Campylobacter jejuni and Campylobacter coli are leading foodborne pathogens, with poultry as a major reservoir. Due to their growth requirements, these Campylobacter spp. may be unable to replicate once excreted by their avian hosts, but their survival in feces and the environment is critical for transmission in the farm ecosystem. Reducing the prevalence of Campylobacter-positive flocks can have major impacts in controlling both contamination of poultry products and environmental dissemination of the pathogens. However, understanding the capacity of these pathogens to survive in transmission-relevant vehicles such as feces and farmhouse water remains poorly understood, and little information is available on species- and strain-associated differences in survival. Here, we employed model conditions to investigate the survival of C. jejuni and C. coli from naturally colonized turkey flocks, and with diverse genotypes and antimicrobial resistance profiles, in turkey feces and in farmhouse water.
KEYWORDS: Campylobacter, Campylobacter coli, Campylobacter jejuni, turkey, antimicrobial resistance, feces, survival, water
ABSTRACT
Campylobacter jejuni and Campylobacter coli are leading causes of human foodborne illness, with poultry as a major vehicle. Turkeys are frequently colonized with Campylobacter, but little is known about Campylobacter survival in turkey feces, even though fecal droppings are major vehicles for Campylobacter within-flock transmission as well as for environmental dissemination. Our objective was to examine survival of Campylobacter, including different strains, in freshly excreted feces from naturally colonized commercial turkey flocks and in suspensions of turkey feces in water from the turkey house. Fecal and water suspensions were stored at 4°C, and Campylobacter populations were enumerated on selective media at 48-h intervals. C. jejuni and C. coli isolates were characterized for resistance to a panel of antibiotics, and a subset was subtyped using multilocus sequence typing. Campylobacter was recovered from feces and water for up to 16 days. Analysis of 548 isolates (218 C. jejuni and 330 C. coli) revealed that C. jejuni survived longer than C. coli in feces (P = 0.0005), while the reverse was observed in water (P < 0.0001). Strain-specific differences in survival were noted. Multidrug-resistant C. jejuni isolates of sequence type 1839 (ST-1839) and the related ST-2935 were among the longest-surviving isolates in feces, being recovered for up to 10 to 16 days, while multidrug-resistant C. coli isolates of ST-1101 were recovered from feces for only up to 4 days. Data on Campylobacter survival upon excretion from the birds can contribute to further understanding of the transmission dynamics of this pathogen in the poultry production ecosystem.
IMPORTANCE Campylobacter jejuni and Campylobacter coli are leading foodborne pathogens, with poultry as a major reservoir. Due to their growth requirements, these Campylobacter spp. may be unable to replicate once excreted by their avian hosts, but their survival in feces and the environment is critical for transmission in the farm ecosystem. Reducing the prevalence of Campylobacter-positive flocks can have major impacts in controlling both contamination of poultry products and environmental dissemination of the pathogens. However, understanding the capacity of these pathogens to survive in transmission-relevant vehicles such as feces and farmhouse water remains poorly understood, and little information is available on species- and strain-associated differences in survival. Here, we employed model conditions to investigate the survival of C. jejuni and C. coli from naturally colonized turkey flocks, and with diverse genotypes and antimicrobial resistance profiles, in turkey feces and in farmhouse water.
INTRODUCTION
Campylobacter spp. are zoonotic bacterial pathogens that are leading agents for human foodborne illness worldwide (1–4), annually resulting in an estimated 0.8 million cases of foodborne illnesses in the United States alone (1). In addition to acute gastroenteritis, human campylobacteriosis can be followed by severe autoimmune sequelae and constitutes the leading antecedent to Guillain-Barré syndrome (5). In the United States and other industrialized nations, Campylobacter jejuni is responsible for the majority (approximately 85%) of human campylobacteriosis cases, with Campylobacter coli being responsible for most of the remainder (4, 6), and contaminated poultry is considered to be a leading vehicle for human campylobacteriosis (7–9). Poultry, including chickens and turkeys, are frequently colonized by C. jejuni and C. coli, which can then be shed in large numbers in the feces (10–15).
Knowledge of Campylobacter survival outside its avian hosts remains poorly characterized. C. jejuni and C. coli are unable to grow below 30°C but can survive for variable lengths of time, with survival markedly better at low temperatures, such as 4°C (16–20). Survival in water can be enhanced by association with other microbes, including other bacteria and waterborne protozoa, such as Acanthamoeba castellanii and Tetrahymena pyriformis (21–24). C. jejuni that had been internalized by protozoa in water from a broiler farm survived longer than C. jejuni that remained extracellular and also exhibited increased tolerance to disinfection (22).
Campylobacter cells are shed, often in high numbers, in the feces of asymptomatic birds (7). Thus, poultry feces constitute a major vehicle for transmission of Campylobacter to the birds within a flock and for subsequent environmental contamination. In addition to coprophagy-mediated transmission within the flock as birds peck on feces-contaminated litter, birds can become infected through water contaminated with the fecal droppings (10, 11, 25). Campylobacter’s capacity to colonize chickens is enhanced upon passage through the birds’ gastrointestinal (GI) tract and shedding in the fecal droppings (26–30). Campylobacter in the poultry feces can be then transmitted to other flocks and farms via insects, such as flies, and other vectors, including farm equipment and human traffic, with potential for downstream dispersal and contamination of the natural environment, e.g., surface water and soil (10, 25, 31–33).
In spite of its clear food safety and public health relevance, Campylobacter survival in poultry feces remains poorly understood. The limited available information is focused on survival in chicken droppings. C. jejuni was found to survive for up to 5 to 6 days in naturally or artificially contaminated laying hen feces at ambient temperature (20°C), with survival significantly higher in naturally contaminated feces (34–36). C. jejuni inoculated into feces and litter from Campylobacter-negative flocks survived significantly longer in feces than in litter, with survival found to be higher at lower temperatures (20°C versus 25 or 30°C) and independent of relative humidity (36). However, major gaps remain in our knowledge of the potential impact of species- (i.e., C. jejuni or C. coli) and strain-specific attributes, including genotype and antimicrobial resistance, on survival. Reports about Campylobacter survival in turkey feces have been lacking, even though turkeys are frequently colonized with C. jejuni and C. coli, including multidrug-resistant strains (12, 15, 37, 38). The objective of the current study was to employ model conditions in order to characterize survival of Campylobacter spp. in turkey feces and in water from turkey farms. To enhance the relevance of the findings to commercial turkey farm systems, we investigated the survival of C. jejuni and C. coli strains of diverse antimicrobial resistance profiles and genotypes in feces excreted by flocks that were already naturally colonized by these species and strains, as well as in water from the turkey farmhouse.
