Ammonia-oxidizing archaea (AOA) and bacteria (AOB) as well as comammox contribute to ammonia oxidation, which plays significant roles in riverine nitrogen cycle and N2O production. Source regions of numerous rivers in the world lie in high-elevation zones, but the abundance, community, and activity of ammonia oxidizers in rivers in high-elevation regions have rarely been investigated. This study revealed distinctive distribution patterns and community structures for ammonia oxidizers in five high-elevation rivers of the Qinghai-Tibet Plateau, and the individual and combined effects of low temperature, low nutrients, and strong solar radiation on ammonia oxidizers were elucidated. The findings of this study are helpful to broaden our knowledge on the biogeography and distribution pattern of ammonia oxidizers in river systems. Moreover, this study provides some implications to predict the performance of ammonia oxidizers in high-elevation rivers and its variations under global climate warming.
Keywords: Qinghai-Tibet Plateau, high-elevation rivers, ammonia-oxidizing archaea (AOA), comammox Nitrospira, suspended sediment, distance-decay pattern
ABSTRACT
Ammonia-oxidizing bacteria (AOB) and archaea (AOA) as well as comammox catalyze ammonia oxidation. The distribution and biogeography of these ammonia oxidizers might be distinctive in high-elevation rivers, which are generally characterized by low temperature and low ammonium concentration but strong solar radiation; however, these characteristics have rarely been documented. This study explored the abundance, community, and activity of ammonia oxidizers in the overlying water of five rivers in the Qinghai-Tibet Plateau (QTP). Potential nitrification rates in these rivers ranged from 5.4 to 38.4 nmol N liter−1 h−1, and they were significantly correlated with ammonium concentration rather than temperature. Comammox were found in 25 of the total 28 samples, and they outnumbered AOA in three samples. Contrary to most studied low-elevation rivers, average AOB amoA gene abundance was significantly higher than that of AOA, and AOB/AOA ratios increased with decreasing water temperature. The Simpson index of the AOA community increased with elevation (P < 0.05), and AOA and AOB communities exhibited high dissimilarities with low-elevation rivers. Cold-adapted (Nitrosospira amoA cluster 1, 33.6%) and oligotrophic (Nitrosomonas amoA cluster 6a, 31.7%) groups accounted for large proportions in the AOB community. Suspended sediment concentration exerted significant effects on ammonia oxidizer abundance (r > 0.56), and owing to their elevational variations in source and concentration, suspended sediments facilitated distance-decay patterns for AOA and AOB community similarities. This study demonstrates distinctive biogeography and distribution patterns for ammonia oxidizers in high-elevation rivers of the QTP. Extensive research should be conducted to explore the role of these microbes in the nitrogen cycle of this zone.
IMPORTANCE Ammonia-oxidizing archaea (AOA) and bacteria (AOB) as well as comammox contribute to ammonia oxidation, which plays significant roles in riverine nitrogen cycle and N2O production. Source regions of numerous rivers in the world lie in high-elevation zones, but the abundance, community, and activity of ammonia oxidizers in rivers in high-elevation regions have rarely been investigated. This study revealed distinctive distribution patterns and community structures for ammonia oxidizers in five high-elevation rivers of the Qinghai-Tibet Plateau, and the individual and combined effects of low temperature, low nutrients, and strong solar radiation on ammonia oxidizers were elucidated. The findings of this study are helpful to broaden our knowledge on the biogeography and distribution pattern of ammonia oxidizers in river systems. Moreover, this study provides some implications to predict the performance of ammonia oxidizers in high-elevation rivers and its variations under global climate warming.
INTRODUCTION
Nitrification is the biological oxidation of ammonia into nitrate (1), and it plays significant roles in the riverine nitrogen cycle and N2O production (2). Aerobic ammonia oxidation (NH3→NO2−) is the first and rate-limiting step in nitrification (1), which has long been thought to be mediated exclusively by beta- and gammaproteobacteria (3). However, during recent decades, ammonia-oxidizing archaea (AOA) of the phylum Thaumarchaeota also have been found to catalyze the ammonia oxidation into nitrite (3). It has been reported that AOA exert primary controls over ammonia oxidation in many terrestrial, marine, and geothermal habitats (4, 5). Recently, some nitrite-oxidizing bacteria within the genus Nitrospira were discovered to be complete ammonia oxidizers (6, 7) that could achieve whole ammonia oxidation into nitrate (NH3→NO3−). The amoA genes of comammox form two monophyletic sister clades, which are referred to as clade A (Com A) and clade B (Com B) (8).
Overlying water of large rivers is believed to have great significance in riverine nitrification (9–11). Some research has been conducted to analyze the distribution and community structure of ammonia oxidizers in riverine overlying water (12, 13). However, these studies focus on low-elevation regions, and high-elevation rivers are still rarely explored. Mountains are the world’s “water towers” (14) and the source region for over 50% of global rivers (15). Compared with low-elevation rivers, the rivers in high-elevation regions are generally characterized by lower temperature and lower ammonium concentration but stronger solar radiation. These factors have profound and differential influences on ammonia oxidizers in low-elevation rivers (13, 16) and other natural systems.
Temperature can significantly affect the diversity, distribution pattern, and community composition of ammonia oxidizers (17). AOA exhibit a stronger tolerance to high temperature than AOB (18); however, their performances in cold environments are condition specific. For example, the prevalence of AOA among ammonia oxidizers and their controlling roles in nitrification were observed in Arctic soils (19). However, in one soil microcosm across a temperature gradient of 4 to 42°C, the minimum temperature for an AOA-supported nitrification rate were estimated to be much higher than those for AOB (18). For the AOB community, Nitrosospira amoA cluster 1, which corresponds to Nitrosospira cluster 0 based on 16S rRNA gene clustering (20), is dominant at low temperature but completely disappears at high temperature (21). Differential responses to temperature are observed among different AOA phylotypes in one soil microcosm (22). Ammonium concentration also drives the community composition variation of ammonia oxidizers (21, 23), and high proportions of Nitrosospira amoA cluster 9 and the Nitrosomonas oligotropha cluster are detected at low ammonium concentrations (21). Moreover, solar radiation has been reported to have more inhibitory effects on AOA growth than on AOB (24). Therefore, under the combined effects of these factors, distinctive community compositions and distribution patterns for ammonia oxidizers could be expected to occur in high-elevation rivers. Cold-adapted and oligotrophic species should account for large proportions in their communities, and AOB might be the dominant ammonia oxidizers in the high-elevation rivers due to the strong solar radiation.
The Qinghai-Tibet Plateau (QTP) is the highest and most extensive highland in the world. The average elevation of the QTP is higher than 4,000 m above sea level, and its mean annual air temperature is −1.7°C (25). To test our hypotheses mentioned above, here, the Yellow River source region, the Yangtze River source region, the Yarlung Zangbo River, the Mekong River source region, and the Salween River source region (Fig. 1) in the QTP were selected to study the abundance, community, and activity of ammonia oxidizers in high-elevation rivers. First, we measured potential nitrification rates of the overlying water samples. Subsequently, distribution patterns of AOA, AOB, and comammox Nitrospira were investigated. Moreover, the community composition and diversity of AOA, AOB, and comammox Nitrospira in the overlying water were determined. Finally, effects of temperature, ammonium concentration, solar radiation intensity, and other factors on the abundance, community diversity, and activity of ammonia oxidizers in the five high-elevation rivers were analyzed.
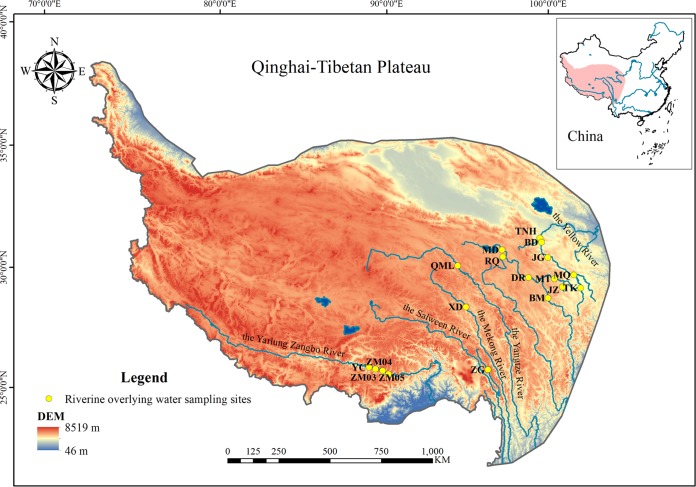
FIG 1.
