S. Heidelberg is a clinically important serovar, linked to foodborne illness and among the top 5 serovars isolated from poultry in the United States and Canada. Acquisition of new genetic material from the microbial flora in the gastrointestinal tract of food animals, including broilers, may contribute to increased fitness of pathogens like S. Heidelberg and may increase their level of antibiotic tolerance. Therefore, it is critical to gain a better understanding of the interactions that occur between important pathogens and the commensals present in the animal gut and other agroecosystems. In this report, we show that the native flora in broiler ceca were capable of transferring mobile genetic elements carrying the AmpC β-lactamase (blaCMY-2) gene to an important foodborne pathogen, S. Heidelberg. The potential role for bacteriophage transduction is also discussed.
KEYWORDS: AMR, chicken, HGT, Salmonella
ABSTRACT
The chicken gastrointestinal tract harbors microorganisms that play a role in the health and disease status of the host. The cecum is the part of the gut that carries the highest microbial densities, has the longest residence time of digesta, and is a vital site for urea recycling and water regulation. Therefore, the cecum provides a rich environment for bacteria to horizontally transfer genes between one another via mobile genetic elements such as plasmids and bacteriophages. In this study, we used broiler chicken cecum as a model to investigate antibiotic resistance genes that can be transferred in vitro from cecal flora to Salmonella enterica serovar Heidelberg. We used whole-genome sequencing and resistome enrichment to decipher the interactions between S. Heidelberg, the gut microbiome, and acquired antibiotic resistance. After 48 h of incubation of ceca under microaerophilic conditions, we recovered one S. Heidelberg isolate with an acquired IncK2 plasmid (88 kb) carrying an extended-spectrum-β-lactamase gene (blaCMY-2). In vitro, this plasmid was transferable between Escherichia coli and S. Heidelberg strains but transfer was unsuccessful between S. Heidelberg strains. An in-depth genetic characterization of transferred plasmids suggests that they share significant homology with P1-like phages. This study contributes to our understanding of horizontal gene transfer between an important foodborne pathogen and the chicken gut microbiome.
IMPORTANCE S. Heidelberg is a clinically important serovar, linked to foodborne illness and among the top 5 serovars isolated from poultry in the United States and Canada. Acquisition of new genetic material from the microbial flora in the gastrointestinal tract of food animals, including broilers, may contribute to increased fitness of pathogens like S. Heidelberg and may increase their level of antibiotic tolerance. Therefore, it is critical to gain a better understanding of the interactions that occur between important pathogens and the commensals present in the animal gut and other agroecosystems. In this report, we show that the native flora in broiler ceca were capable of transferring mobile genetic elements carrying the AmpC β-lactamase (blaCMY-2) gene to an important foodborne pathogen, S. Heidelberg. The potential role for bacteriophage transduction is also discussed.
INTRODUCTION
Numerous studies have demonstrated that horizontal gene transfer (HGT) plays an important role in the spread of antimicrobial resistance (AMR) and virulence genes to foodborne bacterial pathogens (1). The notion that these genes are horizontally transferred from commensal bacteria residing in the animal gut or environment to foodborne pathogens is becoming more recognized (2–4). Likewise, plasmid-mediated antibiotic resistance gene (ARG) transfer is thought to occur between intestinal bacteria and putative pathogens like Salmonella. In an earlier study, we reported that Salmonella enterica serovar Heidelberg strains that acquired Col-like plasmids from poultry litter microbiota survived longer and demonstrated an increased resistance to selected antibiotics (5). Gumpert et al. also demonstrated that Escherichia coli strains present in the gut of infants could acquire multidrug resistance (MDR) plasmids in the absence of antibiotic treatment (6). Salmonella Heidelberg is one of the top Salmonella serovars causing foodborne illness in the United States and Canada (7, 8), and poultry has been reported as the primary source (8, 9).
Enterococcus faecalis, on the other hand, is part of the normal microbiome of poultry and is present in high abundance in day-old chicks (10). However, E. faecalis can be an opportunistic pathogen, causing diseases in poultry and humans (11–13). Only a few studies (14, 15) have investigated their promiscuity to plasmid-mediated ARG of poultry origin; consequently, our knowledge on their fitness factors in the chicken gut is limited.
In this report, we highlight the genetic changes that are associated with acquired antibiotic resistance in S. Heidelberg following in vitro inoculation into broiler ceca. Our primary aim was to determine ARG transfer events that occurred after inoculation of S. Heidelberg into chicken ceca, rather than mimicking what would have happened in vivo. We show that an AmpC β-lactamase gene (blaCMY-2) carried on an IncK2 plasmid could be transferred successfully between E. coli and S. Heidelberg, but transfer between S. Heidelberg strains was not possible.
RESULTS AND DISCUSSION
In vitro bacterial survival in broiler ceca.
To further explain how mobile elements may contribute to the fitness of Salmonella and Enterococcus, we monitored the survival of a nalidixic acid (Nal)-resistant S. Heidelberg (SH-2813nal) and rifampin (Rif)-resistant E. faecalis strain, JH2-2, following microaerophilic incubation in broiler ceca collected from a commercial processing plant (Fig. 1). The average cecum weight on collection was 4.4 ± 0.2 g (n =18), with a range of 2.3 to 6.8 g. SH-2813nal and E. faecalis JH2-2 grew ∼3 and 2 logs after 24 h of incubation in inoculated ceca. No Salmonella was detected in uninoculated ceca; however, the total E. coli organism count increased by ∼1 log after 48 h of incubation (P value < 0.086; Wilcoxon signed-rank test). No significant change was observed in enterococcus populations in uninoculated ceca (Table 1), but low background Rif-resistant populations were detected on Enterococcosel agar supplemented with Rif.
FIG 1.
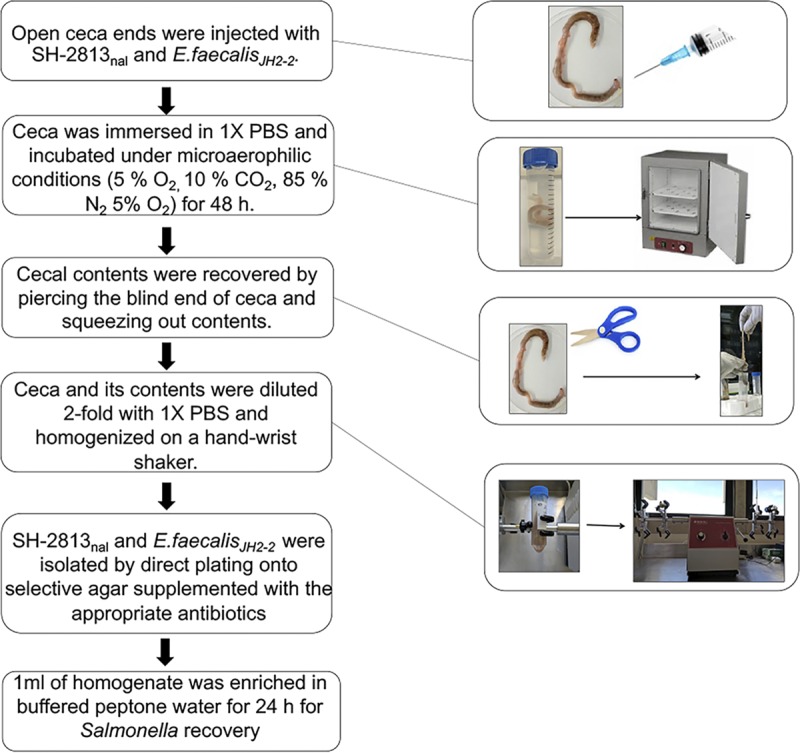
Experimental design. Entire viscera were collected from 18 broiler birds from the evisceration line of a commercial processing plant in Athens, GA. Viscera were put in individual Whirl-Pak bags and transported immediately on ice to the USDA-ARS research unit in Athens. Ceca were removed from viscera and the open end of one cecum/bird was injected with SH-2813nal and E. faecalis JH2-2. Ceca were incubated at 37°C under microaerophilic conditions for 48 h. Three individual ceca were sampled independently at 0.5, 6, 24, and 48 h.
TABLE 1.
Bacterial population in inoculated broiler cecaa
| Time (h)b | Bacterial concn (log CFU/g of cecum), mean (SD) |
||||
|---|---|---|---|---|---|
| SH-2813nalc | E. faecalis JH2-2c |
Total enterococci (control) |
Total E. coli (control) |
Other Salmonella spp. (control) |
|
| 0 | ND | ND | 5.54 (0.60) | 6.10 (0.85) | <LOD |
| 0.5 | 3.62 (0.35) | 3.67 (1.18) | ND | ND | ND |
| 6 | 4.44 (1.19) | 4.57 (1.38) | ND | ND | ND |
| 24 | 6.44 (0.78) | 6.11 (0.62) | ND | ND | ND |
| 48 | 6.15 (0.05) | 5.53 (0.49) | 6.03 (0.42) | 6.94 (0.42) | <LOD |
SH-2813nal is a nalidixic acid-resistant strain of S. Heidelberg; E. faecalis JH2-2 is a rifampin-resistant strain. Abbreviations: ND, not determined; LOD, limit of detection (not detected after 24 h of enrichment).
Concentrations of Enterococcus spp., E. coli, and Salmonella were determined in uninoculated ceca (n = 3) for time points 0 and 48 h only.
Each cecum was inoculated with ∼105 CFU of SH-2813nal and E. faecalis JH2-2 before incubation under microaerophilic conditions.
Although SH-2813nal and E. faecalis JH2-2 were able to grow in vitro in ceca, this does not correspond to colonization, i.e., attachment of bacteria to cecal epithelial cells. For instance, McHan et al. (16) demonstrated in vitro that up to 5 logs of S. Typhimurium could successfully attach to the ceca of 1- and 2-week-old broiler chicks. In our study, the final S. Heidelberg concentration measured in ceca was log 6.15 ± 0.05 CFU/g and likely includes attached and unattached bacterial cells.