RESULTS
Campylobacter spp. in feces and water could be recovered for up to 16 days at 4°C, with a progressive decline during this period.
At time 0, Campylobacter populations in the fecal composite samples ranged from 1.4 × 106 to 3.2 × 106 CFU/g. Due to its growth requirements for high temperature and microaerobic atmosphere, Campylobacter was not expected to grow in these samples, and indeed Campylobacter levels progressively declined with time in all samples. As shown with two representative flocks, population levels in the samples declined slowly (1- to 3-log reduction) over the first 8 days of storage in the feces or in the water suspension (Fig. 1 and data not shown). By day 16, Campylobacter spp. levels were approaching or had already reached the limit of detection, and by day 20, no isolates could be recovered from any of the samples (Fig. 1). The rate of decline of total Campylobacter spp. was not significantly different (P > 0.05) between the fecal composite sample and the suspension of the same composite sample in water; the average number of days at which Campylobacter spp. fell below the limit of detection was 10 and 10.7 for fecal composite samples and water samples, respectively.
FIG 1.
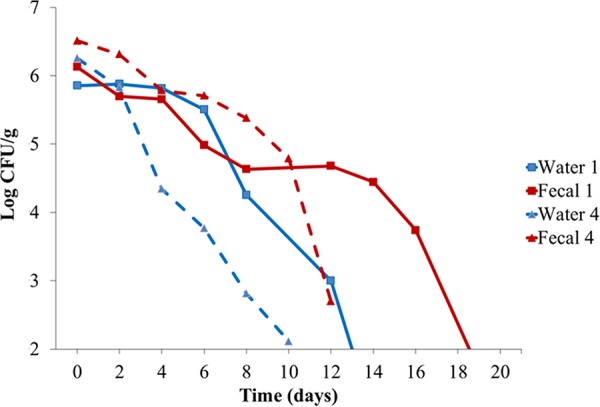
Campylobacter spp. survival in turkey feces and water suspensions. Fecal composite samples (samples 1 and 4) and water suspensions were prepared from two representative Campylobacter-positive turkey flocks as described in Materials and Methods. Total Campylobacter populations were enumerated on selective media (mCCDA) immediately prior to incubation at 4°C (time 0) and at 48-h intervals thereafter, as described in Materials and Methods.
Relative prevalence of C. jejuni increased with time in fecal composite samples, while C. coli predominated at later time points in water suspensions.
Campylobacter species designations (C. jejuni or C. coli) were determined for 548 isolates obtained between time 0 and 16 days from the feces or the water suspensions (Table S1 in the Supplemental Material). Of the 548 isolates, 218 were C. jejuni, with the remaining 330 being C. coli; other Campylobacter species were not detected. The overall relative prevalence of C. jejuni in the feces was approximately 40% at time 0 but increased in the later time points to approximately 90% by day 14 (Fig. 2). The reverse was observed in the water suspensions, where the relative frequency of C. coli increased with time (P ≤ 0.0005). Of the 55 Campylobacter isolates obtained from the water suspensions after 8 days, all but one were C. coli (Fig. 2).
FIG 2.
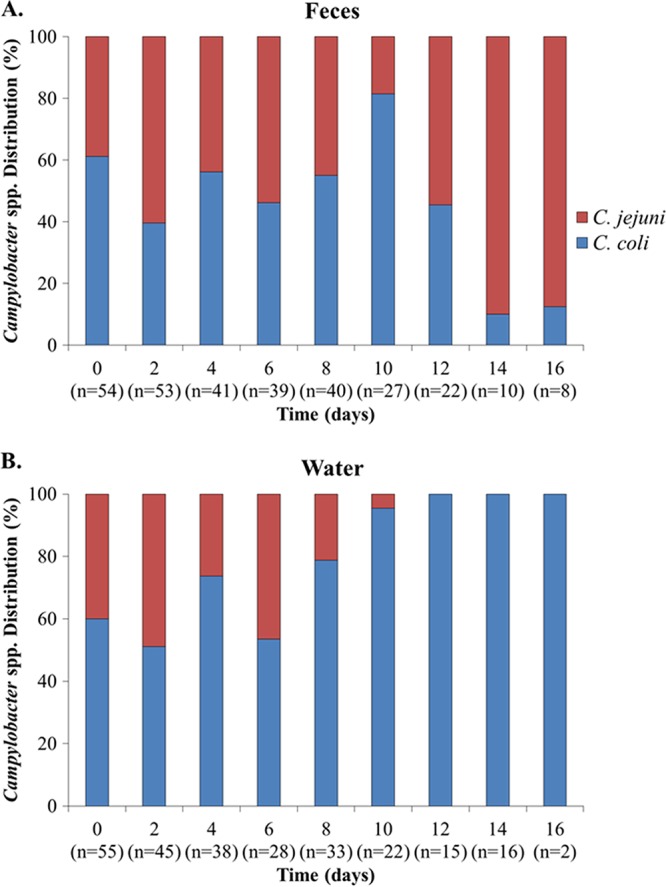
Relative survival of Campylobacter jejuni and C. coli in turkey feces and water suspensions. Campylobacter colonies from the mCCDA plates used for enumerations of total Campylobacter populations from the feces and the water suspensions at different time points (see legend to Fig. 1) were purified and speciated as described in Materials and Methods. C. jejuni and C. coli are shown in red and blue, respectively.
C. jejuni and C. coli with specific antimicrobial resistance profiles differ in their survival in feces and in water suspensions.