Location of sampling sites (yellow circles) in the five high-elevation rivers of the Qinghai-Tibet Plateau. The five rivers consist of the Yellow River source region, the Yangtze River source region, the Mekong River source region, the Salween River source region, and the Yarlung Zangbo River. Ten sites were sampled in the Yellow River source region, with Maduo (MD), Dari (DR), Mentang (MT), Maqu (MQ), Jungong (JG), Banduo (BD), and Tangnaihai (TNH) in the main channel and Requ (RQ), Jiuzhi (JZ), and Tangke (TK) in the tributary. There were two sampling sites in the Yangtze River source region, which were Qumalai (QML) and Banma (BM). Xiangda (XD) was sampled in the Mekong River source region. Two sites were sampled in the Salween River source region, Jiayuqiao (JYQ) and Zuogong (ZG), and Zangmu03 (ZM03), Zangmu04 (ZM04), Zangmu05 (ZM05), and Yangcun (YC) were sampled in the Yarlung Zangbo River.
RESULTS
Physicochemical properties of the overlying water samples.
In our study area, elevation varied from 2,687 to 4,223 m above sea level, and latitude varied from 29.18 to 35.50° N (see Table S1 in the supplemental material). Solar radiation intensity ranged from 21.1 to 25.7 MJ m−2 day−1 and was positively linked to elevation (P < 0.01, n = 28) (Table S2). Dissolved oxygen (DO) concentration significantly declined from 7.97 to 5.57 mg liter−1 with the increase of latitude. Across our sampling sites, water temperature (7.2 to 19.9°C) was negatively correlated to elevation (r = −0.38, P < 0.05, n = 28), and a weaker negative relationship was found between latitude and water temperature (r = −0.34, P = 0.079, n = 28). Suspended sediment concentration varied from 0.001 to 0.711 g liter−1, and it was inversely related to both elevation and latitude (P < 0.05, n = 28). Temperature was significantly correlated to DO (r = −0.63, n = 28) and suspended sediments (r = 0.59, n = 28) (Table S2). Across these five rivers, NH4+ concentration ranged from 0.026 to 0.120 mg N liter−1, and NOx− (NO2− + NO3−)-N concentration was between 0.050 and 0.691 mg N liter−1. The overlying water was under alkaline conditions, with its pH value fluctuating from 7.68 to 9.08.
Potential nitrification rates of the overlying water samples.
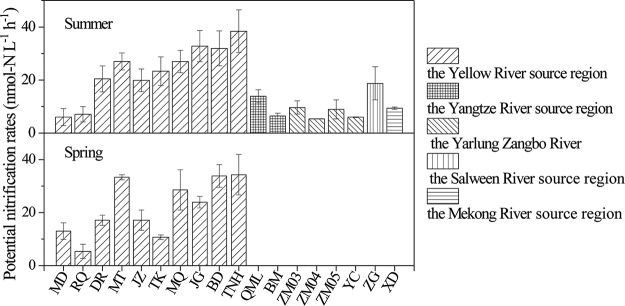
As shown in Fig. 2, potential nitrification rates (PNRs) in the overlying water samples varied from 5.4 to 38.4 nmol N liter−1 h−1, which significantly increased with decreasing elevation (Table S2). Moreover, PNRs were positively linked to ammonium and NOx−-N concentrations (P < 0.05) (Table S2). In contrast, there was no significant relationship between water temperature and PNRs (P > 0.05). In addition, in the Yellow River source region, average PNR in the summer (23.4 nmol N liter−1 h−1) was higher than that in the spring (21.7 nmol N liter−1 h−1), but this difference was not statistically significant (P > 0.05).
FIG 2.
Potential nitrification rates in the overlying water samples. Data represent the mean values ± standard deviations for three replicates.
Abundance of ammonia oxidizers in the overlying water samples.
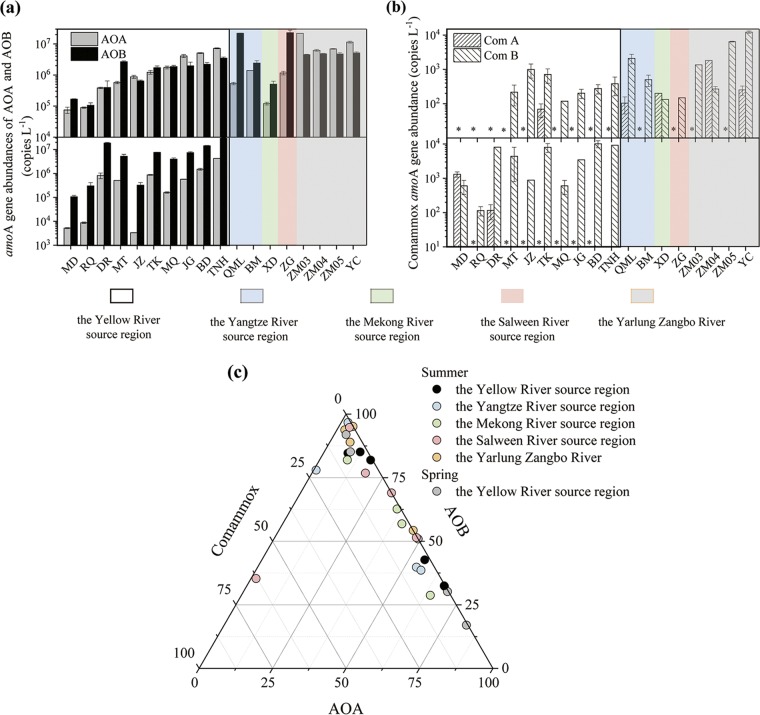
In the overlying water, according to the quantitative real-time PCR (qPCR) method, AOA amoA gene abundance ranged from 3.34 × 103 to 2.18 × 107 copies liter−1 and AOB amoA gene abundance varied between 1.06 × 105 and 2.98 × 107 copies liter−1 (Fig. 3a). AOB amoA gene abundance was higher than that of AOA in 20 of the 28 overlying water samples, with the AOB/AOA ratio ranging from 0.2 to 96.4 (Fig. 3b). The AOB/AOA ratio was negatively correlated with water temperature (P < 0.01) and significantly increased with elevation in the summer. Unlike AOA and AOB, Com A and Com B were found in only 7 and 25 samples, respectively, based on the most probable number PCR (MPN-PCR) method (Fig. 3b), with the highest amoA gene abundance of comammox (Com A plus Com B) being 1.25 × 104 copies liter−1. Comammox abundance was higher than that of AOA in three samples (Fig. 3c) when AOA abundance was corrected through dividing by 99 according to the MPN-PCR method results. No significant variation was found for the comammox/(AOA+AOB) ratio along the elevation or latitude gradient.
FIG 3.
amoA gene abundance of AOA and AOB (a) and Nitrospira comammox (b), as well as their respective proportions in the overlying water samples (c). An asterisk denotes that comammox amoA gene abundance was below the detection limit. AOA abundance (3.34 × 103 copies liter−1) in the JiuZhi station was slightly below the detection limit (3.70 × 103 copies liter−1). Moreover, in qPCR assays for this station, evident amplification was observed and there was no unspecific melt-curve peak. Therefore, 3.34 × 103 copies liter−1 could be accepted to represent its AOA abundance. Data represent the mean values ± SD for three replicates.
