Abstract
Background and Aims
Miscanthus, a C4 perennial grass native to East Asia, is a promising biomass crop. Miscanthus sacchariflorus has a broad geographic range, is used to produce paper in China and is one of the parents (along with Miscanthus sinensis) of the important biomass species Miscanthus × giganteus. The largest study of M. sacchariflorus population genetics to date is reported here.
Methods
Collections included 764 individuals across East Asia. Samples were genotyped with 34 605 single nucleotide polymorphisms (SNPs) derived from restriction site-associated DNA sequencing (RAD-seq) and ten plastid microsatellites, and were subjected to ploidy analysis by flow cytometry.
Key Results
Six major genetic groups within M. sacchariflorus were identified using SNP data: three diploid groups, comprising Yangtze (M. sacchariflorus ssp. lutarioriparius), N China and Korea/NE China/Russia; and three tetraploid groups, comprising N China/Korea/Russia, S Japan and N Japan. Miscanthus sacchariflorus ssp. lutarioriparius was derived from the N China group, with a substantial bottleneck. Japanese and mainland tetraploids originated from independent polyploidization events. Hybrids between diploid M. sacchariflorus and M. sinensis were identified in Korea, but without introgression into either parent species. In contrast, tetraploid M. sacchariflorus in southern Japan and Korea exhibited substantial hybridization and introgression with local diploid M. sinensis.
Conclusions
Genetic data indicated that the land now under the Yellow Sea was a centre of diversity for M. sacchariflorus during the last glacial maximum, followed by a series of migrations as the climate became warmer and wetter. Overall, M. sacchariflorus has greater genetic diversity than M. sinensis, suggesting that breeding and selection within M. sacchariflorus will be important for the development of improved M. × giganteus. Ornamental M. sacchariflorus genotypes in Europe and North America represent a very narrow portion of the species’ genetic diversity, and thus do not well represent the species as a whole.
Keywords: Miscanthus sacchariflorus, Miscanthus × giganteus, Miscanthus sinensis, Miscanthus sacchariflorus ssp, lutarioriparius, restriction site-associated DNA sequencing (RAD-seq), population genetics, polyploidy, plastid haplotype, hybridization, bioenergy
INTRODUCTION
Miscanthus is a genus of C4 perennial grasses in the Andropogoneae tribe that are obligately allogamous and have wind-dispersed seeds. The genus is native to East Asia (especially eastern areas with relatively high precipitation) and Oceania, ranging from tropical to cold temperate climates (Clifton-Brown et al., 2008; Sacks et al., 2013). Miscanthus × giganteus, which is a hybrid between Miscanthus sacchariflorus and Miscanthus sinensis, has recently attracted considerable attention as a feedstock crop for bioenergy and bioproducts (Hodkinson et al., 2002; Clifton-Brown et al., 2008; Dwiyanti et al., 2013b; Sacks et al., 2013). However, nearly all M. × giganteus that is currently grown commercially is a single triploid clone, ‘1993-1780’ (named after the type specimen at Kew Royal Botanic Gardens Herbarium; it is also commonly referred to as M. × giganteus ‘Illinois’ in North America; Hodkinson and Renvoize, 2001; Głowacka et al., 2015), which was collected in southern Japan and imported to Denmark in the 1930s (Nielsen, 1990; Linde-Laursen, 1993). Though biomass yields of M. × giganteus ‘1993-1780’ can exceed those of maize and switchgrass in temperate mid-latitude environments (Heaton et al., 2008; Dohleman and Long, 2009; Somerville et al., 2010), this clone can suffer stand losses during the first winter after planting in the central Midwest USA and locations with similarly cold or colder climates (Greef et al., 1997; Clifton-Brown and Lewandowski, 2000; Farrell et al., 2006; Clark et al., 2016). Yet, in the US southern coastal plain, M. × giganteus ‘1993-1780’ typically flowers too early to achieve the high yields obtained in the Midwest. Thus, breeding efforts are needed to generate improved M. × giganteus cultivars, and these efforts will depend on knowledge of its parent species’ genetic diversity, population structure and adaptation.
Miscanthus sacchariflorus is native to a broad geographic area in China, Korea, Japan and eastern Russia, from 28 to 50 °N and from sea level to approx. 2000 m elevation (Hirayoshi et al., 1957; Lee, 1964; Clifton-Brown et al., 2008; Sun et al., 2010; Sacks et al., 2013). The geographic ranges of M. sacchariflorus and M. sinensis largely overlap, although M. sacchariflorus can be found further north and M. sinensis can be found further south. Especially noteworthy is that M. sacchariflorus probably has the greatest winter hardiness among all the Saccharinae, with populations from northern China and eastern Russia adapted to an average annual minimum air temperature of –40.0 °C (USDA hardiness zone 3; Clark et al., 2016). Miscanthus sacchariflorus also differs from other Miscanthus species in its spreading habit due to long rhizomes and in its preference for wet soil in damp meadows or near the edges of lakes, rivers and streams (Dwiyanti et al., 2013b; Sacks et al., 2013). Miscanthus sacchariflorus ssp. lutarioriparius, which grows along the southern edge of the species’ range near the Yangtze River, has especially tall (3–7 m) and thick (10–20 mm) stems that are harvested to produce paper locally on an industrial scale (Chen and Renvoize, 2005; Sacks et al., 2013); it is adapted to USDA hardiness zones 8 and 9 (average annual minimum temperature of –12.2 to –1.2 °C). Miscanthus sacchariflorus ssp. lutarioriparius is sometimes designated as a separate species, although recent taxonomic evaluation and molecular results favour the subspecies status (Chen and Renvoize, 2005; Sun et al., 2010; Głowacka et al., 2015); a detailed study of its relationship with other populations of M. sacchariflorus has not been conducted previously.
Diploid (2x = 38) and tetraploid (4x = 76) forms of M. sacchariflorus are both common (Hirayoshi et al., 1957; Rayburn et al., 2009; Sun et al., 2010; Moon et al., 2013; Chae et al., 2014), in contrast to M. sinensis, which is nearly always diploid (Clark et al., 2014). However, neither differences in the geographic distribution between diploid and tetraploid M. sacchariflorus, nor the evolutionary relationships between these populations, have been fully described. Diploid M. sacchariflorus has been documented in China, Korea and eastern Russia (Li et al., 2013; Moon et al., 2013; Clark et al., 2016), but is apparently absent from Japan (Hirayoshi et al., 1957; Clark et al., 2015). Tetraploid M. sacchariflorus is common in Japan (Hirayoshi et al., 1957) and Korea (Moon et al., 2013) but less frequent in China (Li et al., 2013) and Russia (Clark et al., 2016). Although early cytogenetic studies suggested an allopolyploid origin for the tetraploids (Adati, 1959; Adati and Shiotani, 1962), recent molecular and cytogenetic studies support an autopolyploid origin from diploid M. sacchariflorus (Takahashi and Shibata, 2002; Dwiyanti et al., 2013a; Clark et al., 2015).
Previous population genetic studies of M. sacchariflorus have sampled from limited areas of the species native range, and no region-wide studies have yet been conducted. Yan et al. (2016) used microsatellite markers to study 644 M. sacchariflorus ssp. lutarioriparius individuals from 25 populations along or near the Yangtze River from 28.9 to 32.5 °N and 111.7 to 120.2 °E, finding high genetic diversity, low clonality and frequent migration among populations, with some anthropogenic influences on population structure. Yook et al. (2014) genotyped 22 M. sacchariflorus accessions using 31 microsatellite markers, and were able to distinguish Korean and non-Korean individuals, but did not find population structure at a finer scale. Using restriction site-associated DNA sequencing (RAD-seq), single nucleotide polymorphisms (SNPs) and plastid microsatellites, our research group has published several population genetic studies of Miscanthus that included two tetraploid M. sacchariflorus individuals from South Korea, two tetraploid and six diploid M. sacchariflorus from China and 11 diploid M. × giganteus from China (Clark et al., 2014), 78 M. sacchariflorus and M. × giganteus (tetraploids and triploids) from Japan (Clark et al., 2015) and 157 diploid and three tetraploid M. sacchariflorus from Russia (Clark et al., 2016). Our previous studies of Miscanthus in China, Korea and Japan lacked sufficient sample size to identify population structure within M. sacchariflorus. Population structure among M. sacchariflorus in Russia was limited to a weak signal of isolation by distance, as well as tetraploid individuals being genetically differentiated from diploids (Clark et al., 2016). The absence of a region-wide assessment of M. sacchariflorus genetic diversity and population structure has been a critical obstacle to using this germplasm efficiently for crop improvement; removing this barrier is a major goal of this study.
Although current knowledge of population structure in M. sacchariflorus is limited, hybridization with M. sinensis is a well-established phenomenon. Here we use the term M. × giganteus to refer to hybrids of any ploidy that have at least 20 % of their ancestry each from M. sacchariflorus and M. sinensis; this cut-off is arbitrary but includes first-generation hybrids (F1) as well as first-generation backcrosses (BC1) of all ploidies. Jiang et al. (2013) demonstrated that M. purpurascens, endemic to China, was a diploid M. × giganteus with equal genetic contributions from M. sinensis and M. sacchariflorus. However, there is no evidence that M. purpurascens introgresses into either parent species beyond the BC1 generation (Jiang et al., 2013; Clark et al., 2014). In Korea, Yook et al. (2014) identified natural M. × giganteus based on morphological data and microsatellite markers, although ploidy and the proportion of ancestry from the two parental species were not assessed. A separate study identified a triploid individual in Korea that was putatively M. × giganteus based on flow cytometry and chromosome counts (Moon et al., 2013). Perhaps owing to the Japanese origin of the high-yielding biomass cultivar M. × giganteus ‘1993-1780’, Japan is the region where Miscanthus hybridization has been most thoroughly studied. Both triploid and tetraploid M. × giganteus have been identified in southern Japan by flow cytometry, chromosome counting, internal transcribed sequence (ITS) sequencing, intron-flanking PCR markers, microsatellites and high-density SNP markers (Hirayoshi et al., 1957; Adati, 1958; Nishiwaki et al., 2011; Dwiyanti et al., 2013a; Clark et al., 2015; Tamura et al., 2016). Variation for agronomic traits such as biomass yield, plant height and spreading habit has been observed among triploid Japanese M. × giganteus accessions (Uwatoko et al., 2016). Unlike the diploid hybrids found in China, tetraploid M. × giganteus in Japan appears to intermate readily with M. sacchariflorus, resulting in substantial introgression of M. sinensis ancestry into tetraploid M. sacchariflorus (Clark et al., 2015). Based on chloroplast data, diploid, triploid and tetraploid M. ×giganteus can have either M. sinensis or M. sacchariflorus as the female parent (Jiang et al., 2013; Clark et al., 2014, 2015). Though interspecific hybridization and cross-ploidy introgression have been well documented in Japan, it is not known if similar processes occur in China and Korea where diploid M. sinensis and tetraploid M. sacchariflorus also grow sympatrically.
In this study, we characterized population structure and its interaction with ploidy for a large collection of M. sacchariflorus and M. × giganteus from China, South Korea, Japan and Russia ranging from 28.6 to 49.3 °N and from 104.5 to 145.2 °E. Our objectives were (1) to better delineate the geographic range and frequency of tetraploid M. sacchariflorus in mainland Asia; (2) to identify distinct genetic clusters and centres of diversity for M. sacchariflorus, and establish their relationship to each other in terms of ancestry and gene flow; (3) to quantify the amount of hybridization with M. sinensis and identify geographic regions in which hybridization is frequent; and (4) to compare M. sacchariflorus (data from the current study) with M. sinensis (data from prior studies) for degree of population structure, genetic diversity and locations of centres of diversity. Using high-density SNP markers obtained from RAD-seq, we provide an in-depth assessment of M. sacchariflorus population genetics throughout most of the species’ native range.