Acquired antibiotic resistance after 48 h of incubation in ceca.
We did not observe any E. faecalis JH2-2 colony on Enterococcosel agar supplemented with rifampin and erythromycin (Ery) after 48 h of incubation; therefore, we performed antimicrobial susceptibility testing (AST) on isolates (n = 9) recovered from Enterococcosel supplemented with Rif. None of these E. faecalis JH2-2 isolates acquired resistance to any antibiotic present on the National Antimicrobial Resistance Monitoring System (NARMS) Gram-positive panel (data not shown); therefore, E. faecalis JH2-2 is not discussed further in this report. To determine if SH-2813nal acquired resistance to β-lactam antibiotics, we used XLT-4 agar supplemented with Nal and ampicillin (Amp) to select for presumptive isolates. In addition, AST was done on presumptive β-lactamase-producing isolates (n = 4) and a selected number of SH-2813nal isolates recovered from XLT-4 agar with Nal only (n = 9). We chose β-lactams due to the reported increase in cephalosporin resistance in S. Heidelberg recovered from chicken and chicken products (8, 17). Five of the thirteen isolates, including the 4 presumptive β-lactamase-producing isolates, acquired increased resistance to multiple antibiotics after 48 h of incubation of ceca. Two isolates displayed complete or intermediate resistance to cefoxitin (a β-lactam), chloramphenicol (a phenicol), and ciprofloxacin (a fluoroquinolones), and 2 other isolates were resistant to tetracyclines (streptomycin and/or tetracycline), sulfizoxazole (a sulfonamide), and cefoxitin (Table 2). One isolate (SH-13A-48h-NalAmp) showed decreased susceptibility or resistance to multiple β-lactams, including cefoxitin, ceftriaxone, cefazolin, cefpodoxime, ceftazidime, cefuroxime, piperacillin, ticarcillin-clavulanic acid, ampicillin, ampicillin-sulbactam, and amoxicillin-clavulanic acid. AST was performed twice on these 5 isolates to ascertain that observed changes in MIC were not reversed after cultivation.
TABLE 2.
Acquired antibiotic resistance profile of S. Heidelberg isolates recovered after cecum incubation
| Isolatea | Antibiotic resistance phenotypeb | Gene(s)/locic |
|---|---|---|
| SH-2813-Parentalphe | Nal | gyrA |
| SH-2813-Parentaltyr | Nal | gyrA |
| SH-4A-0.5h-Nalphe | Nal, Cip (Inter) | gyrA |
| SH-5A-0.5h-Nalphe | Nal, Cip (Inter) | gyrA |
| SH-6A-0.5h-Naltyr | Nal, Cip | gyrA, slyA, mprA |
| SH-10A-24h-Nalphe | Nal, Cip (Inter) | gyrA, 23S-5S rRNA ITS, corA |
| SH-11A-24h-Nalphe | Nal, Cip (Inter) | gyrA |
| SH-12A-24h-Nalphe | Nal, Cip (Inter) | gyrA, 23S-5S rRNA ITS |
| SH-13A-48h-Nalphe | Nal, Cip (Inter) | gyrA, 23S 5S rRNA ITS, envZ |
| SH-14B-48h-Naltyr | Nal, Cip (Inter), Fox (Inter), Chl (Inter), Str, Smx | gyrA, slyA, mprA |
| SH-15A-48h-Naltyr | Nal, Cip (Inter) | gyrA, slyA |
| SH-13A-48h-NalAmpphe | Nal, Cip (Inter), Cro, Amp, Sam, Fox, Caz, Cfz, Cpd, Cro, Cef, Pip, Tim | gyrA, hscA, 23S ribosomal RNA, blaCMY-2 |
| SH-14A-48h-NalAmpphe | Nal, Cip (Inter), Fox, Str, Smx, Tet | gyrA, Int, 23S-5S rRNA ITS, cpxA |
| SH-15A-48h-NalAmpphe | Nal, Cip, Fox, Chl (Inter) | gyrA, Lon, 23S-5S rRNA ITS, ccmF |
| SH-15B-48h-NalAmptyr | Nal, Cip, Fox, Chl (Inter) | gyrA, dinG, slyA, 16S-23S rRNA ITS |
Parental S. Heidelberg strain was made spontaneously resistant to 200 ppm of nalidixic acid. A superscript indicates the Ser83 gyrA mutation associated with the nalidixic acid resistance carried by the isolate.
Abbreviations: Nal, nalidixic acid; Cip, ciprofloxacin; Str, streptomycin; Tet, tetracycline; Smx, sulfamethoxazole; Chl, chloramphenicol; Amc, amoxicillin-clavulanic acid; Amp, ampicillin; Sam, ampicillin-sulbactam; Fox, cefoxitin; Cro, ceftriaxone; Cfz, cefazolin; Cpd, cefpodoxime; Caz, ceftazidime; Cef, cefuroxime; Pip, piperacillin; Tim, ticarcillin-clavulanic acid; Inter, intermediate resistance.
Gene or loci that acquired a mutation. Boldface denotes antibiotic resistance gene carried on a plasmid. ITS, internal transcribed spacer.
To identify the genomic changes underlying the acquisition of antibiotic resistance in these 5 isolates, whole-genome sequencing (WGS) data from all isolates, including sensitive and resistant isolates, were compared using single nucleotide polymorphism (SNP)/indel and resistance gene identification. Each of the resistant isolates carried at least one unique mutation from other sensitive/resistant isolates and the reference genome, except isolate SH-14B-48h-Nal (Table 2; see also Table S2 in the supplemental material). These mutations were in phage integrase (int), sensor histidine kinase (cpxA), ATP-dependent serine endopeptidase La (lon), cytochrome c-type biogenesis protein (ccmF), ATP-dependent DNA helicase (dinG), iron sulfur protein assembly (hscA), and the 16S-23S rRNA internal transcribed spacer (ITS) regions of the genome (Table S2). Among these loci, only the cpxA gene has been reported to confer resistance to cephalosporins via transcriptional regulation of multiple efflux pumps, such as OmpC and OmpF (18, 19). This mutation was identified in only one isolate. The observed phenotypic resistance could not be matched with a known mutation for the other four isolates with acquired resistance. For quinolones, gyrA mutations have been shown to confer cross-resistance to nalidixic acid and ciprofloxacin (20). In this study, the parental strain (SH-2813nal) had either a Ser83Tyr or Ser83Phe substitution in gyrA which could have accounted for the increase in MIC for ciprofloxacin in all isolates tested.
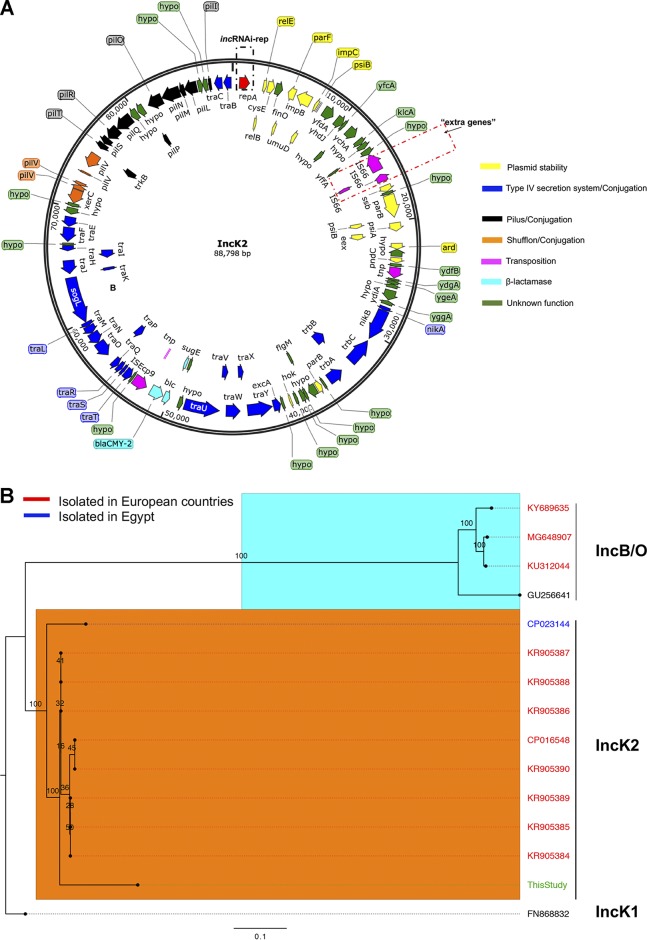
A β-lactamase gene (blaCMY-2) was found in 1 isolate (SH-13A-48h-NalAmp) that was likely responsible for the observed β-lactam resistance phenotype in this isolate. blaCMY-2 was located on a contig totaling 88 kb and predicted as IncB/O/K/Z plasmid using PlasmidFinder. The newly acquired plasmid had a read coverage two times greater than the chromosome and harbored stability, conjugative transfer, and shufflon recombinase genes (Fig. 2A), suggesting a plasmid with ∼2 copies per chromosome. Recently, Roschanski et al. showed that an E. coli isolate carrying an IncB/O/K/Z-like plasmid bearing a blaCMY-2 gene conferred resistance to cephalosporins and carbapenems, and they hypothesized that overexpression of CMY-2 and high plasmid copy numbers (PCNs) (8 per cell) were responsible for decreased susceptibility to carbapenems (21). In our study, this isolate exhibited increased resistance to multiple cephalosporin and penicillin drugs but was susceptible to the carbapenems tested (Table 2), indicative of an extended-spectrum-β-lactamase (ESBL)-like-producing S. Heidelberg strain.
FIG 2.
