Abstract
We present an ultra-high resolution MRI dataset of an ex vivo human brain specimen. The brain specimen was donated by a 58-year-old woman who had no history of neurological disease and died of non-neurological causes. After fixation in 10% formalin, the specimen was imaged on a 7 Tesla MRI scanner at 100 µm isotropic resolution using a custom-built 31-channel receive array coil. Single-echo multi-flip Fast Low-Angle SHot (FLASH) data were acquired over 100 hours of scan time (25 hours per flip angle), allowing derivation of synthesized FLASH volumes. This dataset provides an unprecedented view of the three-dimensional neuroanatomy of the human brain. To optimize the utility of this resource, we warped the dataset into standard stereotactic space. We now distribute the dataset in both native space and stereotactic space to the academic community via multiple platforms. We envision that this dataset will have a broad range of investigational, educational, and clinical applications that will advance understanding of human brain anatomy in health and disease.
Subject terms: Brain, Brain injuries, Magnetic resonance imaging, Brain imaging
| Measurement(s) | nuclear magnetic resonance assay |
| Technology Type(s) | MRI Scanner |
| Factor Type(s) | flip angle |
| Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.9958688
Background & Summary
Postmortem ex vivo MRI provides significant advantages over in vivo MRI for visualizing the microstructural neuroanatomy of the human brain. Whereas in vivo MRI acquisitions are constrained by time (i.e. ~hours) and affected by motion, ex vivo MRI can be performed without time constraints (i.e. ~days) and without cardiorespiratory or head motion. The resultant advantages for characterizing neuroanatomy at microscale are particularly important for identifying cortical layers and subcortical nuclei1–5, which are difficult to visualize even in the highest-resolution in vivo MRI datasets6,7. Ex vivo MRI also provides advantages over histological methods that are associated with distortion and tearing of human brain tissue during fixation, embedding, and slide-mounting.
As the field of ex vivo MRI has developed over the past two decades, several laboratories have focused on imaging blocks of tissue from human brain specimens using small-bore scanners2,8 and specialized receive coils9–11. This approach allows for spatial resolutions of up to 35–75 microns for analyses of specific neuroanatomic regions9,11–13. However, ultra-high resolution imaging of whole human brain specimens at high magnetic field strengths has been far more challenging, due to the need for multi-channel receive coils and large-bore clinical scanners that can accommodate a whole-brain specimen. Whole-brain imaging is required to observe neuroanatomic relationships across distant brain regions, as well as to provide a complete view of human neuroanatomy in standard stereotactic space.
Here, we report the results of a multidisciplinary effort to image a whole human brain specimen ex vivo at an unprecedented spatial resolution of 100 µm isotropic. Central to this effort was the construction of an integrated system consisting of a custom-built 31-channel receive array coil and volume transmit coil, which was designed to accommodate and tightly enclose an ex vivo human brain14. The scans were performed on a 7 Tesla whole-body human MRI scanner using four single-echo spoiled gradient-recalled echo (SPGR/GRE) or Fast Low-Angle SHot (FLASH) sequences. We used varying flip-angles (FA15°, FA20°, FA25°, FA30°) to generate multiple synthesized volumes, each of which is a recomputed image that provides a different tissue contrast. The scans, performed over ~100 hours (~25 hours per FA), generated an ~8 TB dataset (~2 TB per flip angle) that required custom-built computational tools for offline MRI reconstruction and creation of the synthesized volumes. Offline MRI reconstruction considerably reduces the data amount. We release the resulting FA25° acquisition, as well as the synthesized FLASH25 volume here, both in native space and coregistered to standard stereotactic space, for use by the academic community. We envision a broad range of investigational, educational, and clinical applications for this dataset that have the potential to advance understanding of human brain anatomy in health and disease.
Methods
Specimen acquisition and processing
A 58-year-old woman with a history of lymphoma and stem cell transplantation, but no history of neurological or psychiatric disease, died in a medical intensive care unit. She was initially admitted to the hospital for fevers, chills, and fatigue, and then was transferred to the intensive care unit for hypoxic respiratory failure requiring mechanical ventilation. Her hospital course was also notable for a deep venous thrombosis and a pulmonary embolism. The cause of her death on hospital day 15 was determined to be hypoxic respiratory failure due to viral pneumonia. At the time of her death, her surrogate decision-maker provided written informed consent for a clinical autopsy and for donation of her brain for research, as part of a protocol approved by our Institutional Review Board.
At autopsy, her fresh brain weighed 1,210 grams (normal range = 1,200 to 1,500 grams). The brain was fixed in 10% formalin 14 hours after death. Gross examination revealed a normal brain (Fig. 1), without evidence of mass lesions or cerebrovascular disease. To ensure adequate fixation and prevent specimen flattening (which can prevent specimens from fitting into custom ex vivo MRI coils), we followed a series of standard specimen processing procedures, as previously described15.