Antimicrobial resistance profiles were determined for tetracycline (T), streptomycin (S), kanamycin (K), erythromycin (E), and the (fluoro)quinolones nalidixic acid and ciprofloxacin (Q) and are indicated with acronyms that reflect the observed resistance traits. Several antimicrobial resistance (AMR) profiles were identified among the isolates, including five among C. jejuni and nine among C. coli. Of the five AMR profiles in C. jejuni, two clearly predominated both in the feces and in the water suspensions. The majority of the C. jejuni isolates (n = 148, approximately 68%) had AMR profile TSKQ, i.e., resistant to all tested antimicrobials except for erythromycin, while 57 isolates (26% of all C. jejuni) had profile TKQ, i.e., resistant to tetracycline, kanamycin, nalidixic acid, and ciprofloxacin but susceptible to streptomycin and erythromycin (Fig. 3). The remaining three AMR profiles were infrequent; TSQ was detected in seven isolates, while TK and T were detected in three isolates each (Table S1). In C. coli, the leading AMR profiles in both feces and water suspensions included resistance to tetracycline and kanamycin (profile TK), followed by AMR profiles TKQ and TQ (Fig. 4). The multidrug resistance C. coli profile TSEKQ, i.e., resistance to all six tested antimicrobials, was noteworthy in constituting a noticeable (approximately 20%) fraction of the C. coli isolates in the early time points (days 0 to 2) but not thereafter (Fig. 4).
FIG 3.
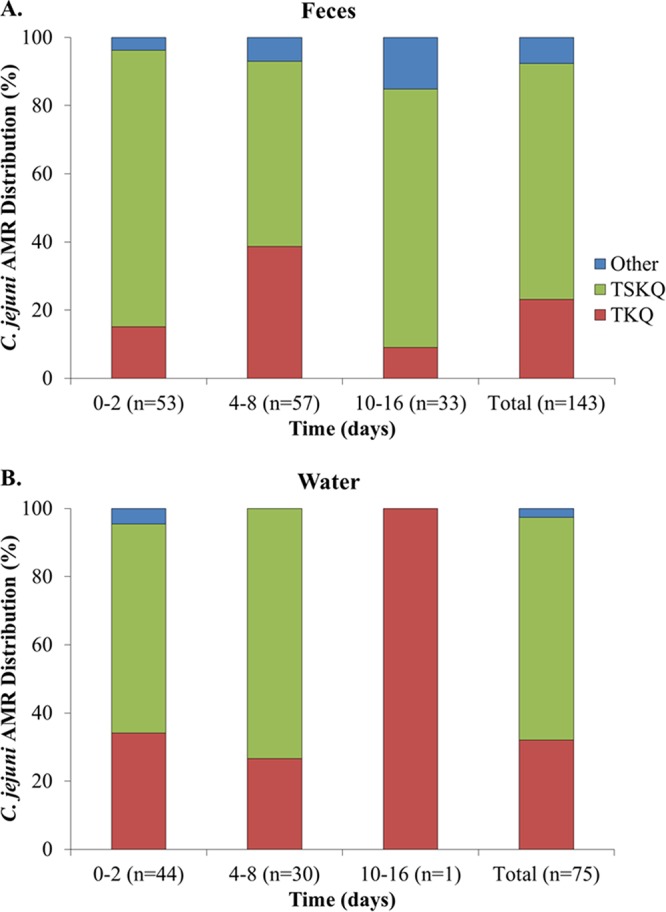
Relative prevalence of Campylobacter jejuni strains with different antimicrobial resistance (AMR) profiles at different time intervals in (A) feces and (B) water suspensions. AMR profiles were determined as described in Materials and Methods. AMR profile acronyms specify resistance to the following antimicrobials: tetracycline (T), streptomycin (S), kanamycin (K), and the (fluoro)quinolones nalidixic acid and ciprofloxacin (Q). The combination of letters indicates the specific AMR profile, e.g., TSEKQ indicates resistance to all tested antimicrobials, while TKQ indicates resistance to only tetracycline, kanamycin, and the quinolones nalidixic acid and ciprofloxacin.
FIG 4.
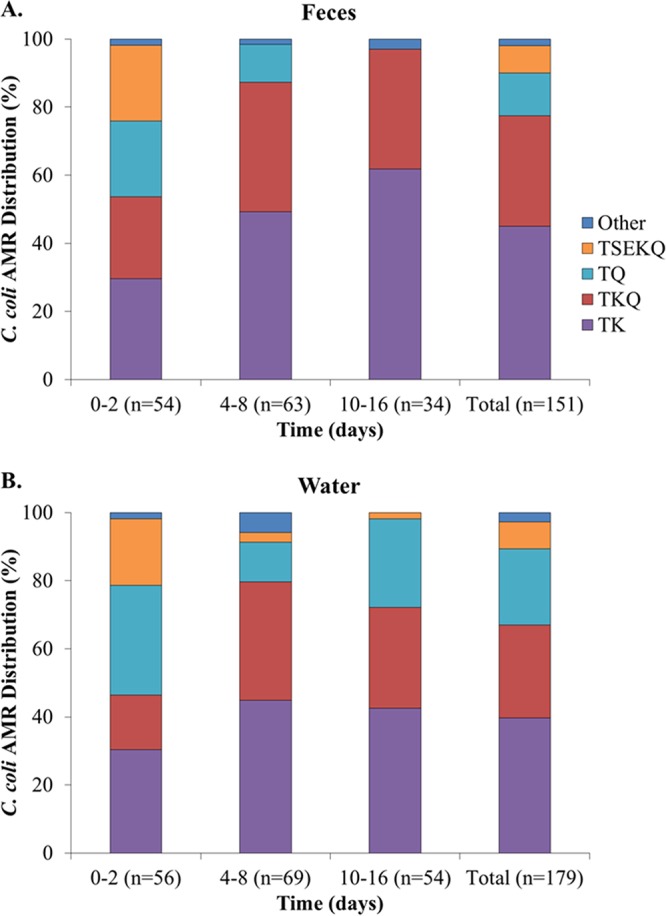
Relative prevalence of Campylobacter coli strains with different antimicrobial resistance (AMR) profiles at different time intervals in (A) feces and (B) water suspensions. AMR profiles were determined as described in Materials and Methods. AMR profile acronyms specify resistance to the following antimicrobials: tetracycline (T), streptomycin (S), erythromycin (E), kanamycin (K), and the (fluoro)quinolones nalidixic acid and ciprofloxacin (Q).