IncK2 acquired by S. Heidelberg. (A) Annotated map of IncK2 plasmid from this study. The dashed red rectangular box shows the extra genes discussed in the text. (B) Maximum likelihood tree of IncB/O/K plasmids based on complete plasmid sequence alignment. (C) Maximum likelihood tree of IncB/O/K plasmids based on incRNAI and rep gene alignment. The TVM model of nucleotide substitution and GAMMA model of rate heterogeneity were used for sequence evolution prediction. Numbers shown next to the branches represent the percentages of replicate trees where associated taxa cluster together based on ∼100 bootstrap replicates. Trees were rooted with reference IncK1 plasmid (GenBank accession number FN868832). The plasmid map was drawn using SnapGene v 4.3.8.1. The tree was viewed using FigTree v 1.4.4. Sources for panel C are Rozwandowicz et al. (25) and Seiffert et al. (26).
For antibiotic-resistant isolates without a known ARG, it is possible that overexpression of multidrug efflux pumps, such as acrAB-tolC, marA sdiA, mdsA, and mdsC, contributed to the higher MICs for cefoxitin, chloramphenicol, and tetracyclines. These genes were present in parental SH-2813nal (Table S3), and up- and downregulation of these efflux pumps have been shown to confer resistance to various antibiotic classes. For instance, Suzuki et al. demonstrated that AR acquisition in E. coli could be quantitatively predicted from the expression changes of a few genes, such as ompF and acrB (18). No resistance gene or mutation was identified to be associated with sulfizoxazole resistance, which agrees with other studies that have reported the lack of sulfonamide resistance genetic elements in some Salmonella enterica isolates (22, 23).
The harsh chicken gut environment may induce proteins that could confer cross protection against antibiotics. In a recent study, the authors exposed chicken breast inoculated with 15 outbreak-linked Salmonella enterica strains to simulated gut conditions of the mouth, stomach, and intestines (24). Acquisition of resistance to ciprofloxacin, ampicillin, ceftriaxone, and sulfamethoxazole-trimethoprim was reported for isolates of serovars Heidelberg, Newport, Albany, and Corvallis after 1 h of exposure.
Comparative genomics of IncB/O/K/Z-like plasmid acquired from ceca.
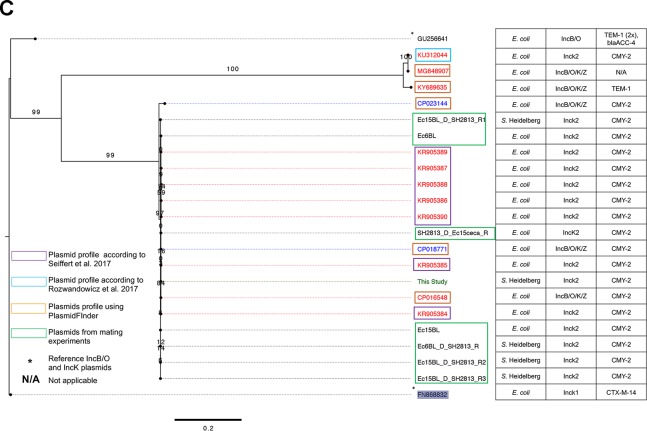
To compare the IncB/O/K/Z-like plasmid found in SH-13A-48h-NalAmp with sequenced IncB/O/K/Z plasmids, we sequenced plasmid DNA extracted from this isolate. An assembly of the sequenced genome with SPAdes resulted in 2 putative plasmid contigs of 88,798 and 37,845 bp with a coverage of >200×. The completeness and circularity of these plasmid contigs were inferred as described previously (5). The 37.8-kb plasmid is an IncX1 plasmid carried by the parental SH-2813nal strain and was present in all S. Heidelberg isolates sequenced for this study. Querying the NCBI nonredundant sequence database with the 88-kb contig yielded plasmids belonging to the IncK2 compatibility group (25) as the closest homologs (data not shown). These plasmids carried the blaCMY-2 gene and ranged in size from 79 to 86 kb. These IncK2 plasmids were found in E. coli strains isolated from clinical and poultry samples collected in European countries (25, 26). A phylogenetic tree reconstructed with selected complete IncK2 and IncB/O DNA sequences showed that the plasmid from this study shared up to 62% pairwise identity with IncK2 plasmids reported by Seiffert et al. (26) and <25% identity with plasmids of the IncB/O compatibility group. Consequently, all IncK2 plasmids were represented by a separate clade on the reconstructed ML tree (Fig. 2B).
The IncB/O, IncK, and IncZ plasmids belong to the I-complex of plasmids and share high homology in their incompability RNA interference (incRNAI) sequences (noncoding antisense RNA that is upstream of ori) (25, 27, 28). Therefore, the use of the incRNAI region as targets in the plasmid-based replicon typing (PBRT) classification scheme of the IncB/O/K plasmids poses challenges with their typing (25). In this study, a phylogenetic tree reconstructed using the incRNAI-rep sequences (i.e., incRNAI and replication protein [RepB] DNA sequences) from plasmids sharing significant incRNAI region homology (E value < 0.0001) with the plasmid from this study showed that this plasmid is more closely related to the IncK2 plasmids (Fig. 2C) than to the IncB/O group. This supports the result from the whole-plasmid DNA comparison. Based on this comparative analysis, this IncB/O/K/Z-like plasmid can be assigned to the IncK2 subgroup of IncI-complex plasmids.
The major difference between the IncK2 plasmids was in the coding DNA sequence (CDS) regions for shufflon proteins (Fig. S1), which has been previously reported (26). However, IncK2 from this study carried three extra IS66 family transposases totaling ∼2.5 kb inserted between the genes for single-stranded DNA-binding protein (ssb) and an uncharacterized protein (yffA) (Fig. 2A and Fig. S1).
In vitro stability, fitness, and plasmid copy number of IncK2 in host.
We serially passed 50 colonies from the SH-2813don on brilliant green sulfur agar (BGS) with or without Amp for 50 generations and used quantitative PCR (qPCR) primers targeting the incRNAI-rep region of IncK2 to confirm their presence. The IncK2 plasmid exhibited 100% stability under Amp selection and 86% stability without selection. In contrast, the CMY-2 gene was stable in only 25/50 colonies (50%) under Amp selection and 23/50 (46%) without selection. Rozwandowicz et al. also reported 100% stability for IncK2 plasmids carried by E. coli isolates from the Netherlands after 30 generations in LB agar without supplementation or supplemented with antibiotics (25). The temporal stability of blaCMY-2 gene has been previously reported for strains of Salmonella enterica (29).
Next, we performed mating experiments in broth culture to simultaneously assess the transfer, fitness cost, and PCN control of IncK2 in SH-2813don. This experiment was done with SH-2813don and IncK2-free SH-2813rec in the presence or absence of ampicillin selection. Transfer of IncK2 to SH-2813rec was not observed in broth experiments. Nevertheless, the IncK2-carrying SH-2813don population had significantly lower fitness than that of the IncK2-free SH-2813rec population during a mating experiment under no selection (P value = 0.0636; Wilcoxon signed-rank test [Fig. 3B]). The PCN of IncK2 was moderate to high during the mating experiment, ranging from 6 to 15 PCN/cell under selection and 21 to 35 PCN/cell without selection (Fig. 3A). This suggests that during ampicillin exposure IncK2 is present at low copy numbers per cell but in the absence of ampicillin IncK2 is present at high copy numbers per cell.
FIG 3.
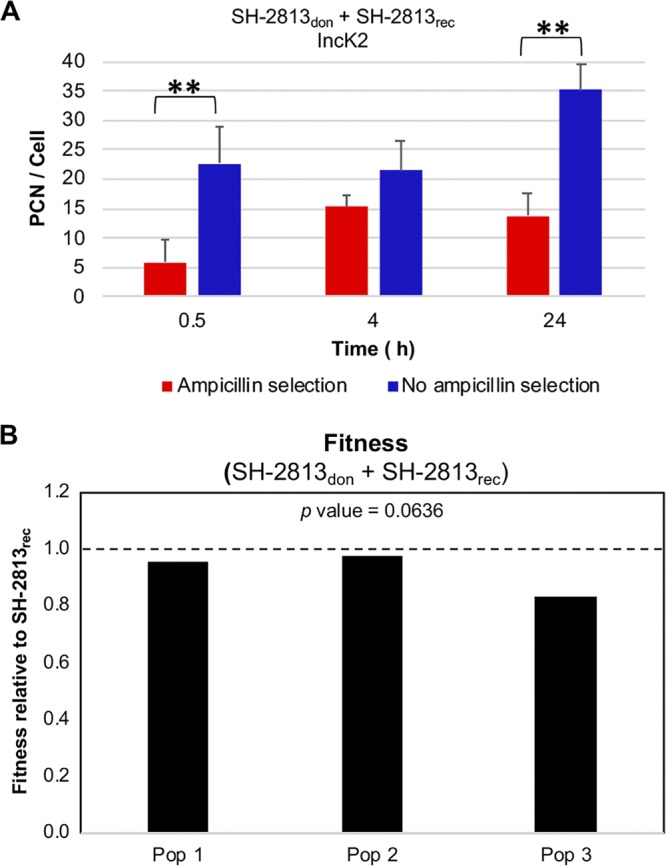
In vitro plasmid copy number (PCN) and fitness of IncK2 in S. Heidelberg host. (A) IncK2 PCN was determined during a mating experiment between IncK2-carrying populations (SH-2813don) and IncK2-free SH-2813rec populations with and without ampicillin selection. (B) Fitness of IncK2-carrying populations relative to IncK2-free populations under no ampicillin selection (**, P < 0.01). Each bar represents the fitness of one population that was established from one single bacterial colony. The horizontal dashed line represents the fitness of SH-2813rec.
Relative abundances of β-lactamase genes and Proteobacteria in ceca.