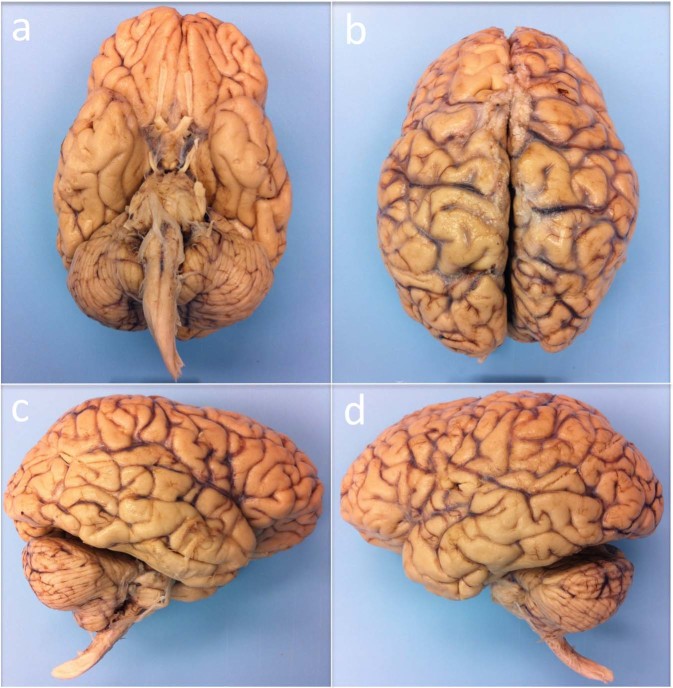
Fig. 1.
Human brain specimen. The human brain specimen that underwent ex vivo MRI is shown from inferior (a), superior (b), right lateral (c) and left lateral (d) perspectives. Gross pathological examination of the brain was normal.
Specimen preparation for scanning
After remaining in fixative for 35 months, the brain specimen was transferred to Fomblin Y LVAC 06/6 (perfluoropolyether, Solvay Specialty Polymers USA, LLC, West Deptford, NJ), which is invisible to MRI and reduces magnetic susceptibility artifacts. The specimen, immersed in Fomblin, was then secured inside a custom-built, air-tight brain holder made of rugged urethane16. The brain holder contains degassing ports for removal of air bubbles, which further reduces magnetic susceptibility artifacts.
Construction of a receive array coil and transmit volume coil for ex vivo imaging of the whole human brain
We built a receive coil apparatus consisting of a 31-channel surface coil loop array with two halves. The apparatus was fabricated using a 3D printer of slightly larger dimensions than the brain holder, which slides inside the single-channel birdcage volume transmit coil (Fig. 2). The brain holder is an oblate spheroid (16 × 19 cm) that conforms to the shape of a whole brain (cerebral hemispheres + cerebellum + brainstem)16 (Fig. 2d). It is made of two separate halves that can be sealed together with a silicone gasket after packing the brain inside. This holder must also withstand the degassing process when under vacuum pressure. Degassing is performed in three steps: 1) introducing vacuum suction into the container with the brain inside, which allows the bubbles to expand under decreased pressure and exit tissue cavities; 2) opening the valve to fill the holder with fomblin and then sealing off the fill valve; and 3) continuation of vacuum suction with low-amplitude vibration of the holder for 2–6 hours. The vibration facilitates the removal of bubbles from tissue cavities. All three steps are performed inside a fume hood.
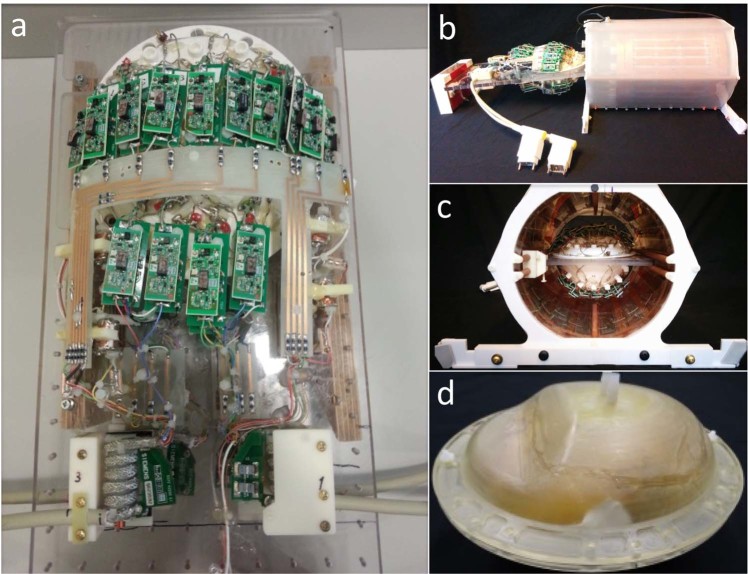
Fig. 2.
Receive array coil and transmit volume coil for ex vivo imaging of the whole human brain. (a) The 31-channel receive array has 15 elements on the top half (with a diameter of 5.5 cm) and 16 on the bottom half (with a diameter of 8.5 cm), each made of 16 AWG wire loops with four or five evenly spaced capacitors. All elements are tuned to 297.2 MHz. (c) The coil former has slightly larger dimensions than the brain holder, which slides inside a volume coil (b). (d) A custom air-tight brain holder was designed to conform to the shape of a whole human brain. The brain holder is an oblate spheroid container (16 × 19 cm) with degassing ports that are used to apply a vacuum suction, thereby reducing air bubbles in the specimen and surrounding fomblin solution.
The coil former (Fig. 2c) consists of two halves and encloses the brain holder. The receive array coil consists of 31 detectors (Fig. 2a), with 15 elements on the top half (diameter = 5.5 cm) and 16 on the bottom half (diameter = 8.5 cm). Coil elements were constructed using 16 AWG wire loops17, each with four or five evenly spaced capacitors (Supplementary Fig. 1). All elements were tuned to 297.2 MHz and matched to a loaded impedance of 75 Ω to minimize preamplifier noise. Preamplifier decoupling was achieved with a cable length of 6 cm. Preamplifiers were placed directly on the coil elements, yielding a substantial reduction in cable losses compared to a previous 30-channel ex vivo brain array18. The active detuning circuit was formed across the match capacitor using an inductor and PIN diode.