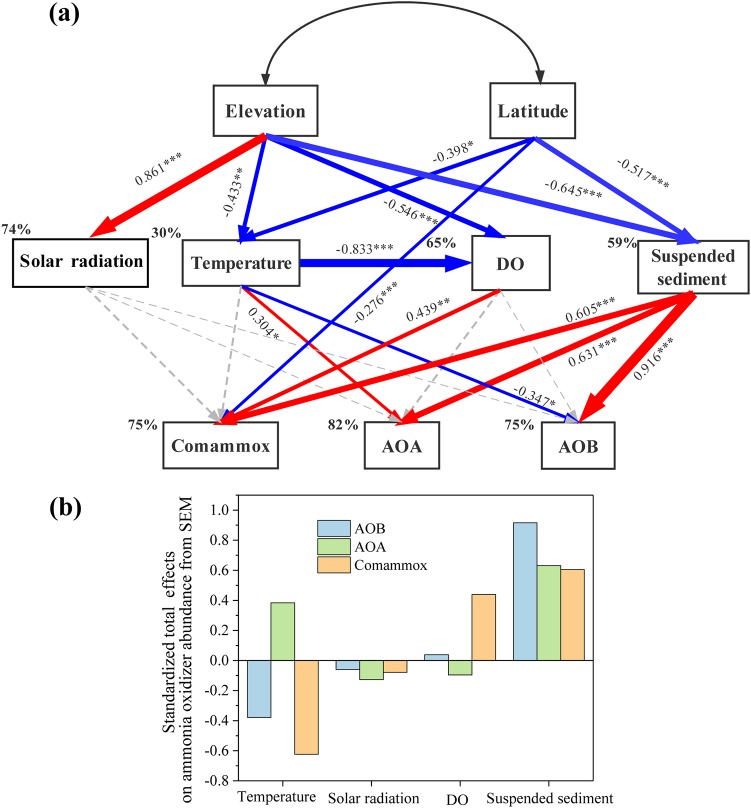
Structural equation models (SEMs) were constructed to reveal the effects of environmental factors on the distribution of ammonia oxidizers in these high-elevation rivers. According to the bootstrap test, the impact of the standard error bias caused by the limited sample size was negligible. A combination of suspended sediment concentration, water temperature, solar radiation, and DO explained 82%, 75%, and 75% of the variance for AOA, AOB, and comammox abundance (Fig. 4a), respectively. According to the total standardized effects (Fig. 4b), suspended sediment concentration was the strongest factor affecting AOA and AOB abundance, followed by water temperature; DO and solar radiation were of much less importance. Suspended sediment and water temperature exerted comparable influences on comammox abundance, with water temperature working mainly through its effects on DO. These findings were supported by (partial) correlation analysis results. After the effects of suspended sediment concentration were controlled, ammonia oxidizer abundance was no longer significantly correlated with elevation (P > 0.1) (Table S3). However, significant relationships still existed between suspended sediment concentration and ammonia oxidizer abundance when controlling for the effects of temperature, DO, ammonium, and solar radiation (Table S3). SEM found that water temperature significantly exerted negative effects on the abundance of AOB and comammox, which was also consistent with the results of partial correlation analysis (Table S3).
FIG 4.
(a) Structural equation models (SEMs) showing the effects of environmental factors on ammonia oxidizer abundance in the overlying water samples. Blue and red arrows indicate significantly negative and positive relationships, respectively, while gray arrows denote insignificant relationships. Numbers close to arrows are path coefficients, and the arrow width is proportional to the strength of these coefficients. Significance levels are indicated: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Percentages adjacent to variables refer to the variance accounted for by the model (R2). The final model showed a satisfactory fit to our data, with a model of χ2 = 16.7, df = 15, P = 0.34, GFI (goodness-of-fit index) = 0.89, and RMSEA (root mean square error of approximation) = 0.066. (b) The standardized total effects of each environmental factor involved in the SEM on the abundance of ammonia oxidizers.
Alpha diversity and taxonomic classification of ammonia oxidizers.
The obtained 423 AOA and 518 AOB sequences in the overlying water were assigned into 52 and 91 operational taxonomic units (OTUs), respectively, based on 95% similarity. Coverage for AOA clone libraries ranged from 70% to 95% and for AOB varied between 67% and 91% (Table 1). There was no significant seasonal fluctuation in AOA and AOB OTU richness of the Yellow River source region (Fig. S1). None of the environmental factors exerted significant effects on the Simpson (Chao1) diversity of both AOA and AOB communities (P > 0.1) according to Spearman correlation analysis. AOA Simpson diversity significantly increased with elevation (Fig. S2a); however, AOB Simpson index experienced unimodal variations along the elevation gradient, which peaked in the mid-elevations of our study area (Fig. S2b).
TABLE 1.
Diversity estimation of archaeal (bacterial) amoA gene clone libraries in the overlying water samplesa
| Season and clone library | Library sizeb | % coverage | Chao1 index | Simpson index |
|---|---|---|---|---|
| Spring | ||||
| MD | 31 (/) | 74 (/) | 21.3 (/) | 0.2043 (/) |
| DR | 15 (30) | 80 (77) | 9.0 (24.5) | 0.2000 (0.0667) |
| MT | 18 (33) | 83 (76) | 8.0 (26.0) | 0.1895 (0.2462) |
| TK | 22 (34) | 73 (91) | 17.5 (10.5) | 0.1255 (0.2210) |
| JG | 20 (32) | 70 (84) | 16.0 (15.0) | 0.1684 (0.2379) |
| TNH | 19 (26) | 79 (54) | 11.2 (37.0) | 0.0760 (0.1446) |
| Summer | ||||
| MD | 14 (27) | 86 (85) | 4.5 (12.0) | 0.5055 (0.1396) |
| DR | 15 (35) | 73 (86) | 13.0 (14.0) | 0.1429 (0.1345) |
| MT | 26 (33) | 81 (73) | 14.4 (28.0) | 0.0738 (0.0625) |
| TK | 25 (33) | 88 (76) | 9.8 (22.3) | 0.1600 (0.1307) |
| JG | 35 (30) | 83 (80) | 17.5 (19.5) | 0.0992 (0.0873) |
| TNH | 27 (31) | 74 (81) | 18.6 (17.0) | 0.0370 (0.0688) |
| ZM03 | 21 (18) | 95 (67) | 7.0 (15.5) | 0.1286 (0.3007) |
| ZM04 | 24 (32) | 78 (94) | 11.3 (12.2) | 0.3877 (0.0827) |
| ZM05 | 21 (28) | 76 (71) | 16.0 (22.0) | 0.5714 (0.0582) |
| XD | 22 (33) | 91 (76) | 7.3 (27.0) | 0.2294 (0.1155) |
| ZG | 23 (34) | 83 (76) | 11.5 (28.0) | 0.1028 (0.1266) |
| QML | 23 (29) | 91 (79) | 7.5 (18.0) | 0.1739 (0.0764) |
| BM | 22 (/) | 82 (/) | 12.5 (/) | 0.0649 (/) |
The numbers in parentheses denote corresponding parameter values for bacterial amoA clone libraries. A slash indicates that corresponding sites were not subjected to sequencing analysis. The shaded zone represents sampling sites in the Yellow River source region.
Number of sequences.