We performed targeted enrichment of ARGs using hybridization capture of the shotgun metagenomic library. Our goal was to sequence ARGs at a higher depth of coverage and determine the relative abundance of blaCMY-2. This enrichment approach has been demonstrated to increase ARG detection severalfold compared to shotgun metagenomics (30). In this study, we validated this method using WGS and phenotypic data on MDR pathogens (see the supplemental material). The average coverage of a de novo-assembled contig carrying an ARG was 803×, with a range of 11 to 32,735×. This method showed a high level of concordance with our WGS data set, and all false positives with a coverage threshold of 10× were removed (Table S4 and Fig. S2).
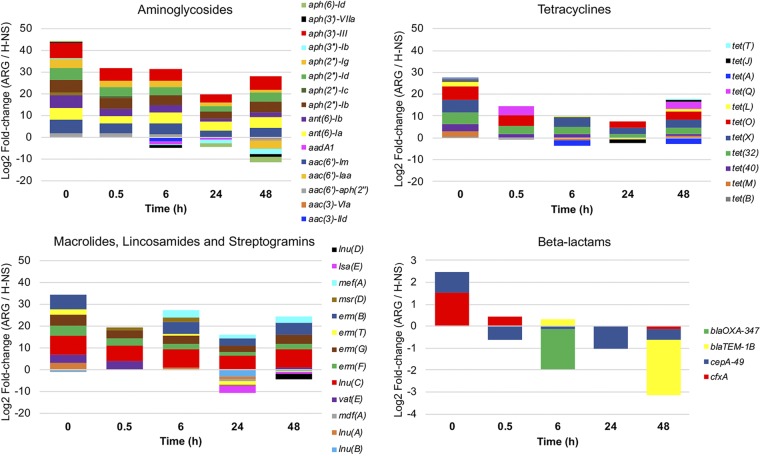
The most abundant ARGs in ceca upon collection conferred resistance to aminoglycosides, tetracyclines, or macrolides, lincosamides, and streptogramins (Fig. 4). Following the inoculation of S. Heidelberg into ceca, ARGs decreased in abundance and a few ARGs that were undetected at time zero were detected at a later time point (Fig. 4). For instance, aadA1 and tet(A) genes were not detected until after 6 h of incubation. Both genes are commonly found on class 1 integrons carried by Enterobacteria (31, 32), suggesting that facultative anaerobes, including E. coli, may bloom in vitro. Four β-lactamase genes were detected at low abundance, but blaCMY-2 was not detected at any time point (Fig. 4). Two genes (cfxA and cepA-49) found mainly in strict anaerobes, including the Bacteroidetes (33, 34), were the β-lactam genes detected at time zero, which is indicative of the microaerophilic conditions of the ceca upon collection. The blaTEM-1B gene that is commonly found in β-lactamase-resistant E. coli isolates (35) was the only other β-lactam gene detected (Fig. 4). However, blaTEM-1B was not detected until 6 h. Collectively, the resistome data suggest that the conditions in the ceca may get less anaerobic in vitro, thus favoring an increase in ARGs harbored by facultative anaerobes. This is further corroborated by our culture data that revealed a log increase in the E. coli population after 48 h of cecum incubation (Table 1).
FIG 4.
Relative abundances of ARGs determined using resistome enrichment. Targeted enrichment of ARGs using hybridization capture of shotgun metagenomic library was done on each cecum. At each time point, three cecum samples were used for resistome determination and ARG abundance was calculated by dividing the ARG contig coverage by the coverage of H-NS. A positive value indicates that ARG abundance is higher than that of H-NS, while a negative value indicates that ARG abundance is lower than that of H-NS.
For the broiler ceca used for this study, the blaCMY-2 gene was below the limit of detection of the metagenomic approach. The blaTEM-1B contig had up to 200× coverage, indicating that the enrichment method should also be sufficient for the detection of blaCMY-2. One explanation for the nondetection could be the instability of blaCMY-2, as we discussed previously. Furthermore, preserving the cecal contents in RNAlater prior to resistome enrichment may have introduced some artifacts. Although biases associated with microbiome and transcriptomic analyses of samples stored in preservatives have been reported (36), we do not know their effects on plasmids or ARG stability.
Retrospective identification of potential IncK2 donors.
The “extra” genes observed in the IncK2 plasmid from this study were part of an ∼9-kb region identified as P1 bacteriophage SJ46 (GenBank accession number NC_031129) by PHAST (Fig. S3). This region included CDS annotated as entry exlusion protein (eex) alleviation of restriction of DNA (ard), plasmid stability genes, and the IS66 family of transposons. This suggests that IncK2 plasmid may be a chimera of P1-like phages with an IncK2 plasmid backbone. In addition, the repL representative gene of the P1 phages is present in two of the IncK2 reference plasmids (Fig. S1). It is unlikely that this IncK2 plasmid was transferred from another Salmonella enterica strain because no Salmonella with IncK2 to our knowledge has been characterized or reported. Also, the cecum samples used in this study were negative for Salmonella even after 24 h of enrichment in buffered peptone water. Additionally, NCBI BLAST search against the nonredundant database found that the closest matches were to plasmids in E. coli strains, suggesting that E. coli was a potential donor of this plasmid. Based on this premise, we retrospectively screened 16 E. coli isolates recovered from 6 cecum samples for IncK2 plasmids.
Only 1 of the 16 isolates (here referred to as Ec15ceca) tested positive for IncK2 via qPCR. However, according to WGS, this isolate harbored no IncK2 plasmid but carried IncFIB and Col440i plasmids and a tet(A) gene that conferred tetracycline resistance (Table 3 and Table S5). This made us question if qPCR primers targeting the incRNAI-rep region of IncK2 plasmids (25, 26) were amplifying homologous regions in the genome of Ec15ceca. To answer this question, we performed a BLAST (tblastx) search against the de novo-assembled genome of Ec15ceca with the incRNAI-rep (∼1,476 bp) sequence from plasmids designated IncK2 in Fig. 2B. The only significant match (E value < 0.02) was an ∼11.7-kb contig with ∼39% pairwise identity. Protein annotation of this contig predicted 11 of 13 CDSs to encode phage tail assembly proteins (Fig. S4).
TABLE 3.
Resistance phenotype, β-lactamases, and plasmids detected in S. Heidelberg and E. coli donor, recipient, and recombinant strains from in vitro mating experimentsa
| Mating IDb | Strain ID | Resistance phenotype | AmpC gene | Plasmid(s) identified | IncK2 contig size (bp) |
|---|---|---|---|---|---|
| Solid plate | |||||
| SH-2813don | SH-13A-48h-NalAmpphe | Amp, Amc, Fox, Cro, Nal | CMY-2 | IncX1, IncK2 | 88,798 (C) |
| SH-2813recc | SH-14A-48h-NalAmpphe | Str, Nal | None | IncX1 | NA |
| SH-2813don | SH-13A-48h-NalAmpphe | Amp, Amc, Fox, Cro, Nal | CMY-2 | IncX1, IncK2 | 88,798 (C) |
| Ec15cecarec | Ec15ceca | Tet | None | IncFIB, Col440i | NA |
| SH2813_D_Ec15ceca_R | NA | Amp, Amc, Fox, Cro, Tet | CMY-2 | IncK2, IncFIB, Col440i | 88,178 |
| SH2813_D_Ec15ceca_Rdon | NA | Amp, Amc, Fox, Cro, Tet | CMY-2 | IncK2, IncFIB, Col440i | 88,178 |
| SH-2813rec | SH-2813-Parentaltyr | Nal | None | IncX1 | NA |
| Ec15ceca_D_SH2813_R | NA | Amp, Amc, Fox, Cro, Nal | ND | ND | ND |
| Ec6BLdon | Ec6BL | Amp, Amc, Fox, Cro, Gen, Str, Smx, Tet | CMY-2 | IncK2, IncF4:A5:B1, IncFII, Col (MG828) | 88,889 (C)e |
| SH-2813rec | SH-2813-Parentaltyr | Nal | None | IncX1 | NA |
| Ec6BL_D_SH2813_R2 | NA | Gen, Str, Smx, Tet, Nal | None | IncFII, IncX1 | NA |
| Ec6BL_D_SH2813_R | NA | Amp, Amc, Fox, Cro, Nal | CMY-2 | IncK2, IncX1 | 85,244 |
| Ec15BLdon | Ec15BL | Amp, Amc, Fox, Cro | CMY-2 | IncK2, IncF4:A-:B1, Col (MG828) | 85,509e |
| SH-2813rec | SH-2813-Parentaltyr | Nal | None | IncX1 | NA |
| Ec15BL_D_SH2813_Rd | NA | Amp, Amc, Fox, Cro, Nal | CMY-2 | IncK2, IncX1 | 84,268, 84,274, 84,273 |
| Broth | |||||
| SH-2813don | SH-13A-48h-NalAmpphe | Amp, Amc, Fox, Cro Nal | CMY-2 | IncX1, IncK2 | 88,798 (C) |
| SH-2813recc | SH-2813 | None | None | IncX1 | NA |
Abbreviations: ID, identifier; Gen, gentamicin; don, donor; rec, recipient; ND, not determined; NA, not applicable; C, complete circular contigs.
Boldface indicates recombinant strains with acquired antibiotic resistance.
Recipient did not acquire β-lactamase resistance from donor.
WGS was done on 3 individual colonies that acquired β-lactamase resistance from the donor.
De novo hybrid assembly was done using Illumina short- and MinION long-read technology (see also Table S5).
To get an improved assembly for the Ec15ceca genome and two E. coli isolates carrying IncK2 collected from broiler litter (BL), we leveraged Illumina short reads with long reads generated using MinION long-read technology (see the supplemental material for the method and Table S5). A rerun of the incRNAI-rep sequence BLAST search produced the same result for Ec15ceca, albeit on a larger contig size, ∼4.7 MB. Further, this incRNAI-rep sequence was identified to be part of a chromosomal region including lambdoid phage mEp460. For the other two E. coli strains the BLAST search produced significant matches (E value < 0.001) for multireplicon IncF plasmids (IncF4:A5:B1 and IncF4:A-:B1) (Table S5).