Survival in feces differed significantly across strains of C. jejuni with different AMR profiles (P < 0.01) (Fig. 3A). However, significant differences were not noted regarding survival in water (P > 0.05), where C. jejuni with profiles TSKQ and TKQ predominated in water suspensions through day 8 (Fig. 3B). As discussed above, total C. jejuni populations declined markedly in the suspensions thereafter (Fig. 2B), and only one C. jejuni isolate was obtained from the water suspensions after 8 days (Fig. 3B). Significant survival differences in feces were also noted across C. coli isolates with different AMR profiles, with the differences remaining significant in water suspensions as well (P ≤ 0.0001). As indicated above, the relative prevalence of C. coli with the multidrug resistance profile TSEKQ diminished markedly after the early time points, both in feces and in the water suspension (Fig. 4). This TSEKQ profile was exclusively encountered among C. coli, in fact, and similar to results from previous studies of Campylobacter in turkeys in this region (14), no erythromycin-resistant C. jejuni isolates were identified in the study.
MLST genotyping of a panel of C. jejuni TSKQ isolates from different time points from the feces and the water suspensions (0, 2, 8, and 10 days) revealed that all shared the same sequence type, ST-1839, which was also detected in all but one of the seven tested C. jejuni isolates with AMR profile TKQ (Fig. 5, Table S2). The closely related ST-2935 (one allele difference from ST-1839) was detected in all three tested isolates of AMR profile TSQ, which were relatively uncommon and identified primarily in the feces and at later time points (days 12, 14, and 16) (Tables S1 and S2 and Fig. 5). One may speculate that C. jejuni ST-1839, of AMR profiles TSKQ or TKQ, and the related ST-2935 (AMR profile TSQ) may have high fitness for survival of Campylobacter after excretion. It is also of interest that ST-2935 isolates were primarily detected at later time points, and only from feces, raising the possibility that they might represent variants of ST-1839 with enhanced capacity to survive in feces. Even though ST-1839 and ST-2935 are genetically similar based on the concatenated allele sequences (Fig. 5), further sequence analysis will be needed to more adequately assess the genetic differences that may account for differences in AMR profiles and potentially in fitness.
FIG 5.
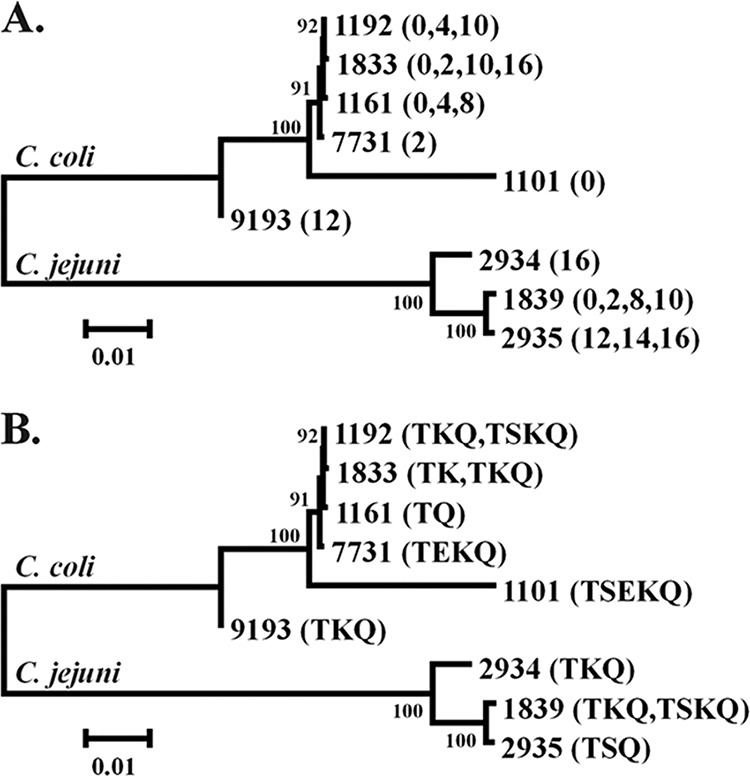
Phylogenetic tree of Campylobacter jejuni and C. coli strains with different sequence types (STs). The tree includes information on (A) time point at which the strains with the specific STs were obtained and (B) the AMR profiles of the same strains. The phylogenetic tree was constructed based on the concatenated sequence of the seven alleles as described in Materials and Methods.
A number of STs were detected among C. coli. ST-1833 and the closely related (one-allele difference) ST-1192 were detected among C. coli isolates of the two leading AMR profiles, TK and TKQ, isolated from feces and water at various time points (0, 2, 10, and 16 days) (Tables S1 and S2 and Fig. 5). C. coli with the third-ranking AMR profile, TQ, had ST-1161, related to both ST-1833 (two-allele differences) and ST-1192 (one-allele difference). Also related (two-allele differences) to these STs was C. coli with ST-7731 and AMR profile TEKQ (Tables S1 and S2 and Fig. 5). Noteworthy was ST-1101, which was encountered among the multidrug-resistant C. coli TSEKQ and was phylogenetically remote, sharing no alleles with the other C. coli STs (Table S2 and Fig. 5). As discussed above, C. coli TSEKQ isolates were primarily obtained from early time points, and their relative prevalence diminished markedly with time.