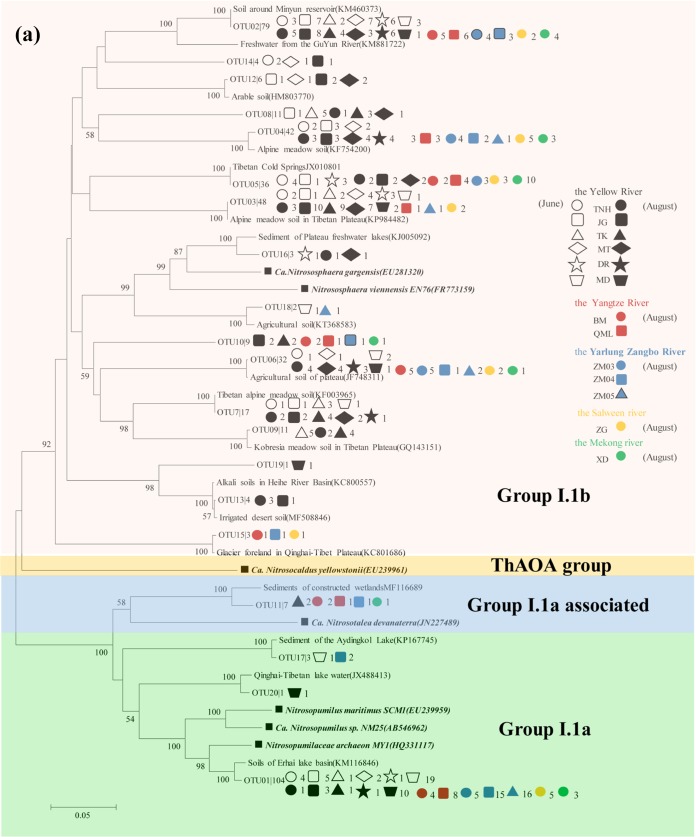
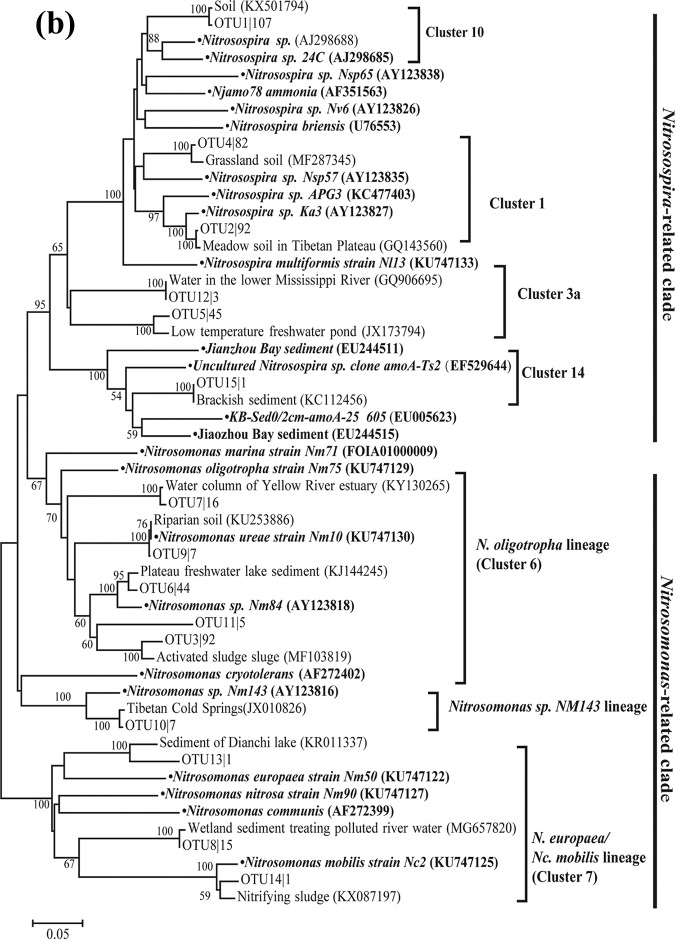
According to the results of phylogenetic analysis (Fig. 5a), most AOA sequences (308) were grouped into the Nitrososphaera cluster, while a smaller quantity (108) belonged to the Nitrosopumilus cluster and the remaining seven sequences were affiliated with the Nitrosotalea cluster. For AOB, their sequences were separately grouped with known species of Betaproteobacteria in the Nitrosomonas and Nitrosospira genera (Fig. 5b). Nitrosospira amoA cluster 1 (Nitrosospira cluster 0 [20]) was the most dominant AOB group in the overlying water, accounting for 33.6% of the total AOB sequences, followed by the N. oligotropha lineage (amoA cluster 6, 31.7%), while 3.3% accounted for the N. europaea/Nc. mobilis lineage (amoA cluster 7). An evident seasonal difference was observed in the AOB community composition of the Yellow River source region (Table S4). The proportion of Nitrosospira amoA cluster 1 was higher in the spring (49.7%) than in the summer (33.3%). Regarding comammox Nitrospira, 39 and 15 sequences were obtained for their clade A and clade B amoA genes, respectively. These sequences were closely matched with the discovered comammox species, and they were distinctly separated from known alpha-, beta-, and gammaproteobacterial amoA sequences according to the phylogenetic analysis (Fig. 5c).
FIG 5.
Phylogenetic trees showing the affiliations of amoA gene sequences of AOA (a), AOB (b), and comammox Nitrospira (c) retrieved from the overlying water samples based on the 85% (AOA and AOB) and 95% (comammox) similarities. The tree was constructed with neighbor-joining methods using maximum composite likelihood with 1,000 bootstraps. The clone names are labeled with OTU number followed by the number of times this OTU was detected in the overlying water samples.
Dissimilarities of AOA and AOB communities in the overlying water.
In the overlying water, weighted UniFrac analysis revealed that there were significant dissimilarities for AOA and AOB community structures among sampling sites (P < 0.01). The first two axes of principal coordinate analysis (PCoA) explained 57.7% and 56.9% of the AOA and AOB community variations, respectively (Fig. 6). No environmental factor was significantly correlated with AOA PCoA axes, and this was supported by the Mantel test results (Table S5). However, the Mantel test found that AOA community weighted UniFrac distances were significantly correlated with geographic distances (Pearson’s r = 0.24, P = 0.02). With respect to AOB, longitude and latitude were the strongest factors closely linking to Axis 2 (20.2%) (r of 0.72 and 0.71, P = 0.001). Similar results were found through the Mantel test, namely, that the geographic distances were significantly related to AOB community weighted UniFrac dissimilarities (r = 0.28, P = 0.04). Unlike the AOA community, the distances of environmental factors (DO, water temperature, suspended sediment, pH, oxidation-reduction potential (ORP), conductivity, and ammonium concentration) were also significantly linked to AOB community dissimilarities (r = 0.27, P = 0.029). However, both spatial and environmental factors failed to work in the partial Mantel test. Geographic distances lost their significant effect on AOB community dissimilarity (P = 0.091) when controlling for environmental factor distances and vice versa (r = 0.19, P = 0.075).
FIG 6.
PCoA plots of AOA (a) and AOB (b) community dissimilarities among sampling sites of five rivers in the QTP using the weighted UniFrac distance.
DISCUSSION
Unique community composition of ammonia oxidizers in the five rivers of the QTP.
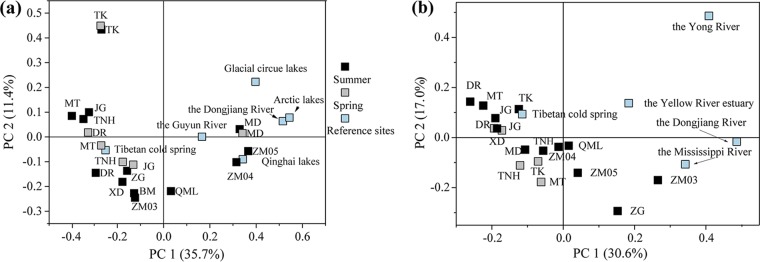
This study reveals the unique community composition of ammonia oxidizers in five high-elevation rivers that are characterized by low temperature and low ammonium concentration but intensive solar radiation. High dissimilarities of AOA and AOB communities were found between our studied rivers and low-elevation ones (Fig. 7). Few of the obtained AOA and AOB sequences matched with those in low-elevation rivers (Fig. 5a and b). Additionally, most AOA communities in the five rivers were also highly divergent from those of the Arctic lake environments (Fig. 7a). These results indicate that the high-elevation environments in the QTP have led to a unique evolution for ammonia oxidizers.
FIG 7.
PCoA plots of AOA (a) and AOB (b) community dissimilarities among sampling sites of this study and other reference sites using the weighted UniFrac distance. Detailed information on these reference sequences is listed in Table S6.
In the five high-elevation rivers, temperature played important roles in shaping the unique AOB community composition, of which Nitrosospira amoA cluster 1 accounted for the highest proportion (33.6%) and was at a much higher level than that of Nitrosospira cluster 3a (9.3%). These amoA cluster 1-type sequences (Fig. 5b) exhibited close phylogenetic relationships with Nitrosospira lacus, which could grow at 4°C but could not grow at 35°C (26). Consistent with our results, in soil microcosms, Nitrosospira amoA cluster 1 has been reported to be prevalent at low temperature (4 to 10°C), while a cluster 3-dominated community is observed at 30°C (21). Additionally, in soils of Mount Everest, cluster 3a predominates in low-elevation soils and disappears at most high-elevation sites (27). Furthermore, the oligotrophic environments also contributed to the distinctive AOB composition in the five high-elevation rivers. The proportion of oligotrophic species (Nitrosomonas amoA cluster 6) was nearly 10 times higher than that for the N. europaea lineage, which generally occurs in eutrophic freshwater environments and wastewater plants (28). However, another oligotrophic cluster (Nitrosospira amoA cluster 9) (21) was not detected in this study, and this might result from the preference of amoA cluster 9 for higher temperature (21). In contrast, most AOB sequences in one low-elevation river (the Yong River) (13) have been clustered with Nitrosomonas communis, which generally occurs in moderately eutrophic environments (28).
