The biological function and the fate of P. luminescens 2° cells were unclear. Here, we performed comparative transcriptomics of P. luminescens 1° and 2° cultures and found several genes, not only those coding for known phenotypic differences of the two cell forms, that are up- or downregulated in 2° cells compared to levels in 1° cells. Our results suggest that when 1° cells convert to 2° cells, they drastically change their way of life. Thus, 2° cells could easily adapt to an alternative environment such as the rhizosphere and live freely, independent of a host, putatively utilizing plant-derived compounds as nutrient sources. Since 2° cells are not able to reassociate with the nematodes, an alternative lifestyle in the rhizosphere would be conceivable.
KEYWORDS: bacterium-host interaction, cell-cell communication, entomopathogenic bacteria, PpyS/PluR
ABSTRACT
Photorhabdus luminescens is a Gram-negative bacterium that lives in symbiosis with soil nematodes and is simultaneously highly pathogenic toward insects. The bacteria exist in two phenotypically different forms, designated primary (1°) and secondary (2°) cells. Yet unknown environmental stimuli as well as global stress conditions induce phenotypic switching of up to 50% of 1° cells to 2° cells. An important difference between the two phenotypic forms is that 2° cells are unable to live in symbiosis with nematodes and are therefore believed to remain in the soil after a successful infection cycle. In this work, we performed a transcriptomic analysis to highlight and better understand the role of 2° cells and their putative ability to adapt to living in soil. We could confirm that the major phenotypic differences between the two cell forms are mediated at the transcriptional level as the corresponding genes were downregulated in 2° cells. Furthermore, 2° cells seem to be adapted to another environment as we found several differentially expressed genes involved in the cells’ metabolism, motility, and chemotaxis as well as stress resistance, which are either up- or downregulated in 2° cells. As 2° cells, in contrast to 1° cells, chemotactically responded to different attractants, including plant root exudates, there is evidence for the rhizosphere being an alternative environment for the 2° cells. Since P. luminescens is biotechnologically used as a bio-insecticide, investigation of a putative interaction of 2° cells with plants is also of great interest for agriculture.
IMPORTANCE The biological function and the fate of P. luminescens 2° cells were unclear. Here, we performed comparative transcriptomics of P. luminescens 1° and 2° cultures and found several genes, not only those coding for known phenotypic differences of the two cell forms, that are up- or downregulated in 2° cells compared to levels in 1° cells. Our results suggest that when 1° cells convert to 2° cells, they drastically change their way of life. Thus, 2° cells could easily adapt to an alternative environment such as the rhizosphere and live freely, independent of a host, putatively utilizing plant-derived compounds as nutrient sources. Since 2° cells are not able to reassociate with the nematodes, an alternative lifestyle in the rhizosphere would be conceivable.
INTRODUCTION
Photorhabdus luminescens is a Gram-negative, entomopathogenic bacterium belonging to the family Enterobacteriaceae (1, 2). The bacteria undergo a dualistic life cycle including mutualistic symbiosis with Heterorhabditidae nematodes and a pathogenic relationship in which they infect and kill insects (1). P. luminescens was first isolated from the gut of Heterorhabditis bacteriophora nematodes, found in temperate climates. The bacteria exist in two phenotypically different forms, which are designated primary (1°) and secondary (2°) cells. After prolonged cultivation, a large portion of single 1° cells undergo phenotypic switching and convert into 2° cells, which differ from 1° cells in various phenotypic traits (3) (Table 1). Most predominant is that 2° cells are less bioluminescent than 1° cells, do not produce red pigments, and are unable to live in symbiosis with the nematode partner (4–7). So far, phenotypic switching of P. luminescens cells has been observed only unidirectionally from the 1° to the 2° cell form (1, 3; our unpublished observations). Previously, phenotypic switching of Photorhabdus has been referred to as phase variation (8). However, this phenomenon differs from classical bacterial phase variation as both variants are genetically identical (1; our own unpublished observations) and has therefore been termed phenotypic heterogeneity (9). The exact regulatory mechanism behind phenotypic switching and the biological role of P. luminescens 2° cells still remain elusive. As 2° cells are known not to be capable of reassociating with nematodes and support their growth and development (6), it has been assumed that they might be better adapted to a life in soil (10, 11). However, 2° cells have thus far not been isolated from soil. The fact that they are found only after prolonged cultivation of 1° cells led to the assumption that the switch occurs as a response to environmental or metabolic stress (12). It was also observed that, after a period of starvation, 2° cells adapted faster to the addition of nutrients and grew faster than 1° cells. Furthermore, proteome analysis demonstrated that 2° cells experience an upregulation of several metabolic enzymes (11). According to this observation, major respiratory enzymes and also the transmembrane proton motive force were found to be upregulated in 2° cells, supporting the assumption that this cell variant might be more adapted for a life in soil (11, 13).
TABLE 1.
Genes corresponding to 1° cell-specific features downregulated in 2° cellsa
| Phenotype and gene | 1° cells | 2° cells | FC by growth phase (2° wt/1° wt)b |
|
|---|---|---|---|---|
| Exp | Stat | |||
| Bioluminescence | +++ | + | ||
| luxC | NS | −11.56 | ||
| luxD | NS | −10.85 | ||
| Pigmentation | + | − | ||
| antA | −19.71 | −25.95 | ||
| antB | −19.52 | −57.15 | ||
| antC | −36.69 | −15.25 | ||
| antD | −35.16 | −13.11 | ||
| antE | −20.61 | NS | ||
| antF | −30.46 | −26.52 | ||
| antG | −20.86 | NS | ||
| antH | −12.11 | −10.52 | ||
| antI | −12.73 | −30.02 | ||
| Crystal proteins | + | − | ||
| cipA | −5.01 | −27.91 | ||
| cipB | NS | −16.21 | ||
| PluDJC_07765 | −4.20 | −47.76 | ||
| Antibiotic production | + | − | ||
| PluDJC_04580 | −11.29 | NS | ||
| PluDJC_045805 | −5.06 | NS | ||
| PluDJC_04590 | −4.90 | NS | ||
| PluDJC_15990 | NS | −5.47 | ||
| PluDJC_16670 | −5.58 | NS | ||
| stlA | −4.95 | NS | ||
| Cell clumping | + | − | ||
| pcfA | NS | −64.84 | ||
| pcfB | NS | −87.19 | ||
| pcfC | NS | −110.61 | ||
| pcfD | NS | −100.05 | ||
| pcfE | NS | −10.98 | ||
| pcfF | NS | −10.52 | ||
| Protease production | ++ | + | ||
| prtA | −8.47 | NS | ||
| Lipase production | + | − | ||
| pdl | NS | −6.33 | ||
Genes were differentially transcribed between 1° and 2° cells in exponentially growing or stationary phase cultures. The presence (+) or absence (−) of the phenotype as it is described in the literature is indicated.
Fold change (FC) was calculated as the level of expression in wild-type 2° cells/expression in wild-type 1° cells. An FC value of less than −3 or greater than 3 was considered significant (P ≤ 0.05). NS, not significant. Exp, exponential growth phase; Stat, stationary growth phase; wt, wild type.
The purpose of the present study was to shed light on the general function of P. luminescens 2° cells and their fate when they are left behind in the soil after an infection cycle. For that reason, we compared the transcriptomes of P. luminescens DJC 1° and 2° cells. Based on the description of the transcriptomic variation observed, we performed various follow-up investigations and bring evidence for an alternative life cycle of 2° cells in soil.
RESULTS AND DISCUSSION
Phenotypic heterogeneity of P. luminescens DJC 1° and 2° cells.