Tuning, matching, and decoupling of neighboring elements was optimized on the bench with a brain sample immersed in periodate-lysine-paraformaldehyde (PLP) solution. Because coil loading varies with the fixative used, the coil must be tuned and matched on the bench using a brain sample with the correct fixative. (For example, testing can be performed with a brain sample immersed in PLP or formalin, but not the regular loading phantom comprised of water and salt). Loops tuned/matched on PLP showed unloaded-to-loaded quality factor ratio (Q-ratio) of QUL/QL = 210/20 = 10.5, corresponding to an equivalent noise resistance of 11 ohms for the loaded coil (Q = wL/R). By contrast, formalin is a less lossy fixative, giving a coil Q-ratio of QUL/QL = 210/60 = 3.5, corresponding to an equivalent noise resistance of 4 ohms.
A shielded detunable volume coil (Fig. 2) was built for excitation, with the following parameters and features: band-pass birdcage, diameter 26.7 cm, and an extended length of 32 cm to accommodate brain samples of larger dimensions. For the detuning circuit we used diodes in every leg of the birdcage. These diodes are powered with the high-power chokes, which can withstand high voltage and short duration inversion pulses.
In summary, this coil system incorporates an improved mechanical design, preamps mounted at the coil detectors, and an extended transmit coil design capable of producing high-power pulses.
7 Tesla MRI data acquisition
The brain specimen was scanned on a whole-body human 7 Tesla (7 T) Siemens Magnetom MRI scanner (Siemens Healthineers, Erlangen, Germany) with the custom-built coil described above. We utilized a GRE sequence19 at 100 µm isotropic spatial resolution with the following acquisition parameters: TR = 40 msec, TE = 14.2 msec, bandwidth = 90 Hz/px, FA = 15°, 20°, 25°, 30°. Total scan time for each FA was 25:01:52 [hh:mm:ss], and each FA acquisition generated 1.98 TB of raw k-space data. To improve the signal-to-noise ratio (SNR) and optimise T1 modelling, we collected FLASH scans at four FAs (Fig. 3). Accounting for localizers, quality assurance (QA) scans, and adjustment scans, the total scan time was 100 hours and 8 minutes, and we collected nearly 7.92 TB of raw k-space data.
Fig. 3.
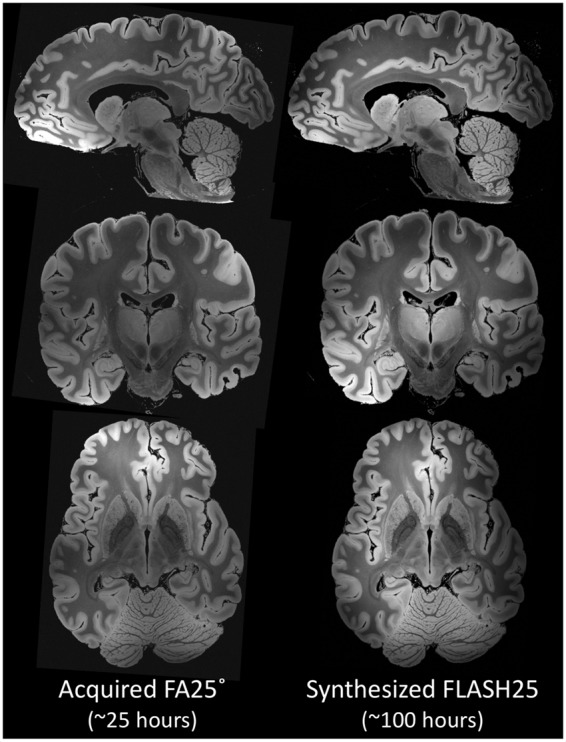
Comparison of FA25° acquisition and synthesized FLASH25 volume. Representative images from the FA25° acquisition (left column) and the synthesized FLASH25 volume (right column) are displayed in the sagittal (top row), coronal (middle row) and axial (bottom row) planes. These images provide a qualitative comparison of the respective signal-to-noise properties of the FA25° acquisition (~25 hours) and the synthesized FLASH25 volume (~100 hours). All images are shown in radiologic convention.
MRI data reconstruction
The size of the k-space data exceeded the storage capacity of the RAID provided by the scanner’s image reconstruction computer. The image reconstruction also required more RAM than what was available. We therefore implemented software on the scanner to stream the data directly via TCP/IP to a server on an external computer added to the scanner network, which saved the data as they were received. Because of additional limitations related to the total size of the raw data for any single scan, as dictated by the imager RAID size, we also divided each acquisition into segments. The server on the external computer stored the data as they were acquired, creating date stamps for every k-space segment.
After the scan was completed, the streamed k-space data were transferred to a computational server where we ran custom software to stitch together the segments, reconstruct the images for each channel (via a 3D FFT on each volume per channel20), and combine the images derived from the 31 channels via the root-sum-of-squares of the signal magnitudes at each voxel. These signal magnitudes were channel-wise decorrelated using a covariance matrix of the channels’ thermal noise. The output from coil combination was the final acquired image (Videos 1, 2 and 3)21.
