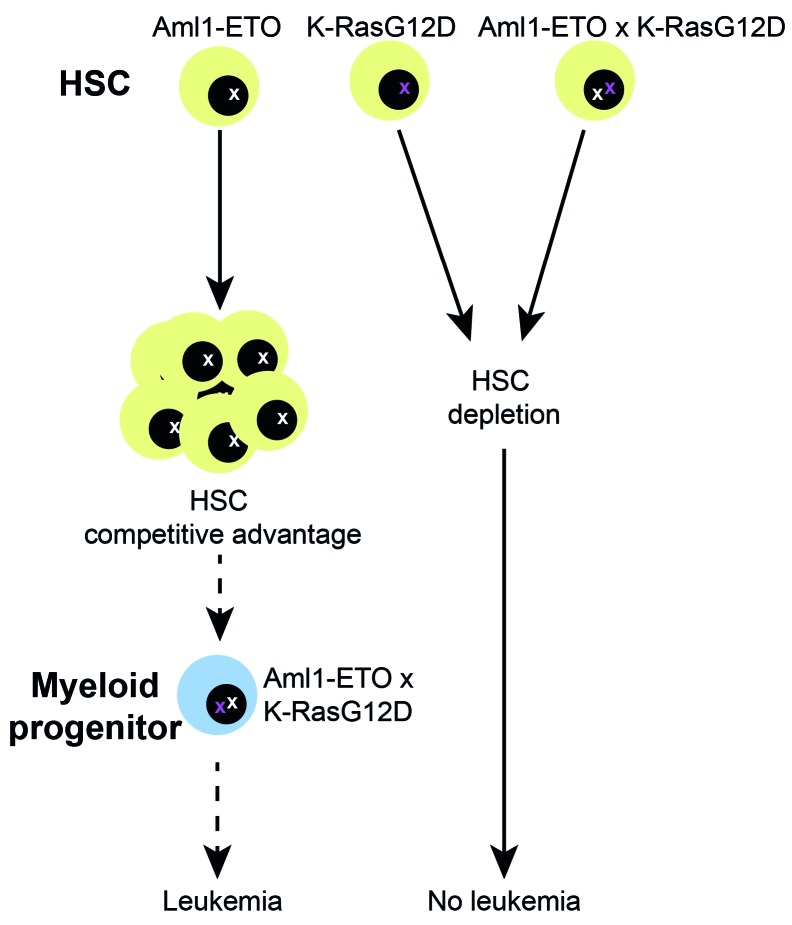
Mutations are acquired in a stepwise manner in AML, and the consequence of the type and order of the mutations acquired will make the HSC more or less likely to facilitate subsequent evolution to leukemia.50 Our findings help to provide a cellular and molecular basis for the observed patterns of clonal evolution during AML development. HSC that acquire Aml1-ETO gain a competitive advantage, leading to an expansion in HSC number, increasing the pool of cells available to acquire additional mutation that could eventually promote leukemia development. HSC that acquire a Kras mutation, either alone or in combination with Aml1-ETO, are depleted. A potential limitation of our study is that the mutations are introduced simultaneously rather than sequentially. New model systems that allow knock-in mutations to be introduced sequentially, and potentially also in specific cellular compartments, warrant further investigation (Figure 6).
Figure 6.
Schematic summarizing the effect of K-RasG12D on pre-leukemic hematopoietic stem cells (HSC). HSC that acquire Aml1-ETO gain a competitive advantage, leading to an expansion in HSC number. Acquisition of K-RasG12D and Aml1-ETO concurrently leads to HSC depletion. It remains to be determined whether sequential acquisition of Aml1-ETO followed-by K-RasG12D might support development of leukemia. As this was not tested in the current study, this is depicted as a dotted arrow.
In summary, our findings help to explain why signaling mutations such as KRAS are not observed within pre-leukemic HSC in AML patients and usually occur as a late event in leukemogenesis. The distinct molecular signatures associated with pre-leukemic mutations in HSC suggest that approaches to target leukemic versus pre-leukemic stem cell expansion are likely to be different.