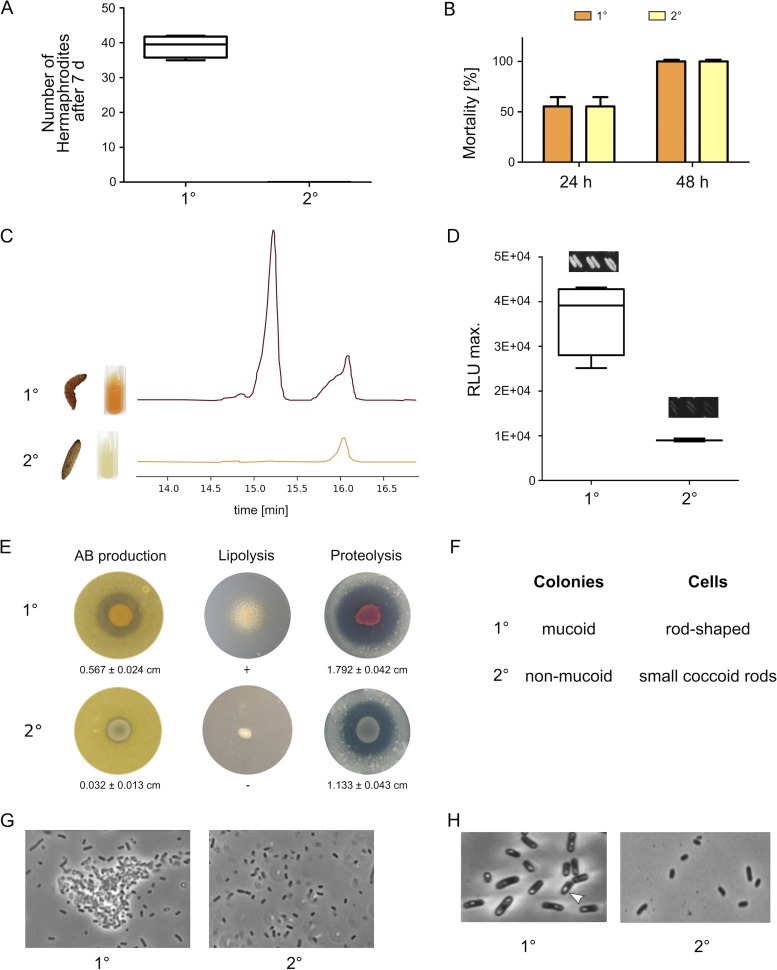
As a first step, we analyzed the phenotypic differences between P. luminescens strain DJC 1° and 2° cells with respect to symbiosis, insect pathogenicity, anthraquinone (pigment) production, and antibiotic, lipase, and protease activities. As also observed for other Photorhabdus strains (6, 14, 15), 2° cells were no longer able to support nematode development (Fig. 1A), whereas insect pathogenicity was comparable to that of 1° cells (Fig. 1B). Furthermore, pigment (anthraquinone) as well as light production was absent from 2° cells (Fig. 1C and D). Antibiotic production and proteolytic activity were strongly decreased while lipase activity, cell clumping, and crystal inclusion proteins were not detectable in 2° cells (Fig. 1E, G, and H). In contrast to the rod-shaped 1° cells that form mucoid colonies, 2° cells are smaller coccoid rods forming nonmucoid colonies (Fig. 1F). The different phenotypes of P. luminescens DJC 1° and 2° cells show that they are comparable to the phenotypic heterogeneity that has been described previously for other Photorhabdus species, such as Photorhabdus temperata (15).
FIG 1.
Phenotypic comparison of P. luminescens DJC 1° and 2° cells. (A) Nematode bioassay. Fifty axenic Heterorhabditis bacteriophora IJs were spotted on 1° or 2° cells grown on lipid agar plates. After 7 days the number of developed hermaphrodites was counted. (B) Pathogenicity assay. Approximately 2,000 of the 1° or 2° cells were injected into 10 G. mellonella larvae each. Mortality was monitored over 48 h. (C) Pigmentation of both phenotypic cell forms was visually monitored over 5 days, and anthraquinone production was quantified from culture supernatant extracts via HPLC. (D) Bioluminescence of 1° and 2° cells was monitored over 24 h using a luminescence plate reader. Additionally, single colonies were streaked, and light production was visually analyzed by taking pictures with 5 min of exposure time. (E) To test for antibiotic production both 1° and 2° cells were spotted onto B. subtilis germ-agar plates. Furthermore, lipolytic or proteolytic activity was tested by spotting both phenotypic cell forms onto Tween agar or skim milk agar plates, respectively. (F) The colony morphology of both cell forms was analyzed by streaking single colonies with a toothpick. The shape of the cells as well as formation of cell clumps (G) and crystal inclusion proteins (H) was investigated via phase-contrast microscopy. Error bars represent standard deviations of three independently performed experiments.
Comparative transcriptome analysis of P. luminescens 1° and 2° cells.
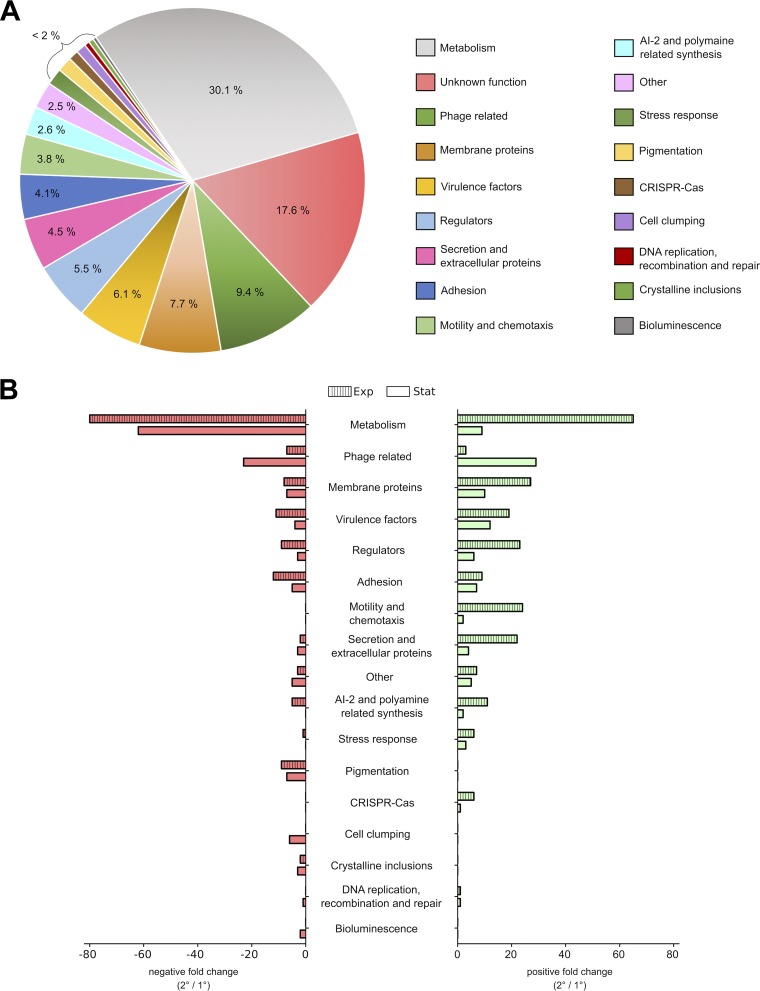
To gain more insights into the differences between P. luminescens 1° and 2° cells, we performed transcriptome sequencing (RNA-Seq) analysis. Thereby, 638 differentially expressed genes (DEGs) were found in 1° and 2° cells, including 373 genes present during exponential growth phase, 178 in early stationary phase, and 87 in both growth phases (see Table S1 in the supplemental material). Ignoring the genes whose function is unclear, the remaining DEGs were divided into 18 subgroups corresponding to their specific functions (Fig. 2A). The subgroup referred to as “others” contains genes that were predicted to be truncated or even pseudogenes, together with genes not yet classified.
FIG 2.
Overview of genes differentially expressed in 2° cells compared to levels in 1° cells. (A) Division of the 638 DEGs according to their functionality into 19 subgroups. (B) Negative (red) and positive (green) FCs of all DEGs obtained at the exponential (Exp; striped bars) as well as stationary (Stat; plain bars) growth phase.
First, we looked for genes that correlate with the distinct phenotypic differences of 1° and 2° cells described above. We found genes responsible for all phenotypic traits mentioned above, such as bioluminescence (luxCD), pigmentation (antABCDEFGHI), crystal inclusion proteins (e.g., cipA), cell clumping (pcfABCDEF), antibiotic production (e.g., PluDJC_04580), proteases (prtA), and lipases (pdl), to be downregulated in 2° cells (Table 1).
2° cells of P. luminescens DJC are unable to reassociate with the nematodes and are therefore left behind in the soil. Thus, phenotypic switching has to be tightly regulated as a switching frequency of 100% would lead to a breakdown of the bacterium's life cycle. However, the exact mechanism is still unclear. Our transcriptome analysis revealed 35 DEGs encoding transcriptional regulators, of which two-thirds are of unknown function (Table S1). Consequently, one or more of these regulatory genes could be involved in the regulation of phenotypic heterogeneity in P. luminescens DJC cell populations.
2° cells of Photorhabdus sp. are commonly described as cell variants that lack several phenotypes. However, our transcriptome analysis revealed that several of the DEGs were upregulated in 2° cells, including genes involved in the cells’ metabolism, stress response, motility, and chemotaxis (Fig. 2B). This indicates that 2° cells are adapted to living in an environment other than that of the symbiotic host. Due to the incapability of 2° cells to reassociate with the nematodes, it seems likely that they are adapted to a free life in soil or the rhizosphere.