DISCUSSION
The turkey flocks employed for materials in the current study were from a region (eastern North Carolina) dense in turkey production, and turkey flocks from this region were previously found to be frequently colonized with both C. jejuni and C. coli (14, 38). Similarly, both species were encountered in the current study. Interestingly, C. jejuni survived significantly longer than C. coli in the feces, while the opposite was noted when feces were suspended in water from the turkey house. Such species-specific differences in survival in poultry feces versus water suspensions have not been reported before, and the underlying mechanisms remain to be elucidated. One possibility is that C. coli might have higher fitness than C. jejuni in protozoa present in the farm water. Protozoa were commonly found in the water systems of broiler farms, and the survival of C. jejuni and C. coli was shown to be significantly enhanced when cocultured with protozoa from the water systems (22). The presence of protozoa was not assessed in the turkey-house water in the current study, and the extent to which C. coli may have a fitness advantage in waterborne protozoa remains to be demonstrated, e.g., via cocultures with protozoa. Furthermore, studies using feces from flocks colonized with C. jejuni and C. coli strains different from those prevailing here will be valuable to determine whether the findings that we observed here pertain generally to C. coli versus C. jejuni.
Strain-specific differences were noted in survival, supporting previous data which were obtained with broiler feces naturally colonized with C. jejuni and stored at 20°C. In that study, genotyping employed fla typing, and of the five detected fla types, some tended to predominate in later time points after shedding (34). Our findings also strongly suggested that multidrug-resistant C. coli isolates with resistance profile TSEKQ had impaired capacity to persist in feces or water compared to other C. coli strains. Traits that may compromise the fitness of these strains have yet to be elucidated. In another study with commercial turkeys in the same region (eastern North Carolina), we discovered that C. coli TSEKQ isolates of ST-1161, ST-1149, and ST-906, related (one- to three-allele differences) to ST-1101 of C. coli TSEKQ in the current study, were frequently isolated from the cecal contents of young turkeys in the first 5 weeks of life, but their frequency declined significantly as birds aged (39). Such data had led to the hypothesis that these strains may have relatively low fitness in regard to their capacity to compete with other Campylobacter strains and other microbes in the gut as birds age (39). The current data suggest that C. coli TSEKQ with ST-1101 may also have relatively impaired fitness after excretion. It would be of interest to identify potential linkages between the low fitness of these strains in the feces from the relatively old birds in the current study (flocks were, on average, 12.8 weeks old when feces were collected) and their previously observed significant decrease in cecal prevalence with increasing age of the flocks (39).
In the current study, Campylobacter could be recovered from most fecal samples for up to 12 to 16 days at 4°C. This was noticeably longer than the observed survival of C. jejuni either naturally present or spiked in chicken feces which were then incubated at higher temperatures than we employed (20 to 30°C); in those studies, C. jejuni could only be recovered from the chicken feces for up to 5 to 6 days (34–36). Storage temperature may be a key reason for the observed differences, since Campylobacter survival is enhanced at low temperature (17, 19, 40). The low temperature (4°C) employed in the current study was intentionally chosen to enhance the potential to detect differences in survival among different strains of C. jejuni and C. coli shed by the birds in the feces. Campylobacter strains can differ in their cold tolerance (41–44), and such differences may contribute to the current findings. In addition to temperature, other attributes, including the avian source (chicken versus turkey) and the Campylobacter species and strains involved, may contribute to the observed differences in survival between our study and others.
Previous studies of survival of Campylobacter shed by colonized poultry involved only C. jejuni (34–36), whereas both C. jejuni and C. coli were shed in the feces and monitored in the current study. Previous studies also did not compare survival in feces and farm water and did not provide information on AMR profiles or STs of the C. jejuni strains that were investigated (34–36). Inclusion of ST or other readily cross-comparable genotypic designations, as well as antimicrobial resistance data, will be increasingly useful for meaningful comparisons of the findings from different studies. It will, in addition, facilitate placement of findings from diverse regions and laboratories in the wider context of Campylobacter strain diversity and antimicrobial resistance.
In the current study, we characterized AMR profiles and genotypes of C. jejuni and C. coli strains isolated following different lengths of time in the feces or the water suspensions. Even though strains with certain AMR profiles and MLST-based genotypes differed in their capacity to survive in these materials, the underlying factors remain to be identified. Current data permitted the identification of associations between strain-specific traits such as resistance and genotype and relative survival capacity but do not constitute evidence of causality. The potential contribution of AMR profiles to persistence cannot be assessed with the current design of the study, which specifically investigated strains that had naturally colonized the flocks and were excreted in the feces in the farm environment. Such strains are genotypically diverse, and multiple attributes can contribute to their performance in feces and water. Genetic characterization of these strains via whole-genome sequence analysis is expected to be valuable in identifying potential genome content associations with strain-specific differences in survival in transmission-relevant vehicles that we observed in the current study.
The finding that C. jejuni survived significantly longer in feces than C. coli is important from a public health perspective. C. jejuni accounts for over 85% of human campylobacteriosis. Thus, higher relative fitness of C. jejuni in the feces may suggest greater potential for preharvest colonization of turkey flocks with this species. As mentioned earlier, Campylobacter can spread within a turkey house directly via coprophagy or among different turkey houses and farms via vectors such as flies, equipment, and transport crates as well as via human traffic, e.g., through footwear, clothing, and vehicles (10, 25, 31–33). During slaughter and processing, birds that became colonized with Campylobacter at the farm are more likely to lead to contamination of the carcasses with Campylobacter from the birds’ intestinal contents (11, 45).
The current study monitored levels of Campylobacter populations naturally shed in the feces of Campylobacter-positive turkeys. This alleviates potential pitfalls associated with inoculation of feces or water with laboratory-grown strains of C. jejuni or C. coli and enhances the real-life relevance of the survival assessments. Further characterization of Campylobacter survival outside the poultry host will enhance our understanding of Campylobacter transmission within the farm ecosystem, as well as potential dissemination of the pathogen to the natural environment. Findings from such studies can inform assessments of the food safety and public health risk posed by different Campylobacter species and strains in the poultry production ecosystem, contributing to science-based interventions to reduce the disease burden of campylobacteriosis.
MATERIALS AND METHODS
Sample collection.
Fecal droppings and water samples were obtained from seven turkey flocks grown conventionally at six different commercial turkey farms in eastern North Carolina between 29 July 2012 and 11 April 2013. Flock surveillance for a different project at our laboratory had indicated that these turkey flocks were Campylobacter positive and colonized with different Campylobacter species (C. jejuni and C. coli) and strains. All turkeys were males (toms) with an average age of 90 days (approximately 12.8 weeks) at the time that fecal droppings were collected for the present investigation.