The largest homologous regions identified by PHAST to have the required modules for P1 (37) and mEp460 phage (GenBank accession number NC_019716) virion assembly were ∼113 kb and 90 kb, respectively. The P1 phage encoded 2 of the 3 “extra” IS66 family transposases present in the IncK2 plasmid from this study (Fig. S5). The mEp460 carried toxin/antitoxin systems, including relE, hicAB, and hokD (data not shown). Together, these findings suggest that a phage may have packaged plasmid DNA during exit from a bacterial cell carrying IncK2 (38). This form of recombination has been reported for P1-like bacteriophages carrying blaCTX-M-27, blaCTX-M-15, blaSHV-2, and mcr-1 present in Salmonella and E. coli strains (39–42). Otherwise, the tail module in mEp460 may include variable regions that determine the specificity of phage attachment to different bacterial hosts (43).
IncK2 plasmid is transferable between E. coli and S. Heidelberg.
We made attempts to recreate the recombination event between Ec15ceca and SH-2813don in vitro using solid plate mating. Our goal was to determine if transfer was possible, rather than determining the rate of transfer. First, we used Ec15ceca as the recipient of IncK2 and SH-2813don as the donor. After 24 h of mating, we observed multiple E. coli colonies on CHROMagar plates supplemented with ampicillin. Whole-genome sequencing and AST on a selected colony confirmed the successful transfer of IncK2 carrying blaCMY-2 to Ec15ceca (Table 3). Next, we performed the same experiment with the Ec15ceca strain that acquired IncK2 (Ec15cecadon) serving as the donor to SH-2813rec. We observed two SH-2813rec colonies that acquired β-lactam resistance. In addition, we performed a mating experiment with SH-2813don and SH-2813rec but observed no transfer of β-lactam resistance.
Finally, we selected two E. coli strains isolated from the broiler litter of a grow-out house carrying blaCMY-2 on an IncK2 plasmid. Strain Ec15BL carries only the blaCMY-2 gene, whereas Ec6BL carries additional ARGs on other type of plasmids (Table 3 and Table S5). The IncK2 in Ec6BL also carries three extra IS66 family transposases (data not shown). We performed the same solid mating experiment with these two strains acting as the donor of IncK2 to SH-2813rec. For both strains, we observed transfer of the IncK2 to SH-2813rec after 24 h of mating (Table 3). A plasmid designated IncFII carrying multidrug resistance and virulence genes was also transferred from Ec6BL to SH-2813rec (Fig. S6).
Although de novo assembly of plasmids from recombinants resulted in incomplete contigs, protein annotation revealed that the core genome of IncK2 did not change from donor to recipient (data not shown). Also, the blaCMY-2 backbone (ISEcp9-blaCMY-2-blc-sugE) was conserved in all recombinants. The IncFII contig (∼119 kb) carried ARGs that conferred resistance to aminoglycosides, tetracyclines, and sulfonamides (Fig. S6) and was calculated to be present at ∼4 copies/cell. In addition, this contig carried genes for metal resistance (mer operon), colicin production, and multiple toxin-antitoxin systems. Interestingly, an ∼38.8-kb segment of the IncFII contig was identified as phage SJ46, and it included a class 1 integron (intI1), transposases, and the aforementioned ARGs. Also, this IncFII plasmid shared significant homology (E value < 0.0001) with the incRNAI-rep region of IncK2 plasmids.
Determining the mode of recombination responsible for the transfer of these plasmids between E. coli and S. Heidelberg was beyond the scope of the study, but our analysis suggests that phage-mediated transfer cannot be ignored. The detection of P1-like phage DNA in IncK2 and IncF genomes and in the core genome of E. coli strains suggests that the interactions between phage-plasmid and host are complex. Our hypothesis is that P1-like phages, including SJ46, transduce plasmids between permissive hosts (generalized transduction). This does not preclude plasmid transfer that has been shown to occur through the uptake of free DNA (transformation) made possible after viral lysis of host cell (44). However, Salmonella has not been demonstrated to be naturally competent.
For conjugation to occur, cell-to-cell contact is required, i.e., the donor bacterium and recipient bacterium will need to meet. Although this is achievable in vitro, it is a bigger challenge in the cecum. One limiting factor to finding a mate carrying IncK2 is the low relative abundance of Proteobacteria, including E. coli and Salmonella residing in the chicken gut (10, 45). In contrast, lysis of an E. coli strain carrying a P1 phage can produce up 200 infective phage particles per cell (37) that are capable of generalized transduction (46). The rate of transduction will depend nonlinearly on the turnover and replication rates of E. coli and Salmonella in the cecum (46).
The inability to transfer IncK2 between Heidelberg strains in vitro was unexpected. Zhang and LeJeune (47) demonstrated that phage-mediated transfer of blaCMY-2, tet(A), and tet(B) genes from S. Heidelberg to S. Typhimurium was possible. One explanation is that the conditions used in vitro for mating experiments in our study are not reflective of those in the cecum. Otherwise, transfer may require phage or plasmid genes that are available only when E. coli is present. Further, it is possible that this transfer event is strain specific due to restriction modification systems, plasmid incompatibility, and the clustered regularly interspaced short palindromic repeat (CRISPR) repertoire (48). In other Salmonella strains or serovars, such HGT defense systems may serve to limit the acquisition of Inck2 plasmids. Lastly, this event maybe unique to the microbiome of the ceca used for this study.
Increasing resistance to cephalosporins has been attributed to the production of plasmid-mediated extended-spectrum or AmpC β-lactamases. The majority of these β-lactamase genes (blaCMY-2 and blaCTX-M) have been found on the IncI complex plasmids (17, 49–51). In this study, we showed that S. Heidelberg could acquire blaCMY-2 carried on an IncK2 plasmid from E. coli. Our analysis of the IncK2 plasmids suggests that PBRT is not sufficient for their detection or discrimination. Consequently, PCR primers targeting the inc/ori sites of IncK2 are likely to detect homologous regions present in phages or IncF plasmids. These results highlight the complex nature of interactions between plasmid-phage and ARG transfer between commensals and pathogens in the gut and support the need for a more targeted approach.
Conclusion.
Although it is important that S. Heidelberg acquired IncK2 carrying blaCMY-2 in vitro, this result must be appraised within the context of the experimental design employed. For the same event to occur in vivo, a permissive Heidelberg strain will have to evade the physical and biological barriers of the gut to get to the cecum, where a bacterial strain carrying blaCMY-2 must be a coresident. Furthermore, the inability to transfer IncK2 between Heidelberg strains may result in a bottleneck of IncK2 plasmid populations in vivo. The latter hypothesis still needs to be tested. The results presented here should be viewed as new information on HGT and acquisition of multidrug resistance in S. Heidelberg.
MATERIALS AND METHODS
Bacterial isolates.
A strain of Salmonella enterica serovar Heidelberg (SH-2813) isolated from chicken carcass rinsate was spontaneously made resistant to 200 ppm of nalidixic acid (Nal) (SH-2813nal) (5). This resulted in two isogenic S. Heidelberg strains with different gyrA substitutions conferring Nal resistance. One strain had a Ser83Tyr substitution, while the other had a Ser83Phe substitution. These strains are referred to as SH-2813-Parentaltyr and SH-2813-Parentalphe, respectively. The parental SH-2813 strain carries an ∼37-kb IncX1 conjugative plasmid (5). Enterococcus faecalis strain JH2-2 with chromosomally conferred resistance to rifampin (Rif), fusidic acid, and lincosamides was obtained from Charlene Jackson (USDA-ARS, Athens, GA). The JH2-2 strain harbors no plasmid.
Bacterial inoculum preparation.
Frozen bacterial stocks of SH-2813nal (SH-2813-Parentaltyr and SH-2813-Parentalphe) and E. faecalis JH2-2 were recovered on XLT-4 and Enterococcosel (BD Difco, Sparks, MD) agar supplemented with 200 ppm and 512 ppm of Nal and Rif (Sigma-Aldrich Corp., St. Louis, MO), respectively. Plates were incubated at 37°C for 48 h, and 6 colonies were randomly selected for inoculation into brain heart infusion broth (BHIB) with no antibiotics. After inoculation, BHIB tubes were incubated in a water bath overnight at 37°C and 85 rpm. Following overnight growth, BHIB culture was diluted 20-fold with fresh BHIB and incubated for ∼3 h in a water bath shaker at 37°C and 200 rpm. Afterwards, bacterial cells were centrifuged at 4,600 × g for 5 min and washed in 1× phosphate-buffered saline (PBS). Washed cells were used as the inoculum.
Cecum inoculation.
Entire viscera were collected randomly from 18 broiler birds from the evisceration line of a commercial processing plant. Viscera were put in individual Whirl-Pak bags and transported immediately on ice for processing. Ceca were removed from viscera, and the open end of one cecum/bird was injected with ∼105 CFU of SH-2813nal and E. faecalis JH2-2 (Fig. 1). Following inoculation, the cecum was immersed in an equal weight volume of sterile 1× PBS and incubated at 37°C under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) for 48 h. We chose these conditions after Aviv et al. (4) showed that a plasmid (pESI) present in multidrug-resistant Salmonella Infantis had its highest rate of transfer to E. coli and Salmonella Typhimurium at 37°C rather than 41°C and under microaerophilic rather than aerobic or anaerobic conditions.
Microbiological analysis.