MRI data processing
The acquired data underwent a series of processing steps, culminating in the creation of synthesized FLASH volumes, which are recomputed images incorporating all acquired measurements (Fig. 3; Videos 4, 5, 6, and 7)21. The volumes were estimated directly from the four FLASH acquisitions using the DESPOT1 algorithm19,22 with the program ‘mri_ms_fitparms’ distributed in FreeSurfer (http://surfer.nmr.mgh.harvard.edu)23 to quantify tissue properties independent of scanner and sequence types. This algorithm fits the tissue parameters (i.e. T1 and proton density) of the signal equation for the FLASH scan at each voxel using multiple input volumes. The volumes at the originally acquired TRs and FAs were then regenerated from the parameter maps by evaluating the FLASH signal equation. Of note, in this analysis we did not correct for B1 transmit inhomogeneities as we did not acquire the B1 transmit field map. Nevertheless, this method represents a disciplined approach to combining the volumes acquired with different FAs, while also providing a way to generate a variety of contrasts for distinguishing between multiple tissue classes.
In principle, a volume with any TR and FA combination could be synthesized. These synthesized volumes are created from all the acquired data and therefore have better SNR than the individually acquired input volumes. We choose to release the 25 degree synthetic volume (FLASH25) as it has, on average, maximal SNR and the best apparent contrast for cortical and subcortical structures9. Representative images from the FLASH25 volume are shown in Figs 4, 5, and Supplementary Fig. 2.
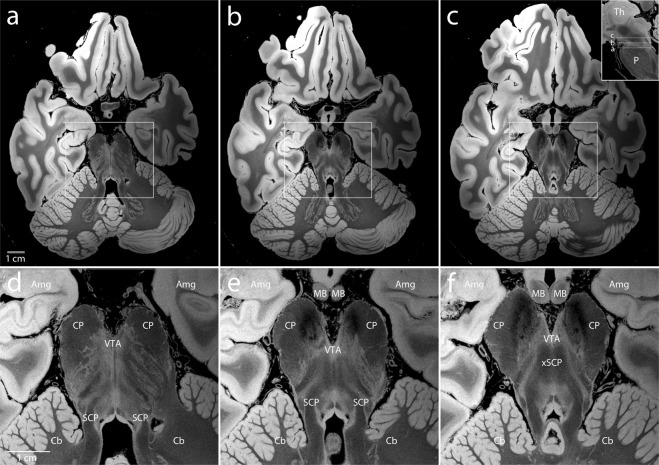
Fig. 4.
Delineation of brainstem neuroanatomy. Representative axial sections from the synthesized FLASH25 volume are shown at the level of the rostral pons and caudal midbrain (a-c, see inset in panel c). Zoomed views of the brainstem, medial temporal lobe, and anterior cerebellum (within the white rectangles in a–c) are shown in the bottom row (d–f). The anatomic detail that can be visualized in this ex vivo 100 μm resolution MRI dataset is beyond that which can be seen in typical in vivo MRI datasets. All images are shown in radiologic convention. Neuroanatomic abbreviations: Amg = amygdala; Cb = cerebellum; CP = cerebral peduncle; MB = mammillary body; P = pons; SCP = superior cerebellar peduncle; VTA = ventral tegmental area; xSCP = decussation of the superior cerebellar peduncle; Th = thalamus.
Fig. 5.
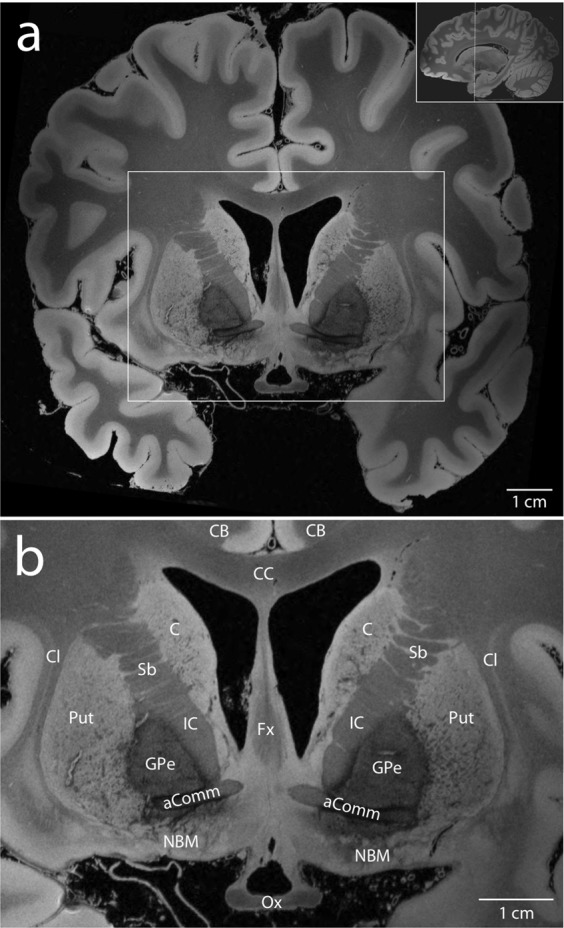
Delineation of basal ganglia and basal forebrain neuroanatomy. A representative coronal section from the synthesized FLASH25 volume is shown in the plane of the anterior commissure (aComm; see inset in a). A zoomed view of the basal ganglia and basal forebrain (within the white rectangle in a) is shown in (b). The anatomic detail that can be visualized in this ex vivo 100 μm resolution MRI dataset is beyond that which can be seen in standard in vivo MRI datasets. Neuroanatomic abbreviations: C = caudate; CB = cingulum bundle; CC = corpus callosum; Cl = claustrum; Fx = fornix; GPe = globus pallidus externa; IC = internal capsule; NBM = nucleus basalis of Meynert; Ox = optic chiasm; Put = putamen; Sb = striatal bridges.