The Nitrososphaera cluster (also referred to as the group I.1b lineage) (29) contributed to 72.8% of the AOA community in the five high-elevation rivers. However, it was the Nitrosopumilus cluster (also called group I.1a) (29) that dominated the Dongjiang River, a low-elevation river (12). In the Yellow River source region, Nitrosopumilus abundance was higher in spring than in summer (Fig. 5a). This is contrary to their increase with elevated temperature in one soil microcosm (22), which suggests different mechanisms. Compared with AOB, temperature appeared to have less significant influences on AOA community composition (Fig. 6; also see Table S5 in the supplemental material). This might be because that Nitrososphaera cluster, the principal AOA component in this study, has been reported to be insensitive to temperature variations over a temperature range of 10 to 30°C (22).
Abundance and activity of ammonia oxidizers in the five rivers of the QTP.
Average amoA gene abundances of AOA and AOB were 1.42 × 106 and 3.04 × 106 copies liter−1, respectively, which were comparable to those in low-elevation rivers (Fig. S3). In this study, NanoDrop was used to quantify qPCR standards, which makes qPCR result comparison among different works of research less reliable at any precision finer than an order of magnitude. Therefore, here the comparison was only made for the general trends. Contrary to findings of most studies on low-elevation rivers (16), average AOB amoA gene abundance was significantly higher than that of AOA in this study. In the slightly eutrophic and cold lake sediments (10°C), AOB amoA gene abundance has also been found to be greater than that of AOA (30).
In this study, the dominance of AOB benefited from their community composition. As mentioned above, a large proportion of AOB community has adapted to the low-ammonium environment of the five high-elevation rivers. These oligotrophic AOB species (Nitrosomonas amoA cluster 6) possess ammonium inhibitory concentrations comparable to those of Nitrososphaera viennensis EN76 (31), with which most AOA sequences in this study shared close phylogenetic relationships (Fig. 5a). The AOB/AOA ratio significantly decreased with increasing temperature (P < 0.05), and the Nitrosospira amoA cluster 1 proportion was higher in spring than summer. This suggests the significant roles of low temperature in shaping the predominance of AOB in this zone. Consistent results have been found in soils of Mount Everest (27) and several freshwater lakes on the Yunnan Plateau, China (32). However, AOA are the predominant ammonia oxidizers in Arctic soils (19), and such a discrepancy could be partially incurred by the intensive solar radiation in high-elevation regions, which exerts more inhibitory effects on AOA growth than AOB growth (24).
PNRs in this study were measured with the chlorate inhibition method and, given the limitations of this method (33), only those studies using similar methods were selected for PNR comparison. The PNRs in the Yong River ranged from 642 to 1,428 nmol N liter−1 h−1 (13), and the average rate (1,100 nmol N liter−1 h−1) was approximately 60 times greater than that of this research (19 nmol N liter−1 h−1). Such a difference should be slightly larger, because our PNRs were measured in darkness. However, the bias between in situ and our determined results would be less than 14% (24), because the current photoinhibition effects on AOA and AOB are evaluated by laboratory culture experiments, and moreover AOA would not recover from photoinhibition (24). In the five high-elevation rivers, PNRs were positively correlated with ammonium concentration (P < 0.05) rather than temperature (P > 0.05), which suggests the key role of ammonium concentration in constraining ammonia oxidizer activity. Ammonium concentration in this study was less than 120 μg N liter−1, while it was 1,490 to 9,850 μg N liter−1 in the Yong River (13). Such a difference is within the same range as their PNRs. In contrast, the average temperature in this study was 14.9°C, which is merely ∼2-fold lower than the 25°C in the Yong River. However, this needs to be confirmed by an extended survey.
Effects of suspended sediments on community structure variations for AOA and AOB.
Different from soil and sediment, overlying water of one river has high hydrological connectivity. Neighboring sites are expected to share high similarity in their microbial composition. However, this was not observed in the Yellow River source region for AOA and AOB communities (Fig. S4). Instead, weighted UniFrac distances of its AOA community were significantly correlated with corresponding geographic distances (Pearson r = 0.69, P = 0.001). After controlling for the geographic distances, AOB community UniFrac distances were significantly correlated with elevation variations during summer (r = 0.47, P = 0.051). This means that the community similarity for AOA and AOB in the overlying water samples was closely linked to the geographic proximity instead of their hydrological connectivity in the Yellow River source region.
This contradictory result might arise from the community coalescence in river systems (34). In the overlying water, relative significance of dispersal-facilitated (“mass effects”) and environmental condition-based sorting (“species sorting”) shapes microbial community composition (35, 36). In this study, ammonia oxidizer abundance was greatly affected by suspended sediment concentration (Fig. 4), which significantly increased with the decrease of elevation in the Yellow River source region (Fig. S5). This indicates that bed sediment and ambient soil could continuously supply overlying water with ammonia oxidizers via solid-particle transportation. Both AOA and AOB have relatively long generation times. It has been reported that the doubling time of Nitrososphaera viennensis is between 27.5 h and 46 h (37), and minimum doubling times for AOB are typically in the range of 12 h to several days (38). This allows AOA and AOB to preserve some of their original characteristics in community composition and structure for a period. According to our study, nearly 79% of suspended sediment came from terrestrial sources in the Yellow River source region (data not shown here). Significant distance-decay patterns for ammonia oxidizer community similarities have been observed in soil systems (39, 40). AOA and AOB community structures greatly shift along the elevation gradient in the soils of Mount Everest (27). Under such circumstances, the presence of suspended sediment facilitated the distance-decay patterns for ammonia oxidizer community similarities in the overlying water of the Yellow River source region.
Previous studies have reported that nitrifier abundance increases with suspended sediment concentration (11, 16), and this relationship was also observed in this study. Moreover, we found that the presence of suspended sediment facilitated the decrease of community similarity for AOA and AOB with increasing spatial distances in the Yellow River source region, which might affect their functional roles. Many studies have found links between microbial community structures and the corresponding functions (41), and different AOA phylotypes are functionally heterogeneous in Arctic soils (19).
Conclusions.
This study investigated the abundance, community, and activity of ammonia oxidizers in the five rivers of the QTP. The unique conditions (low temperature, low ammonium concentration, and intensive solar radiation) in the five high-elevation rivers shaped distinctive communities for AOA and AOB. AOB were the dominant ammonia oxidizers, and cold-adapted (Nitrososphaera amoA cluster 1, 33.6%) and oligotrophic (Nitrososphaera amoA cluster 6a, 31.7%) groups accounted for large proportions in the AOB community. PNRs of these rivers were much lower than those of low-elevation rivers, which could be attributed mainly to their low ammonium concentration rather than low temperature. Additionally, the presence of suspended sediment facilitated the distance-decay pattern for AOA and AOB community similarity in the overlying water. In addition, Nitrospira comammox were found in 25 of the 28 total samples, and they outnumbered AOA in three samples. This study demonstrates the distinctive communities and distribution patterns for ammonia oxidizers in high-elevation rivers of the QTP. Extensive research should be conducted to explore the role of these microbes in the nitrogen cycle of this zone. Moreover, the community and function of ammonia oxidizers, especially Nitrosospira amoA cluster 1, in the high-elevation rivers of the QTP might change under global climate warming, which deserves further study.
MATERIALS AND METHODS
Sample collection and physiochemical factor analysis.
In this study, the sampling work in the Yellow River source region was conducted twice during July 2016 (summer) and May 2017 (spring), and the other four rivers were sampled during July 2016. Ten sampling sites were selected in the Yellow River source region. There were four and two sampling sites in the Yarlung Zangbo River and the Yangtze River source regions, respectively, and one sampling site was selected in each of the remaining two rivers. A total of 28 samples were collected from these five rivers. During each sampling period, triplicate overlying water samples (20 cm below the surface) were collected at each site using presterilized Nalgene bottles. For each sample, approximately 400 ml to 1,000 ml water was filtered with 0.20-μm-pore-size polycarbonate filters (Millipore, USA) in situ. These filters were temporarily stored at –15°C in a vehicle-mounted refrigerator (FYL-YS-30L; Fuyilian, China) during the whole sampling process and stored at −80°C in the laboratory for further DNA extraction. Incubation samples for nitrification rate analysis were transported to the laboratory on ice within 72 h.