After cecum inoculation, three individual ceca were sampled independently at 0.5, 6, 24, and 48 h. Cecal contents were removed by piercing the blind end of the cecum with sterile scissors and squeezing out contents with sterile gloves. Afterwards, the cecum and its contents were diluted 2-fold with 1× PBS and homogenized in a hand-wrist shaker at 450 rpm for 5 min. Nalidixic acid-resistant S. Heidelberg organisms were recovered by direct plating of serial dilutions on XLT-4 agar supplemented with 200 ppm of Nal (XLT-4nal). Serial dilutions were also performed on XLT-4 agar supplemented with 200 ppm of Nal and 32 ppm of ampicillin (Amp) for isolation of S. Heidelberg clones with acquired resistance to Amp. For E. faecalis JH2-2, Rif-resistant isolates were recovered on Enterococcosel agar supplemented with 512 ppm of Rif. For selection of JH2-2 clones with acquired resistance to erythromycin (Ery), serial dilutions were screened on Enterococcosel agar supplemented with 512 ppm of Rif and 8 ppm of Ery. Ampicillin and erythromycin were chosen to represent the common classes of broad-spectrum β-lactam and macrolide drug resistance found in Salmonella serovars and enterococcus strains. We also quantified the total population of E. coli and enterococci in uninoculated cecal preinoculation (n = 3) and at 48 h (n = 3) using CHROMagar ECC and Enterococcosel agar with no antibiotics. All bacterial incubations were carried out at 37°C, unless otherwise noted. The contents of the ceca were saved at a 1:1 ratio in RNAlater (Thermo Fisher, Waltham, MA) at –80°C before DNA was extracted for further analysis.
Antibiotic susceptibility testing.
Antimicrobial susceptibility testing was performed on selected SH-2813nal (n = 13) and E. faecalis JH2-2 (n = 9) isolates recovered during the 48 h of incubation of ceca and on isolates from in vitro mating experiments (n = 14) using the National Antimicrobial Resistance Monitoring System (NARMS) protocol (52). In addition, a Gram-negative panel (GN2F) (Thermo Fisher Scientific, Waltham, MA) was used to determine the MICs of one isolate to β-lactam drugs that are not available on the NARMS panel. MICs for isolates were determined by broth microdilution using the Sensititre semiautomated antimicrobial susceptibility system (Thermo Fisher Scientific). Results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) guidelines when available (53); otherwise, breakpoints established by NARMS were used (52).
Whole-genome sequencing and single nucleotide polymorphism identification.
Sequencing was performed on SH-2813nal (n =27) isolates recovered from cecal contents after incubation of ceca and on selected SH-2813rec (n = 6) (recipients) and ECrec (n = 1) strains that acquired antibiotic resistance during in vitro mating experiments. WGS was performed as described previously (5). A single run of sequencing on the MiSeq platform resulted in low-read depth coverage for some samples, so a second set of technical replicates were sequenced using the same methodology for each sample.
Single nucleotide polymorphisms (SNPs) and indels present in sequenced isolates were determined by aligning raw sequence reads to the chromosome of S. Heidelberg strain AMR588-04-00435 (GenBank accession number NC_CP016573) using Burrows-Wheeler Aligner (BWA) (54). To avoid false-positive SNPs from sequencing or batch effect error, only the sequence reads from the second technical replicates were used in this analysis. SAM file sorting and removal of PCR duplicates were done using SAMtools (55). Genome Analysis ToolKit (56) v 3.6 with a minimum mapping quality of 30 and a minimum base quality of 30 was used for SNP identification. Manipulation of generated variant call format files (VCF) was done using vcftools v 0.1.12b (57).
Resistome analysis.
Cecal DNA was used for resistome analysis. DNA was extracted and purified from 250 mg of cecal plus RNAlater homogenate using a previously described method for poultry litter (5). This DNA was used for target enrichment of shotgun libraries to characterize the antibiotic resistance genes (i.e., the resistome) present in ceca. Shotgun metagenomic libraries were prepared using New England BioLabs Ultra FS II library kits (New England BioLabs, Ipswich, MA), ligated to iTru adapter stubs (58), cleaned with a 0.8 ratio of speedbeads after ligation, and amplified with iTru dual-indexing primers. Following library preparation, samples were cleaned with a 1:1 ratio of speedbeads, quantified with a Qubit 2.0 fluorometer DNA high-sensitivity assay kit (Thermo Fisher, Waltham, MA), and pooled in sets of 8 samples for target enrichment by hybridization capture. Baits were designed and synthesized by Arbor Bioscience (Ann Arbor, MI) for all ARGs present in the Comprehensive Antibiotic Resistance Database (CARD) (59) database on November 2016. Sequencing was done on the Illumina MiSeq platform with 250-bp paired-end reads using the MiSeq reagent V2 (500 cycles).
Bioinformatics.
Whole-genome sequence reads from technical replicates were combined and assembled de novo into contigs using SPAdes and plasmidSPAdes (60, 61). The quality of assembled genomes of combined runs was assessed using Quality Assessment Tool for Genome Assemblies (QUAST) (59) (File S1). Assembled contigs were submitted to the Center for Genomic Epidemiology’s ResFinder (62) and CARD for the identification of resistance genes carried on plasmids or chromosome (63). Contigs were also submitted to PlasmidFinder (64) to determine existing plasmid replicon types.
Prophage-like regions were identified using PHAST (65) and PHASTER (66), and putative sequences were extracted for annotation. Annotation was done with Prokka (67) and by performing a PSI-BLAST search against the NCBI nonredundant database for viruses. Phylogenetic tree reconstructions of plasmids were performed using the maximum likelihood method implemented in RAxML-NG v 0.6.0 (68), with the number of bootstrap replicates criteria set to 100. We used the best model of sequence evolution predicted by jModelTest for tree reconstruction (69). ProgressiveMAUVE (70) and MAFFT (71) implemented in Geneious Prime v 2019.1.1 was used for the comparative analysis of plasmids and phages.
Fastq reads for resistome libraries were assembled using SPAdes (--meta). Contigs with ≤10× coverage or shorter than ∼500 bp were not used for ARG identification. Contigs were queried against ResFinder for acquired ARG determination and against CARD for global transcriptional regulator H-NS. The relative abundance of an ARG (log2 fold change) was determined from the coverage of the contig carrying the ARG, normalized against the coverage of H-NS. Prior to using this method on the resistome data set, we benchmarked it against WGS data of susceptible and MDR strains of S. Heidelberg (n = 1), S. Kentucky (n = 4), S. Enteritidis (n = 1), Campylobacter jejuni (n = 3), Campylobacter coli (n = 4), and E. faecalis (n = 4) (see the supplemental material).
Identification of E. coli strains carrying IncK2 plasmids.
One hundred microliters of cecal plus RNAlater homogenate from time point 48 h (n =6) was spread onto CHROMagar and incubated overnight. Afterwards, 16 E. coli colonies were selected randomly and restreaked on fresh CHROMagar plates. DNA extraction by boiling (5) was done on pure colonies before screening with two qPCR primers targeting IncK2 (Table S1).
Solid plate mating experiments.
Bacterial strains used for mating experiments are listed in Table 3. Five colonies from relevant recipient and donor strains (∼108 CFU/ml) were selected from overnight cultures grown on sheep blood agar (Remel Inc, San Diego, CA) and resuspended in 900 μl of 1× PBS. Recipient and donor were diluted 10,000- and 100,000-fold, respectively, using 1× PBS. Thereafter, 100 μl from the 1:10,000 recipient dilution was spread onto CHROMagar and incubated for 6 h. Following incubation, 100 μl from the 1:100,000 donor dilution was spread onto the recipient bacterial lawn and incubated for 18 to 24 h. After incubation, two areas of equal sizes (∼0.3 cm) were stamped from the middle and edge of the agar and added to 900 μl of 1× PBS. Vigorous shaking at 1,800 rpm was performed with a Fastprep-96 bead beater (MP Biomedicals, Solon, Ohio) for 1 min to suspend the cells from the agar plug. Serial dilutions were then spread onto brilliant green sulfur agar (BGS; BD Difco, Sparks, MD) or CHROMagar containing relevant antibiotics to distinguish the recipient, donor, and “recombinant population” that acquired antibiotic resistance. Agar plates were supplemented with antibiotics at the following concentrations, unless otherwise noted: 8 ppm of ampicillin, 1 ppm of gentamicin, 16 ppm of tetracycline, and 16 ppm of streptomycin, all purchased from Sigma (St. Louis, MO).
Mating/competition in liquid culture.
Mating between SH-13A-48h-NalAmp (SH-2813don) (donor) and IncK2-free SH-2813 (SH-2813rec) was performed in Mueller-Hinton broth (MHB) (BD Difco, Sparks, MD). The donor-recipient mixture was made as previously described (15). Mixtures were diluted 10,000-fold in 50-ml centrifuge tubes containing 20 ml of MHB without supplementation or supplemented with 32 ppm of Amp and incubated under the same microaerophilic conditions as the ceca for 24 h without shaking. To identify SH-2813rec that acquired IncK2 carrying the CMY-2 gene, we screened 56 colonies from each population at 24 h on BGS supplemented with Nal and/or Amp. Colonies that grew on BGS supplemented with Amp but showing no growth on BGS with Nal were designated presumptive recipients of IncK2.
The in vitro fitness cost associated with acquiring the IncK2 plasmid was tested with a 24-h population of SH-2813don and SH-2813rec under no Amp selection. The fitness of the population carrying IncK2 plasmids relative to the IncK2-free population was determined as described by San Millan et al. (72):
| (1) |
where Wstrain is the fitness of the population carrying IncK2 plasmids (SH-2813don), Ninitial,don and Nfinal,don are the numbers of cells (in CFU) in the population carrying the IncK2 plasmids before and after competition, and Ninitial,rec and Nfinal,rec are the numbers of cells of the IncK2-free (SH-2813rec) population before and after competition. To determine Nfinal,don and Nfinal,rec, we screened 56 colonies per population from BGS containing no antibiotics onto BGS with Nal and Amp. From here, the fraction of IncK2 plasmid-carrying cells (isolates resistant to both Nal and Amp) within each population was estimated by scoring the presence of growth on both BGS plates with Nal and Amp (Nfinal,don). Isolates that were susceptible to Amp were used for estimation of the IncK2-free population (Nfinal,rec).
Plasmid copy number determination.