As the fate of 2° cells is a crucial missing piece to understanding phenotypic heterogeneity of P. luminescens, we therefore focused on genes that could support 2° cells to deal with alternative environmental conditions such as those of the soil and the rhizosphere.
Changes in signaling and cell-cell communication.
Among the genes with affected expression in 2° cells were various genes encoding regulators involved in signaling and cell-cell communication. Two of these are pluR and ppyS, which code for the LuxR solo (16) and the photopyrone synthase, respectively, were also downregulated in 2° cells. PpyS/PluR is the quorum sensing system used by P. luminescens to control expression of the pcfABCDEF operon and, therefore, cell clumping via PluR (17). This explains the diminished pcfABCDEF transcription and therefore the absence of cell clumps in 2° cells, since PluR positively regulates expression of the pcf operon. However, downregulation of pluR would also affect the cells’ ability to communicate with each other. Since P. luminescens harbors 40 LuxR solo receptors, which are supposed to be involved in cell-to-cell communication as well as interkingdom signaling (18, 19), it is likely that 2° cells use an alternative to the PpyS/PluR communication system.
Transcriptome analysis revealed upregulation of 12 LuxR solo genes in 2° cells: the 8 genes of the PluDJC_10415-PluDJC_10460 operon, which are part of the largest PAS4-LuxR solo cluster of P. luminescens; two single PAS4-LuxR solos (PluDJC_04850 and PluDJC_18380); and the only two LuxR solos with a yet undefined signal binding domain (SBD) (PluDJC_09555 and PluDJC_21150). The LuxR solos of P. luminescens can be divided into four subgroups corresponding to their SBDs. The largest group comprises 34 LuxR solos harboring a PAS4 signal binding domain (19). PAS4 domains of P. luminescens are homologous to the PAS3 domain of the fruit fly Drosophila melanogaster, in which it has been shown that this domain acts as a juvenile hormone (JH) receptor (20). Therefore, it is suggested that PAS4 domains of P. luminescens play an important role in interkingdom signaling and also bind hormone-like molecules (21). Moreover, it has also been shown that LuxR solos of plant-associated bacteria can respond to plant signaling molecules (22, 23), which might also be true for one or more LuxR solos upregulated in 2° cells. However, no specific signal sensed by the PAS4-LuxR solos of P. luminescens has been identified yet.
In summary, the DEGs encoding LuxR receptors strongly suggest that 2° cells utilize other cell-cell communication systems for intra- as well as interkingdom signaling than 1° cells and thereby are able to adapt to an alternative lifestyle. Future work will investigate to which signals the LuxR solos respond and if they support the adaptation of 2° cells to a life in the soil and the rhizosphere.
Differences in LPS composition.
We observed an alteration in expression of six wbl genes, which were either up- or downregulated, that play a role in the O-antigen biosynthesis of lipopolysaccharide (LPS) in the cells (24) (Table S1). For host-associated microbes, changes in LPS composition have previously been associated with differences in host niche (25, 26). Therefore, we hypothesize that the change in LPS composition in 2° cells strongly indicates a specificity for environmental conditions other than those to which 1° cells are adapted. Whether the differences in LPS composition could support the idea that the 2° cells live free in soil that is in contact with plants remains to be tested.
Metabolic changes.
Our transcriptome analysis of 1° and 2° cells revealed a large set of DEGs involved in the cells’ metabolism, which already gives hints of an adaption of 2° cells to alternative nutrients. Among these DEGs were, e.g., genes playing a role in cobalamin biosynthesis or fumarate degradation (Table S1). The complete set of genes involved in hydroxyphenylacetate (HPA) metabolism were expressed at higher levels in 2° cells. 4-HPA is a common fermentation product of aromatic amino acids. Several bacteria, such as Escherichia coli, are able to degrade 4-HPA over several converting steps to finally metabolize it to pyruvate and succinate. Furthermore, it is also often found in soil as a result of plant material degradation by animals (27). Therefore, an enhanced capability to degrade 4-HPA could help 2° cells to grow in soil as it can be used as a carbon source.
In contrast, 2° cells seem to have less affinity for phenylpropanoid compounds than 1° cells as we found the respective cluster (hcaCFE, hcaB, and hcaD) (28) to be downregulated. However, as phenylpropanoids most commonly originate from proteins (28), which are the main nutrient source inside the larvae, reorientation of 2° cells after leaving the cadaver would be obligatory.
Furthermore, the genes astABDE and PluDJC_15875, encoding enzymes for arginine degradation (29), are upregulated in 2° cells. In E. coli the arginine succinyltransferase (AST) pathway is induced when nitrogen is limited and aspartate and arginine are present (30). Again, this could be a mechanism allowing 2° cells to overcome starvation in soil as in the rhizosphere large amounts of amino acids, which are secreted, e.g., from plant roots, are present (31).
As the bacterium-nematode complex, which comprises only 1° cells, emerges from the cadaver when all nutrients of the larvae are depleted, 2° cells might be exposed to starvation. An increase in motility and a higher sensitivity to nutrients and, therefore, enhanced chemotaxis would be an essential strategy for the bacteria to overcome nutrient limitation.
Increased motility and chemotaxis of 2° cells.
The general function of P. luminescens 2° cells is still unclear, but it is assumed that they might be better adapted to a life in soil (10, 11). Since the nutrients present in the rhizosphere differ from the those present in the bioconverted insect cadaver and may not always be easily available, an increase in motility and a higher sensitivity to alternative nutrients could therefore be of great advantage for the whole cell population.
As flagellum formation and directed or nondirected motility are highly complex, including many different operons, we evaluated this group of data considering fold change (FC) values above 1.5 or below −1.5 to include all DEGs involved in these processes. Indeed, we found several DEGs involved in motility and chemotaxis that were upregulated in 2° cells.
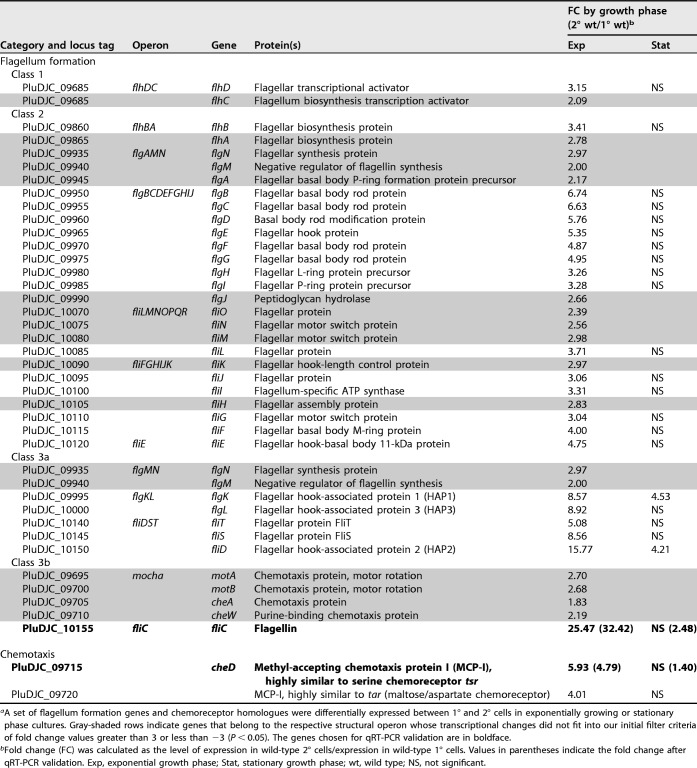
(i) Motility. The transcriptome analysis demonstrated increased expression of 22 genes involved in flagellum formation with an FC of >3 and an additional 13 genes with fold changes ranging from 2.0 to 2.98. We found flhD and flhC, the two parts of the transcriptional activator complex FlhDC (32), to be upregulated in 2° cells (Table 2). Furthermore, we found that several structural genes involved in flagellar hook-basal body complex assembly, which are designated class 2 flagellar genes, (32) were upregulated. In detail, expression levels of either parts of or the complete operons flgBCDEFGHIJ, flhBAE, fliFGHIJK, and fliLMNOPQR as well as the gene encoding FliE were higher in 2° cells. Furthermore, two class 3a structural gene clusters, fliDST and flgKL, as well as fliC (class 3b), which encodes flagellin, exhibited increased expression in exponentially growing 2° cells (Table 2) (32).
TABLE 2.