MATERIALS AND METHODS
Plant material, DNA extraction and genotyping
Plant material consisted primarily of M. sacchariflorus collected in the wild across East Asia, including Russia, China, South Korea and Japan (Table 1; Fig. 1). Collection sites were targeted based on herbarium records as well as the tendency of M. sacchariflorus to grow near water. With the goal of broad regional sampling, between one and five individuals were sampled at each collection site. In total, 764 individual Miscanthus genotypes were studied, comprising 722 M. sacchariflorus, 37 M. × giganteus and five M. sinensis (Tables 1 and 2; Fig. 1; Supplementary Data S1). Of the individuals analysed in the current study, 255 were included in previous studies that examined population structure and genetic relationships at smaller geographic scales (Clark et al., 2014, 2015, 2016; Głowacka et al., 2015). For the new individuals, rhizomes were collected in the wild (primarily in 2014; Supplementary Data S1) and propagated in greenhouses. DNA was extracted from lyophilized, ground leaf tissue using a CTAB (cetyltrimethylammonium bromide) method with minor modifications (Fulton et al., 1995; Clark et al., 2016). All individuals were genotyped at ten plastid microsatellites (de Cesare et al., 2010; Jiang et al., 2012) as previously described (Clark et al., 2014). Size separation of the PCR products was accomplished by capillary electrophoresis on a 3730 × l DNA Analyzer (Applied Biosystems, Foster City, CA, USA) with the GeneScan 500 LIZ size standard at the Roy J. Carver Biotechnology Center at the University of Illinois. RAD-seq using PstI and MspI with size selection from 200 to 500 bp was performed as previously described (Clark et al., 2014). A total of seven new RAD-seq libraries with 95 samples each were generated for the study, with data available on the NCBI Sequence Read Archive, accession SRP087645. RAD-seq data from our previous studies of M. sacchariflorus and M. × giganteus are available under accessions SRP026347, SRP048207 and SRP063572, and were also included in the current analysis.
Table 1.
Summary of provenance and ploidy for Miscanthus spp. individuals included in the study
| Country of origin | Total no. of individuals | No. of individuals from previous publications | Diploid | Triploid | Tetraploid | Not determined |
|---|---|---|---|---|---|---|
| M. sacchariflorus | ||||||
| Russia | 150 | 150* | 147 | 0 | 3 | 0 |
| South Korea | 106 | 1† | 27 | 1 | 78 | 0 |
| China | 262 | 8†,‡ | 206 | 2 | 52 | 2 |
| Japan | 194 | 82‡,§ | 0 | 0 | 184 | 10 |
| Unknown | 10 | 8†,‡ | 7 | 0 | 3 | 0 |
| Total | 722 | 249 | 387 | 3 | 320 | 12 |
| M. × giganteus | ||||||
| South Korea | 24 | 0 | 9 | 5 | 10 | 0 |
| Japan | 13 | 5§ | 0 | 4 | 9 | 0 |
| Total | 37 | 5 | 9 | 9 | 19 | 0 |
| M. sinensis | ||||||
| China | 3 | 0 | 3 | 0 | 0 | 0 |
| Japan | 1 | 0 | 0 | 0 | 0 | 1 |
| Unknown | 1 | 1‡ | 1 | 0 | 0 | 0 |
| Total | 5 | 1 | 4 | 0 | 0 | 1 |
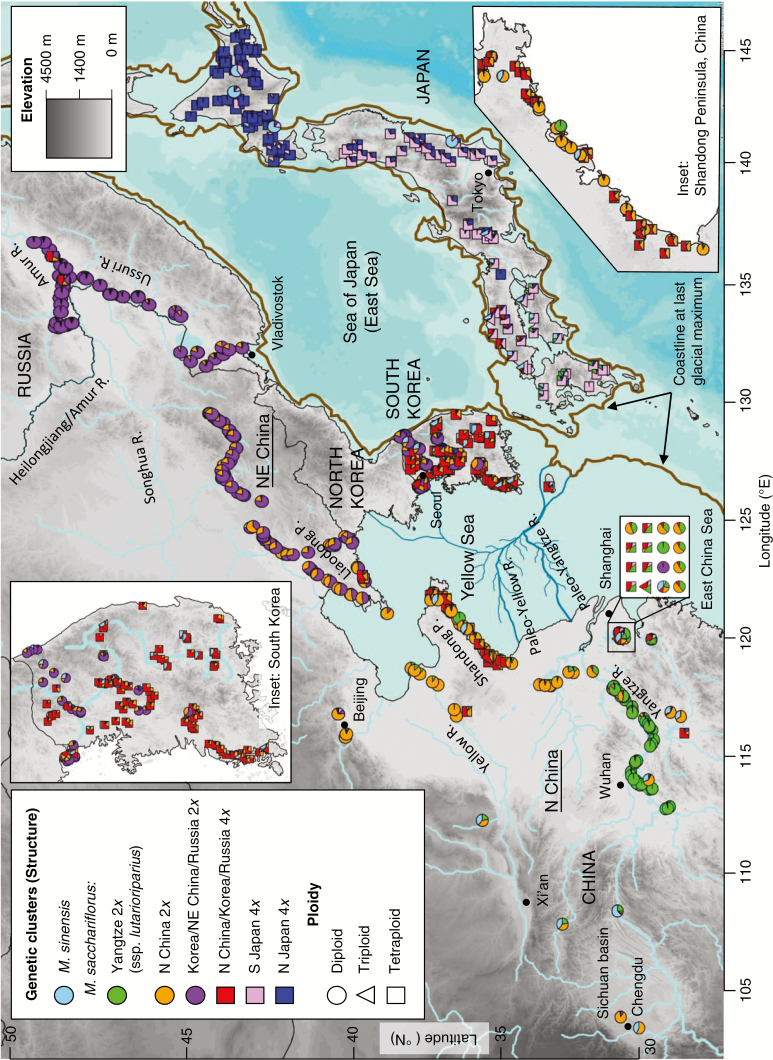
Fig. 1.
Map of M. sacchariflorus and M. × giganteus individuals indicating collection sites, ploidy and genetic relationships. Each accession is represented by a pie chart showing ancestry (Q) among seven genetic clusters as determined by analysis with the software Structure using 34 605 RAD-seq SNPs. All individuals from the current study with known collection site and ploidy are plotted, including 699 M. sacchariflorus and 38 M. × giganteus. Fourteen diploid M. × giganteus found in China and Japan in previous studies (Clark et al., 2014, 2015) are also included. Elevation is shown as a grey scale. The coastline at the last glacial maximum is indicated by a brown line drawn according to Lee et al. (2008). Paleo rivers on the land that is now under the Yellow Sea are drawn according to Song et al. (2016) and Yoo et al. (2016). High-resolution maps of Structure results are available in Supplementary Data S3.
Table 2.
Genetic diversity within Miscanthus sacchariflorus and M. × giganteus collected in eastern Asia
| DAPC group | No. of individuals | SNP diversity (D) | Mean extrapolated allelic richness* | Number of SNPs with minor allele frequency >0.05 | N P | N P unique | Plastid Gini–Simpson index | Plastid Gini–Simpson index excluding Msi haplotypes |
|---|---|---|---|---|---|---|---|---|
| Yangtze diploids (ssp. lutarioriparius) | 67 | 0.115 | 1.71 | 11 377 | 17 | 5 | 0.69 ± 0.06 | 0.69 ± 0.06 |
| N China diploids | 64 | 0.139 | 1.82 | 15 087 | 18 | 3 | 0.85 ± 0.03 | 0.85 ± 0.03 |
| Korea/NE China/Russia diploids | 256 | 0.141 | 1.89 | 14 778 | 20 | 9 | 0.85 ± 0.01 | 0.85 ± 0.01 |
| N China/Korea/Russia tetraploids | 139 | 0.133 | 1.94 | 14 648 | 19 | 7 | 0.79 ± 0.03 | 0.79 ± 0.03 |
| S Japan tetraploids | 120 | 0.139 | 1.89 | 15 509 | 20 | 6 | 0.75 ± 0.04 | 0.74 ± 0.04 |
| N Japan tetraploids | 76 | 0.134 | 1.80 | 14 249 | 9 | 2 | 0.46 ± 0.07 | 0.45 ± 0.07 |
| Mxg (2x × 2x) | 7 | 0.123 | 1.82 | 12 616 | 5 | 0 | 0.86 ± 0.10 | 0.86 ± 0.10 |
| Mxg (4x × 2x) | 30 | 0.187 | 1.86 | 18 897 | 9 | 0 | 0.77 ± 0.06 | 0.53 ± 0.13 |
Groupings are based on discriminant analysis of principal components (DAPC) for 34 605 RAD-seq SNPs (Fig. 2).
Msi, M. sinensis; Mxg, M. × giganteus; D, diversity, as calculated from expected heterozygosity of RAD-seq SNPs (allele frequencies were estimated from read count data using the R package polyfreqs for tetraploid and triploid groups, and directly from genotypes called with UNEAK for diploid groups; standard error of D = 0.001 for each group); NP, number of plastid haplotypes; NP unique, number of plastid haplotypes unique to each group.
*Standard error of 0.001 or 0.002 for each group for allelic richness.
Ploidy analysis
Flow cytometry, for determining DNA content and inferring ploidy, was performed using previously described protocols (Clark et al., 2015, 2016). In brief, leaves were co-chopped in buffer with Sorghum bicolor (1.74 pg/2C) or M. sinensis (5.38 pg/2C) as an internal standard, then stained with propidium iodide or 4’,6-diamidino-2-phenylindole (DAPI), before analysis on an LSR II Flow Cytometry Analyzer (BD Biosciences, San Jose, CA, USA) at Zheijiang University or at the Roy J. Carver Biotechnology Center at the University of Illinois or on a Partec PA II Flow Cytometer (Sysmex Partec, Görlitz, Germany) at Aarhus University, respectively. Expected genome sizes were 4.4 pg/2C for diploid M. sacchariflorus, 8.5 pg/2C for tetraploid M. sacchariflorus and 6.9 pg/2C for triploid M. × giganteus (Rayburn et al., 2009; Li et al., 2013; Chae et al., 2014). Out of 764 individuals included in the population structure analysis, ploidy was inferred from genome sizes determined by flow cytometry for 694 individuals with available leaf tissue. Out of the remaining 70 individuals, we were able to infer ploidy of 57 based on heterozygosity of RAD-seq data (Supplementary Data Fig. S1), leaving only 13 individuals with undetermined ploidy.
RAD-seq data analysis
The UNEAK pipeline (Lu et al., 2013) was used to identify SNPs from RAD-seq data and to genotype individuals. Tags with fewer than five reads were excluded from tag pair identification. After filtering for SNPs with <50 % missing data, a minimum minor allele frequency of 0.01 and no apparent heterozygous genotypes in any of three doubled haploid M. sinensis accessions (Głowacka et al. 2012), 34 605 SNPs remained (Supplementary Data S2). In M. sinensis, we previously found improved resolution of population structure by using these filtering parameters as compared with more stringent parameters (Clark et al., 2014), which we confirmed in M. sacchariflorus (Supplementary Data Fig. S2).
Structure 2.3.4 (Falush et al. 2003), discriminant analysis of principal components (DAPC) implemented in the R package adegenet (Jombart et al., 2010) and TESS3 implemented in the tess3r R package (Caye et al., 2016) were used to assess population structure, hybridization and major groupings of M. sacchariflorus. Structure was run with 50 000 Markov chain Monte Carlo (MCMC) reps after a 10 000 rep burn-in using the admixture model with correlated allele frequencies. A preliminary Structure run at K = 1 was performed to infer lambda (the parameter for determining allele frequency priors), which was estimated at 0.4686. This value of lambda was then fixed, and six Structure runs each at K = 1–10 were performed. The Evanno method (Evanno et al., 2005) was used to explore possible values of K (Supplementary Data Fig. S3). Principal components analysis for DAPC was performed using the ‘glPca’ function of adegenet, with parameters set so that each marker would be centred and scaled, and all principal components would be retained. The ‘find.clusters’ function was then used to make initial groupings, with 1000 randomly chosen centroids to ensure convergence, and all principal components included. The Bayesian information criterion was used as a guide for selection of the number of clusters (Supplementary Data Fig. S4). DAPC was then performed with the ‘dapc’ function using the groupings from ‘find.clusters’, the first 250 principal components and all discriminant axes for K = 2–10. TESS3 was run in six replicates at K = 1–10, with the optimal replicate at each K being selected by the software. The cross-validation score at each value of K was plotted in order to help identify the optimal K value (Supplementary Data Fig. S5). The K value that was ultimately chosen for further analysis was the lowest K that resulted in consistency between groups identified by Structure, DAPC and TESS3, and was also biologically and geographically meaningful (Supplementary Data Fig. S6).