Of note, ex vivo MRI of the fixed human brain yields a different contrast than in vivo MRI, mainly from a shortened T1, but also from a decrease in T2*, both of which are related to formalin fixation24. The predominant source of signal contrast in ex vivo MRI is likely myelin25 and/or iron26. Specifically, myelin appears to be a source of T1 contrast, while cortical iron appears to be a source of T2* contrast27.
Coregistration of the dataset to standard stereotactic space
The dataset was spatially normalized into the MNI ICBM 2009b NLIN ASYM template28 (Supplementary Fig. 3a). This template constitutes the newest version of the “MNI space” and is considered a high-resolution version of MNI space because it is available at 0.5 mm isotropic resolution. To combine structural information present on T1 and T2 versions of the template, we created a joint template using PCA, as previously described29. The four synthesized FLASH volumes (FLASH15, FLASH20, FLASH25, and FLASH30) were downsampled to isotropic voxel-sizes of 0.5 mm for spatial normalization and initially registered into template space in a multispectral approach using Advanced Normalization Tools (ANTs; http://stnava.github.io/ANTs/)30. This multispectral approach simultaneously accounts for intensity data in all four volumes. The initial normalization was performed in four stages (rigid body, affine, whole brain SyN and subcortically focused SyN) as defined in the “effective: low variance + subcortical refine” preset implemented in Lead-DBS v2.0 software (www.lead-dbs.org)31. Lead-DBS is a MATLAB-based, version-controlled software package whose code is available at GitHub (www.github.com/netstim/leaddbs).
To refine the warp, we introduced fiducial regions of interest (ROI) iteratively using a tool developed for this task (available within Lead-DBS). Specifically, we manually drew line and point fiducial markers in both native and template spaces (Supplementary Fig. 3b). In addition, we manually segmented four structures in native space (subthalamic nucleus, internal and external pallidum and red nucleus). The three types of fiducials (line ROI, spherical ROI and manual segmentations of key structures) were then added as “spectra” in subsequent registration refinements (Supplementary Fig. 3c). Thus, the final registration consisted of a large number of pairings between native and template space (the first four being the actual anatomical volumes, the subsequent ones being manual segmentations and paired helper fiducials). To achieve maximal registration precision, the warp was refined in 132 iterations with extensive manual expert interaction, each refinement continuing directly from the last saved state. We used linear interpolation to create the normalized files in the data release21. Because the registration required manual segmentations and fiducial markers created by a neuroanatomy expert (A.H.), the registration is not replicable using code. Nevertheless, segmentations and fiducial markers, as well as final warp-files (which can be applied to the dataset using the ANTs package), are available upon request.
Data Records
The native space FA25° acquisition and synthesized FLASH25 volume are available for download at the Dryad Digital Repository21 and OpenNeuro32. Additional synthesized volumes are available upon request to the corresponding author. Axial, coronal, and sagittal videos of the native space FA25° acquisition (Videos 1, 2, and 3) and synthesized FLASH25 volume (Videos 4, 5, 6, and 7) are also available at the Dryad Digital Repository21 and OpenNeuro32. The synthesized FLASH25 volume is available for interactive, online viewing at https://histopath.nmr.mgh.harvard.edu/100micronMRI. The normalized FLASH25 volume in standard stereotactic space is available at the Dryad Digital Repository21, OpenNeuro32, and is hosted on www.lead-dbs.org (preinstalled as part of the Lead-DBS software package).
To view the nifti volumes released with this dataset, we recommend using the FreeView viewer, which can be downloaded at: https://surfer.nmr.mgh.harvard.edu/fswiki/UpdateFreeview. FreeView is compatible with Linux, Mac, and Windows (via VirtualBox) operating systems. FreeView provides an opportunity to visualize the data in 3-dimensions, while also performing basic measurements and segmentations, and taking screenshots. With respect to system requirements, we recommend at least 8 GB of RAM and a processor speed of 2 GHz. For users whose computing hardware does not support visualization of the 100 micron resolution volumes, we downsampled the acquired FA25° volume, synthesized FLASH25 volume, and MNI-coregistered FLASH25 volume to 200 micron and 500 micron resolution. All downsampled volumes are available at Dyrad Digital Repository21 and OpenNeuro32. Users can also downsample the 100 micron volumes to their preferred spatial resolution using commands provided in the “README_code” file on OpenNeuro32.
To optimize the potential applications of this dataset, we also release the acquired volumes for all four flip angles (FA15°, FA20°, FA25°, FA30°) as source data on Open Neuro32, along with code to process the source data. With this code, users can create derivative volumes according to their preferences. We also provide the full set of sequence parameters from this scan on Dryad Digital Repository21 and OpenNeuro32.
Technical Validation
Coil signal-to-noise ratio (SNR) measurements
The receive coil has a QUL/QL ratio that ranged from 6 in the top half elements to 8 in the bottom half elements due to larger coil diameter. The S12 coupling between neighbouring elements, measured with all other coils active detuned, ranged from −10.9 to −24 dB. All individual elemnts had S11 < −20 dB and active detuning of >30 dB. We evaluated the performance of the transmit coil by examining the B1+ profile14, which shows the efficiency throughout the entire spatial distribution of the brain specimen. The efficiency was greatest in the center of the specimen and fell off gradually towards the edges, as expected for a whole brain specimen at 7 T.