Motility- and chemotaxis-related genes transcribed at higher levels in 2° cells than in 1° cells in exponential or stationary growth phasea
A setof flagellum formation genes and chemoreceptor homologues were differentially expressed between 1° and 2° cells in exponentially growing or stationary phase cultures. Gray-shaded rows indicate genes that belong to the respective structural operon whose transcriptional changes did not fit into our initial filter criteria of fold change values greater than 3 or less than −3 (P < 0.05). The genes chosen for qRT-PCR validation are in boldface.
Fold change (FC) was calculated as the level of expression in wild-type 2° cells/expression in wild-type 1° cells. Values in parentheses indicate the fold change after qRT-PCR validation. Exp, exponential growth phase; Stat, stationary growth phase; wt, wild type; NS, not significant.
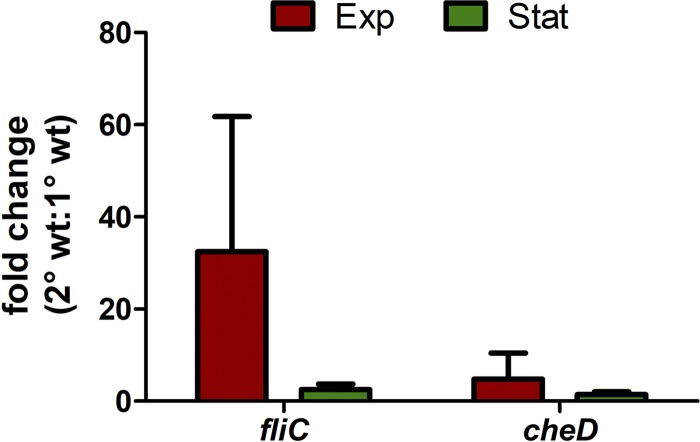
As a representative for motility genes, fliC, the major driving force for flagellum formation, was chosen for RNA-Seq data validation via reverse transcription-quantitative PCR (RT-qPCR). Thereby, we could confirm upregulation of fliC in 2° cells during the exponential growth phase (Fig. 3).
FIG 3.
RT-qPCR data on fliC and cheD displaying higher transcription in 2° than in 1° cells. RT-qPCR revealed a higher level of transcription of fliC and cheD in 2° cells than in 1° cells either in the exponential growth phase (red) or in the stationary phase (green); the fold change is significantly higher in the exponential growth phase for both genes. The data are presented as the fold change ratio of 2° cells to 1° cells with recA used as the housekeeping gene. Values are means of three independent biological replicates and were calculated using the Pfaffl method. wt, wild type.
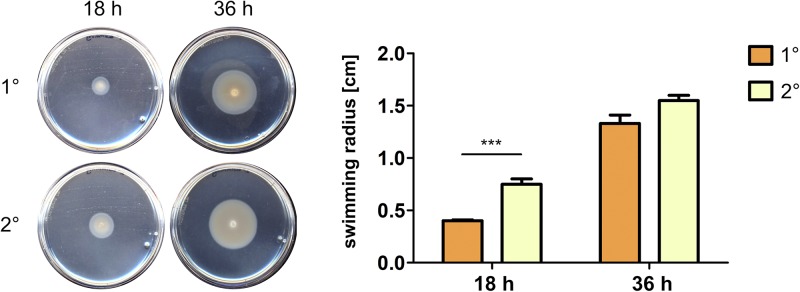
Previously, for Xenorhabdus nematophila and Photorhabdus temperata strains, motility was described to be a specific feature of 1° cells (33). However, we found upregulation of motility-related genes in P. luminescens 2° cells and therefore analyzed whether motility is truly increased in 2° cells. For that purpose, we performed swimming assays by spotting the respective cell forms onto soft-agar swimming plates and measuring the zone of colonization at two different time points. Previously, growth rates of 1° and 2° cells were confirmed to be similar in the medium that was used for the swimming assays (data not shown). In fact, 2° cells exhibited a significantly increased swimming motility compared to that of 1° cells after 18 h of incubation. However, after 36 h the difference between the two cell forms decreased to a nonsignificant level (Fig. 4). This is in accordance with the transcriptome data, which showed that changes in expression of almost all motility-related genes occurred only in the exponential growth phase and were not significant during the stationary growth phase (Table 2).
FIG 4.
Enhanced swimming motility of 2° cells in comparison to that of 1° cells. Upon spotting 5 × 106 1° or 2° cells onto semisolid swimming agar plates, 2° cells showed significantly increased swimming activity compared to that of 1° cells after 18 h of incubation. Error bars represent standard deviations of three independently performed experiments. ***, P < 0.001.
As the transcriptome analysis was performed under noninducing conditions, increased motility seems to be a specific feature of 2° cells of the P. luminescens DJC strain. In E. coli the master activator of flagella formation, flhDC, is directly repressed by lrhA (34). P. luminescens harbors a homologue of this LysR-type transcriptional regulator, HexA, which was identified to act as a master repressor of 1°-cell-specific genes and is highly upregulated in 2° cells of P. temperata (15). However, in X. nematophila, which is closely related to P. luminescens, lrhA positively regulates motility (35). Thus, the flhDC operon might also be activated by hexA in P. luminescens 2° cells. High levels of flhDC, in turn, could cause the increased swarming of 2° cells as positive regulation of swarming motility via FlhDC was observed for X. nematophila (36). We also found hexA upregulated in P. luminescens DJC 2° cells. However, due to the strong cutoff criteria we used, it is not listed.
(ii) Chemotaxis. As motility and chemotaxis go hand in hand, we next analyzed if increased motility in 2° cells subsequently leads to an enhanced chemotactic behavior of the cells. We found upregulation of the complete mocha operon described for E. coli (37) with fold changes in 2° cells of between 1.83 and 2.7 (Table 2). This operon (class 3b flagellar genes) comprises four genes, motA, motB, cheA, and cheW, and is an important part of the chemotaxis systems as it drives motor rotation and attractant sensing (38, 39).
In E. coli the last part of the chemotaxis system is the meche or tar operon, which consists of four sensory (cheRBYZ) and two receptor (tar and tap) genes (40, 41). Transcriptome analysis of P. luminescens DJC 1° and 2° cells revealed one homologue of tar, PluDJC_09720, as upregulated in 2° cells. Despite that, PluDJC_09715, which is highly similar to tsr of E. coli, was also expressed at a higher level in 2° cells (Table 2). Tsr, a type I methyl-accepting chemotaxis protein (MCP-I), is a primary chemoreceptor for the transduction of the attractant serine, while Tar, a type II MCP, is a chemoreceptor for the transduction of aspartate and maltose in E. coli (42). Gene expression of PluDJC_09715 was exemplarily verified via RT-qPCR (Fig. 3).
In order to investigate the difference in the chemotaxis-driven motilities of P. luminescens 1° and 2° cells, swarming assays were performed. For that purpose, a single bacterial colony was spotted onto the center of a semisolid agar plate containing 1 mM or 10 mM serine or maltose, respectively. E. coli MG1655 wild type served as a positive control for chemotactic swarming, while the nonmotile P. luminescens 2° ΔfliC strain was used as a negative control.
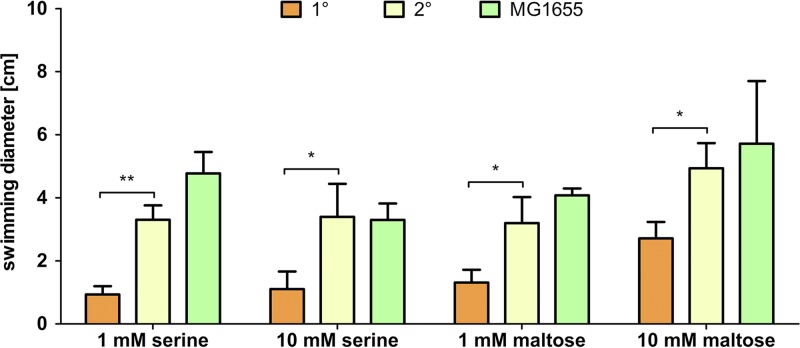
P. luminescens 1° cells showed only a low response to both concentrations of serine as well as 1 mM maltose. However, there was increased movement on the soft-agar plates containing 10 mM maltose. In contrast, 2° cells showed a significantly stronger response to both serine and maltose. Here, a higher sensibility to serine was observed as the swarming diameter on serine plates was significantly bigger than the diameters on plates supplemented with maltose. E. coli MG1655 cells were slightly more motile than P. luminescens 2° cells with 1 mM serine as well as with both concentrations of maltose (Fig. 5 and Fig. S1).