Allele frequencies within DAPC groups were estimated directly from the sampled genotypes for the diploid groups, and with the R package polyfreqs (Blischak et al., 2016) for the tetraploid and triploid groups. SNP diversity (D) was estimated as the expected heterozygosity (probability of drawing two different alleles from the population) at each SNP, averaged across all SNPs (Nei, 1973). Allelic richness was estimated using the extrapolation method of Foulley and Ollivier (2006). Pairwise Jost’s D (Jost, 2008) among DAPC groups was estimated from genotypes and allele frequencies using a custom R function (available at doi:10.5281/zenodo.58614). Pairwise Jost’s D values were then used as a distance matrix for the calculation of a Neighbor–Joining tree among populations using the R package ape (Paradis et al., 2004). The software TreeMix (Pickrell and Pritchard, 2012) was used to infer evolutionary relationships and gene flow among the groups identified by DAPC, including one group for M. sinensis. The number of migration edges selected was the highest number at which each edge connected a unique pair of tree edges.
A second Neighbor–Joining tree was calculated to show relationships among individuals. To reduce the confounding effect of hybridization on the calculation of a Neighbor–Joining tree between Miscanthus individuals, individuals in M. × giganteus DAPC groups were excluded, as were SNPs that were highly differentiated between M. sinensis and M. sacchariflorus according to Structure results, leaving 731 individuals and 31 743 SNPs, respectively. Manhattan distances were calculated in R between each pair of individual genotypes; the distance between two genotypes homozygous for different alleles was 2, the distance between a homozygous and heterozygous genotype was 1 and the distance between identical genotypes was 0; these distances were then summed across all SNPs and scaled by the proportion of non-missing data (Black, 2006). Manhattan distances were used for calculating the Neighbor–Joining tree using the R package ape (Paradis et al., 2004). This same genetic distance matrix was used for conducting Mantel tests using the R package ade4 (Chessel et al., 2004) utilizing geographic distances calculated with the R package geosphere (Hijmans, 2017).
To test hypotheses about the history of population divergence and admixture, scenarios were tested using DIYABC 2.1.0 (Cornuet et al., 2014). The origins of tetraploid populations and the relationships among diploid populations were examined separately. In both cases, six scenarios with uniform prior probabilities were tested using a total of 600 000 simulations. Posterior probabilities of scenarios were estimated using the logistic regression approach.
Plastid haplotype analysis
Each unique combination of amplicon sizes across ten microsatellite loci was considered to be one plastid haplotype. Individuals with any missing data were excluded from plastid analysis, leaving 759 individuals. In our previous studies of Miscanthus (Clark et al., 2014, 2015, 2016), distances between haplotypes were calculated simply as the number of markers at which alleles differed (i.e. with ten microsatellites, distances ranged from zero to ten). However, due to the large number of closely related haplotypes in this study, greater resolution was needed so distances between haplotypes were calculated as the sum of differences in amplicon size across all ten loci. A plastid haplotype network was then calculated using a modified source code from pegas (Paradis, 2010). Pairwise Jost’s D using plastid haplotype frequencies among the DAPC groups identified from RAD-seq SNPs was estimated using the R package mmod (Winter, 2012) then used for generating a Neighbor–Joining tree among populations using the R package ape (Paradis et al., 2004).
All data sets and scripts used for both SNP and plastid analysis are available at the Illinois Data Bank (https://doi.org/10.13012/B2IDB-0170190_V3).
RESULTS
Ploidy types and their geographic distribution
The distribution of ploidy types among species and among geographies within species was not uniform. Among all the M. sacchariflorus and M. × giganteus studied, we identified 396 diploids, 12 triploids and 339 tetraploids; ploidy was not determined for 12 individuals (Table 1). Four out of the five M. sinensis included in the study were diploid, as expected (Clark et al., 2014), and ploidy was not determined for one individual. From mainland Asia, 380 diploid, three triploid and 133 tetraploid M. sacchariflorus individuals were observed (Table 1). Among the mainland Asian M. × giganteus, we observed nine diploids, five triploids and ten tetraploids (Table 1). In China, tetraploid M. sacchariflorus individuals were found primarily in coastal regions, especially on the Shandong Peninsula, Liaodong Peninsula and near Taihu Lake (near Shanghai), whereas tetraploids were common throughout South Korea (Fig. 1). In Russia, we found only three M. sacchariflorus tetraploids, which were collected near the Amur River. Diploid M. sacchariflorus predominated in inland regions of China and Russia. In South Korea, M. sacchariflorus diploids were frequently found in the north but were uncommon south of 36.5 °N. Three triploid M. sacchariflorus were collected at Taihu Lake, the Shandong Peninsula and in north-western South Korea, all regions where both diploid and tetraploid M. sacchariflorus were common. We also identified five triploid M. × giganteus from geographically diverse areas of South Korea, which was consistent with previous reports (Moon et al., 2013). No triploid M. × giganteus individuals were found in China.
All 184 Japanese M. sacchariflorus individuals that we tested were tetraploid (Table 1; Fig. 1). In addition to three triploid Japanese M. × giganteus that we identified previously (Clark et al., 2015), one new triploid M. × giganteus from the Japanese island of Shikoku was identified (JM2014-S-4). To the best of our knowledge, this is the first discovery of triploid M. × giganteus on Shikoku Island.
Population structure inferred from RAD-Seq SNPs
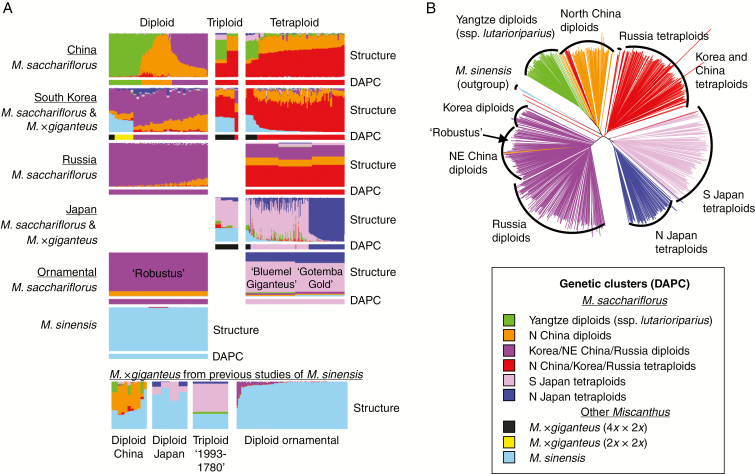
We identified seven genetic groups via Structure and TESS3 (K = 7; six M. sacchariflorus and one M. sinensis) and nine groups via DAPC (K = 9; six M. sacchariflorus, one M. sinensis and two M. × giganteus); given that Structure and TESS3 can detect hybrids between groups as an admixture but DAPC cannot, this represented a consistent result among all three analyses. Cluster assignments of individuals for all values of K tested are shown in Supplementary Data Fig. S6. Out of the 34 605 SNPs used in analysis, 34 564 had call rates above zero in all six M. sacchariflorus genetic groups. Cluster assignments of each individual for the selected values of K are also provided in Supplementary Data S1. Structure results at K = 7 are mapped in Fig. 1, with higher resolution maps provided in Supplementary Data S3.
Three of the M. sacchariflorus groups were primarily diploid, including Yangtze (ssp. lutarioriparius), N China and Korea/NE China/Russia (Figs 1 and 2A; green, orange and purple, respectively). The other three M. sacchariflorus groups were primarily tetraploid, including N China/Korea/Russia, S Japan and N Japan (Figs 1 and 2A; red, pink and dark blue, respectively). One M. × giganteus DAPC group consisted only of diploids from Korea (yellow), whereas the other consisted primarily of triploids and tetraploids from Korea and Japan, plus two diploids from Korea (black) (Fig. 2A; Supplementary Data S1). Structure results indicated that all diploid M. × giganteus were derived from hybridization between M. sinensis (diploid; sky blue) and diploid M. sacchariflorus, whereas all triploid and tetraploid M. × giganteus were derived from hybridization between M. sinensis (diploid) and tetraploid M. sacchariflorus; in all cases, the M. sacchariflorus parent originated from the same geographic region as where the M. × giganteus genotype was collected (Fig. 2A). The three triploid M. sacchariflorus from China and Korea appeared to be the result of hybridization between diploid and tetraploid M. sacchariflorus (Figs 1 and 2A).
Fig. 2.
Individual-based analysis of population structure of Miscanthus sacchariflorus and M. × giganteus using RAD-seq SNP data. Individuals that had ≥20 % M. sinensis admixed with M. sacchariflorus were considered to be M. × giganteus. (A) Bar charts showing Q values from Structure analysis (top of each set by provenance) or assignment to groups based on discriminant analysis of principal components (DAPC, bottom of each set) using 34 605 RAD-seq SNPs. Shown are nine DAPC groups, comprising six M. sacchariflorus, one M. sinensis and two M. ×giganteus; in the Structure analysis, seven groups are shown with colour as for DAPC groupings except that the M. × giganteus individuals are represented as an admixture of M. sinensis and M. sacchariflorus. All 764 individuals from the current study are plotted. The bottom portion includes an additional 93 individuals from previous studies (Clark et al., 2014, 2015). ‘Ornamental’ indicates cultivars available from the horticultural nursery trade in North America. (B) Neighbor–Joining tree of 722 Miscanthus sacchariflorus individuals using 31 743 RAD-seq SNPs. Five M. sinensis individuals were included as an outgroup. Branches are coloured based on DAPC groupings (A). To reduce the signal of hybridization, 37 early generation hybrids were excluded, as were 2862 SNPs that were highly differentiated between M. sinensis and M. sacchariflorus.
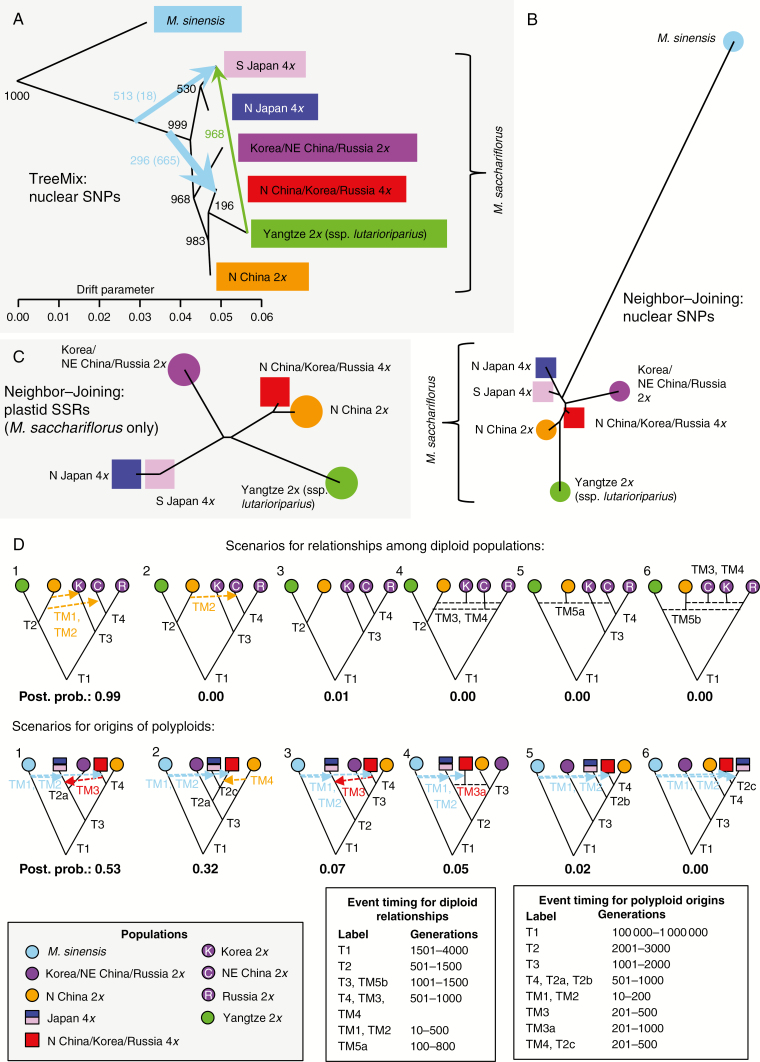
The divergence of M. sinensis and M. sacchariflorus, as indicated by TreeMix analysis, was followed chronologically by the Japan–mainland split within M. sacchariflorus, then by the split of the Korea/NE China/Russia diploids from the other mainland groups (Fig. 3A). Divergence of N China diploids, Yangtze diploids and N China/Korea/Russia M. sacchariflorus tetraploids occurred approximately simultaneously based on TreeMix. In the Neighbor–Joining tree of the consolidated groups (Fig. 3B), M. sinensis branched off the tree between the Japan and mainland M. sacchariflorus groups, indicating that the most basal division of M. sacchariflorus was Japan vs. mainland, consistent with the TreeMix results. Moreover, in TreeMix, the clade containing all mainland Asian M. sacchariflorus was found in 968 out of 1000 bootstrap replicates, strongly supporting an early Japan–mainland split (Fig. 3A).