Elevation, latitude, and longitude of each sampling site were recorded. Some physicochemical properties of the overlying water (temperature, DO, pH, ORP, and conductivity) were measured in situ with portable meters (SevenGo Duo SG23 and Seven2Go S4; Mettler Toledo). The solar radiation intensity is estimated based on observations from meteorological stations on the QTP according to the China Meteorological Administration. In the laboratory, one part of each water sample was filtered through 0.45-μm-pore-size polycarbonate filters (Millipore, USA). These filters then were dried in a freeze drier to constant weight and weighed to determine the suspended sediment concentration. Subsequently, concentrations of NH4+-N and NOx− (NO2− + NO3−)-N in the obtained filtrate were analyzed colorimetrically with an AutoAnalyzer 3 (Bran & Luebbe, France).
Determination of PNRs for the overlying water samples.
After overlying water samples arrived at our laboratory, PNRs were determined within 24 h with a chlorate inhibition method (42). Unfiltered water samples (total of 100 ml) from each site were transferred into a series of 300-ml serum vials. Potassium chlorate (KClO3) was added into the vials to a final concentration of 10 mM to inhibit the oxidation of nitrite into nitrate (42, 43). Incubations with sterile water samples were conducted to evaluate abiotic ammonia oxidation. These vials were capped by silicone rubber septa and crimp sealed with aluminum closures. A specific volume of headspace air was drawn out from the serum vials with syringes to simulate the corresponding in situ pressure (detailed information can be seen in the supplemental material). The vials then were incubated in the dark at in situ temperature for 0.25 h, 4 h, 6 h, 12 h, and 24 h with a shaking speed of 190 rpm, and each incubation was conducted in triplicate. At each sampling point, a 3-ml water sample was collected for nitrite analysis with the AutoAnalyzer 3. Subsequently, these vials were sealed again with silicone rubber septa and aluminum closures, followed by the extraction of air from the vials to simulate in situ air pressure. The subsamples then were incubated in the dark again for 24 h. PNRs were determined through the linear accumulation of NO2− concentration during the incubation period, as shown in Fig. S6 in the supplemental material.
DNA extraction, cloning, and sequence analysis for ammonia oxidizers.
For each sample, triplicate DNA was extracted from the 0.20-μm filters, obtained by filtration of the overlying water samples, with a FastDNA Spin kit for soil (QBIOgene, Carlsbad, CA, USA) according to the manufacturer’s instructions. The concentration and quality of obtained DNA were roughly determined with a NanoDrop 2000 UV-visible spectrophotometer (Thermo Scientific) and 1% agarose gel electrophoresis. Primer pairs Arch-amoAF/Arch-amoAR (44) and amoA-1F/amoA-1R (45) were used to amplify archaeal and bacterial ammonia monooxygenase subunit A (amoA) genes, respectively, in the overlying water samples. Archaeal and bacterial amoA genes were sequenced in 19 and 17 samples, respectively. Detailed information is shown in Table 1. Primer pairs comaA-244F/comaA-659R and comaB-244F/comaB-659R were employed to retrieve comammox Nitrospira clade A (Com A) and clade B (Com B) sequences, respectively (8). Specific reaction mixtures and thermal profiles for these PCR amplifications are listed in Table 2. Triplicate amoA amplicons were pooled before sequencing analysis to diminish within-sample heterogeneity. These amplicons then were purified and ligated into a pMD18-T vector (TaKaRa, Dalian, China). The inserts with correct patterns were sequenced with an ABI3730 DNA sequencer (Applied Biosystems, USA).
TABLE 2.
Primer pairs used in this study and corresponding amplification conditions
| Reference | Specificity | Primer | Sequence (5′–3′) | Thermal profile |
Reaction mixturea (25 μl total) |
|---|---|---|---|---|---|
| 44 | AOA amoA (MPN) PCR | Arch-amoAF | STAATGGTCTGGCTTAGACG | 5 min at 95°C; 30 cycles consisting of 94°C for 45 s, 53°C for 60 s, and 72°C for 60 s; 72°C for 15 min | 2.5 μl 10× PCR buffer (Mg2+ plus), 2 μl dNTPs (2.5 mM), 0.2 μl Ex Taq polymerase (5 U μl−1; Takara, Dalian, China), 1 μl of each primer (10 mM), 0.25 μl BSA (20 mg ml−1), and 2 μl DNA template |
| Arch-amoAR | GCGGCCATCCATCTGTATGT | ||||
| 45 | AOB amoA (MPN) PCR | amoA 1F | GGGGTTTCTACTGGTGGT | 5 min at 95°C; 35 cycles consisting of 94°C for 45 s, 55°C for 45 s, and 72°C for 60 s; 72°C for 10 min | 2.5 μl 10× PCR buffer (Mg2+ plus), 2 μl dNTPs (2.5 mM), 0.2 μl Ex Taq polymerase (5 U μl−1; Takara, Dalian, China), 1 μl of each primer (10 mM), 0.25 μl BSA (20 mg ml−1), and 2 μl DNA template |
| amoA 2R | CCCCTCKGSAAAGCCTTCTTC | ||||
| 8 | Com A amoA (MPN) PCR | comaA-244F | TAYAAYTGGGTSAAYTA | 5 min at 95°C; 35 cycles consisting of 94°C for 45 s, 53°C for 45 s, and 72°C for 60 s; 72°C for 10 min | 2.5 μl 10× PCR buffer (Mg2+ plus), 2 μl dNTPs (2.5 mM), 0.2 μl Ex Taq polymerase (5 U μl−1; Takara, Dalian, China), 1 μl of each primer (10 mM), 0.25 μl BSA (20 mg ml−1), and 3 μl DNA template |
| comaA-659R | ARATCATSGTGCTRTG | ||||
| Com B amoA (MPN) PCR | comaB-244F | TAYTTCTGGACRTTYTA | |||
| comaB-659R | ARATCCARACDGTGTG |
dNTPs, deoxynucleoside triphosphates.
Quantification of amoA genes by qPCR and most probable number PCR (MPN-PCR) methods.
Primer pairs Arch-amoAF/Arch-amoAR and amoA-1F/amoA-1R were also used to estimate AOA and AOB amoA gene abundance, respectively, through qPCR assays. The amplification was performed in triplicate on a C1000 thermal cycler (Bio-Rad, CA, USA). Each quantitative real-time PCR (qPCR) assay was conducted with a 25-μl reaction mixture containing 12.5 μl SYBR Ex Taq premix (TaKaRa Bio, Inc., Shiga, Japan), 400 nM each amoA gene primer, 0.25 μl bovine serum albumin (BSA; 20 mg ml−1) for AOA, and 3 μl DNA (1 to 10 ng) as the qPCR template. qPCR thermal programs were 95°C for 1 min, 40 cycles of 10 s at 95°C, 30 s at 53°C for AOA or 55°C for AOB, 1 min at 72°C, and then plate reads at 83°C (46). Melting curve analysis was executed at the end of each qPCR run to check the specificity of amplification products, followed by confirmation with agarose gel electrophoresis. Negative controls without DNA template were subjected to the same qPCR procedures in all experiments to check and exclude any potential DNA contamination. To construct the standard curve, a 10-fold dilution series of a known amount of plasmid containing the amoA gene was used as the standard. The standard curves for archaeal and bacterial amoA spanned from 1.05 × 102 to 1.05 × 107 copies and from 1.24 × 102 to 1.24 × 107 copies, respectively, per assay. Detection limits for the AOA and AOB amoA gene abundance in the overlying water samples were 3.70 × 103 and 7.60 × 103 copies liter−1, respectively. Amplification efficiencies ranged from 85.2 to 90.3% for archaeal amoA and from 90.2 to 90.4% for bacterial amoA, and r2 values for all amplifications were higher than 98%.
Primer pairs comaA-244F/comaA-659R and comaB-244F/comaB-659R in our experiments produced nonspecific products during qPCR amplifications. To avoid such quantification errors, the MPN-PCR method was employed to estimate Com A and Com B amoA gene abundance in this study. Briefly, extracted DNA was serially diluted by a factor of 10 and then amplified in triplicate. Reaction mixtures and amplification thermal profiles are shown in Table 2. Five microliters of the amplicons was visualized on agarose gels (1% Tris-acetate-EDTA) stained with Goldview nucleic acid stain (SBS Genetech, China). The bands of about 415 bp, corresponding to the expected size, were calculated as positive results, which was further verified by the simultaneously amplified comammox plasmids. Negative controls without DNA template were added to exclude potential DNA contamination. Finally, comammox abundance was enumerated with the Cochran tables (47). The AOA and AOB abundances of three randomly selected samples were also determined through MPN-PCR methods. It was found that the abundances of AOA and AOB determined through qPCR were approximately 100 (99) times higher than those through MPN-PCR. Therefore, compared with that of comammox, AOA and AOB abundance was divided by 99.