FIG 5.
Swimming diameters after addition of different putative attractants. Attractant-dependent motility of P. luminescens DJC 1° and 2° cells and E. coli MG1655 cells, as indicated, was determined. Error bars represent standard deviations of at least three independently performed experiments. *, P < 0.05; **, P < 0.01.
By increasing the serine concentration, a negative effect could be perceived for E. coli. Here, supplementing the plates with 10 mM instead of 1 mM serine led to a 30.9% shrinkage of the swimming diameter. This effect could be observed only for E. coli and has been reported before as a result of saturation of the serine-sensing transducer Tsr in E. coli (43). However, the swimming diameter of 2° cells did not increase by raising the serine concentration from 1 mM to 10 mM but was similar to the value obtained with the lower serine concentration (Fig. 5 and Fig. S1). Therefore, the Tsr homolog of P. luminescens PluDJC_09715 might be able to cope with a higher concentration of serine. The 2° cells of the ΔfliC strain, which does not produce any flagellin, served as a negative control and were nonmotile upon addition of any putative attractant (data not shown).
The putative role of plants in the life cycle of 2° cells.
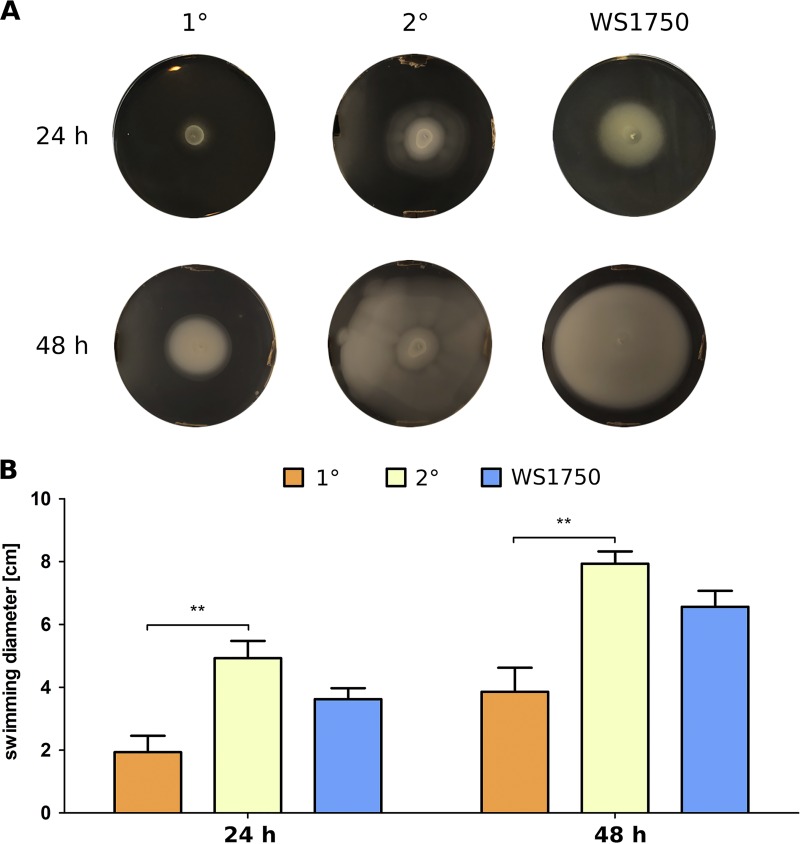
The main producers of nutrients in the soil are plants, as the majority of compounds in the rhizosphere, such as amino acids or sugars as organic acids peptides, proteins, or lipids, derive from root exudates (44–46). Therefore, we investigated whether P. luminescens cells also respond to plant root exudates. For that purpose, we used soft-agar swimming plates supplemented with root exudates of the pea plant Pisum sativum extracted in methanol (MeOH-Ex) and spotted P. luminescens 1° and 2° cells on the plates. The plant-pathogenic strain Pseudomonas fluorescens WS1750 served as a positive control. Effects of methanol on swimming activity were excluded by solely adding the solvent (data not shown). Analysis of the swimming diameters after 24 h or 48 h revealed a significantly higher response of 2° cells to MeOH-Ex than that of 1° cells (Fig. 6A and B). The compositions of compounds contained in the root exudates are unknown. Comparing the swimming activities in the presence of MeOH-Ex to those in the presence of serine and maltose showed them to be comparable or even higher for 2° cells. However, we already applied serine and maltose in excess, as this amino acid and sugar are usually excreted from plants in micromolar or nanomolar amounts (47, 48). Thus, a stronger response of 2° than 1° cells toward other compounds derived from the plant seems likely. Here, further evaluation of the exudate ingredients to resolve the structure and thus the specific signal to which 2° cells respond is needed.
FIG 6.
Effects of plant root exudates on swimming motility of P. luminescens 1° and 2° cells. On plates containing MeOH-Ex, 2° cells showed a significantly stronger response in terms of increased swimming activity than 1° cells. The recorded swimming diameters were even bigger than those observed with the positive-control P. fluorescens WS1750. (A) Pictures of soft-agar swimming plates supplemented with MeOH-Ex after 24 h and 48 h. (B) Graphical depiction of swimming diameters of 1° and 2° cells as well as the WS1750 strain. Error bars represent standard deviations of three independently performed experiments. **, P < 0.01.
The sensing of plant root exudates by 2° cells might be attributable to PluDJC_09715 and PluDJC_09720, as they are MCPs not only for serine and maltose but also for the amino acids alanine/glycine and aspartic acid/glycine, respectively. Furthermore, fruAB was upregulated in 2° cells, which indicates a higher affinity for taking up and utilizing fructose. In addition to galactose, arabinose, raffinose, rhamnose, xylose, and sucrose, fructose and also maltose are the dominant sugars found in root exudates (49). Therefore, a higher-level response of 2° cells than of 1° cells to maltose underlines the suggestion of an increased affinity of 2° cells toward compounds primarily derived from plants.
However, in addition to sugars, vitamins, and amino acids, plants also secrete a wide variety of organic acids that are known to attract bacteria and serve as a nutrient source (50). Thus, additional, as-yet-unknown MCPs involved in the response of 2° cells to plant root exudates might be present in P. luminescens.
Increased temperature tolerance of 2° cells.
Our findings that P. luminescens 2° cells are better adapted to different nutrients than 1° cells support the theory of free-living 2° cells in soil. Additionally, although cultures were grown in rich medium, our transcriptome analysis revealed that several genes involved in the stress response were upregulated in 2° cells (Fig. 1B). Among them, the majority of genes we found are usually induced upon starvation (e.g., dppABCDF, phoH, cstA, or cspD).
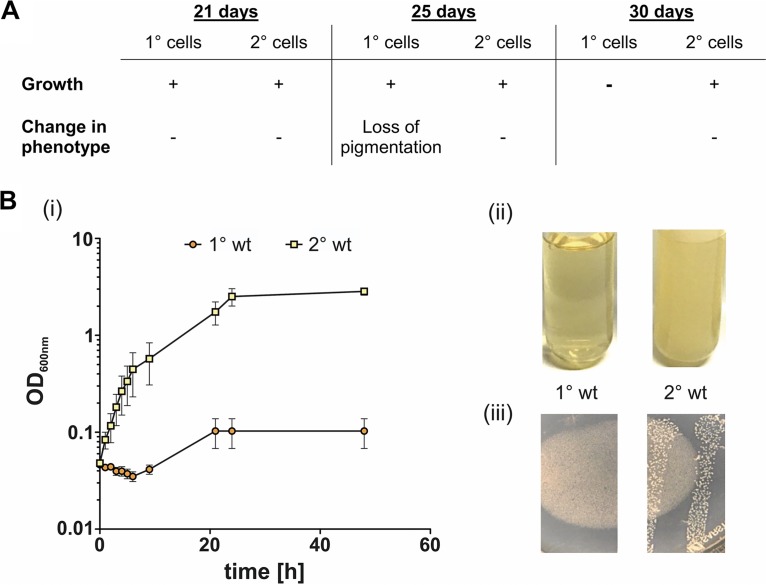
However, it has already been described that 2° cells recover faster from periods of starvation than 1° cells (11), although outside the host, 2° cells would also be more exposed to changing temperatures. Therefore, we attempted to examine whether 2° cells show a higher tolerance to low and high temperatures. As we performed the RNA-Seq analysis under noninducing conditions, no relevant genes were found. For that purpose, we cultivated both cell forms at low temperatures. Here, neither 1° nor 2° cells showed growth when cultivated at 4°C (data not shown). However, we observed an advantage for 2° cells upon storing LB plates with colonies of each cell form at 4°C for 30 days. Every 4 to 5 days, a single colony was inoculated into LB medium and cultivated at 30°C to determine whether the cells were able to recover and to restart growth. While 2° cells grew perfectly well at all tested time points (Fig. 7A), 1° cells were not able to grow after 30 days. Furthermore, although the 1° cells grew after 25 days of incubation at 4°C, we observed a loss of pigmentation (Fig. 7A), which indicates decreased fitness of the cells, as they remained 1° cells with respect to all other phenotypes (data not shown). Even though we did not find upregulation of any genes encoding heat shock proteins, we also tested the capability of both cell forms to deal with higher temperatures. We found that 2° cells grew significantly better in terms of reaching higher cell densities than 1° cells when they were cultivated at 37°C (Fig. 7B, panels i and ii). Furthermore, only 2° and not 1° cells formed colonies when plated onto LB plates and incubated at 37°C (Fig. 7B, panel iii). Growth at different temperatures is much more important for a free life in soil than for a life inside a host. Night and day as well as the different seasons have a great impact on soil temperature. Therefore, the larger temperature tolerance of 2° cells further supports the idea that they are better adapted for a life in soil than 1° cells.