Fig. 3.
Relationships among genetic groups of Miscanthus sacchariflorus based on RAD-seq SNPs and plastid microsatellites. Groups were identified using discriminant analysis of principal components (DAPC; Fig. 2A). (A) Relationships among six M. sacchariflorus groups, and one M. sinensis outgroup, as determined by the software TreeMix based on 34 605 RAD-seq SNPs and 727 individuals. The black tree indicates population divergence. Coloured arrows indicate subsequent migration, colour coded by population of origin. Arrow width signifies the magnitude of migration. The numbers of bootstrap replicates out of 1000 supporting each clade or migration event are indicated. Numbers in parentheses indicate the number of replicates in which there was a migration event from M. sinensis to the indicated population. (B) Neighbor–Joining tree calculated from pairwise Jost’s D statistics using allele frequencies at 34 605 RAD-seq SNPs across 727 individuals. (C) Neighbor–Joining tree calculated from pairwise Jost’s D statistics using frequencies of plastid haplotypes across ten microsatellite markers, across 712 M. sacchariflorus individuals with complete plastid data (M. sinensis is not shown due to the lack of shared haplotypes with M sacchariflorus). (D) Scenarios tested by DIYABC and their estimated posterior probabilities. Arrows indicate migration events, and dashed horizontal lines indicate admixture events. Model constraints upon divergence (T) and migration times (TM) are indicated in terms of number of generations before the present.
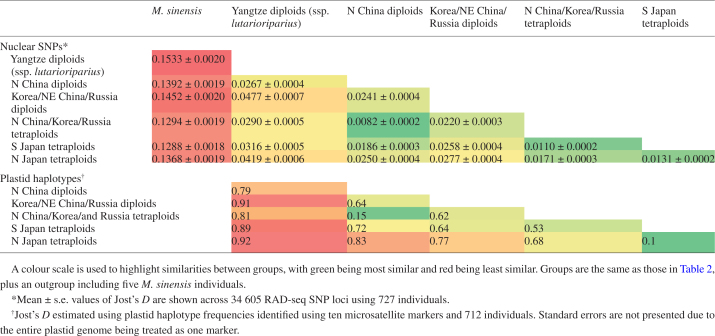
Population sub-structure of M. sacchariflorus was further elucidated via Neighbor–Joining trees and estimates of genetic differentiation (Figs 2B and 3B, C; Table 3). Neighbor–Joining trees indicated that the tetraploid S Japan group was basal to the tetraploid N Japan group (Figs 2B and 3C) which, in combination with S Japan being the group least differentiated from N Japan (Table 3), suggested that N Japan was derived from S Japan. Similarly, the diploid N China group was basal to the diploid Yangtze group (Fig. 2B) and the Yangtze diploids were most closely related to the N China diploids (Table 3), suggesting that the Yangtze diploids were derived from the N China diploids. Within the Korea/NE China/Russia diploid group, Korea was closest to the centre of the tree, followed by NE China, then Russia as the most derived group (Fig. 2B). Eight individuals from the N China/Korea/Russia tetraploid DAPC group also appeared in the N China clade of the Neighbor–Joining tree; these were tetraploid and triploid hybrids with the Yangtze diploid group (Fig. 2). Moreover, the two most closely related M. sacchariflorus groups were the N China diploids and N China/Korea/Russia tetraploids, suggesting recent derivation of the latter group from the former (Table 3). Although the two M. sacchariflorus Japan groups were more closely related to the N China/Korea/Russia tetraploids than to any other mainland group (Table 3), the Neighbor–Joining trees positioned the Japanese M. sacchariflorus as approximately equally related to all mainland groups (Figs 2B and 3B). Jost’s D statistic from SNP data indicated that S Japan tetraploids and N China/Korea/Russia tetraploids were the M. sacchariflorus groups least diverged from M. sinensis, whereas Yangtze diploids were the most diverged from M. sinensis (Table 3). Mantel tests indicated significant isolation by distance within all M. sacchariflorus groups except for N Japan tetraploids and Yangtze diploids (Supplementary Data Fig. S7).
Table 3.
Differentiation among Miscanthus sacchariflorus genetic groups using Jost’s D statistic
High intraspecific admixture within and across ploidies of M. sacchariflorus was indicated by the Structure results (Figs 1 and 2A). The observed admixture was typically consistent with geographical proximity of groups, and probably reflects some combination of isolation by distance and true admixture. For example, tetraploid M. sacchariflorus from S Japan (pink) frequently had some ancestry from the adjacent tetraploid populations in northern Japan (blue) or South Korea (red). Similarly, a mostly continuous gradient of admixture was observed among diploid M. sacchariflorus in mainland Asia from the Yangtze River to north China and from north-east China to Russia (Figs 1 and 2A; green to orange to purple). Other admixtures suggested gene flow over long distances and across ploidy barriers; for example, ancestry from the Yangtze diploid (ssp. lutarioriparius; green) group was found not only in nearby tetraploids in China, but also in tetraploids in S Japan. TreeMix analysis also identified gene flow from the Yangtze diploid (ssp. lutarioriparius) group to the S Japan tetraploids (Fig 3A). The rare tetraploids in Russia that were primarily part of the N China/Korea/Russia tetraploid group appeared to be admixed with diploids from Korea/NE China/Russia (purple). Korean diploids (purple) had some ancestry from the Japan tetraploids, and notably it appeared to be more from N Japan (blue) than from S Japan (pink).
Hybridization between tetraploid M. sacchariflorus and diploid M. sinensis resulted in considerable introgression of M. sinensis DNA into M. sacchariflorus in both Korea and southern Japan, but curiously evidence of such cross-ploidy introgression was absent from China, as indicated in the Structure results (Fig. 2A). TreeMix indicated gene flow from ancestral M. sacchariflorus to the S Japan and N China/Korea/Russia tetraploid M. sacchariflorus groups (Fig. 3A); given that it is biologically impossible for an ancestral and extant population to interbreed, this is likely to be an artefact of TreeMix not being designed for interspecies comparisons, but rather represents interspecific gene flow from M. sinensis to the tetraploid groups, as identified by Structure (Fig. 2A). In contrast to the tetraploids, diploid M. × giganteus hybrids were observed in Korea, but without introgression. No M. × giganteus hybrids were identified in China; however, diploid hybrids have been identified in previous studies (Jiang et al., 2013; Clark et al., 2014; Figs 1 and 2A) but probably were not collected for this study because they morphologically resemble M. sinensis. The M. × giganteus DAPC group that consisted of triploids and tetraploids from Korea and Japan had substantially greater SNP diversity than all the other groups studied (Table 2), probably the result of combining genomes that had evolved in isolation over considerable time.
The SNP diversity was high and similar among five of the six M. sacchariflorus DAPC groups. However, the Yangtze diploid (ssp. lutarioriparius) group had approx. 10–20 % lower SNP diversity based on three different estimates [expected heterozygosity (D), allelic richness and number of SNPs with a minor allele frequency >0.05; Table 2]. Among the tetraploid M. sacchariflorus groups, S Japan had slightly higher SNP diversity than N Japan or N China/Korea/Russia (Table 2). The low relative diversity estimates for the Yangtze diploid group and for the N Japan group (Table 2) are consistent with genetic bottlenecks associated with sub-groups that formed via migration from larger initial groups, as indicated in the Neighbor–Joining trees (Figs 2B and 3C).
Model testing with DIYABC was performed to test hypotheses about the relationships among populations based on the results of Structure, Neighbor–Joining and TreeMix analyses (Fig. 3D). Of six scenarios that tested relationships among the diploid M. sacchariflorus populations, the most probable (0.99) involved all extant populations radiating from a common ancestor, with subsequent gene flow from N China to Korea and NE China. Hypotheses involving independent refugia in the north and south of mainland Asia, followed by admixture to produce extant diploid populations surrounding the Yellow Sea, were not supported. Of six scenarios that tested the origins of the M. sacchariflorus polyploids, the most probable (0.53) supported multiple polyploidization events, with the Japan tetraploids being most closely related to the Korea/NE China/Russia diploids, and the N China/Korea/Russia tetraploids being most closely related to the N China diploids. Among scenarios that tested the hypothesis of a single M. sacchariflorus polyploidization event, the most probable (0.32) involved all tetraploids being derived from the Korea/NE China/Russia diploid group, with subsequent gene flow from the N China diploids to the N China/Korea/Russia tetraploids.
Plastid haplotype analysis
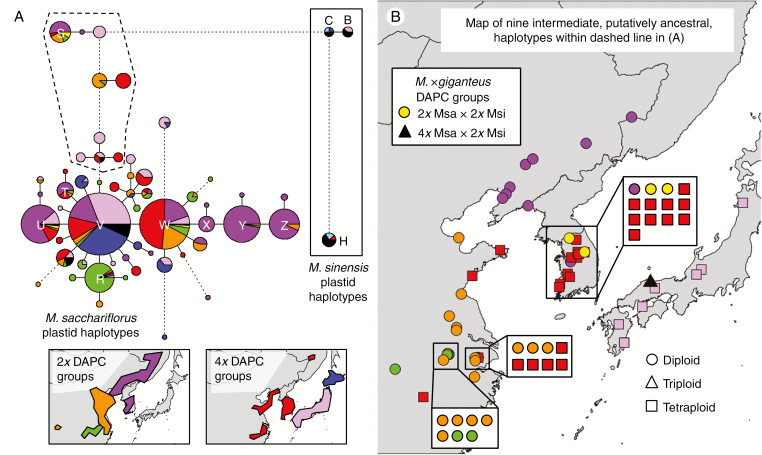
Among all M. sacchariflorus and M. sinensis entries studied, 56 unique plastid haplotypes were identified. Eight (S–Z) were identified previously in a study of Russian M. sacchariflorus (Clark et al., 2016). The most common M. sacchariflorus haplotype (V) was widely distributed across the sampling range, and the second most common haplotype (W) was common on the mainland but rare in Japan (Fig. 4). Several common haplotypes (U, X, Y and Z) were found primarily in the diploid Korea/NE China/Russia DAPC group, and one (R) was found primarily in the Yangtze diploid DAPC group. Eight newly observed M. sacchariflorus haplotypes plus the previously observed haplotype S (dashed line, Fig. 4A) were intermediate between the most common M. sacchariflorus and M. sinensis haplotypes; these haplotypes could be found throughout most of the sampling range in China, Korea and southern Japan, but were notably absent from Hokkaido (N Japan) and Russia (except for one individual near the Chinese border; Fig. 4B). Geographic distributions of common haplotypes are shown in Supplementary Data Fig. S8.
Fig. 4.
Plastid haplotypes for 759 Miscanthus spp. individuals using ten microsatellite markers. Colours from Fig. 2 indicate discriminant analysis of principal components (DAPC) groups identified with nuclear SNPs, with six M. sacchariflorus, one M. sinensis (light blue) and two M. × giganteus (yellow and black) shown; geographical distributions of M. sacchariflorus groups are shown in the inset. (A) Haplotype network. Each circle or pie chart represents one haplotype, and the area is proportional to the number of individuals with the haplotype. Letters indicate haplotypes referred to in the text. Solid lines connecting circles and pie charts indicate single mutational steps (one nucleotide difference in amplicon size at one microsatellite marker). Dotted lines indicate multiple mutational steps, proportional in number to the length of the line. Nine haplotypes surrounded by a dashed line are intermediate between common M. sacchariflorus and common M. sinensis haplotypes. (B) Map of individuals possessing any of the nine intermediate haplotypes surrounded by the dashed line in (A). Shapes indicate ploidy.
Three plastid haplotypes (B, C and H) that were previously found to be common in M. sinensis (Clark et al., 2014, 2015) were observed but infrequent in M. sacchariflorus (Fig. 4A). The M. sinensis haplotype C was found in M. sacchariflorus from N Japan and M. × giganteus in S Japan and Korea, haplotype B was found in M. sacchariflorus and M. × giganteus from S Japan, and haplotype H was found in M. sacchariflorus and M. × giganteus from Korea. Haplotypes B, C and H were common in M. sinensis from these same regions (Clark et al., 2014, 2015), supporting the hypothesis that sympatric hybridization between M. sinensis and M. sacchariflorus was common in Japan and Korea. These three haplotypes are highly differentiated from all other M. sacchariflorus haplotypes (Fig. 4A), and therefore the alternative hypothesis that they were found in M. sacchariflorus due to shared ancestry with M. sinensis is unlikely. Miscanthus × giganteus individuals from Japan and Korea also possessed haplotypes typical of M. sacchariflorus, indicating that hybridization occurred in both directions but perhaps with preference for M. sacchariflorus as the female parent, which may be due to M. sacchariflorus being more common at our sampling sites. Out of 30 individuals in the triploid and tetraploid (4x × 2x) M. × giganteus DAPC group, only 11 had plastid haplotypes from M. sinensis. Among the seven individuals in the Korean diploid (2x × 2x) M. × giganteus DAPC group, all had haplotypes characteristic of M. sacchariflorus.