We compared the SNR of the 31-channel ex vivo array to that of a standard 31-channel 7 T head coil and a 64-channel 3 T head coil. SNR maps were computed following the method of Kellman & McVeigh33. We calibrated the voltage required for a 180° pulse using a B1+ map (estimated with the AFI method)34 with an ROI of 3-cm diameter at the center of the brain. We estimated array noise covariance from thermal noise data acquired without RF excitation. The SNR gain with the 31-channel ex vivo array was 1.6-fold versus the 31-channel 7 T standard coil and 3.3-fold versus the 64-channel 3 T head array (Fig. 6). The noise coupling between channels was 11% for the 31-channel ex vivo array, a 2-fold improvement relative to our previous array18.
Fig. 6.
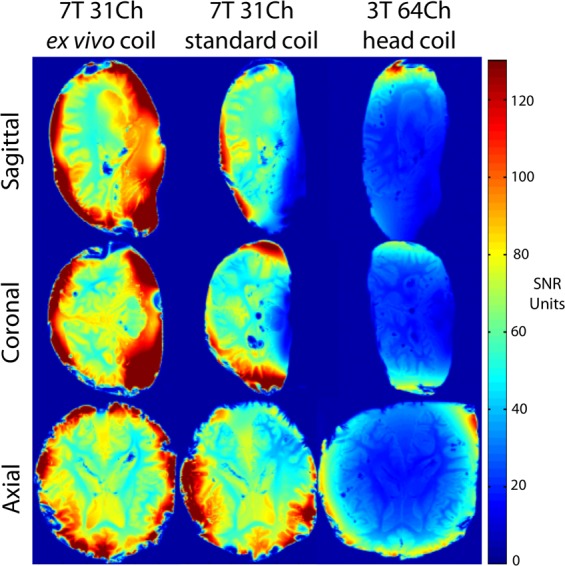
Signal-to-noise ratio (SNR) analysis of coil performance. Representative SNR maps are shown in the sagittal (top row), coronal (middle row) and axial (bottom row) planes for a test brain sample immersed in periodate-lysine-paraformaldehyde. The maps demonstrate an SNR gain of 1.6-fold for the 31-channel 7 Tesla (7 T) ex vivo coil (left column) compared to the 31-channel 7 T standard coil (middle column), and a gain of 3.3-fold compared to the 64-channel 3 T head coil (right column). The noise coupling between channels was 11% for the 31-channel ex vivo coil array, a 2-fold improvement relative to our previous array18.
Coregistration accuracy
We assessed the neuroanatomic accuracy of the final registration results (i.e. the fit between structures on the normalized FLASH volumes versus the high-resolution MNI template) by visual inspection using a tool specifically designed for this task (implemented in Lead-DBS). An example of this visual inspection assessment for the subthalamic nucleus and globus pallidus interna is provided in Supplementary Fig. 4. The normalized FLASH25 volume is distributed pre-installed within Lead-DBS software and can be selected for visualization in the 3D viewer (www.lead-dbs.org). Figure 7 shows an example of deep brain stimulation electrode reconstructions in a hypothetical patient being treated for Parkinson’s Disease.
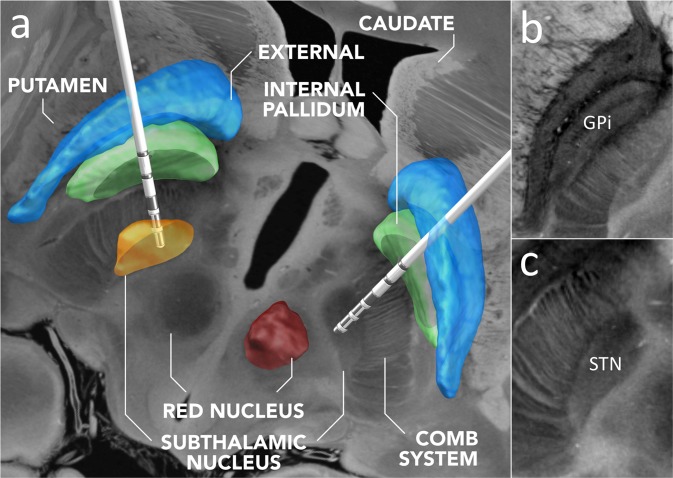
Fig. 7.
Normalization of the ex vivo MRI dataset into standard stereotactic space and integration into the Lead-DBS software platform. (a) Exemplary use-case of the normalized FLASH25 volume in deep brain stimulation (DBS). DBS electrodes are visualized for a hypothetical patient using Lead-DBS software (https://www.lead-dbs.org)31. An axial image from the normalized scan, at the level of the rostral midbrain, is shown as a backdrop, with 3D-structures defined by the DISTAL atlas35 (right subthalamic and left red nucleus hidden for optimal visualization of the underlying anatomy). Panels (b) and (c) show zoomed views of key DBS target regions: the left globus pallidus interna (GPi in b) and subthalamic nucleus (STN in c).