Statistical analysis.
Operational taxonomic unit (OTU) richness and the Chao1 and Simpson indices of AOA and AOB communities were calculated with Mothur software, v1.41.3, at 95% similarity (48). The taxonomic identities of the representative OTU sequences of AOA, AOB, and comammox were determined by constructing phylogenetic trees with reference sequences obtained from the National Centre for Biotechnology Information (NCBI) database using MEGA v7.0 (49). Weighted (unweighted) UniFrac and principal coordinate analyses (PCoA) for AOA and AOB communities were also executed with the Mothur software. Geographic distances among sampling sites were calculated in R, v3.4 (50), with RStudio, v1.1.463 (Boston, MA), by the geosphere package, v1.5-7 (51). Environmental factors were prefiltered through the vif.cca function of the vegan package, v2.5-4 (52), and the variables with vif of >3 were removed to avoid collinearity issues (53). The distance matrixes for environmental factors and elevation were calculated with the Euclidean method, and (partial) Mantel test was used to compare the relationship between ammonia oxidizer communities and factors, all of which were analyzed using the vegan package.
The abundance of AOA, AOB, and comammox Nitrospira, AOB/AOA ratio, as well as suspended sediment concentration were natural logarithm transformed to meet normality assumptions before further statistical analysis. Pearson (Spearman) correlation and partial correlation analyses, paired-sample t test, and other general statistical analyses were conducted under the Statistical Product and Service Solutions 21.0 software (SPSS) at a significance level of 0.05. Paired sample t test was used to identify the seasonal differences in the abundance, community diversity, and activity of ammonia oxidizers, as well as the physiochemical factors, of the Yellow River source region. It was also used to compare the abundance differences between AOA and AOB and between Com A and Com B.
Structural equation models (SEMs) were constructed to identify the factors controlling the distribution of ammonia oxidizers in the overlying water. Based on our prior knowledge (11, 24) and correlation analysis results, DO, water temperature, solar radiation intensity, and suspended sediment concentration were adopted into SEMs. The data matrix was fit to the model using the maximum-likelihood estimation method. A bootstrap test was conducted with 1,000 bootstrap samples to correct the bias of standard error due to the small sample size of our study, and then we recalculated relevant statistical indices. Moreover, the SEM results were further validated by other statistical methods. SEM analysis was performed using AMOS 20.0 (Amos Development Corporation, Meadville, PA, USA).
Data availability.
Sequences have been deposited in GenBank under accession numbers MH179343 to MH179467 and MK987213 to MK987510 for the archaeal amoA gene and MK987511 to MK988028 for the bacterial amoA gene. The GenBank accession numbers for comammox sequences are MN011812 to MN011865.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the National Key R&D Program of China (2017YFA0605001), the National Natural Science Foundation of China (91547207), the Fund for Innovative Research Group of the National Natural Science Foundation of China (51721093), and the National Natural Science Foundation of China (91547109).
We sincerely appreciate Petra Pjevac for her help in the phylogenetic analysis of comammox Nitrospira.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01701-19.
REFERENCES
- 1.Stein LY. 2019. Insights into the physiology of ammonia-oxidizing microorganisms. Curr Opin Chem Biol 49:9–15. doi: 10.1016/j.cbpa.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Beaulieu JJ, Tank JL, Hamilton SK, Wollheim WM, Hall RO, Mulholland PJ, Peterson BJ, Ashkenas LR, Cooper LW, Dahm CN, Dodds WK, Grimm NB, Johnson SL, McDowell WH, Poole GC, Valett HM, Arango CP, Bernot MJ, Burgin AJ, Crenshaw CL, Helton AM, Johnson LT, O'Brien JM, Potter JD, Sheibley RW, Sobota DJ, Thomas SM. 2011. Nitrous oxide emission from denitrification in stream and river networks. Proc Natl Acad Sci USA 108:214–219. doi: 10.1073/pnas.1011464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 4.Stahl DA, de la Torre JR. 2012. Physiology and diversity of ammonia-oxidizing archaea. Annu Rev Microbiol 66:83–101. doi: 10.1146/annurev-micro-092611-150128. [DOI] [PubMed] [Google Scholar]
- 5.Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. 2009. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev 33:855–869. doi: 10.1111/j.1574-6976.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- 6.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M. 2015. Complete nitrification by Nitrospira bacteria. Nature 528:504. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Kessel M, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B, Jetten MSM, Lücker S. 2015. Complete nitrification by a single microorganism. Nature 528:555. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pjevac P, Schauberger C, Poghosyan L, Herbold CW, van Kessel M, Daebeler A, Steinberger M, Jetten MSM, Lucker S, Wagner M, Daims H. 2017. amoA-targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment. Front Microbiol 8:1508. doi: 10.3389/fmicb.2017.01508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laanbroek HJ, Bollmann A. 2011. Nitrification in inland waters, p 385–403. In Ward BB, Arp DJ, Klotz MG (ed), Nitrification, 1st ed ASM Press, Washington, DC. [Google Scholar]
- 10.Xia X, Liu T, Yang Z, Michalski G, Liu S, Jia Z, Zhang S. 2017. Enhanced nitrogen loss from rivers through coupled nitrification-denitrification caused by suspended sediment. Sci Total Environ 579:47–59. doi: 10.1016/j.scitotenv.2016.10.181. [DOI] [PubMed] [Google Scholar]
- 11.Xia X, Yang Z, Zhang X. 2009. Effect of suspended-sediment concentration on nitrification in river water: importance of suspended sediment–water interface. Environ Sci Technol 43:3681–3687. doi: 10.1021/es8036675. [DOI] [PubMed] [Google Scholar]
- 12.Liu Z, Huang S, Sun G, Xu Z, Xu M. 2011. Diversity and abundance of ammonia-oxidizing archaea in the Dongjiang River, China. Microbiol Res 166:337–345. doi: 10.1016/j.micres.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q, Tang F, Zhou Y, Xu J, Chen H, Wang M, Laanbroek HJ. 2015. Shifts in the pelagic ammonia-oxidizing microbial communities along the eutrophic estuary of Yong River in Ningbo City, China. Front Microbiol 6:1180. doi: 10.3389/fmicb.2015.01180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Immerzeel WW, Van Beek LP, Bierkens MF. 2010. Climate change will affect the Asian water towers. Science 328:1382–1385. doi: 10.1126/science.1183188. [DOI] [PubMed] [Google Scholar]
- 15.Beniston M. 2003. Climatic change in mountain regions: a review of possible impacts, p 5–31. In Diaz HF. (ed), Climate variability and change in high elevation regions: past, present & future. Springer, Dordrecht, Netherlands. [Google Scholar]
- 16.Xia X, Zhang S, Li S, Zhang L, Wang G, Zhang L, Wang J, Li Z. 2018. The cycle of nitrogen in river systems: sources, transformation, and flux. Environ Sci Process Impacts 20:863–891. doi: 10.1039/c8em00042e. [DOI] [PubMed] [Google Scholar]
- 17.Urakawa H, Tajima Y, Numata Y, Tsuneda S. 2008. Low temperature decreases the phylogenetic diversity of ammonia-oxidizing archaea and bacteria in aquarium biofiltration systems. Appl Environ Microbiol 74:894–900. doi: 10.1128/AEM.01529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor AE, Giguere AT, Zoebelein CM, Myrold DD, Bottomley PJ. 2017. Modeling of soil nitrification responses to temperature reveals thermodynamic differences between ammonia-oxidizing activity of archaea and bacteria. ISME J 11:896–908. doi: 10.1038/ismej.2016.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alves RJE, Wanek W, Zappe A, Richter A, Svenning MM, Schleper C, Urich T. 2013. Nitrification rates in Arctic soils are associated with functionally distinct populations of ammonia-oxidizing archaea. ISME J 7:1620. doi: 10.1038/ismej.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purkhold U, Pommerening-Roser A, Juretschko S, Schmid MC, Koops HP, Wagner M. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol 66:5368–5382. doi: 10.1128/aem.66.12.5368-5382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avrahami S, Liesack W, Conrad R. 2003. Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers. Environ Microbiol 5:691–705. doi: 10.1046/j.1462-2920.2003.00457.x. [DOI] [PubMed] [Google Scholar]