Collection and processing of fecal droppings and water.
During the visit to each turkey flock, eight fresh individual turkey droppings (two from each quarter of the turkey house) were collected. Droppings were considered to be fresh upon visual observation of voiding. During the same visit, 50 ml of water was collected from the same turkey house from drinking water available for the birds. Fecal and water samples were brought to the laboratory on ice within 3 hours of collection and processed on the same day.
In the laboratory, a fecal composite sample was prepared in a 50-ml sterile plastic Falcon tube (Becton, Dickinson and Co., San Jose, CA) by combining all eight individual turkey droppings and mixing them using a sterile metal spatula. Water suspensions were made in 15-ml sterile plastic Falcon tubes (Becton, Dickinson and Co.) by adding 1 g of the fecal composite sample into 9 ml of the water from the turkey house and vortexing. The fecal composite and the water suspension were stored in the dark at 4°C.
Isolation and enumeration of Campylobacter spp.
Beginning on day 0 (immediately prior to placement of the samples at 4°C), dilutions of both the fecal composite sample and the water suspension were plated on the selective medium modified charcoal-cefoperazone-deoxycholate agar (mCCDA) (Oxoid, Ogdensburg, NY) for isolation and enumeration of Campylobacter spp. Plates were incubated microaerobically using Campy GasPaks (Becton, Dickinson and Co.) at 42°C, and colonies were enumerated after 48 h of incubation. Enumerations were done at 48-h intervals until the number of Campylobacter colonies declined to near or below the detection limit (100 CFU/g feces or 100/ml of the water suspension). At every time point, 10 to 16 presumptive Campylobacter colonies were isolated and purified on Mueller-Hinton agar (MHA) (Becton, Dickinson and Co.) following microaerobic incubation at 42°C for 48 h as described above. Purified cultures were preserved at –80°C as described (14).
Species and antimicrobial resistance profile determinations.
Campylobacter species designations (C. jejuni or C. coli) of a panel of 545 isolates from four flocks, each grown on a different farm, are summarized in Table S1. Designations were determined by multiplex PCR using primers specific for hip and ceu, as described (46). Isolates were tested as described (37) for susceptibility to a panel of six antibiotics, tetracycline (T), streptomycin (S), erythromycin (E), kanamycin (K), and the (fluoro)quinolones nalidixic acid and ciprofloxacin (Q). The pan-sensitive strain C. jejuni ATCC 33560 (American Type Culture Collection) was used as a quality control strain. All isolates were simultaneously grown on MHA plates to ensure viability. Antimicrobial resistance profiles are presented with acronyms that reflect the encountered resistance traits from among the tested antimicrobials. For instance, T represents an isolate with resistance only to tetracycline, while TKQ indicates an isolate with resistance to tetracycline, kanamycin, nalidixic acid, and ciprofloxacin but susceptible to the remaining two tested antimicrobials, and isolates with profile TSEKQ were resistant to all six tested antimicrobials.
Multilocus sequence typing and phylogenetic tree construction.
A subset of isolates was characterized by multilocus sequencing typing (MLST) as described (47). The isolates were chosen to represent the two Campylobacter species that were encountered, i.e., C. jejuni and C. coli, the different time points, as well as the different antimicrobial resistance profiles. Thus, efforts were made to include isolates representing each specific species/antimicrobial resistance profile from each time point. All sequence data were deposited in PubMLST (https://pubmlst.org/campylobacter/) (48), and the MLST profiles identified in this study were downloaded from PubMLST as in-frame concatenated allele sequences. The concatenated sequences were aligned using ClustalX, and a dendrogram was constructed within MEGA v. 6 (49) using the neighbor-joining method with the Kimura 2-parameter distance estimation method.
Statistical analysis.
Statistical analyses employed Chi-square and Fisher’s exact tests and were conducted using SAS v. 9.3 (SAS Institute, Cary, NC). Linear models for mean log CFU were developed treating time points either as a factorial effect or as a continuously valued regressor with effect quantified by a slope. Sample type (feces or water) was included as a factorial effect, along with a possible interaction term with time point to allow for the possibility that the change over time depended on sample type. Fisher’s exact test was used to determine whether there were differences among strains with different antimicrobial resistance (AMR) profiles in terms of how long they survived in feces or water. Two-way contingency tables were constructed between time and strain (AMR profile), separately for each species and sample type (feces and water). Time was dichotomized according to t < 4 days or t ≥ 4 days.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by USDA-NIFA grant 2011-51110-31050.
We are grateful to turkey industry collaborators for their support throughout the project. We are grateful to Michael Mann and Rusty Romine for assistance in sample collection and to all turkey industry staff for access to farm samples. We thank all members of our laboratory for their support in this study.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01579-19.