We determined the plasmid copy number (PCN) of IncK2 present in each population during liquid culture mating. PCN was determined as described previously (5). Primer sets targeting gapA of S. Heidelberg and the incRNAI-rep of IncK2 were used for PCN determination (Table S1). Real-time qPCR amplification and analysis were performed on a CFX96 Touch real-time PCR detection system (Bio-Rad Inc., Hercules, CA) and carried out as previously described (5). PCN was determined as the copy ratio of plasmid-borne genes to gapA (73).
IncK2 and blaCMY-2 stability in S. Heidelberg.
The stability of IncK2 plasmid present in SH-2813don was tested over 50 generations. We serially passaged 50 individual colonies of SH-2813don consecutively for 5 days on brilliant green sulfur agar without supplementation or supplemented with Amp. On the 6th day, hot-boil DNA extraction was performed on 10 μl of each bacterial colony/culture. Afterwards, qPCR was performed on extracted DNA with primers targeting the incRNAI-rep (region upstream of repA) of IncK2 and blaCMY-2 (Fig. 2A and Table S1).
Statistical analyses.
Continuous variables did not meet the assumption of a normal distribution; therefore, comparisons between strains or treatment were performed using Wilcoxon signed-rank test. Analyses were performed using R (v 3.4.1).
Ethics statement.
We did not physically interact with the chickens before viscera collection at the processing plant; therefore, we are exempt from university guidelines and USDA/NIH regulations regarding animal use.
Data availability.
Whole-genome sequence and resistome fastq files are available under NCBI Sequence Read Archive (SRA) BioProject number PRJNA509629.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Troy Kierant, Denice Cudnik, Mark Berrang, Steven Knapp, Carolina Hall, Mary Katherine Crews, and Anna-Marie Bosch for their logistical and technical assistance.
Any opinions expressed in this paper are those of the authors and do not necessarily reflect the official positions and policies of the USDA, and any mention of products or trade names does not constitute recommendation for use.
We declare no competing commercial interests in relation to this work.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01903-19.
REFERENCES
- 1.Colavecchio A, Cadieux B, Lo A, Goodridge LD. 2017. Bacteriophages contribute to the spread of antibiotic resistance genes among foodborne pathogens of the Enterobacteriaceae family—a review. Front Microbiol 8:1108. doi: 10.3389/fmicb.2017.01108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Card RM, Cawthraw SA, Nunez-Garcia J, Ellis RJ, Kay G, Pallen MJ, Woodward MJ, Anjum MF. 2017. An in vitro chicken gut model demonstrates transfer of a multidrug resistance plasmid from Salmonella to commensal Escherichia coli. mBio 8:e00777-17. doi: 10.1128/mBio.00777-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, Pickard DJ, Barthel M, Westendorf AM, Krogfelt KA, Walker AW, Ackermann M, Dobrindt U, Thomson NR, Hardt W-D. 2012. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci U S A 109:1269–1274. doi: 10.1073/pnas.1113246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aviv G, Rahav G, Gal-Mor O. 2016. Horizontal transfer of the Salmonella enterica serovar Infantis resistance and virulence plasmid pESI to the gut microbiota of warm-blooded hosts. mBio 7:e01395-16. doi: 10.1128/mBio.01395-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oladeinde A, Cook K, Orlek A, Zock G, Herrington K, Cox N, Lawrence JP, Hall C. 2018. Hotspot mutations and ColE1 plasmids contribute to the fitness of Salmonella Heidelberg in poultry litter. PLoS One 13:e0202286. doi: 10.1371/journal.pone.0202286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gumpert H, Kubicek-Sutherland JZ, Porse A, Karami N, Munck C, Linkevicius M, Adlerberth I, Wold AE, Andersson DI, Sommer MO. 2017. Transfer and persistence of a multi-drug resistance plasmid in situ of the infant gut microbiota in the absence of antibiotic treatment. Front Microbiol 8:1852. doi: 10.3389/fmicb.2017.01852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans PS, Luo Y, Muruvanda T, Ayers S, Hiatt B, Hoffman M, Zhao S, Allard MW, Brown EW. 2014. Complete genome sequences of Salmonella enterica serovar Heidelberg strains associated with a multistate food-borne illness investigation. Genome Announc 2:e01154-13. doi: 10.1128/genomeA.01154-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edirmanasinghe R, Finley R, Parmley EJ, Avery BP, Carson C, Bekal S, Golding G, Mulvey MR. 2017. A whole-genome sequencing approach to study cefoxitin-resistant Salmonella enterica serovar Heidelberg isolates from various sources. Antimicrob Agents Chemother 61:e01919-16. doi: 10.1128/AAC.01919-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gieraltowski L, Higa J, Peralta V, Green A, Schwensohn C, Rosen H, Libby T, Kissler B, Marsden-Haug N, Booth H, Kimura A, Grass J, Bicknese A, Tolar B, Defibaugh-Chávez S, Williams I, Wise M. 2016. National outbreak of multidrug resistant Salmonella Heidelberg infections linked to a single poultry company. PLoS One 11:e0162369. doi: 10.1371/journal.pone.0162369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oakley BB, Buhr RJ, Ritz CW, Kiepper BH, Berrang ME, Seal BS, Cox NA. 2014. Successional changes in the chicken cecal microbiome during 42 days of growth are independent of organic acid feed additives. BMC Vet Res 10:282. doi: 10.1186/s12917-014-0282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregersen RH, Petersen A, Christensen H, Bisgaard M. 2010. Multilocus sequence typing of Enterococcus faecalis isolates demonstrating different lesion types in broiler breeders. Avian Pathol 39:435–440. doi: 10.1080/03079457.2010.517250. [DOI] [PubMed] [Google Scholar]
- 12.Hammerum AM. 2012. Enterococci of animal origin and their significance for public health. Clin Microbiol Infect 18:619–625. doi: 10.1111/j.1469-0691.2012.03829.x. [DOI] [PubMed] [Google Scholar]
- 13.Olsen RH, Schonheyder HC, Christensen H, Bisgaard M. 2012. Enterococcus faecalis of human and poultry origin share virulence genes supporting the zoonotic potential of E. faecalis. Zoonoses Public Health 59:256–263. doi: 10.1111/j.1863-2378.2011.01442.x. [DOI] [PubMed] [Google Scholar]
- 14.Cavaco LM, Bernal JF, Zankari E, Léon M, Hendriksen RS, Perez-Gutierrez E, Aarestrup FM, Donado-Godoy P. 2017. Detection of linezolid resistance due to the optrA gene in Enterococcus faecalis from poultry meat from the American continent (Colombia). J Antimicrob Chemother 72:678–683. doi: 10.1093/jac/dkw490. [DOI] [PubMed] [Google Scholar]
- 15.Hasan KA, Ali SA, Rehman M, Bin-Asif H, Zahid S. 2018. The unravelled Enterococcus faecalis zoonotic superbugs: emerging multiple resistant and virulent lineages isolated from poultry environment. Zoonoses Public Health 65:921–935. doi: 10.1111/zph.12512. [DOI] [PubMed] [Google Scholar]
- 16.McHan F, Cox NA, Blankenship LC, Bailey JS. 1989. In vitro attachment of Salmonella Typhimurium to chick ceca exposed to selected carbohydrates. Avian Dis 33:340–344. doi: 10.2307/1590853. [DOI] [PubMed] [Google Scholar]
- 17.Liakopoulos A, Geurts Y, Dierikx CM, Brouwer MS, Kant A, Wit B, Heymans R, Van Pelt W, Mevius DJ. 2016. Extended-spectrum cephalosporin-resistant Salmonella enterica serovar Heidelberg strains, the Netherlands. Emerg Infect Dis 22:1257–1261. doi: 10.3201/eid2207.151377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki S, Horinouchi T, Furusawa C. 2014. Prediction of antibiotic resistance by gene expression profiles. Nat Commun 5:5792. doi: 10.1038/ncomms6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun S, Berg OG, Roth JR, Andersson DI. 2009. Contribution of gene amplification to evolution of increased antibiotic resistance in Salmonella Typhimurium. Genetics 182:1183–1195. doi: 10.1534/genetics.109.103028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vila J, Ruiz J, Marco F, Barcelo A, Goñi P, Giralt E, De Anta TJ. 1994. Association between double mutation in gyrA gene of ciprofloxacin-resistant clinical isolates of Escherichia coli and MICs. Antimicrob Agents Chemother 38:2477–2479. doi: 10.1128/AAC.38.10.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roschanski N, Fischer J, Falgenhauer L, Pietsch M, Guenther S, Kreienbrock L, Chakraborty T, Pfeifer Y, Guerra B, Roesler UH. 2018. Retrospective analysis of bacterial cultures sampled in German chicken-fattening farms during the years 2011–2012 revealed additional vim-1 carbapenemase-producing Escherichia coli and a serologically rough Salmonella enterica serovar Infantis. Front Microbiol 9:538. doi: 10.3389/fmicb.2018.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maka L, Mackiw E, Sciezynska H, Modzelewska M, Popowska M. 2015. Resistance to sulfonamides and dissemination of sul genes among Salmonella spp. isolated from food in Poland. Foodborne Pathog Dis 12:383–389. doi: 10.1089/fpd.2014.1825. [DOI] [PubMed] [Google Scholar]
- 23.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. 2016. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 60:5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sales C, Melo A, Niedzwiedzka K, Souza E, Schaffner D, Magnani M. 2018. Changes of antibiotic resistance phenotype in outbreak-linked Salmonella enterica strains after exposure to human simulated gastrointestinal conditions in chicken meat. J Food Prot 81:1844–1850. doi: 10.4315/0362-028X.JFP-18-213. [DOI] [PubMed] [Google Scholar]