Assessment of plastid haplotype diversity indicated a strong bottleneck in the N Japan DAPC group, and a weaker bottleneck in the Yangtze diploid DAPC group (Table 2). Plastid haplotype diversity was highest among the N China diploids and Korea/NE China/Russia diploids (Table 2). Pairwise Jost’s D using plastid haplotype frequencies between pairs of DAPC groups indicated a very close relationship between the N China/Korea/Russia tetraploids and the N China diploids, as well as between N Japan and S Japan (Table 3). Although the smallest Jost’s D between Japan and mainland Asia was between S Japan and the N China/Korea/Russia tetraploid group (Table 3), a Neighbor–Joining tree constructed from the Jost’s D matrix indicated that Japan was similarly distant from all mainland groups, and thus not derived from the mainland tetraploids (Fig. 3C).
DISCUSSION
Centre of radiation for M. sacchariflorus
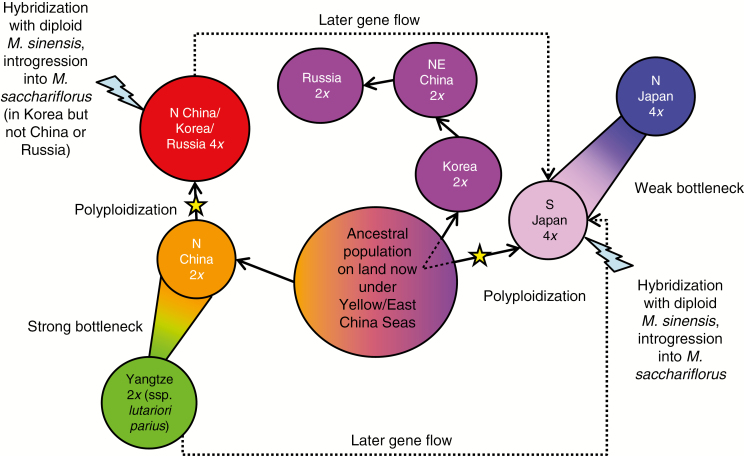
Multiple lines of evidence indicate that current populations of M. sacchariflorus were derived from an ancestral population that refuged during the last glacial maximum (LGM) on land that is now under the Yellow Sea and East China Sea (Figs 1 and 5). Perhaps most compelling is the observation that the four most genetically diverse extant M. sacchariflorus groups currently inhabit land surrounding these seas, whereas derived sub-groups are found more distant, indicating a radiation from this region. For example, the N China diploid group is found on the western edge of the Yellow and East China Seas, and was the parent population of the substantially less genetically diverse Yangtze group found further south (Figs. 1, 3B and 5; Tables 2 and 3). To the east, the S Japan tetraploid group gave rise to the less diverse N Japan tetraploids (Figs 2B and 5; Table 2). Similarly, the basal clade of the Korea/NE China/Russia diploid group in the RAD-seq SNP Neighbor–Joining tree was from Korea, followed by NE China, then Russia (i.e. from south to north; Fig. 2B). Additionally, the N China/Korea/Russia tetraploid group is common in coastal areas adjacent to both the western and eastern sides of the Yellow Sea, especially on the Shandong Peninsula in the west and on the Korean Peninsula in the east, but is infrequent in the interior of the continent (Fig. 1A), suggesting that it arose on a land mass between the Shandong and Korean Peninsulas. Moreover, none of the M. sacchariflorus groups surrounding the Yellow and East China Seas had substantially greater genetic diversity than the rest (Table 2), suggesting that they each similarly represent the diversity of the ancestral population.
Fig. 5.
Model depicting relationships among Miscanthus sacchariflorus populations based on RAD-seq SNP and plastid microsatellite data. Colours are based on discriminant analysis of principal components (DAPC) in Fig. 1.
Model testing with DIYABC strongly supported a scenario with a single refugium for all diploid populations, as opposed to multiple refugia followed by admixture (Fig. 3D). In both the TreeMix (Fig. 3A) and Neighbor–Joining (Fig. 3B,C) analyses, Japanese and mainland Asian M. sacchariflorus appeared to be derived from an extinct common ancestor, with Japan vs. mainland Asia being the first split, although DIYABC results suggest that an ancestor of both the Japanese M. sacchariflorus (tetraploids) and the diploid Korea/NE China/Russia group diverged from all other M. sacchariflorus before Japan became a distinct population (Fig. 3D).
Plastid haplotypes that were intermediate between common M. sacchariflorus and M. sinensis haplotypes, and therefore putatively ancestral, were absent from N Japan and Russia, the regions of our sampling range that are furthest from the Yellow Sea and East China Sea (Fig. 4B). Consistent with the RAD-seq SNP data, the N China diploid and Korea/NE China/Russia diploid DAPC groups had similarly high plastid diversity (Table 2), but did not share all of their plastid haplotypes (Fig. 4A), suggesting that they were independently derived from a nearby centre of diversity.
During the LGM, the climate of East Asia was too cold and dry to support populations of M. sacchariflorus throughout much of its current range, but the environment in the southern Yellow Sea Basin would have been favourable. Contemporary populations of M. sacchariflorus are found primarily in open riparian areas of climatic regions that predominantly support cool temperate forests and have average annual low temperatures warmer than –40 °C (USDA hardiness zone ≥3). In the northern portion of M. sacchariflorus’ current range in north-east China, eastern Russia and northern Japan, vegetation during the LGM was steppe–tundra (Adams and Faure, 1997; Ray and Adams, 2001) and thus unsuitably cold and dry for this species. However, during the LGM, a steppe-dominated ecosystem covered a wide area from the present day Korean Peninsula in the east to the mountains surrounding the Sichuan Basin in the west, including all of the Yellow Sea Basin and the north-western edge of the East China Sea Basin (Adams and Faure, 1997; Ray and Adams, 2001). Moreover, the two largest rivers in East Asia, the Yellow River and the Yangtze River, flowed over the southern portion of the Yellow Sea Basin during the LGM (Ryu et al., 2008; Song et al., 2016; Yoo et al., 2016). Song et al. (2016) reported that the main paleo-channel of the Yellow River was at approx. 35 °N, south of the present day Shandong Peninsula (in contrast to its present position north of the Shandong Peninsula) and flowed to the east and south-east; whereas the paleo-channel of the Yangtze River entered the Yellow Sea Basin approx. 2 °N north of its current location, flowed from east to north-east where it merged with the Yellow River at approx. 35 °N and 124.5 °E, and ultimately formed a large delta near present day Jeju Island (Fig. 1). During the LGM, average flow rates of the Yellow River (460 m3 s–1) and Yangtze River (730 m3 s–1) were approx. six and 41 times less than current flow rates, respectively (Song et al., 2016); however, the flow rate of the merged paleo-Yellow and Yangtze River was similar to that of the present day Ussuri River in eastern Russia, which currently supports large populations of M. sacchariflorus (Clark et al., 2016). Thus the climate and hydrology of the southern Yellow Sea Basin during the LGM would have been a conducive refugium for M. sacchariflorus, and this is consistent with inferences from our population genetics analyses. Additional sampling of mud cores from the southern Yellow Sea to enable analysis of phytoliths and pollen deposited during the LGM would help test the hypothesis that this region was key to the survival and subsequent expansion of M. sacchariflorus during the last ice age.
Origins of M. sacchariflorus ssp. lutarioriparius
Our genetic data support the conclusion that M. sacchariflorus ssp. lutarioriparius is a subspecies of M. sacchariflorus, and not a separate species. Miscanthus sacchariflorus ssp. lutarioriparius, which has been described as Triarrhena lutarioriparia (Liu, 1997; Liu et al., 2001; Liu and Yu, 2004) or M. lutarioriparius (Chen and Renvoize, 2005, 2006), is reported to be endemic to the Yangtze River watershed, has tall (3–7 m) and thick (15–30 mm) stems, yields about 30 Mg ha–1 of dry biomass, and is used to manufacture paper on a commercial scale in China. Subspecies status of lutarioriparius was the conclusion of Sun et al. (2010) based on trait comparisons and of Chen et al. (2007) based on ITS nuclear ribosomal DNA sequence data. The Yangtze diploid DAPC group in the current study corresponded to M. sacchariflorus ssp. lutarioriparius, given its geographical location and great height (Supplementary Data S1). Our population genetic analyses clearly indicated that the Yangtze diploid group was derived from the N China diploid M. sacchariflorus group via a substantial genetic bottleneck (Figs 2B, 3 and 5; Tables 2 and 3). Moreover, all plastid haplotypes found in the Yangtze diploid group were also found in or closely related to plastid haplotypes found in other M. sacchariflorus DAPC groups (Fig. 4A). If M. sacchariflorus ssp. lutarioriparius had truly been a different species, then we would have expected the Neighbor–Joining and TreeMix results to show it forming a basal branch separate from M. sacchariflorus, before the Japan–mainland split, but this was not observed.
Previous studies have found that M. sacchariflorus ssp. lutarioriparius included both diploid and tetraploid individuals, with the latter observed primarily along the most eastern portion of the Yangtze River and around nearby Lakes Taihu and Hung-tse (Li et al., 2013; Sheng et al., 2016). In contrast, our DAPC analysis identified only diploid individuals in the Yangtze group. However, our Structure analysis identified seven tetraploid individuals that had 41–45 % of their ancestry from the Yangtze diploid group (in some cases, this was the single greatest genetic contribution to the admixture), with most of the remainder being from the N China/Korea/Russia tetraploid group (Fig. 2A; Supplementary Data S1). We collected six of these tetraploids on the shores of Lake Taihu along with a triploid individual of similar ancestry, and the seventh was found 131 km south (Fig. 1; Supplementary Data S1 and S3). It is likely that the tetraploid M. sacchariflorus ssp. lutarioriparius identified by Li et al. (2013) and Sheng et al. (2016) were similar to those that we collected near Lake Taihu given that all three studies sampled there, although we did not have any samples from Lake Hung-tse to compare with the previous studies. Notably, our assessment of admixture indicated a small genetic contribution of M. sacchariflorus ssp. lutarioriparius to S Japan M. sacchariflorus tetraploids from Kyushu and Shikoku Islands (Figs 1 and 2A), though it is unknown if the most direct source of lutarioriparius was diploid or tetraploid. Further study of the eastern tetraploid population of M. sacchariflorus ssp. lutarioriparius and its gene flow with other M. sacchariflorus is warranted given the breeding potential of M. sacchariflorus ssp. lutarioriparius for high biomass yield.
Origins of tetraploid M. sacchariflorus and subsequent gene flow
Our results confirm and add resolution to the known geographic range of tetraploid M. sacchariflorus. In Japan, we found M. sacchariflorus to be exclusively tetraploid, consistent with previous surveys of ploidy (Hirayoshi et al., 1957; Clark et al., 2015). In a previous study, we found rare diploid M. sinensis × M. sacchariflorus hybrids in Japan (Clark et al., 2015), suggesting that diploid M. sacchariflorus might exist in Japan. New genetic analysis of these hybrid individuals in the context of the current study indicates that the M. sacchariflorus component of their ancestry corresponds to the S Japan and N Japan tetraploid populations (Figs 1 and 2A), suggesting that gene flow may occur from tetraploid M. sacchariflorus to diploid M. sinensis, perhaps by rare haploid gametes produced by triploid M. × giganteus.
In mainland Asia, we found both diploid and tetraploid M. sacchariflorus. No mainland region was exclusively tetraploid, although tetraploids were concentrated in the Korean and Shandong Peninsulas. Tetraploids were found as far north as the Amur River, 1400 km away from the nearest mainland Asia tetraploids, which were on the Liaodong Peninsula. The Amur River tetraploids might be a relict population, indicating that tetraploids were once much more widespread north of the Liaodong Peninsula or, alternatively, they might be recent anthropogenic introductions. The only tetraploids found south of the Shandong Peninsula were the hybrids with M. sacchariflorus ssp. lutarioriparius mentioned above.