- 22.Tourna M, Freitag TE, Nicol GW, Prosser JI. 2008. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ Microbiol 10:1357–1364. doi: 10.1111/j.1462-2920.2007.01563.x. [DOI] [PubMed] [Google Scholar]
- 23.Verhamme DT, Prosser JI, Nicol GW. 2011. Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J 5:1067. doi: 10.1038/ismej.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merbt SN, Stahl DA, Casamayor EO, Martí E, Nicol GW, Prosser JI. 2012. Differential photoinhibition of bacterial and archaeal ammonia oxidation. FEMS Microbiol Lett 327:41–46. doi: 10.1111/j.1574-6968.2011.02457.x. [DOI] [PubMed] [Google Scholar]
- 25.Kang S, Xu Y, You Q, Flügel W-A, Pepin N, Yao T. 2010. Review of climate and cryospheric change in the Tibetan Plateau. Environ Res Lett 5:015101. doi: 10.1088/1748-9326/5/1/015101. [DOI] [Google Scholar]
- 26.Urakawa H, Garcia JC, Nielsen JL, Le VQ, Kozlowski JA, Stein LY, Lim CK, Pommerening-Röser A, Martens-Habbena W, Stahl DA, Klotz MG. 2015. Nitrosospira lacus sp. nov., a psychrotolerant, ammonia-oxidizing bacterium from sandy lake sediment. Int J Syst Evol Microbiol 65:242–250. doi: 10.1099/ijs.0.070789-0. [DOI] [PubMed] [Google Scholar]
- 27.Zhang LM, Wang M, Prosser JI, Zheng YM, He JZ. 2009. Altitude ammonia-oxidizing bacteria and archaea in soils of Mount Everest. FEMS Microbiol Ecol 70:52–61. doi: 10.1111/j.1574-6941.2009.00775.x. [DOI] [PubMed] [Google Scholar]
- 28.Koops HP, Purkhold U, Pommerening-Röser A, Timmermann G, Wagner M. 2006. The lithoautotrophic ammonia-oxidizing bacteria, p 778–811. In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (ed), The prokaryotes, 3th ed, vol 5 Springer Science & Business Media, New York, NY. [Google Scholar]
- 29.Pester M, Rattei T, Flechl S, Gröngröft A, Richter A, Overmann J, Reinhold-Hurek B, Loy A, Wagner M. 2012. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol 14:525–539. doi: 10.1111/j.1462-2920.2011.02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu L, Han C, Zhu G, Zhong W. 29 August 2019. Responses of active ammonia-oxidizers and nitrification activity in eutrophic lake sediments to nitrogen and temperature. Appl Environ Microbiol doi: 10.1128/AEM.00258-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sedlacek CJ, McGowan B, Suwa Y, Sayavedra-Soto L, Laanbroek HJ, Stein LY, Norton JM, Klotz MG, Bollmann A. 11 April 2019. A physiological and genomic comparison of Nitrosomonas cluster 6a and 7 ammonia-oxidizing bacteria. Microb Ecol doi: 10.1007/s00248-019-01378-8. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y, Zhang J, Zhao Q, Zhou Q, Li N, Wang Y, Xie S, Liu Y. 2016. Sediment ammonia-oxidizing microorganisms in two plateau freshwater lakes at different trophic states. Microb Ecol 71:257–265. doi: 10.1007/s00248-015-0642-3. [DOI] [PubMed] [Google Scholar]
- 33.Belser LW, Mays EL. 1980. Specific inhibition of nitrite oxidation by chlorate and its use in assessing nitrification in soils and sediments. Appl Environ Microbiol 39:505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mansour I, Heppell CM, Ryo M, Rillig MC. 2018. Application of the microbial community coalescence concept to riverine networks. Biol Rev Camb Philos Soc 93:1832–1845. doi: 10.1111/brv.12422. [DOI] [PubMed] [Google Scholar]
- 35.Leibold MA, Holyoak M, Mouquet N, Amarasekare P, Chase JM, Hoopes MF, Holt RD, Shurin JB, Law R, Tilman D, Loreau M, Gonzalez A. 2004. The metacommunity concept: a framework for multi‐scale community ecology. Ecol Lett 7:601–613. doi: 10.1111/j.1461-0248.2004.00608.x. [DOI] [Google Scholar]
- 36.Savio D, Sinclair L, Ijaz UZ, Parajka J, Reischer GH, Stadler P, Blaschke AP, Blöschl G, Mach RL, Kirschner AKT, Farnleitner AH, Eiler A. 2015. Bacterial diversity along a 2600 km river continuum. Environ Microbiol 17:4994–5007. doi: 10.1111/1462-2920.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauder LA, Albertsen M, Engel K, Schwarz J, Nielsen PH, Wagner M, Neufeld JD. 2017. Cultivation and characterization of Candidatus Nitrosocosmicus exaquare, an ammonia-oxidizing archaeon from a municipal wastewater treatment system. ISME J 11:1142. doi: 10.1038/ismej.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webster G, Embley TM, Freitag TE, Smith Z, Prosser JI. 2005. Links between ammonia oxidizer species composition, functional diversity and nitrification kinetics in grassland soils. Environ Microbiol 7:676–684. doi: 10.1111/j.1462-2920.2005.00740.x. [DOI] [PubMed] [Google Scholar]
- 39.Hu H, Zhang L-m, Yuan C, Zheng Y, Wang J, Chen D, He J. 2015. The large-scale distribution of ammonia oxidizers in paddy soils is driven by soil pH, geographic distance, and climatic factors. Front Microbiol 6:938. doi: 10.3389/fmicb.2015.00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang H, Huang L, Deng Y, Wang S, Zhou Y, Liu L, Dong H. 2014. Latitudinal distribution of ammonia-oxidizing bacteria and archaea in the agricultural soils of eastern China. Appl Environ Microbiol 80:5593–5602. doi: 10.1128/AEM.01617-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bier RL, Bernhardt ES, Boot CM, Graham EB, Hall EK, Lennon JT, Nemergut DR, Osborne BB, Ruiz-Gonzalez C, Schimel JP, Waldrop MP, Wallenstein MD. 2015. Linking microbial community structure and microbial processes: an empirical and conceptual overview. FEMS Microbiol Ecol 91:fiv113. doi: 10.1093/femsec/fiv113. [DOI] [PubMed] [Google Scholar]
- 42.Kurola J, Salkinoja-Salonen M, Aarnio T, Hultman J, Romantschuk M. 2005. Activity, diversity and population size of ammonia-oxidising bacteria in oil-contaminated landfarming soil. FEMS Microbiol Lett 250:33–38. doi: 10.1016/j.femsle.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 43.Berg P, Rosswall T. 1985. Ammonium oxidizer numbers, potential and actual oxidation rates in two swedish arable soils. Biol Fert Soils 1:131–140. doi: 10.1007/BF00301780. [DOI] [Google Scholar]
- 44.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rotthauwe JH, Witzel KP, Liesack W. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang LM, Hu HW, Shen JP, He JZ. 2012. Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J 6:1032–1045. doi: 10.1038/ismej.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cochran WG. 1950. Estimation of bacterial densities by means of the most probable number. Biometrics 6:105–116. doi: 10.2307/3001491. [DOI] [PubMed] [Google Scholar]
- 48.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stecher G, Kumar S, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 51.Hijmans RJ, Williams E, Vennes C, Hijmans M. 2017. geosphere: spherical trigonometry. https://cran.r-project.org/web/packages/geosphere/index.html. Accessed 28 October 2018.
- 52.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’hara R, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2013. vegan: community ecology package, version 2. https://cran.r-project.org/web/packages/vegan/index.html. Accessed 28 October 2018.
- 53.Zuur AF, Ieno EN, Elphick CS. 2010. A protocol for data exploration to avoid common statistical problems. Methods Ecol Evol 1:3–14. doi: 10.1111/j.2041-210X.2009.00001.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences have been deposited in GenBank under accession numbers MH179343 to MH179467 and MK987213 to MK987510 for the archaeal amoA gene and MK987511 to MK988028 for the bacterial amoA gene. The GenBank accession numbers for comammox sequences are MN011812 to MN011865.