Acknowledgments
A.J.M received funding from a Medical Research Council Senior Clinical Fellowship (MR/L006340/1). This work was supported by the Medical Research Council (MC_UU_12009, and G0701761, G0900892 and MC_UU_12009/7 to CN). The authors acknowledge the contributions of the WIMM Flow Cytometry Facility, supported by the MRC HIU; MRC MHU (MC_UU_12009); NIHR Oxford BRC and John Fell Fund (131/030 and 101/517), the EPA fund (CF182 and CF170) and by the WIMM Strategic Alliance awards G0902418 and MC_UU_12025.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/11/2215
References
- 1.Sykes SM, Kokkaliaris KD, Milsom MD, Levine RL, Majeti R. Clonal Evolution of Preleukemic hematopoietic stem cells in acute myeloid leukemia. Exp Hematol. 2015; 43(12):989–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368(9550):1894–1907. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013; 368(22):2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reckzeh K, Bereshchenko O, Mead A, et al. Molecular and cellular effects of oncogene cooperation in a genetically accurate AML mouse model. Leukemia. 2012;26(7):1527–1536. [DOI] [PubMed] [Google Scholar]
- 5.Shih AH, Jiang Y, Meydan C, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell. 2015; 27(4):502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L, Rodriguez B, Mayle A, et al. DNMT3A Loss Drives Enhancer Hypomethylation in FLT3-ITD-Associated Leukemias. Cancer Cell. 2016;29(6):922–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374(23):2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–737. [DOI] [PubMed] [Google Scholar]
- 9.Miyamoto T, Weissman IL, Akashi K. AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc Natl Acad Sci U S A. 2000;97(13):7521–7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jan M, Snyder TM, Corces-Zimmerman MR, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med. 2012;4(149):149ra118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shlush LI, Zandi S, Mitchell A, et al. Identification of pre-leukaemic haematopoi-etic stem cells in acute leukaemia. Nature. 2014;506(7488):328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci U S A. 2014;111(7):2548–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyamoto T, Nagafuji K, Akashi K, et al. Persistence of multipotent progenitors expressing AML1/ETO transcripts in long-term remission patients with t(8;21) acute myelogenous leukemia. Blood. 1996;87(11):4789–4796. [PubMed] [Google Scholar]
- 15.Krauth MT, Eder C, Alpermann T, et al. High number of additional genetic lesions in acute myeloid leukemia with t(8;21)/RUNX1-RUNX1T1: frequency and impact on clinical outcome. Leukemia. 2014;28(7):1449–1458. [DOI] [PubMed] [Google Scholar]
- 16.Higuchi M, O’Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1(1):63–74. [DOI] [PubMed] [Google Scholar]
- 17.Guryanova OA, Shank K, Spitzer B, et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med. 2016;22(12):1488–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braun BS, Tuveson DA, Kong N, et al. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc Natl Acad Sci U S A. 2004;101(2):597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan IT, Kutok JL, Williams IR, et al. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J Clin Invest. 2004;113(4):528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabnis AJ, Cheung LS, Dail M, et al. Oncogenic Kras initiates leukemia in hematopoietic stem cells. PLoS Biol. 2009;7(3):e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu SH, Heiser D, Li L, et al. FLT3-ITD knockin impairs hematopoietic stem cell quiescence/homeostasis, leading to myeloproliferative neoplasm. Cell Stem Cell. 2012;11(3):346–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan G, Kalaitzidis D, Usenko T, et al. Leukemogenic Ptpn11 causes fatal myeloproliferative disorder via cell-autonomous effects on multiple stages of hematopoiesis. Blood. 2009;113(18):4414–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q, Bohin N, Wen T, et al. Oncogenic Nras has bimodal effects on stem cells that sustainably increase competitiveness. Nature. 2013;504(7478):143–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mead AJ, Neo WH, Barkas N, et al. Niche-mediated depletion of the normal hematopoietic stem cell reservoir by Flt3-ITD-induced myeloproliferation. J Exp Med. 2017;214(7):2005–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson EL, Willis N, Mercer K, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15(24):3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269(5229):1427–1429. [DOI] [PubMed] [Google Scholar]