Our genetic results are most consistent with the hypothesis that the Japanese and mainland Asian populations of tetraploid M. sacchariflorus originated from distinct polyploidization events (Fig. 5). In Neighbor–Joining and TreeMix analyses (Fig. 3), the mainland tetraploids (N China/Korea/Russia tetraploids) appeared to be closely related to the N China and Yangtze diploid groups, suggesting that the mainland tetraploids were derived from a common ancestor of these diploid groups. Derivation of the mainland tetraploids exclusively from the Yangtze diploid group is unlikely given that the genetic diversity of the mainland tetraploids is higher than that of the Yangtze diploids. The Japan tetraploids (S Japan and N Japan), on the other hand, were similarly related to all extant diploid groups that we identified, suggesting that they were derived either from the common ancestor of all diploid M. sacchariflorus or from a diploid population that once existed in Japan but has become extinct. The DIYABC results indicated that the Japan tetraploids and the Korea/NE China/Russia diploids instead shared a common ancestor that had already diverged from all other M. sacchariflorus (Fig. 3D), and in this scenario the ancestor was probably located in Korea. Notably, the Korean diploids have apparent admixture from Japan according to Structure (Figs 1 and 2A), which could in reality reflect shared ancestry. Some combination of the two hypotheses is possible, for example if there was a gradient of isolation by distance in the ancestral Yellow Sea population, and the eastern segment of the population gave rise to both the Korean diploids and Japanese tetraploids (Fig. 5). After the LGM, coastal regions in the east warmed sooner than more interior areas to the west (Winkler and Wang, 1993), and this warming probably drove the initial migration and differentiation of populations out of the Yellow Sea basin. It should be noted that the Japan tetraploids were more closely related to the mainland tetraploids than to any diploid group; however, our analysis suggests that this similarity is due to gene flow between the tetraploid groups and hybridization with M. sinensis in both groups, as opposed to shared ancestry.
Structure results indicated that all tetraploid M. sacchariflorus arose by autopolyploidization (doubling of the diploid M. sacchariflorus genome) rather than allopolyploidization (hybridization with M. sinensis followed by genome doubling). It should also be noted that we did not observe any tetraploid M. × giganteus derived from diploid M. sacchariflorus, indicating that allotetraploid formation via hybridization of diploid M. sinensis and diploid M. sacchariflorus is rare to non-existent. However, polyploidy in M. sacchariflorus appears to have facilitated considerable introgression from M. sinensis. In Korea and S Japan, where both triploid and tetraploid M. × giganteus were found, we observed a gradient of introgression of M. sinensis into tetraploid M. sacchariflorus (Fig. 2A), similar to what we previously observed in Japan (Clark et al., 2015). Intriguingly, however, no hybridization or introgression from M. sinensis was observed among tetraploid M. sacchariflorus in China or Russia; we do not know what has caused this notable difference in cross-ploidy interspecific hybridization and introgression. Interestingly, we also identified many diploid M. × giganteus in Korea, but did not observe any introgression of M. sinensis into diploid M. sacchariflorus. Therefore, it appears that there exists a barrier to introgression of M. sinensis into diploid M. sacchariflorus that is broken by polyploidy, allowing introgression of M. sinensis into tetraploid M. sacchariflorus. Moreover, polyploidy has broken the barrier to introgression independently in both Korea and Japan.
It is worthwhile to consider what mechanisms might permit the generation and normal development of interspecific F1 hybrids from a diploid species crossed with diploid or tetraploid forms of another species, but allow introgression only or highly preferentially into the tetraploid form. Sterility of diploid F1M. × giganteus is an unlikely mechanism, because we have observed more than a dozen such populations in crossing experiments, and the progeny were typically fertile (unpublished data); moreover, many ornamental cultivars that are predominantly M. sinensis in genetic composition have a substantial portion of their genome introgressed from M. sacchariflorus (Clark et al., 2014, 2015). Lack of fitness in the diploid F1 is also unlikely to be the cause, given that we have observed many diploid M. × giganteus F1 hybrids to be vigorous and high yielding in field trials (Dong et al., 2018). Perhaps then, a decrease in fitness in subsequent generations (F2 or backcross) may occur to a greater extent for the interspecific diploids than for the tetraploids. For any given backcross generation, the overall proportion of alleles from M. sinensis would be the same regardless of whether the M. sacchariflorus was diploid or tetraploid, but tetraploid introgressed M. sacchariflorus would have M. sinensis alleles distributed across a greater number of loci than would a diploid introgressed M. sacchariflorus. Moreover, most loci with an M. sinensis allele would have a 1:3 ratio of M. sinensis alleles to M. sacchariflorus alleles in an introgressed tetraploid, whereas any locus with an M. sinensis allele would necessarily have a 1:1 ratio of M. sinensis to M. sacchariflorus alleles in a diploid. These differences in distribution of M. sinensis alleles could affect fitness via dominance and epistasis effects. Additionally, polyploidy in plants is associated with an increased rate of meiotic recombination (Melamed-Bessudo et al., 2016), which could facilitate introgression by breaking linkage between deleterious alleles and neutral or advantageous alleles.
Intraspecific cross-ploidy gene flow primarily from diploid to tetraploid forms of M. sacchariflorus was also indicated by the Structure and TreeMix results. We identified six tetraploid individuals in China that appeared to have half of their ancestry from the Yangtze diploid group, and all three tetraploid individuals from Russia had approximately half of their ancestry from the Korea/NE China/Russia and N China diploid groups (Figs 1 and 2A). Both TreeMix and Structure also identified gene flow from the Yangtze diploid group into the S Japan tetraploid group (Figs 1, 2A, 3A and 5). Given that M. sacchariflorus ssp. lutarioriparius (Yangtze diploids) are exceptionally tall and high yielding, ancestry from this group may have contributed to the high yield potential of the biomass cultivar M. × giganteus ‘1993-1780’, which had approx. 4 % ancestry from M. sacchariflorus ssp. lutarioriparius (Fig. 2A). Also, diploid M. sacchariflorus in Korea notably appeared to have some ancestry from the N Japan tetraploids (Figs 1 and 2A). The N Japan group, which is more geographically isolated and less impacted by admixture and hybridization than the S Japan group, may more closely resemble an ancient population that existed in S Japan and in the area that is now the Yellow Sea. Thus, the Korea diploids might not truly be admixed with N Japan, but may appear that way because they are also closely related to the ancestral population from that area (Figs 2B and 5).
Provenance of cultivated Miscanthus
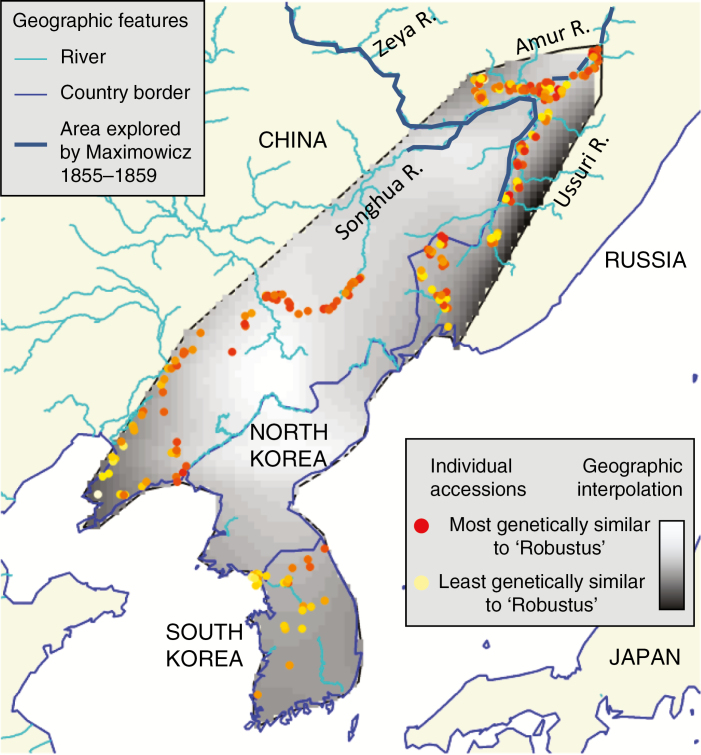
By taking into account both historical records and new genomic data from our study, we determined that the ornamental and much-studied M. sacchariflorus cultivar ‘Robustus’ was probably derived from a wild population along the southern Amur River between the Songhua and Zeya Rivers, or on the lower (northern) reaches of the Songhua River, in either case near the border of Russia and China. In the summers of 1855 and 1856, Carl Maximowicz, Conservator of the Imperial Botanical Garden at St. Petersburg, became the first Western botanist to collect M. sacchariflorus, which he named Imperata (Triarrhena) sacchariflora. By his own account, Maximowicz observed this species and collected specimens along the southern Amur River between the mouths of the Songhua River and Zeya River (near the city of Blagoveshchensk), and along the lower (northern) reaches of the Ussuri River (Maximowicz, 1859). Maximowicz returned to the Amur Region in 1859 and was able to travel about 270 km up the Songhua River in China before returning to Russia (Ravenstein, 1861; Stapf, 1891; Bretschneider, 1898). By 1862, the M. sacchariflorus that Maximowicz introduced from the Amur region to St. Petersburg was found to be winter-hardy there, and it was promoted as a beautiful ornamental as well as a valuable forage crop for cattle (Regel, 1862). Miscanthus sacchariflorus seeds and divisions were subsequently distributed from St. Petersburg to other botanical gardens in Europe (Regel, 1862). Karl Foerster, an influential horticulturalist with a nursery in Potsdam, Germany and who led the mid-twentieth century revival of using perennial grasses in Western gardens (Foerster, 1988; Darke, 1994, 2007), obtained germplasm of M. sacchariflorus from a botanical garden (M. Deuter, pers. comm. from K. Naeser, E. Pagels, R. Simon and H. Hagemann). Foerster selected the ‘Robustus’ cultivar in around 1950 (Simon, 1952), which like all ornamental Miscanthus was propagated vegetatively for distribution. Genomic data from our current study indicated that ‘Robustus’ is closely related to populations of M. sacchariflorus found along the southern Amur and northern Songhua Rivers, which is consistent with the locations from which Maximowicz reported collecting, and less related to populations from the Ussuri River (Figs 2B and 6). The analysis presented in Fig. 6 also indicates that ‘Robustus’ could have originated from the upper (southern) Songhua River, but we know of no historical botanical introductions from this area of China to Europe. Thus, ‘Robustus’ was probably derived from the material Maximowicz introduced from the north-western portion of his collection expedition.
Fig. 6.
Genetic relationship between the ornamental Miscanthus sacchariflorus ‘Robustus’ and wild populations of M. sacchariflorus in eastern Russia, north-east China and Korea, based on 34 605 RAD-seq SNPs. A total of 249 individuals in the Korea/NE China/Russia diploid group with known provenance are plotted. The thick blue line in the upper right indicates river segments known to have been explored in the mid to late 1850s by botanist Carl Maximowicz, who collected and brought back to St. Petersburg seed and/or rhizomes of M. sacchariflorus from which we believe ‘Robustus’ is derived (Maximowicz, 1859; Simon, 1952).
In contrast to the diploid ‘Robustus’, the ornamental tetraploid M. sacchariflorus cultivars ‘Gotemba Gold’ and ‘Bluemel Giganteus’ were both in the S Japan group, with Structure results similar to individuals from the Kanto region of Japan (Fig. 2A). ‘Gotemba Gold’, a variegated cultivar also known as ‘Gotemba’, originated from Kenji Watanabe’s nursery in 20th century Japan (Darke, 2007) and is most probably named after the city of Gotemba in the Kanto region. The biomass cultivar M. × giganteus ‘1993-1780’ had ancestry matching M. sacchariflorus from southern Japan (Fig. 2A), consistent with its origins as a wild collection from that region (Greef et al. 1997).
We previously determined that about half of ornamental Miscanthus cultivars sold as M. sinensis in North America and Europe were actually interspecific hybrids with M. sacchariflorus ancestry introgressed into a central or southern Japan M. sinensis genetic background (about BC2, typically; Clark et al., 2014, 2015). The results presented here indicate that the M. sacchariflorus ancestry of these ornamental cultivars came from the Korea/NE China/Russia diploid M. sacchariflorus group (Fig. 2A). Given the different geographic origins of the M. sinensis and M. sacchariflorus portions of the genomes of these ornamentals, this introgression almost certainly did not occur in nature, but instead was likely to be the result of European breeding efforts for early flowering and winter hardiness (Darke, 1994; Clark et al., 2014). Results from our current study indicated that the ornamental M. × giganteus ‘Purpurascens’ was also derived from the Korea/NE China/Russia diploid M. sacchariflorus group, consistent with our previous hypothesis that ‘Purpurascens’ was the source of the M. sacchariflorus ancestry in ornamental M. sinensis (Clark et al., 2015). ‘Purpurascens’ in cultivation is itself likely to be a naturally occurring hybrid given that its M. sinensis ancestry also came from Korea, NE China or Russia (Clark et al., 2015, 2016).