FIG 7.
Growth and phenotype of 1° and 2° cells at high and low temperatures. (A) 1° cells do not recover growth after being incubated for 30 days at 4°C and already show loss of pigmentation after 25 days at 4°C. In contrast, 2° cells restart growth after 30 days of exposure to cold and are not affected at all in their fitness or phenotype. (B) 2° cells were capable of growing at 37°C when cultivated in liquid culture while growth of 1° cells was highly decreased under this condition (i and ii). Upon streaking both cell forms onto agar plates and incubating them at 37°C, only 2° cells were able to form colonies (iii). All experiments were independently performed three times. Error bars represent standard deviation.
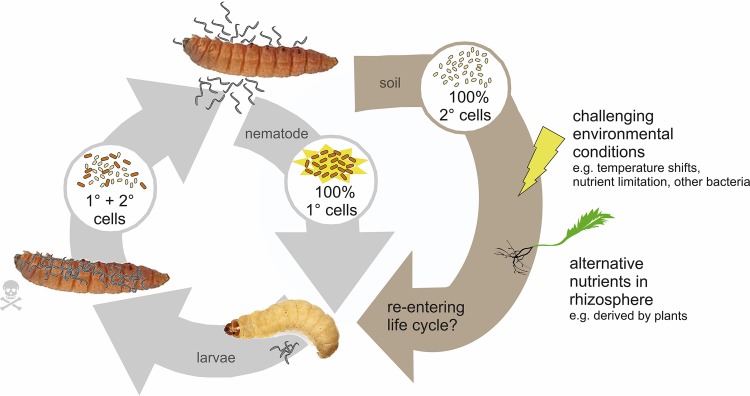
Conclusion.
We could confirm that the most prominent phenotypic traits of P. luminescens DJC 1° and 2° cells are mediated at the transcriptional level. Furthermore, our transcriptome data support the idea that 2° cells are better adapted to an alternative environment outside insect hosts. We found evidence that 2° cells change their metabolism in order to be better adapted to alternative nutrients. Furthermore, 2° cells highly express genes that deal with stress situations, and we could show that they are less sensitive to high or low temperatures than 1° cells. These data thereby strongly support the theory of free-living 2° cells in soil where they withstand challenging environmental conditions and feed from nutrients present in the soil (Fig. 8). Furthermore, we found evidence that 2° cells might somehow be associated with plants or feed on plant-derived nutrients in the rhizosphere.
FIG 8.
Model of extended life cycle of 2° cells in soil. As only 1° cells are able to reassociate with the nematodes and emerge from the cadaver, 2° cells are left behind in the soil. Based on our transcriptome data, it seems likely that 2° cells are better adapted to free living in soil and thereby are able to survive changing and challenging environmental conditions but also develop strategies to utilize alternative nutrients which are present in soil and which are most likely derived from plants. Eventually, they may find a yet unknown way to reenter the life cycle of P. luminescens.
If and how 2° cells can reenter the pathogenic life cycle or can convert to the 1° phenotype again still remain elusive. However, since the bacteria are already used as a bio-insecticide in agriculture, further investigation of a putative interaction of Photorhabdus sp. 2° cells with plant roots is of great importance for biotechnology and agriculture.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strains MG1655 and DH5α λpir were used in this study. They were routinely grown at 37°C in LB medium [1% (wt/vol) NaCl, 1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract]. If necessary, 50 μg/ml antibiotic was added into the medium. P. luminescens DJC (2) 1° and 2° cells were obtained from the lab of David Clarke (University College Cork, Ireland) and were cultivated aerobically in either LB medium or CASO medium (0.5% [wt/vol] NaCl, 0.5% [wt/vol] peptone from soy, 1.5% [wt/vol] tryptone) at 30°C. If necessary, the growth medium was supplemented with 50 μg/ml rifampin (Sigma-Aldrich). For preparation of agar plates, 1.5% (wt/vol) agar was added to the respective medium.
Bioluminescence bioassays.
Luminescence measurements were performed by cultivation of P. luminescens DJC 1° and 2° cells in black 96-well plates with transparent bottoms (Corning, Bodenheim, Germany) and recording of optical density (OD) as well as luminescence using an Infinite-500 reader (Tecan, Salzburg, Austria). Additionally, single colonies of the respective P. luminescens variants were streaked onto LB plates and incubated at 30°C for 48 h. Subsequently, bioluminescence was monitored using a chemiluminescence imager (Peqlab, Erlangen, Germany) with a 5-min exposure time.
Pathogenicity bioassays.
Fifth-instar larvae of Galleria mellonella (reared in our lab) were incubated on ice for 10 min to reduce movement and surface sterilized in a 70% (vol/vol) ethanol bath, followed by a bath of sterile water. Larvae were infected via subcutaneous injection of approximately 2,000 P. luminescens DJC 1° or 2° cells using a sterilized microsyringe (1702 RN, 25 μl; Hamilton). The infected larvae were then incubated at 30°C, and the mortality rate was determined by counting dead and live animals after 24 h and 48 h.
Protease bioassays.
P. luminescens DJC 1° and 2° cells were grown overnight in LB medium at 30°C. Then, an aliquot of 50 μl (OD at 600 nm [OD600] of 1.0) was dropped onto the middle of a skim-milk agar plate (1% [wt/vol] skim milk, 0.3% [wt/vol] yeast extract, 1.2% [wt/vol)] agar), and the plates were incubated for 2 days at 30°C.
Lipase activity bioassays.
P. luminescens DJC 1° and 2° cells were grown overnight in LB medium at 30°C. Then, an aliquot of 50 μl (OD600 of 1.0) was dropped onto the middle of a Tween 20 agar plate (1% Tween 20 [vol/vol)], 1% [wt/vol] tryptone, 0.5% [wt/vol] NaCl2, 0.1% [wt/vol] CaCl2·2 H2O, 2% [wt/vol] agar], and the plates were incubated for 2 days at 30°C. The precipitation of the calcium salt was visually monitored.
Antibiotic bioassays.
For testing antibiotic activity, we used soft-agar plates supplemented with Bacillus subtilis as a test strain. For that purpose, an overnight culture of B. subtilis at an OD600 of 2 to 3 in a 1:100 dilution was added to liquid hand-warm LB agar medium (0.8% [wt/vol] agar). After the plates were polymerized, an aliquot of 30 μl (OD600 of 1.0) of the respective P. luminescens DJC 1° or 2° cells was dropped onto the middle of the agar plate and incubated for 48 h at 30°C.
Symbiosis bioassays.
An aliquot of 50 μl of an overnight culture of P. luminescens DJC 1° and 2° cells, diluted to an OD600 of 1.0, was spread in a Z pattern onto the surface of a lipid agar plate (1% [vol/vol] corn syrup, 0.5% [wt/vol] yeast extract, 5% [vol/vol] cod liver oil, 2% [wt/vol] MgCl2·6 H2O, 2.5% [wt/vol] Difco nutrient agar [Becton, Dickinson, Heidelberg, Germany]) using an inoculating loop. The plates were incubated at 30°C for 3 days before addition of 50 surface-sterilized axenic Heterorhabditis bacteriophora infective juvenile (IJ) nematodes to the bacterial biomass. Nematodes were surface sterilized by washing in a solution (0.4% [wt/vol]) of hyamine (Sigma-Aldrich, Deisenhofen, Germany). The plates were kept at room temperature. Nematode recovery was assessed 7 to 8 days after addition of IJ nematodes by counting the number of hermaphrodites on the lipid agar plate.