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.p11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaakoush NO, Castaño-Rodríguez N, Mitchell HM, Man SM. 2015. Global epidemiology of Campylobacter infection. Clin Microbiol Rev 28:687–720. doi: 10.1128/CMR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Döpfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. 2015. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12:e1001921. doi: 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman CR, Neimann J, Wegener HC, Tauxe RV. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p 121–138. In Nachamkin I, Blaser MJ (ed), Campylobacter, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 5.Nyati KK, Nyati R. 2013. Role of Campylobacter jejuni infection in the pathogenesis of Guillain-Barré syndrome: an update. Biomed Res Int 2013:852195. doi: 10.1155/2013/852195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzgerald C. 2015. Campylobacter. Clin Lab Med 35:289–298. doi: 10.1016/j.cll.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Andreoletti O, Budka H, Buncic S, Collins JD, Griffin J, Hald T, Hendrik Havelaar A, Hope J, Klein G, McLauchlin J, Messens W, Müller-Graf C, Nguyen-The C, Noerrung B, Peixe L, Prieto Maradona M, Ricci A, Sofos J, Threlfall J, Vågsholm I, Vanopdenbosch E, Hofshagen M, Kruse H, McCarthy N, Monteiro Pires S, Newell D, Strachan N. 2010. Scientific opinion on quantification of the risk posed by broiler meat to human campylobacteriosis in the EU. EFSA J 8:1437. [Google Scholar]
- 8.Domingues AR, Pires SM, Halasa T, Hald T. 2012. Source attribution of human campylobacteriosis using a meta-analysis of case-control studies of sporadic infections. Epidemiol Infect 140:970–981. doi: 10.1017/S0950268811002676. [DOI] [PubMed] [Google Scholar]
- 9.Lindmark H, Boqvist S, Ljungström M, Agren P, Björkholm B, Engstrand L. 2009. Risk factors for campylobacteriosis: an epidemiological surveillance study of patients and retail poultry. J Clin Microbiol 47:2616–2619. doi: 10.1128/JCM.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs‐Reitsma WF. 1997. Aspects of epidemiology of Campylobacter in poultry. Vet Q 19:113–117. doi: 10.1080/01652176.1997.9694753. [DOI] [PubMed] [Google Scholar]
- 11.Herman L, Heyndrickx M, Grijspeerdt K, Vandekerchove D, Rollier I, De Zutter L. 2003. Routes for Campylobacter contamination of poultry meat: epidemiological study from hatchery to slaughterhouse. Epidemiol Infect 131:1169–1180. doi: 10.1017/s0950268803001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallace J, Stanley K, Jones K. 1998. The colonization of turkeys by thermophilic campylobacters. J Appl Microbiol 85:224–230. doi: 10.1046/j.1365-2672.1998.00470.x. [DOI] [PubMed] [Google Scholar]
- 13.Miller WG, Mandrell R. 2005. Prevalence of Campylobacter in the food and water supply: incidence, outbreaks, isolation and detection, p 101–163. In Ketley J, Konkel ME (ed), Campylobacter jejuni: new perspectives in molecular and cellular biology. Horizon Bioscience, Wymondham, Norfolk, United Kingdom. [Google Scholar]
- 14.Wright SL, Carver DK, Siletzky RM, Romine S, Morrow WEM, Kathariou S. 2008. Longitudinal study of prevalence of Campylobacter jejuni and Campylobacter coli from turkeys and swine grown in close proximity. J Food Prot 71:1791–1796. doi: 10.4315/0362-028X-71.9.1791. [DOI] [PubMed] [Google Scholar]
- 15.Luangtongkum T, Jeon B, Han J, Plummer P, Logue CM, Zhang Q. 2009. Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol 4:189–200. doi: 10.2217/17460913.4.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SF. 2002. The physiology of Campylobacter species and its relevance to their role as foodborne pathogens. Int J Food Microbiol 74:177–188. doi: 10.1016/s0168-1605(01)00678-x. [DOI] [PubMed] [Google Scholar]
- 17.Hazeleger WC, Janse JD, Koenraad PM, Beumer RR, Rombouts FM, Abee T. 1995. Temperature-dependent membrane fatty acid and cell physiology changes in coccoid forms of Campylobacter jejuni. Appl Environ Microbiol 61:2713–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodgers JD, Clifton-Hadley FA, Marin C, Vidal AB. 2010. An evaluation of survival and detection of Campylobacter jejuni and C. coli in broiler caecal contents using culture-based methods. J Appl Microbiol 109:1244–1252. doi: 10.1111/j.1365-2672.2010.04748.x. [DOI] [PubMed] [Google Scholar]
- 19.Bronowski C, James CE, Winstanley C. 2014. Role of environmental survival in transmission of Campylobacter jejuni. FEMS Microbiol Lett 356:8–19. doi: 10.1111/1574-6968.12488. [DOI] [PubMed] [Google Scholar]
- 20.Hilbert F, Scherwitzel M, Paulsen P, Szostak MP. 2010. Survival of Campylobacter jejuni under conditions of atmospheric oxygen tension with the support of Pseudomonas spp. Appl Environ Microbiol 76:5911–5917. doi: 10.1128/AEM.01532-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young KT, Davis LM, DiRita VJ. 2007. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol 5:665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- 22.Snelling WJ, McKenna JP, Lecky DM, Dooley J. 2005. Survival of Campylobacter jejuni in waterborne protozoa. Appl Environ Microbiol 71:5560–5571. doi: 10.1128/AEM.71.9.5560-5571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buswell CM, Herlihy YM, Lawrence LM, McGuiggan JT, Marsh PD, Keevil CW, Leach SA. 1998. Extended survival and persistence of Campylobacter spp. in water and aquatic biofilms and their detection by immunofluorescent-antibody and -rRNA staining. Appl Environ Microbiol 64:733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vieira A, Karlyshev AV, Seddon AM. 2015. Campylobacter-Acanthamoeba interactions. Microbiology 161:933–947. doi: 10.1099/mic.0.000075. [DOI] [PubMed] [Google Scholar]
- 25.Ellis-Iversen J, Ridley A, Morris V, Sowa A, Harris J, Atterbury R, Sparks N, Allen V. 2012. Persistent environmental reservoirs on farms as risk factors for Campylobacter in commercial poultry. Epidemiol Infect 140:916–924. doi: 10.1017/S095026881100118X. [DOI] [PubMed] [Google Scholar]