Differences in genetic diversity and population structure between M. sinensis and M. sacchariflorus
Higher genetic diversity and weaker population structure were observed for M. sacchariflorus in comparison with M. sinensis, which suggests differences in their demographic history. We previously hypothesized, based on a comparison between population genetics of M. sacchariflorus in Russia with M. sinensis across East Asia, that M. sacchariflorus underwent less of a genetic bottleneck during the LGM than M. sinensis (Clark et al., 2016), and our new results also support this hypothesis. Genetic diversity, measured as the number of SNPs identified with a minor allele frequency >0.05, ranged from 11 337 to 15 509 among DAPC groups in M. sacchariflorus (Table 2) but was only 6942–9262 in M. sinensis (Clark et al., 2016). Similarly, pairwise Jost’s D for the six M. sacchariflorus genetic groups ranged from only 0.008 to 0.048 (Table 3), whereas for six M. sinensis genetic groups that we identified previously it ranged from 0.019 to 0.076 (Clark et al., 2014), indicating about half as much within-species differentiation for M. sacchariflorus compared with M. sinensis.
Differences between the plastid haplotype networks of M. sacchariflorus and M. sinensis were also consistent with our hypothesis of a greater genetic bottleneck for M. sinensis during the LGM. In contrast to M. sinensis, in which the most common haplotypes consisted of two groups separated by multiple mutational steps (Clark et al., 2014), the most common haplotypes in M. sacchariflorus formed a continuous network of single mutational steps (Fig. 4A). This indicates that extinction of haplotypes due to genetic drift has occurred less frequently in M. sacchariflorus than in M. sinensis, which was probably due to a greater effective population size over time. Moreover, the most common haplotype in M. sacchariflorus (V) was distributed very broadly across East Asia, whereas the three most common haplotypes in M. sinensis (B, D and H; Clark et al., 2014) all had distinctly restricted geographies, indicating large changes in haplotype frequency due to founder effects as M. sinensis colonized Asia. Lastly, we found rare haplotypes in M. sacchariflorus that potentially belonged to the common ancestor of M. sinensis and M. sacchariflorus, or at least an ancient population of M. sacchariflorus, given that they were intermediate between the common M. sinensis haplotypes and common M. sacchariflorus haplotypes (Fig. 4A). Such ancestral haplotypes appear to have become extinct in M. sinensis (Clark et al., 2014, 2015).
Conclusions
Here we provide the most extensive analysis of M. sacchariflorus population structure to date, revealing high genetic diversity, geographical differentiation of six genetic groups, multiple origins of polyploidy and a history of introgression from diploid M. sinensis into tetraploid M. sacchariflorus in Japan and Korea but not in China or Russia (Fig. 5). Over half of the SNP markers identified in this study (18 555 out of 34 605) had not been identified in any of our previous studies of Miscanthus (Supplementary Data S2), highlighting the importance of broad sampling for capturing the diversity of this genus. These results provide important context for physiological and agronomic studies of M. sacchariflorus.
Given the great genetic diversity and population structure in M. sacchariflorus, we caution against using any single genotype to draw conclusions about the species as a whole. Because the ornamental cultivar ‘Robustus’ has been one of only two M. sacchariflorus clones readily available in North America and Europe, considerable research and breeding have been conducted on this genotype, including development of the ‘Amuri’ clones that were released by Tinplant (Deuter and Abraham, 2004; Anonymous, 2006; Pude, 2011; Supplementary Data S4). However, ‘Robustus’ is exceptionally early flowering and adapted to higher latitudes and colder winter temperatures than many other M. sacchariflorus because it originated near the north-eastern edge of the species’ range (Figs 2B and 6). For example, if studies were to include representatives of the Yangtze group (ssp. lutarioriparius) or the S Japan group, we would expect very different conclusions about yield potential, height, flowering time and winter-hardiness than from studies that include only ‘Robustus’ or its progeny. Thus, for studies that wish to make broad conclusions about M. sacchariflorus as a species, it would be advisable to include multiple representatives from each of the six genetic groups that we identified.
To develop new triploid M. × giganteus biomass cultivars, plant breeders should especially note that China and Korea offer tetraploid germplasm that is genetically distinct from the Japanese tetraploid germplasm that has predominantly been used for breeding thus far. The two tetraploid groups of M. sacchariflorus should be evaluated for differences in yield potential and stress tolerance, and used in controlled crosses with M. sinensis to test for heterosis and their potential to improve yield of M. × giganteus. For experiments seeking to examine the consequences of polyploidy in M. sacchariflorus, we recommend comparing the diploids and tetraploids from the Shandong Peninsula, since these possess the closest relationship between diploid and tetraploid M. sacchariflorus populations, and hybridization with M. sinensis is rare or non-existent in that region (Figs 1, 2A and 5).
Because of the greater amount of M. sinensis germplasm imported to North America and Europe during the last century and a half in comparison with all other Miscanthus species, a disproportionate amount of Miscanthus research to date has focused on M. sinensis (Hodkinson et al., 2015). However, an understanding of the biology and genetic diversity of M. sacchariflorus will be equally important, if not more so, for breeding improved M. × giganteus cultivars. The tetraploid M. sacchariflorus parent not only contributes two-thirds of the genetic material of triploid M. × giganteus (Rayburn et al., 2009), but is also thought to make a large contribution to its high yield via chilling-tolerant photosynthesis, which effectively extends its growing season (Głowacka et al., 2014). Given that M. sacchariflorus also has substantially more genetic diversity than M. sinensis, we expect that selection within M. sacchariflorus will provide the greatest opportunity to make genetic gains for breeding improved M. × giganteus.
SUPPLEMENTARY DATA
Data S1: list of all accessions in the study, including collection locations, plastid haplotypes, and DAPC, Structure and TESS3 results. Data S2: list of SNP markers used in the study, including tag sequences and allele frequencies within genetic groups. Data S3: GeoTIFF images showing Structure Q values on high-resolution maps. Data S4: flyer from Tinplant describing the Miscanthus × giganteus ‘Amuri’ clones. Figure S1: heterozygosity vs. missing data rate, by individual, across 34 605 RAD-seq SNPs. Figure S2: comparison of population structure identified with different SNP filtering criteria. Figure S3: Delta K values calculated using the method of Evanno et al. (2005) across six Structure runs each at K = 1–10. Figure S4: Bayesian information criterion for selection of the optimal number of genetic groups using ‘find.clusters’ in adegenet. Figure S5: cross-validation score for selection of the optimal number of groups by TESS3. Figure S6: barplots indicating clustering result of 768 Miscanthus individuals using 34 605 SNP markers and the TESS3, Structure and DAPC methods. Figure S7: genetic vs. geographic distance in Miscanthus sacchariflorus. Figure S8: geographic distributions of common plastid haplotypes.
ACKNOWLEDGEMENTS
This work was supported by the DOE Office of Science, Office of Biological and Environmental Research (BER) [grant nos DE-SC0012379 and DE-SC0006634], the Plant Exploration Program of the National Plant Germplasm System at the US Department of Agriculture via Cooperative Agreement #59-1275-1-338, and the Energy Biosciences Institute. New Energy Farms provided in-kind support. Myling Luu and Melina Salgado performed DNA extractions. Ben Baechle, Colten Maertens and Helen Gapsis assisted with plant propagation. Ursula Reuter-Carlson translated German texts.
LITERATURE CITED
- Adams JM, Faure H. 1997. Preliminary vegetation maps of the world since the last glacial maximum: an aid to archaeological understanding. Journal of Archaeological Science 24: 623–647. [Google Scholar]
- Adati S. 1958. Studies on the genus Miscanthus with special reference to the Japanese species suitable for breeding purposes as fodder crops. Bulletin of the Faculty of Agriculture, Mie University 17: 1–112. [Google Scholar]
- Adati S. 1959. Cytogenetics of Japanese wild forage Miscanthus species. Proceedings of the X International Congress of Genetics, August 20–27, 1958, vol. 2 McGill University, Montreal, Canada Toronto, Ontario, Canada: University of Toronto Press, 1–2. [Google Scholar]
- Adati S, Shiotani I. 1962. The cytotaxonomy of the genus Miscanthus and its phylogenic status. Bulletin of the Faculty of Agriculture, Mie University 25: 1–24. [Google Scholar]
- Anonymous 2006. TINPLANT bringt neue Miscanthus-Mehrklonsorte ‘Amuri’ auf den Markt. Bio-based News. [Google Scholar]
- Black PE. 2006. Manhattan distance. In: Pieterse V, Black PE, eds. Dictionary of algorithms and data structures. Gaithersburg, MD: National Institute of Standards and Technology. [Google Scholar]
- Blischak PD, Kubatko LS, Wolfe AD. 2016. Accounting for genotype uncertainty in the estimation of allele frequencies in autopolyploids. Molecular Ecology Resources 16: 742–754. [DOI] [PubMed] [Google Scholar]
- Bretschneider EV. 1898. History of European botanical discoveries in China. Hamburg: Severus Verlag. [Google Scholar]
- Caye K, Deist TM, Martins H, Michel O, François O. 2016. TESS3: fast inference of spatial population structure and genome scans for selection. Molecular Ecology Resources 16: 540–548. [DOI] [PubMed] [Google Scholar]
- de Cesare M, Hodkinson TR, Barth S. 2010. Chloroplast DNA markers (cpSSRs, SNPs) for Miscanthus, Saccharum and related grasses (Panicoideae, Poaceae). Molecular Breeding 26: 539–544. [Google Scholar]
- Chae WB, Hong SJ, Gifford JM, Lane Rayburn A, Sacks EJ, Juvik JA. 2014. Plant morphology, genome size, and SSR markers differentiate five distinct taxonomic groups among accessions in the genus Miscanthus. GCB Bioenergy 6: 646–660. [Google Scholar]
- Chen S-F, Dong S-S, Wu W, Shi S-H, Zhao P-H. 2007. Phylogenetics of Triarrhena and related genera based on ITS sequence data. Journal of Wuhan Botanical Research 25: 239–244. [Google Scholar]
- Chen SL, Renvoize SA. 2005. A new species and a new combination of Miscanthus (Poaceae) from China. Kew Bulletin 60: 605–607. [Google Scholar]
- Chen S, Renvoize SA. 2006. Miscanthus Andersson, Öfvers. Kongl. Vetensk.-Akad. Förh. 12: 165. 1855. In: Wu Z, Raven, Hong D, eds. Flora of China, Vol. 22 Beijing: Science Press, 581–583. [Google Scholar]
- Chessel D, Dufour AB, Thioulouse J. 2004. The ade4 package – I : one-table methods. R News 4: 5–10. [Google Scholar]
- Clark LV, Brummer JE, Głowacka K, et al. 2014. A footprint of past climate change on the diversity and population structure of Miscanthus sinensis. Annals of Botany 114: 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LV, Dzyubenko E, Dzyubenko N, et al. 2016. Ecological characteristics and in situ genetic associations for yield-component traits of wild Miscanthus from eastern Russia. Annals of Botany 118: 941–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LV, Stewart JR, Nishiwaki A, et al. 2015. Genetic structure of Miscanthus sinensis and Miscanthus sacchariflorus in Japan indicates a gradient of bidirectional but asymmetric introgression. Journal of Experimental Botany 66: 4213–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton-Brown J, Chiang Y-C, Hodkinson TR. 2008. Miscanthus: genetic resources and breeding potential to enhance bioenergy production. In: Vermerris W, ed. Genetic improvement of bioenergy crops. New York: Springer, 273–294. [Google Scholar]