- 27.Bereshchenko O, Mancini E, Moore S, et al. Hematopoietic stem cell expansion precedes the generation of committed myeloid leukemia-initiating cells in C/EBPalpha mutant AML. Cancer Cell. 2009;16(5):390–400. [DOI] [PubMed] [Google Scholar]
- 28.Pronk CJ, Rossi DJ, Mansson R, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1(4):428–442. [DOI] [PubMed] [Google Scholar]
- 29.Velasco-Hernandez T, Sawen P, Bryder D, Cammenga J. Potential pitfalls of the Mx1-Cre system: implications for experimental modeling of normal and malignant hematopoiesis. Stem Cell Reports. 2016; 7(1):11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ichikawa M, Asai T, Saito T, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004; 10(3):299–304. [DOI] [PubMed] [Google Scholar]
- 31.Growney JD, Shigematsu H, Li Z, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106(2):494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai X, Gaudet JJ, Mangan JK, et al. Runx1 loss minimally impacts long-term hematopoietic stem cells. PloS One. 2011;6(12):e28430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drissen R, Buza-Vidas N, Woll P, et al. Distinct myeloid progenitor-differentiation pathways identified through single-cell RNA sequencing. Nat Immunol. 2016;17(6):666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pratilas CA, Taylor BS, Ye Q, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A. 2009; 106(11):4519–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carnevalli LS, Scognamiglio R, Cabezas-Wallscheid N, et al. Improved HSC reconstitution and protection from inflammatory stress and chemotherapy in mice lacking granzyme B. J Exp Med. 2014;211(5):769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aguilo JI, Anel A, Catalan E, et al. Granzyme B of cytotoxic T cells induces extramitochondrial reactive oxygen species production via caspasedependent NADPH oxidase activation. Immunol Cell Biol. 2010;88(5):545–554. [DOI] [PubMed] [Google Scholar]
- 37.Taniguchi Ishikawa E, Gonzalez-Nieto D, Ghiaur G, et al. Connexin-43 prevents hematopoietic stem cell senescence through transfer of reactive oxygen species to bone marrow stromal cells. Proc Natl Acad Sci U S A. 2012;109(23):9071–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito K, Ito K. Hematopoietic stem cell fate through metabolic control. Exp Hematol. 2018;64:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tonks A, Pearn L, Musson M, et al. Transcriptional dysregulation mediated by RUNX1-RUNX1T1 in normal human progenitor cells and in acute myeloid leukaemia. Leukemia. 2007;21(12):2495–2505. [DOI] [PubMed] [Google Scholar]
- 40.Zhao S, Zhang Y, Sha K, et al. KRAS (G12D) cooperates with AML1/ETO to initiate a mouse model mimicking human acute myeloid leukemia. Cell Physiol Biochem. 2014;33(1):78–87. [DOI] [PubMed] [Google Scholar]
- 41.Chou FS, Wunderlich M, Griesinger A, Mulloy JC. N-Ras(G12D) induces features of stepwise transformation in preleukemic human umbilical cord blood cultures expressing the AML1-ETO fusion gene. Blood. 2011;117(7):2237–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adolfsson J, Borge OJ, Bryder D, et al. Upregulation of Flt3 expression within the bone marrow Lin(−)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15(4):659–669. [DOI] [PubMed] [Google Scholar]
- 43.Chang YI, You X, Kong G, et al. Loss of Dnmt3a and endogenous Kras(G12D/+) cooperate to regulate hematopoietic stem and progenitor cell functions in leukemogenesis. Leukemia. 2015;29(9):1847–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue L, Pulikkan JA, Valk PJ, Castilla LH. NrasG12D oncoprotein inhibits apoptosis of preleukemic cells expressing Cbfbeta-SMMHC via activation of MEK/ERK axis. Blood. 2014;124(3):426–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewandowski D, Barroca V, Duconge F, et al. In vivo cellular imaging pinpoints the role of reactive oxygen species in the early steps of adult hematopoietic reconstitution. Blood. 2010;115(3):443–452. [DOI] [PubMed] [Google Scholar]
- 46.El-Hadya SBM, Almasryc E, ElHefneyb AM. Detection of Cx43 and P27 in acute myeloid leukemia patients with t(8;21). Egypt J Haematol. 2012;37(2):1110–1067. [Google Scholar]
- 47.Ng SW, Mitchell A, Kennedy JA, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540(7633):433–437. [DOI] [PubMed] [Google Scholar]
- 48.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818–822. [DOI] [PubMed] [Google Scholar]
- 49.Kirstetter P, Schuster MB, Bereshchenko O, et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13(4):299–310. [DOI] [PubMed] [Google Scholar]
- 50.Grove CS, Vassiliou GS. Acute myeloid leukaemia: a paradigm for the clonal evolution of cancer¿ Dis Model Mech. 2014;7(8):941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]