Pigmentation.
The development of red pigments was visually noted after 3 days of growth of P. luminescens DJC 1° and 2° cells on LB plates at 30°C or 3 days after injection of the bacteria into G. mellonella larvae. Additionally, pigmentation was quantified by determining the anthraquinone (AQ) production via high-performance liquid chromatography (HPLC). To this end, 100 ml of LB medium was inoculated to an OD600 of 0.1 using overnight cultures of P. luminescens DJC 1° and 2° cells. After 72 h of growth at 30°C, 15 ml of each culture was centrifuged for 5 min at 5,000 rpm (at room temperature [RT]). Then, 10 ml of the resulting supernatant was transferred into a new reaction tube and mixed with 10 ml of ethyl acetate plus 0.1% (vol/vol) formic acid (FA) and shaken for 1 h at RT. Subsequently, the reaction tube was kept standing for 1 h and briefly centrifuged in order to separate the organic (upper phase) from the hydrophilic phase. The latter was removed with a vacuum evaporator (Heidolph) at 240 × 105 Pa at 42°C. The extracts were resuspended in 750 μl of methanol and analyzed by HPLC-UV (Thermo Scientific) using a C18 Hypersil Gold column (particle size, 5 μm; 250 by 4.6 mm), with detection achieved by measuring UV absorbance at 430 nm. Acetonitrile (ACN) plus 0.1% (vol/vol) FA was used as the mobile phase. With that, a gradient from 5% (vol/vol) to 95% (vol/vol) ACN–0.1% (vol/vol) FA in a period of 25 min was followed by an isocratic step (95% [vol/vol] ACN plus 0.1% FA) with a flow rate of 0.5 ml/min. The column temperature was set at 30°C. The resulting peak areas were normalized against the optical density of the culture measured at the harvesting step.
RNA preparation.
Total RNA from three independent cultures of DJC 1° or DJC 2° cells in the exponential growth phase (6-h culture, 3 × 109 CFU/ml) and early stationary phase (18-h culture, 10 × 109 CFU/ml) grown at 30°C was extracted. The pellets of harvested cells were resuspended in 500 μl of ice-cold AE buffer (20 mM NaAc, pH 5.2; 1 mM EDTA, pH 8.0), and then 500 μl of Roti-Aqua-P/C/I (where P/C/I is phenol, chloroform, and isoamyl alcohol) (Roth) and 25 μl of 10% SDS were added. After vortexing, the mixture was incubated for 30 min at 60°C with shaking. Subsequently, the samples were placed into a refrigerator for one night. On the next day, the samples were centrifuged at 16,100 relative centrifugal force units (rcf) for 20 min at 0°C. Afterwards, the supernatant was transferred into 5PRIME Phase Lock gel tubes (Quantabio), supplemented with 500 μl of P/C/I and 50 μl of 3 M NaAc, pH 5.2, and after mixing the tubes were centrifuged at 16,100 rcf for 10 min at 0°C. Then the supernatants were mixed with 1 ml of 96% ethanol (EtOH) and held at −80°C for overnight precipitation. On day 3 samples were again centrifuged at 16,100 rcf for 30 min at 0°C, but this time the supernatant was discarded. To wash the pellet, 1 ml of 80% EtOH was added and subsequently removed by centrifugation at 16,100 rcf for 10 min at 0°C. This washing step was repeated two times. Then the pellet was air dried for 60 min with an open lid and resolved in 100 μl of diethyl pyrocarbonate (DEPC)-treated water. Five micrograms of RNA was then treated with DNase I (ThermoFisher) to remove genomic DNA. Integrity and quantity of total RNA samples were tested with an Agilent 2100 Bioanalyzer system. To eliminate rRNA, a Ribo-Zero rRNA removal kit for Gram-negative bacteria was used according to the protocol provided by the manufacturer (Illumina). Afterwards, an additional quality check with the Agilent 2100 Bioanalyzer system was performed.
Transcriptome analysis.
To sequence RNA samples, cDNA libraries were generated using an NEBNext Ultra II RNA Library Prep kit for Illumina (New England Biolabs [NEB]), according to the manufacturer’s instructions, starting from 50 ng of rRNA-depleted RNA. The libraries were quality controlled by analysis on an Agilent 2000 Bioanalyzer with an Agilent High Sensitivity DNA kit (Agilent Technologies) for fragment sizes of around 200 to 500 bp. Libraries were pooled, and sequencing on a MiSeq sequencer (2- by 75-bp paired-end sequencing; version 3 chemistry [Illumina]) was performed at the Genomics Service Unit (Ludwig-Maximilians-Universität [LMU] Biocenter, Martinsried, Germany). CLC Genomics Workbench (version 11.0.0; Qiagen) was used to analyze the data. Raw reads were trimmed for quality and adapter sequences, mapped to the reference genome (P. luminescens DJC; GenBank accession number NZ_CP024900.1), and analyzed using an RNA-Seq analysis tool. We selected differentially expressed genes having a P value of ≤0.05, and the filter for the fold change was set to values of less than −3 or greater than 3. To exclude single outliers, the limit for the maximum group mean was set to ≥20. The functions of the genes of interest were extracted from the UniProt (https://www.uniprot.org) and NCBI (https://www.ncbi.nlm.nih.gov) databases.
RT-qPCR.
To validate the whole-transcriptome data, reverse transcription-quantitative PCR (RT-qPCR) was carried out on three independent total RNA preparations, in each case in triplicate. cDNAs were synthesized during the run using a Luna Universal One-Step RT-qPCR kit (NEB), and the reactions were performed according to the protocol provided by the manufacturer. Reactions and melting curves were monitored in a LightCycler (Bio-Rad). Differences in gene expression levels were calculated using the Pfaffl method (51) with recA serving as a housekeeping gene. All data are presented as a ratio of three independent biological replicates. Values are means ± the standard deviations.
Generation of knockout mutants.
The fliC gene was deleted in P. luminescens 2° cells as described previously (52). In brief, 500 bp upstream and downstream of genomic fliC (PluDJC_10155) were amplified by PCR using the primer pair BamHI_fliC FA fwd (ACGGGATCCGGCAACGAATGCATCATG) and FliC FA ovl FB rev (CCCTAGCTGAGCGATTAACGTGCCATAGTTAGAGTTCC) and the pair FliC FB ovl FA fwd (GGAACTCTAACTATGGCACGTTAATCGCTCAGCTAGGG) and fliC FB_EagI rev (ACTCGGCCGCAATCACGGCTCCTTAAC), introducing BamHI and EagI restriction sites (underlined) into the 5′ end of the upstream fragment and the 3′ end of the downstream fragment, respectively. Overlap extension PCR was used to fuse the two PCR products, which were then cloned into pNPTs138-R6KT using the BamHI and EagI restriction sites, resulting in pNPTS-FAB ΔfliC. Correctness of the plasmid was confirmed by PCR using primers Check pNPTS-FA FB FWD (TGCTTCCGGCTCGTATG) and Check pNPTS-FA FB REV (GTAAAACGACGGCCAGTCC). This plasmid was then conjugated from E. coli S17-1 λpir into 2° cells, and exconjugants were selected as Rifr Kmr colonies. The pNPTs138-R6KT plasmid contains the sacB gene, and after growth in LB broth (with no selection), putative mutants were identified by screening for Rifr Sucr Kms colonies. The deletion of fliC was confirmed by PCR using the primer pair BamHI_fliC FA fwd/fliC FB_EagI rev, followed by DNA sequencing.
Swimming assays.
Swimming assays were performed using soft-agar plates containing 0.3% (wt/vol) agar, 1% tryptone (wt/vol), and 0.5% NaCl (wt/vol). Overnight cultures of 1° and 2° cells were set to an OD600 of 1, and 5 μl was spotted into the center of a soft-agar swimming plate. Without any further movement, the plates were incubated at RT. After 18 and 36 h the diameters of the colonies representing swimming were documented and evaluated using the ImageJ tool for measuring distances. The data were obtained from three independently performed biological and technical replicates.
Chemotaxis movement assays.