- 25.Rozwandowicz M, Brouwer MS, Zomer AL, Bossers A, Harders F, Mevius DJ, Wagenaar JA, Hordijk J. 2017. Plasmids of distinct Inck lineages show compatible phenotypes. Antimicrob Agents Chemother 61:e01954-16. doi: 10.1128/AAC.01954-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seiffert SN, Carattoli A, Schwendener S, Collaud A, Endimiani A, Perreten V. 2017. Plasmids carrying Blacmy-2/4 in Escherichia coli from poultry, poultry meat, and humans belong to a novel Inck subgroup designated Inck2. Front Microbiol 8:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Praszkier J, Wei T, Siemering K, Pittard J. 1991. Comparative analysis of the replication regions of IncB, IncK, and IncZ plasmids. J Bacteriol 173:2393–2397. doi: 10.1128/jb.173.7.2393-2397.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang D, Zhao Y, Feng J, Hu L, Jiang X, Zhan Z, Yang H, Yang W, Gao B, Wang J, Li J, Yin Z, Zhou D. 2019. Replicon-based typing of Inci-complex plasmids, and comparative genomics analysis of IncIgamma/k1 plasmids. Front Microbiol 10:48. doi: 10.3389/fmicb.2019.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Toro M, Garcia P, Rodriguez I, Rojo-Bezares B, Helmuth R, Saenz Y, Rodicio MR, Guerra B, Torres C. 2013. Characterisation of plasmids implicated in the mobilisation of extended-spectrum and AmpC beta-lactamase genes in clinical Salmonella enterica isolates and temporal stability of the resistance genotype. Int J Antimicrob Agents 42:167–172. doi: 10.1016/j.ijantimicag.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 30.Noyes NR, Weinroth ME, Parker JK, Dean CJ, Lakin SM, Raymond RA, Rovira P, Doster E, Abdo Z, Martin JN, Jones KL, Ruiz J, Boucher CA, Belk KE, Morley PS. 2017. Enrichment allows identification of diverse, rare elements in metagenomic resistome-virulome sequencing. Microbiome 5:142. doi: 10.1186/s40168-017-0361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dessie HK, Bae DH, Lee YJ. 2013. Characterization of integrons and their cassettes in Escherichia coli and Salmonella isolates from poultry in Korea. Poultry Sci 92:3036–3043. doi: 10.3382/ps.2013-03312. [DOI] [PubMed] [Google Scholar]
- 32.Sunde M, Simonsen GS, Slettemeas JS, Bockerman I, Norstrom M. 2015. Integron, plasmid and host strain characteristics of Escherichia coli from humans and food included in the Norwegian antimicrobial resistance monitoring programs. PLoS One 10:e0128797. doi: 10.1371/journal.pone.0128797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veloo ACM, Baas WH, Haan FJ, Coco J, Rossen JW. 2019. Prevalence of antimicrobial resistance genes in Bacteroides spp. and Prevotella spp. Dutch clinical isolates. Clin Microbiol Infect 25:1156.e9–1156.e13. doi: 10.1016/j.cmi.2019.02.017. [DOI] [PubMed] [Google Scholar]
- 34.Sydenham TV, Soki J, Hasman H, Wang M, Justesen US, ESGAI (ESCMID Study Group on Anaerobic Infections) . 2015. Identification of antimicrobial resistance genes in multidrug-resistant clinical Bacteroides fragilis isolates by whole genome shotgun sequencing. Anaerobe 31:59–64. doi: 10.1016/j.anaerobe.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 35.Brinas L, Zarazaga M, Saenz Y, Ruiz-Larrea F, Torres C. 2002. Beta-lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrob Agents Chemother 46:3156–3163. doi: 10.1128/aac.46.10.3156-3163.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarthy A, Chiang E, Schmidt ML, Denef VJ. 2015. RNA preservation agents and nucleic acid extraction method bias perceived bacterial community composition. PLoS One 10:e0121659. doi: 10.1371/journal.pone.0121659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Łobocka MB, Rose DJ, Plunkett G, Rusin M, Samojedny A, Lehnherr H, Yarmolinsky MB, Blattner FR. 2004. Genome of bacteriophage P1. J Bacteriol 186:7032–7068. doi: 10.1128/JB.186.21.7032-7068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mann BA, Slauch JM. 1997. Transduction of low-copy number plasmids by bacteriophage P22. Genetics 146:447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L, Li W, Jiang GZ, Zhang WH, Ding HZ, Liu YH, Zeng ZL, Jiang HX. 2017. Characterization of a P1-like bacteriophage carrying CTX-M-27 in Salmonella spp. resistant to third generation cephalosporins isolated from pork in China. Sci Rep 7:40710. doi: 10.1038/srep40710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin J, Ko KS. 2015. A plasmid bearing the bla(CTX-M-15) gene and phage P1-like sequences from a sequence type 11 Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 59:6608–6610. doi: 10.1128/AAC.00265-15. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Billard-Pomares T, Fouteau S, Jacquet ME, Roche D, Barbe V, Castellanos M, Bouet JY, Cruveiller S, Médigue C, Blanco J, Clermont O, Denamur E, Branger C. 2014. Characterization of a P1-like bacteriophage carrying an SHV-2 extended-spectrum beta-lactamase from an Escherichia coli strain. Antimicrob Agents Chemother 58:6550–6557. doi: 10.1128/AAC.03183-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai L, Wang J, Hurley D, Yu Z, Wang L, Chen Q, Li J, Li F, Fanning S. 2017. A novel disrupted mcr-1 gene and a lysogenized phage P1-like sequence detected from a large conjugative plasmid, cultured from a human atypical enteropathogenic Escherichia coli (aEPEC) recovered in China. J Antimicrob Chemother 72:1531–1533. [DOI] [PubMed] [Google Scholar]
- 43.Sandmeier H, Iida S, Arber W. 1992. DNA inversion regions Min of plasmid p15B and Cin of bacteriophage P1: evolution of bacteriophage tail fiber genes. J Bacteriol 174:3936–3944. doi: 10.1128/jb.174.12.3936-3944.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keen EC, Bliskovsky VV, Malagon F, Baker JD, Prince JS, Klaus JS, Adhya SL. 2017. Novel “superspreader” bacteriophages promote horizontal gene transfer by transformation. mBio 8:e02115-16. doi: 10.1128/mBio.02115-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oakley BB, Lillehoj HS, Kogut MH, Kim WK, Maurer JJ, Pedroso A, Lee MD, Collett SR, Johnson TJ, Cox NA. 2014. The chicken gastrointestinal microbiome. FEMS Microbiol Lett 360:100–112. doi: 10.1111/1574-6968.12608. [DOI] [PubMed] [Google Scholar]
- 46.Volkova VV, Lu Z, Besser T, Grohn YT. 2014. Modeling the infection dynamics of bacteriophages in enteric Escherichia coli: estimating the contribution of transduction to antimicrobial gene spread. Appl Environ Microbiol 80:4350–4362. doi: 10.1128/AEM.00446-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, LeJeune JT. 2008. Transduction of blaCMY-2, tet (A), and tet (B) from Salmonella enterica subspecies enterica serovar Heidelberg to S. Typhimurium. Vet Microbiol 129:418–425. doi: 10.1016/j.vetmic.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 48.Forsberg KJ, Malik HS. 2018. Microbial genomics: the expanding universe of bacterial defense systems. Curr Biol 28:R361–R364. doi: 10.1016/j.cub.2018.02.053. [DOI] [PubMed] [Google Scholar]
- 49.Voets GM, Fluit AC, Scharringa J, Schapendonk C, van den Munckhof T, Leverstein-van Hall MA, Stuart JC. 2013. Identical plasmid AmpC beta-lactamase genes and plasmid types in E. coli isolates from patients and poultry meat in the Netherlands. Int J Food Microbiol 167:359–362. doi: 10.1016/j.ijfoodmicro.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 50.Vogt D, Overesch G, Endimiani A, Collaud A, Thomann A, Perreten V. 2014. Occurrence and genetic characteristics of third-generation cephalosporin-resistant Escherichia coli in Swiss retail meat. Microb Drug Resist 20:485–494. doi: 10.1089/mdr.2013.0210. [DOI] [PubMed] [Google Scholar]
- 51.Smith H, Bossers A, Harders F, Wu G, Woodford N, Schwarz S, Guerra B, Rodríguez I, van Essen-Zandbergen A, Brouwer M, Mevius D. 2015. Characterization of epidemic IncI1-Igamma plasmids harboring Ambler class A and C genes in Escherichia coli and Salmonella enterica from animals and humans. Antimicrob Agents Chemother 59:5357–5365. doi: 10.1128/AAC.05006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Food and Drug Administration. 2015. The National Antimicrobial Resistance Monitoring System manual of laboratory methods, 2nd ed. https://www.fda.gov/downloads/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/UCM453381.pdf.
- 53.CLSI. 2017. Performance standards for antimicrobial susceptibility testing, 27th ed CLSI document M100 CLSI, Wayne, PA. [Google Scholar]
- 54.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glenn TC, Pierson TW, Bayona-Vásquez NJ, Kieran TJ, Hoffberg SL, Thomas JC, Lefever DE, Finger JW, Gao B, Bian X, Louha S. 2019. Adapterama II: universal amplicon sequencing on Illumina platforms (TaggiMatrix). bioRxiv 10.1101/619544. [DOI] [PMC free article] [PubMed]
- 59.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA. 2016. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32:3380–3387. [DOI] [PubMed] [Google Scholar]
- 62.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FSL, Wright GD, McArthur AG. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 68.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019:btz305. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 70.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.San Millan A, Heilbron K, MacLean RC. 2014. Positive epistasis between co-infecting plasmids promotes plasmid survival in bacterial populations. ISME J 8:601–612. doi: 10.1038/ismej.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee C, Kim J, Shin SG, Hwang S. 2006. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J Biotechnol 123:273–280. doi: 10.1016/j.jbiotec.2005.11.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole-genome sequence and resistome fastq files are available under NCBI Sequence Read Archive (SRA) BioProject number PRJNA509629.