Supplementary information
Acknowledgements
We thank Michelle Siciliano and Terrence Ott for assistance in obtaining and processing the brain specimen; Simon Sigalovsky for assistance with coil construction; Gunjan Madan for assistance with coil testing and evaluation; L. Daniel Bridgers for constructing the brain container and coil array housing; Andrew Hoopes for assistance with creation of visual media; and Paul Raines for expert technical support in implementing the online Biolucida viewer for this dataset. This work was supported by the National Institutes of Health (NIH) National Institute of Neurological Disorders and Stroke (K23-NS094538, R01-NS052585, R21-NS072652, R01-NS070963, R01-NS083534, U01-NS086625, R21-NS109627), National Institute for Biomedical Imaging and Bioengineering (P41-EB015896, R01-EB006758, R21-EB018907, R01-EB019956, R01-EB023281, R00-EB021349), National Institute on Aging (R01-AG057672, R01-AG022381, R01-AG008122, R01-AG016495, R01-AG008122, U01-AG006781, R21-AG046657, P41-RR014075, P50-AG005136, R56-AG064027), National Center for Alternative Medicine (RC1-AT005728), Eunice Kennedy Shriver National Institute of Child Health and Human Development (K01-HD074651, R01-HD071664, R00-HD074649), NIH Director’s Office (DP2-HD101400) and the Centers for Disease Control and Prevention (R49-CE001171). Support for this research was also provided by the BRAIN Initiative Cell Census Network (U01-MH117023) and the NIH Blueprint for Neuroscience Research (U01-MH093765) as part of the multi-institutional Human Connectome Project. This research utilized resources provided by the National Center for Research Resources (U24-RR021382) and by NIH shared instrumentation grants S10-RR023401, S10-RR019307, and S10-RR023043. Additional support was provided by the James S. McDonnell Foundation, Rappaport Foundation, Tiny Blue Dot Foundation, and the German Research Foundation (Emmy Noether Grant 410169619).
Author contributions
B.L.E. designed the study, analyzed the data, and prepared the manuscript. A.M. built the coil, acquired and analyzed the data, and contributed to the manuscript. A.H. created the warp from native space to standard stereotactic space, performed the coregistration for Lead-DBS implementation, and contributed to the manuscript. J.R.P. designed the study, acquired and analyzed the data, and contributed to the manuscript. T.W. acquired the data and contributed to the manuscript. M.D.T. acquired and analyzed the data, and contributed to the manuscript. J.A. designed the study, acquired and analyzed the data, and contributed to the manuscript. J.P.S. advised on the building and testing of the coil, and contributed to the manuscript. B.R.D. analyzed the data and contributed to the manuscript. A.S. acquired and analyzed the data, and contributed to the manuscript. L.S.T. processed and analyzed the data, and contributed to the manuscript. R.D.F. performed the pathological assessment and contributed to the manuscript. L.L.W. supervised the building of the coil and contributed to the manuscript. B.F. supervised and designed the study, analyzed the data, and contributed to the manuscript. A.v.d.K. supervised and designed the study, acquired and analyzed the data, and contributed to the manuscript.
Code availability
Neuroimaging data were processed using standard processing pipelines in FreeSurfer v6.023 (http://surfer.nmr.mgh.harvard.edu, https://github.com/freesurfer/freesurfer). All code used for registration of volumes into standard stereotactic space is available within the open-source Lead-DBS v2.0 software31 (https://github.com/leaddbs/leaddbs). Because registration involved multiple manual user interface steps, no ready-made code is provided, but the process can be readily reproduced with the provided data and software.
Competing interests
None of the authors has a conflicting financial interest. Dr. Fischl and Mr. Tirrell have financial interest in CorticoMetrics, a company whose medical pursuits focus on brain imaging and measurement technologies. Their interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Bruce Fischl and Andre van der Kouwe.
Supplementary information
is available for this paper at 10.1038/s41597-019-0254-8.
References
- 1.Augustinack JC, van der Kouwe AJ, Fischl B. Medial temporal cortices in ex vivo magnetic resonance imaging. J Comp Neurol. 2013;521:4177–4188. doi: 10.1002/cne.23432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edlow BL, et al. Neuroanatomic connectivity of the human ascending arousal system critical to consciousness and its disorders. J Neuropathol Exp Neurol. 2012;71:531–546. doi: 10.1097/NEN.0b013e3182588293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McNab JA, et al. The Human Connectome Project and beyond: Initial applications of 300mT/m gradients. Neuroimage. 2013;80:234–245. doi: 10.1016/j.neuroimage.2013.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roebroeck A, Miller KL, Aggarwal M. Ex vivo diffusion MRI of the human brain: Technical challenges and recent advances. NMR Biomed. 2019;32:e3941. doi: 10.1002/nbm.3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aggarwal M, et al. Feasibility of creating a high-resolution 3D diffusion tensor imaging based atlas of the human brainstem: a case study at 11.7 T. Neuroimage. 2013;74:117–127. doi: 10.1016/j.neuroimage.2013.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lusebrink F, Sciarra A, Mattern H, Yakupov R, Speck O. T1-weighted in vivo human whole brain MRI dataset with an ultrahigh isotropic resolution of 250 mum. Sci Data. 2017;4:170032. doi: 10.1038/sdata.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horn A, et al. Teaching NeuroImages: In vivo visualization of Edinger comb and Wilson pencils. Neurology. 2019;92:e1663–e1664. doi: 10.1212/WNL.0000000000007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi E, Song JW, Folkerth RD, Grant PE, Schmahmann JD. Detection of postmortem human cerebellar cortex and white matter pathways using high angular resolution diffusion tractography: a feasibility study. Neuroimage. 2013;68:105–111. doi: 10.1016/j.neuroimage.2012.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Augustinack JC, et al. Detection of entorhinal layer II using 7Tesla [corrected] magnetic resonance imaging. Ann Neurol. 2005;57:489–494. doi: 10.1002/ana.20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gangolli M, et al. Quantitative validation of a nonlinear histology-MRI coregistration method using generalized Q-sampling imaging in complex human cortical white matter. Neuroimage. 2017;153:152–167. doi: 10.1016/j.neuroimage.2017.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Veluw SJ, et al. Microbleed and microinfarct detection in amyloid angiopathy: a high-resolution MRI-histopathology study. Brain. 2016;139:3151–3162. doi: 10.1093/brain/aww229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sengupta S, et al. High resolution anatomical and quantitative MRI of the entire human occipital lobe ex vivo at 9.4T. Neuroimage. 2018;168:162–171. doi: 10.1016/j.neuroimage.2017.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Kouwe, A. et al. High Resolution Structural and Diffusion MRI of Ex Vivo Human Motor Cortex. Organization for Human Brain Mapping. Abstract #4427 (2011).