- 26.Cawthraw SA, Wassenaar TM, Ayling R, Newell DG. 1996. Increased colonization potential of Campylobacter jejuni strain 81116 after passage through chickens and its implication on the rate of transmission within flocks. Epidemiol Infect 117:213–215. doi: 10.1017/s0950268800001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ringoir DD, Korolik V. 2003. Colonisation phenotype and colonisation potential differences in Campylobacter jejuni strains in chickens before and after passage in vivo. Vet Microbiol 92:225–235. doi: 10.1016/s0378-1135(02)00378-4. [DOI] [PubMed] [Google Scholar]
- 28.Jones MA, Marston KL, Woodall CA, Maskell DJ, Linton D, Karlyshev AV, Dorrell N, Wren BW, Barrow PA. 2004. Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect Immun 72:3769–3776. doi: 10.1128/IAI.72.7.3769-3776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, Geys H, Cawthraw S, Havelaar A, Teunis P. 2006. Dose response for infectivity of several strains of Campylobacter jejuni in chickens. Risk Anal 26:1613–1621. doi: 10.1111/j.1539-6924.2006.00850.x. [DOI] [PubMed] [Google Scholar]
- 30.Conlan AJK, Line JE, Hiett K, Coward C, Van Diemen PM, Stevens MP, Jones MA, Gog JR, Maskell DJ. 2011. Transmission and dose-response experiments for social animals: a reappraisal of the colonization biology of Campylobacter jejuni in chickens. J R Soc Interface 8:1720–1735. doi: 10.1098/rsif.2011.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hald B, Skovgard H, Pedersen K, Bunkenborg H. 2008. Influxed insects as vectors for Campylobacter jejuni and Campylobacter coli in Danish broiler houses. Poult Sci 87:1428–1434. doi: 10.3382/ps.2007-00301. [DOI] [PubMed] [Google Scholar]
- 32.Hald B, Skovgård H, Bang DD, Pedersen K, Dybdahl J, Jespersen JB, Madsen M. 2004. Flies and Campylobacter infection of broiler flocks. Emerg Infect Dis 10:1490–1492. doi: 10.3201/eid1008.040129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones NR, Millman C, van der Es M, Hukelova M, Forbes KJ, Glover C, Haldenby S, Hunter PR, Jackson K, O’Brien SJ, Rigby D, Strachan NJC, Williams N, Lake IR, ENIGMA Consortium . 2017. Novel sampling method for assessing human-pathogen interactions in the natural environment using boot socks and citizen scientists, with application to Campylobacter seasonality. Appl Environ Microbiol 83:e00162-17. doi: 10.1128/AEM.00162-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmed MFM, Schulz J, Hartung J. 2013. Survival of Campylobacter jejuni in naturally and artificially contaminated laying hen feces. Poult Sci 92:364–369. doi: 10.3382/ps.2012-02496. [DOI] [PubMed] [Google Scholar]
- 35.Bui XT, Wolff A, Madsen M, Bang DD. 2012. Reverse transcriptase real-time PCR for detection and quantification of viable Campylobacter jejuni directly from poultry faecal samples. Res Microbiol 163:64–72. doi: 10.1016/j.resmic.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 36.Smith S, Meade J, Gibbons J, McGill K, Bolton D, Whyte P. 2016. The impact of environmental conditions on Campylobacter jejuni survival in broiler faeces and litter. Infect Ecol Epidemiol 6:31685. doi: 10.3402/iee.v6.31685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu W, Siletzky RM, Wright S, Islam M, Kathariou S. 2009. Antimicrobial susceptibility profiles and strain type diversity of Campylobacter jejuni isolates from turkeys in eastern North Carolina. Appl Environ Microbiol 75:474–482. doi: 10.1128/AEM.02012-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee BC, Reimers N, Barnes HJ, D’Lima C, Carver D, Kathariou S. 2005. Strain persistence and fluctuation of multiple-antibiotic resistant Campylobacter coli colonizing turkeys over successive production cycles. Foodborne Pathog Dis 2:103–110. doi: 10.1089/fpd.2005.2.103. [DOI] [PubMed] [Google Scholar]
- 39.Niedermeyer JA, Ring L, Miller WG, Genger S, Parr Lindsey C, Osborne J, Kathariou S. 2018. Proximity to other commercial turkey farms affects colonization onset, genotypes and antimicrobial resistance profiles of Campylobacter in turkeys: suggestive evidence from a paired-farm model. Appl Environ Microbiol 84:e01212-18. doi: 10.1128/AEM.01212-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González M, Hänninen M-L. 2012. Effect of temperature and antimicrobial resistance on survival of Campylobacter jejuni in well water: application of the Weibull model. J Appl Microbiol 113:284–293. doi: 10.1111/j.1365-2672.2012.05342.x. [DOI] [PubMed] [Google Scholar]
- 41.Chan KF, Le Tran H, Kanenaka RY, Kathariou S. 2001. Survival of clinical and poultry-derived isolates of Campylobacter jejuni at a low temperature (4°C). Appl Environ Microbiol 67:4186–4191. doi: 10.1128/AEM.67.9.4186-4191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cools I, Uyttendaele M, Caro C, D’Haese E, Nelis HJ, Debevere J. 2003. Survival of Campylobacter jejuni strains of different origin in drinking water. J Appl Microbiol 94:886–892. doi: 10.1046/j.1365-2672.2003.01916.x. [DOI] [PubMed] [Google Scholar]
- 43.Bronowski C, Mustafa K, Goodhead I, James CE, Nelson C, Lucaci A, Wigley P, Humphrey TJ, Williams NJ, Winstanley C, ENIGMA Consortium . 2017. Campylobacter jejuni transcriptome changes during loss of culturability in water. PLoS One 12:e0188936. doi: 10.1371/journal.pone.0188936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oh E, Chui L, Bae J, Li V, Ma A, Mutschall SK, Taboada EN, McMullen LM, Jeon B. 2018. Frequent implication of multistress-tolerant Campylobacter jejuni in human infections. Emerg Infect Dis 24:1037–1044. doi: 10.3201/eid2406.171587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berndtson E, Danielsson-Tham M-L, Engvall A. 1996. Campylobacter incidence on a chicken farm and the spread of Campylobacter during the slaughter process. Int J Food Microbiol 32:35–47. doi: 10.1016/0168-1605(96)01102-6. [DOI] [PubMed] [Google Scholar]
- 46.Smith K, Reimers N, Barnes HJ, Lee BC, Siletzky R, Kathariou S. 2004. Campylobacter colonization of sibling turkey flocks reared under different management conditions. J Food Prot 67:1463–1468. doi: 10.4315/0362-028x-67.7.1463. [DOI] [PubMed] [Google Scholar]
- 47.Miller WG, Englen MD, Kathariou S, Wesley IV, Wang G, Pittenger-Alley L, Siletz RM, Muraoka W, Fedorka-Cray PJ, Mandrell RE. 2006. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology 152:245–255. doi: 10.1099/mic.0.28348-0. [DOI] [PubMed] [Google Scholar]
- 48.Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