- Clifton-Brown JC, Lewandowski I. 2000. Overwintering problems of newly established Miscanthus plantations can be overcome by identifying genotypes with improved rhizome cold tolerance. New Phytologist 148: 287–294. [Google Scholar]
- Cornuet JM, Pudlo P, Veyssier J, et al. 2014. DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30: 1187–1189. [DOI] [PubMed] [Google Scholar]
- Darke R. 1994. A century of grasses. Arnoldia 54: 3–11. [Google Scholar]
- Darke R. 2007. The encyclopedia of grasses for livable landscapes. Portland, Oregon: Timber Press, Inc. [Google Scholar]
- Deuter M, Abraham J. 2004. Miscanthus-Züchtung. Kreuzungen zwischen tagneutralen M. sacchariflorus und M. sinensis. In: Pude R, ed. Anbau, Verwertung und Erfolgsaussichten von Miscanthus in Europa, Beitrage zu Agrarwissenschaften, Band 28. Bad Neuenahr: Wehle, 7–11. [Google Scholar]
- Dohleman FG, Long SP. 2009. More productive than maize in the Midwest: how does Miscanthus do it?Plant Physiology 150: 2104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Liu S, Clark LV, et al. 2018. Genetic mapping of biomass yield in three interconnected Miscanthus populations. GCB Bioenergy 10: 165–185. [Google Scholar]
- Dwiyanti MS, Rudolph A, Swaminathan K, et al. 2013a. Genetic analysis of putative triploid Miscanthus hybrids and tetraploid M. sacchariflorus collected from sympatric populations of Kushima, Japan. BioEnergy Research 6: 486–493. [Google Scholar]
- Dwiyanti MS, Stewart JR, Yamada T. 2013b. Germplasm resources of Miscanthus and their application in breeding. In: Saha MC, Bhandari HS, Bouton JH, eds. Bioenergy feedstocks: breeding and genetics. Oxford: John Wiley & Sons, Inc, 49–66. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell AD, Clifton-Brown JC, Lewandowski I, Jones MB. 2006. Genotypic variation in cold tolerance influences the yield of Miscanthus. Annals of Applied Biology 149: 337–345. [Google Scholar]
- Foerster K. 1988. Einzug der Gräser und Farne in die Gärten. Stuttgart: Eugen Ulmer. [Google Scholar]
- Foulley JL, Ollivier L. 2006. Estimating allelic richness and its diversity. Livestock Science 101: 150–158. [Google Scholar]
- Fulton TM, Chunwongse J, Tanksley SD. 1995. Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Molecular Biology Reporter 13: 207–209. [Google Scholar]
- Głowacka K, Kaczmarek Z, Jeżowski S. 2012. Androgenesis in the bioenergy plant Miscanthus sinensis: from calli induction to plant regeneration. Crop Science 52: 2659–2673. [Google Scholar]
- Głowacka K, Adhikari S, Peng J, et al. 2014. Variation in chilling tolerance for photosynthesis and leaf extension growth among genotypes related to the C4 grass Miscanthus × giganteus. Journal of Experimental Botany 65: 5267–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Głowacka K, Clark LV, Adhikari S, et al. 2015. Genetic variation in Miscanthus × giganteus and the importance of estimating genetic distance thresholds for differentiating clones. GCB Bioenergy 7: 386–404. [Google Scholar]
- Greef J, Deuter M, Jung C, Schondelmaier J. 1997. Genetic diversity of European Miscanthus species revealed by AFLP fingerprinting. Genetic Resources and Crop Evolution 44: 185–195. [Google Scholar]
- Heaton EA, Dohleman FG, Long SP. 2008. Meeting US biofuel goals with less land: the potential of Miscanthus. Global Change Biology 14: 2000–2014. [Google Scholar]
- Hijmans RJ. 2017. geosphere: spherical trigonometry. R package version 1.5–7. [Google Scholar]
- Hirayoshi I, Nishikawa K, Kubono M, Murase T. 1957. Cyto-genetical studies on forage plants (VI): On the chromosome number of Ogi (Miscanthus sacchariflorus). Research Bulletin of the Faculty of Agriculture, Gifu University 8: 8–13. [Google Scholar]
- Hodkinson TR, Renvoize S. 2001. Nomenclature of Miscanthus × giganteus (Poaceae). Kew Bulletin 56: 759–760. [Google Scholar]
- Hodkinson TR, Chase MW, Takahashi C, Leitch IJ, Bennet MD, Renvoize SA. 2002. The use of DNA sequencing (ITS and trnL–F), AFLP, and fluorescent in situ hybridization to study allopolyploid Miscanthus (Poaceae). American Journal of Botany 89: 279–286. [DOI] [PubMed] [Google Scholar]
- Hodkinson TR, Klaas M, Jones MB, Prickett R, Barth S. 2015. Miscanthus: a case study for the utilization of natural genetic variation. Plant Genetic Resources 13: 219–237. [Google Scholar]
- Jiang J, Zhu M, Ai X, Xiao L, Deng G, Yi Z. 2013. Molecular evidence for a natural diploid hybrid between Miscanthus sinensis (Poaceae) and M. sacchariflorus. Plant Systematics and Evolution 299: 1367–1377. [Google Scholar]
- Jiang J-X, Wang Z-H, Tang B-R, Xiao L, Ai X, Yi Z-L. 2012. Development of novel chloroplast microsatellite markers for Miscanthus species (Poaceae). American Journal of Botany 99: e230–e233. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics 11: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L. 2008. GST and its relatives do not measure differentiation. Molecular Ecology 17: 4015–4026. [DOI] [PubMed] [Google Scholar]
- Lee E, Kim S, Nam S. 2008. Paleo-Tsushima Water and its effect on surface water properties in the East Sea during the last glacial maximum: revisited. Quaternary International 176–177: 3–12. [Google Scholar]
- Lee Y. 1964. Taxonomic studies on the genus Miscanthus: relationships among the section, subsection, and species, part 2: enumeration of species and varieties. Journal of Japanese Botany 39: 257–265. [Google Scholar]
- Li X, Hu D, Luo M, et al. 2013. Nuclear DNA content variation of three Miscanthus species in China. Genes & Genomics 35: 13–20. [Google Scholar]
- Linde-Laursen I. 1993. Cytogenetic analysis of Miscanthus ‘Giganteus’, an interspecific hybrid. Hereditas 119: 297–300. [Google Scholar]
- Liu J, Yu X. 2004. The exploitation and utilization of Triarrhena lutarioriparia resources. Journal of Zhongkai Agrotechnology 7: 63–67. [Google Scholar]
- Liu L. 1997. Miscanthus, Diandranthus, Triarrhena. In: Chen SL, ed. Flora reipublicae popularis sinicae, Vol. 10 Beijing: Science Press, 4–26. [Google Scholar]
- Liu L, Zhu M, Zhu T. 2001. Exploitation and utilization of Miscanthus & Triarrhena. Journal of Natural Resources 16: 562–563. [Google Scholar]
- Lu F, Lipka AE, Glaubitz J, et al. 2013. Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS Genetics 9: e1003215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximowicz CJ. 1859. Primitiae florae Amurensis. St. Petersburg, Russia: Buchdruckerei der kaiserlichen Akademie der Wissenschaften. [Google Scholar]
- Melamed-Bessudo C, Shilo S, Levy AA. 2016. Meiotic recombination and genome evolution in plants. Current Opinion in Plant Biology 30: 82–87. [DOI] [PubMed] [Google Scholar]
- Moon Y-H, Cha Y-L, Choi Y-H, et al. 2013. Diversity in ploidy levels and nuclear DNA amounts in Korean Miscanthus species. Euphytica 193: 317–326. [Google Scholar]
- Nei M. 1973. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences, USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PN. 1990. Elefantengrassanbau in Dänemark – Praktikerbericht. Pflug und Spaten 3: 1–4. [Google Scholar]
- Nishiwaki A, Mizuguti A, Kuwabara S, et al. 2011. Discovery of natural Miscanthus (Poaceae) triploid plants in sympatric populations of Miscanthus sacchariflorus and Miscanthus sinensis in southern Japan. American Journal of Botany 98: 154–159. [DOI] [PubMed] [Google Scholar]
- Paradis E. 2010. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26: 419–420. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genetics 8: e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pude R. 2011. Mehrklonsorte ‘Amuri’ – eine neue Züchtung der Tinplant-GmbH Available at http://www.miscanthus.de/zuechtung/mehrklonsorte.htm. Last accessed 18 June 2018.
- Ravenstein EG. 1861. The Russians on the Amur. London: Trübner and Co. [Google Scholar]
- Ray N, Adams J. 2001. A GIS-based vegetation map of the world at the last glacial maximum (25,000–15,000 BP). Internet Archaeology 11. [Google Scholar]
- Rayburn AL, Crawford J, Rayburn CM, Juvik JA. 2009. Genome size of three Miscanthus species. Plant Molecular Biology Reporter 27: 184–188. [Google Scholar]
- Regel E. 1862. Imperata sacchariflora Maxim. In: Gartenflora: allgemeine monatsschrift fur deutsche, russische und schweizerische garten- und blumenkunde, Vol. 11 Erlangen: Verlag von Ferdinand Enke, 92–93. [Google Scholar]
- Ryu E, Lee SJ, Yang DY, Kim JY. 2008. Paleoenvironmental studies of the Korean peninsula inferred from diatom assemblages. Quaternary International 176–177: 36–45. [Google Scholar]
- Sacks EJ, Juvik JA, Lin Q, Stewart JR, Yamada T. 2013. The gene pool of Miscanthus species and its improvement. In: Paterson AH, ed. Genomics of the Saccharinae. New York: Springer New York, 73–101. [Google Scholar]
- Sheng J, Hu X, Zeng X, et al. 2016. Nuclear DNA content in Miscanthus sp. and the geographical variation pattern in Miscanthus lutarioriparius. Scientific Reports 6: 34342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon H. 1952. Die Gartengräser. Ein Beitrag zur Oekologie der Gartenstauden. Thesis, Technische Hochschule München, Fakultät für Landwirtschaft, Freising Weihenstephan. [Google Scholar]
- Somerville C, Youngs H, Taylor C, Davis SC, Long SP. 2010. Feedstocks for lignocellulosic biofuels. Science 329: 790–792. [DOI] [PubMed] [Google Scholar]
- Song Z, Li J, Gu Z, Tang W, Yu J, Gao L. 2016. Characteristics of buried paleo-channels in the western South Yellow sea during the late last glaciation. Tehnicki Vjesnik 23: 835–842. [Google Scholar]
- Stapf O. 1891. Carl Johann Maximowicz. Nature 43: 449. [Google Scholar]
- Sun Q, Lin Q, Yi Z-L, Yang Z-R, Zhou F-S. 2010. A taxonomic revision of Miscanthus s.l. (Poaceae) from China. Botanical Journal of the Linnean Society 164: 178–220. [Google Scholar]
- Takahashi C, Shibata F. 2002. Analysis of Miscanthus sacchariflorus and M. sinensis chromosomes by fluorescence in situ hybridization using rDNA and total genomic DNA probes. Chromosome Science 6: 7–11. [Google Scholar]
- Tamura K, Uwatoko N, Yamashita H, et al. 2016. Discovery of natural interspecific hybrids between Miscanthus sacchariflorus and Miscanthus sinensis in southern Japan: morphological characterization, genetic structure, and origin. BioEnergy Research 9: 315–325. [Google Scholar]
- Uwatoko N, Tamura K, Yamashita H, Gau M. 2016. Naturally occurring triploid hybrids between Miscanthus sacchariflorus and M. sinensis in Southern Japan, show phenotypic variation in agronomic and morphological traits. Euphytica 212: 355–370. [Google Scholar]
- Winkler MG, Wang PK. 1993. The late-quaternary vegetation and climate of China, global climates since the last glacial maximum. In: Wright HE, Kutzbach JE Jr, Webb T, Ruddiman WF, Street-Perrott FA, Bartlein PJ eds. Global climates since the last glacial maximum, Minneapolis, MN: University of Minnesota Press, 221–261. [Google Scholar]
- Winter DJ. 2012. MMOD: an R library for the calculation of population differentiation statistics. Molecular Ecology Resources 12: 1158–1160. [DOI] [PubMed] [Google Scholar]
- Yan J, Zhu M, Liu W, et al. 2016. Genetic variation and bidirectional gene flow in the riparian plant Miscanthus lutarioriparius, across its endemic range: implications for adaptive potential. GCB Bioenergy 8: 764–776. [Google Scholar]
- Yoo DG, Lee GS, Kim GY, et al. 2016. Seismic stratigraphy and depositional history of late Quaternary deposits in a tide-dominated setting: an example from the eastern Yellow Sea. Marine and Petroleum Geology 73: 212–227. [Google Scholar]
- Yook MJ, Lim S-H, Song J-S, et al. 2014. Assessment of genetic diversity of Korean Miscanthus using morphological traits and SSR markers. Biomass and Bioenergy 66: 81–92. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.