- 14.Mareyam, A. et al. Array coil and sample preparation and support system for whole brain ex vivo imaging at 100 μm. International Society for Magnetic Resonance in Medicine. Abstract #3130 (2015).
- 15.Edlow BL, et al. Multimodal Characterization of the Late Effects of Traumatic Brain Injury: A Methodological Overview of the Late Effects of Traumatic Brain Injury Project. J Neurotrauma. 2018;35:1604–1619. doi: 10.1089/neu.2017.5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bridgers, L. D. Design and Manufacture of an Ultra-High Field Ex Vivo Coil Assembly, Massachusetts Institute of Technology (2012).
- 17.Keil B, et al. Size-optimized 32-channel brain arrays for 3 T pediatric imaging. Magn Reson Med. 2011;66:1777–1787. doi: 10.1002/mrm.22961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mareyam, A., Polimeni, J. R., Alagappan, V., Fischl, B. & Wald, L. L. A 30 channel receive-only 7 T array for ex vivo brain hemisphere imaging. International Society for Magnetic Resonance in Medicine. Abstract #106 (2009).
- 19.Fischl B, et al. Sequence-independent segmentation of magnetic resonance images. Neuroimage. 2004;23(Suppl 1):S69–84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 20.Frigo M, Johnson SG. The Design and Implementation of FFTW3. Proceedings of the IEEE. 2005;93:216–231. doi: 10.1109/JPROC.2004.840301. [DOI] [Google Scholar]
- 21.Edlow BL, 2019. Data from: 7 Tesla MRI of the ex vivo human brain at 100 micron resolution. Dryad Digital Repository. [DOI] [PMC free article] [PubMed]
- 22.Deoni SC, Peters TM, Rutt BK. High-resolution T1 and T2 mapping of the brain in a clinically acceptable time with DESPOT1 and DESPOT2. Magn Reson Med. 2005;53:237–241. doi: 10.1002/mrm.20314. [DOI] [PubMed] [Google Scholar]
- 23.Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tovi M, Ericsson A. Measurements of T1 and T2 over time in formalin-fixed human whole-brain specimens. Acta Radiol. 1992;33:400–404. doi: 10.1177/028418519203300503. [DOI] [PubMed] [Google Scholar]
- 25.Eickhoff S, et al. High-resolution MRI reflects myeloarchitecture and cytoarchitecture of human cerebral cortex. Hum Brain Mapp. 2005;24:206–215. doi: 10.1002/hbm.20082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukunaga M, et al. Layer-specific variation of iron content in cerebral cortex as a source of MRI contrast. Proc Natl Acad Sci U S A. 2010;107:3834–3839. doi: 10.1073/pnas.0911177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuber C, et al. Myelin and iron concentration in the human brain: a quantitative study of MRI contrast. Neuroimage. 2014;93(Pt 1):95–106. doi: 10.1016/j.neuroimage.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 28.Fonov VS, Evans AC, McKinstry RC, Almli CR, Collins D. Unbiased nonlinear average age-appropriate brain templates from birth to adulthood. Neuroimage. 2009;47:S102. doi: 10.1016/S1053-8119(09)70884-5. [DOI] [Google Scholar]
- 29.Horn A, 2017. PCA MNI 2009b NLIN template. figshare. [DOI]
- 30.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical image analysis. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horn A, et al. Lead-DBSv2: Towards a comprehensive pipeline for deep brain stimulation imaging. Neuroimage. 2019;184:293–316. doi: 10.1016/j.neuroimage.2018.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edlow BL, 2019. 7 Tesla MRI of the ex vivo human brain at 100 micron resolution. OpenNeuro. [DOI] [PMC free article] [PubMed]
- 33.Kellman P, McVeigh ER. Image reconstruction in SNR units: a general method for SNR measurement. Magn Reson Med. 2005;54:1439–1447. doi: 10.1002/mrm.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yarnykh VL. Actual flip-angle imaging in the pulsed steady state: a method for rapid three-dimensional mapping of the transmitted radiofrequency field. Magn Reson Med. 2007;57:192–200. doi: 10.1002/mrm.21120. [DOI] [PubMed] [Google Scholar]
- 35.Ewert S, et al. Toward defining deep brain stimulation targets in MNI space: A subcortical atlas based on multimodal MRI, histology and structural connectivity. Neuroimage. 2018;170:271–282. doi: 10.1016/j.neuroimage.2017.05.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Edlow BL, 2019. Data from: 7 Tesla MRI of the ex vivo human brain at 100 micron resolution. Dryad Digital Repository. [DOI] [PMC free article] [PubMed]
- Horn A, 2017. PCA MNI 2009b NLIN template. figshare. [DOI]
- Edlow BL, 2019. 7 Tesla MRI of the ex vivo human brain at 100 micron resolution. OpenNeuro. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Neuroimaging data were processed using standard processing pipelines in FreeSurfer v6.023 (http://surfer.nmr.mgh.harvard.edu, https://github.com/freesurfer/freesurfer). All code used for registration of volumes into standard stereotactic space is available within the open-source Lead-DBS v2.0 software31 (https://github.com/leaddbs/leaddbs). Because registration involved multiple manual user interface steps, no ready-made code is provided, but the process can be readily reproduced with the provided data and software.