Soft-agar swarming assays were performed using agar plates containing 0.3% (wt/vol) agar, 1% tryptone (wt/vol), 1% NaCl (wt/vol), and the putative attractant. After autoclaving, the soft agar was kept at 60°C. Right before use, 20 ml of soft agar was supplemented with either 1 mM or 10 mM l-serine or maltose. As the concentration of the plant root exudate was unknown, 600 μl of exudate dissolved in methanol (MeOH-Ex) was added to 20 ml of 0.3% soft agar. After the plates were polymerized, 10 μl of P. luminescens DJC 1° and 2° wild-type (WT), DJC 2° ΔfliC, and E. coli MG1655 cells at an OD600 of 0.1 were spotted into the center of the soft-agar plates. Swarming plates were incubated for 24 h and at 30°C without motion. The swimming diameters, representing chemotaxis-dependent movement, were documented and analyzed via the ImageJ tool for measuring distances. The data were obtained from three independently performed biological and technical replicates.
Extraction of plant root exudates.
To extract plant root exudates, 75 Pisum sativum plants were put in flasks containing 250 ml of methanol. After 16 h of shaking at RT, the liquid was collected, filter sterilized, and stored at 4°C until further use.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alice Regaiolo (Mainz, Germany) for helpful discussions and proofreading of the manuscript. RNA-Seq was performed in the Genomics core facility of the LMU Biocenter.
S.E. and A.B. performed and evaluated RNA-Seq analysis. S.E. conducted the phenotypic comparison of the two cell forms and performed the swimming assays as well as RT-qPCR analysis. N.D. generated the P. luminescens 2° ΔfliC mutant and performed the growth and chemotaxis assays. S.E. generated the figures. S.E. and R.H. designed the study and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Research was funded from the Deutsche Forschungsgemeinschaft, priority program SPP 1617 (HE 5247/5-2).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01910-19.
REFERENCES
- 1.Forst S, Dowds B, Boemare N, Stackebrandt E. 1997. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu Rev Microbiol 51:47–72. doi: 10.1146/annurev.micro.51.1.47. [DOI] [PubMed] [Google Scholar]
- 2.Zamora-Lagos M-A, Eckstein S, Langer A, Gazanis A, Pfeiffer F, Habermann B, Heermann R. 2018. Phenotypic and genomic comparison of Photorhabdus luminescens subsp. laumondii TT01 and a widely used rifampicin-resistant Photorhabdus luminescens laboratory strain. BMC Genomics 19:19:854. doi: 10.1186/s12864-018-5121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forst S, Clarke DJ. 2002. Nematode-bacterium symbiosis. CABI Publishing, Oxford, United Kingdom. [Google Scholar]
- 4.Akhurst RJ. 1980. Morphological and functional dimorphism in Xenorhabdus spp, bacteria symbiotically associated with the insect pathogenic nematodes Neoplectana and Heterorhabditis. J Gen Microbiol 121:303–309. doi: 10.1099/00221287-121-2-303. [DOI] [PubMed] [Google Scholar]
- 5.Heinrich AK, Glaeser A, Tobias NJ, Heermann R, Bode HB. 2016. Heterogeneous regulation of bacterial natural product biosynthesis via a novel transcription factor. Heliyon 2:e00197. doi: 10.1016/j.heliyon.2016.e00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han R, Ehlers RU. 2001. Effect of Photorhabdus luminescens phase variants on the in vivo and in vitro development and reproduction of the entomopathogenic nematodes Heterorhabditis bacteriophora and Steinernema carpocapsae. FEMS Microbiol Ecol 35:239–247. doi: 10.1016/S0168-6496(01)00097-6. [DOI] [PubMed] [Google Scholar]
- 7.You J, Liang S, Cao L, Liu X, Han R. 2006. Nutritive significance of crystalline inclusion proteins of Photorhabdus luminescens in Steinernema nematodes. FEMS Microbiol Ecol 55:178–185. doi: 10.1111/j.1574-6941.2005.00015.x. [DOI] [PubMed] [Google Scholar]
- 8.Boemare NE, Akhurst RJ. 1988. Biochemical and physiological characterization of colony form variants in Xenorhabdus spp. (Enterobacteriaceae). J Gen Microbiol 134:751–761. doi: 10.1099/00221287-134-3-751. [DOI] [PubMed] [Google Scholar]
- 9.Davidson CJ, Surette MG. 2008. Individuality in bacteria. Annu Rev Genet 42:253–268. doi: 10.1146/annurev.genet.42.110807.091601. [DOI] [PubMed] [Google Scholar]
- 10.Joyce SA, Lango L, Clarke DJ. 2011. The regulation of secondary metabolism and mutualism in the insect pathogenic bacterium Photorhabdus luminescens. Adv Appl Microbiol 76:1–25. doi: 10.1016/B978-0-12-387048-3.00001-5. [DOI] [PubMed] [Google Scholar]
- 11.Smigielski AJ, Akhurst RJ, Boemare NE. 1994. Phase variation in Xenorhabdus nematophilus and Photorhabdus luminescens: differences in respiratory activity and membrane energization. Appl Environ Microbiol 60:120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joyce SA, Watson RJ, Clarke DJ. 2006. The regulation of pathogenicity and mutualism in Photorhabdus. Curr Opin Microbiol 9:127–132. doi: 10.1016/j.mib.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Turlin E, Pascal G, Rousselle J-C, Lenormand P, Ngo S, Danchin A, Derzelle S. 2006. Proteome analysis of the phenotypic variation process in Photorhabdus luminescens. Proteomics 6:2705–2725. doi: 10.1002/pmic.200500646. [DOI] [PubMed] [Google Scholar]
- 14.Han RR, Ehlers R. 2000. Pathogenicity, development, and reproduction of Heterorhabditis bacteriophora and Steinernema carpocapsae under axenic in vivo conditions. J Invertebr Pathol 75:55–58. doi: 10.1006/jipa.1999.4900. [DOI] [PubMed] [Google Scholar]
- 15.Joyce SA, Clarke DJ. 2003. A hexA homologue from Photorhabdus regulates pathogenicity, symbiosis and phenotypic variation. Mol Microbiol 47:1445–1457. doi: 10.1046/j.1365-2958.2003.03389.x. [DOI] [PubMed] [Google Scholar]
- 16.Subramoni S, Venturi V. 2009. LuxR-family “solos”: bachelor sensors/regulators of signalling molecules. Microbiology 155:1377–1385. doi: 10.1099/mic.0.026849-0. [DOI] [PubMed] [Google Scholar]
- 17.Brachmann AO, Brameyer S, Kresovic D, Hitkova I, Kopp Y, Manske C, Schubert K, Bode HB, Heermann R. 2013. Pyrones as bacterial signaling molecules. Nat Chem Biol 9:573–578. doi: 10.1038/nchembio.1295. [DOI] [PubMed] [Google Scholar]
- 18.Brameyer S, Heermann R. 2016. Quorum sensing and LuxR solos in Photorhabdus, p 103–119. In ffrench-Constant RH. (ed), The molecular biology of Photorhabdus bacteria. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 19.Brameyer S, Kresovic D, Bode HB, Heermann R. 2014. LuxR solos in Photorhabdus species. Front Cell Infect Microbiol 4:166. doi: 10.3389/fcimb.2014.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubrovsky EB. 2005. Hormonal cross talk in insect development. Trends Endocrinol Metab 16:6–11. doi: 10.1016/j.tem.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Heermann R, Fuchs TM. 2008. Comparative analysis of the Photorhabdus luminescens and the Yersinia enterocolitica genomes: uncovering candidate genes involved in insect pathogenicity. BMC Genomics 9:40. doi: 10.1186/1471-2164-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Covaceuszach S, Degrassi G, Venturi V, Lamba D. 2013. Structural insights into a novel interkingdom signaling circuit by cartography of the ligand-binding sites of the homologous quorum sensing LuxR-family. Int J Mol Sci 14:20578–20596. doi: 10.3390/ijms141020578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venturi V, Fuqua C. 2013. Chemical signaling between plants and plant-pathogenic bacteria. Annu Rev Phytopathol 51:17–37. doi: 10.1146/annurev-phyto-082712-102239. [DOI] [PubMed] [Google Scholar]
- 24.Derzelle S, Hallet B, Ferain T, Delcour J, Hols P. 2003. Improved adaptation to cold-shock, stationary-phase, and freezing stresses in Lactobacillus plantarum overproducing cold-shock proteins. Appl Environ Microbiol 69:4285–4290. doi: 10.1128/AEM.69.7.4285-4290.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGowan CC, Necheva A, Thompson SA, Cover TL, Blaser MJ. 1998. Acid-induced expression of an LPS-associated gene in Helicobacter pylori. Mol Microbiol 30:19–31. doi: 10.1046/j.1365-2958.1998.t01-1-01079.x. [DOI] [PubMed] [Google Scholar]
- 26.Duelli DM, Tobin A, Box JM, Kolli VS, Carlson RW, Noel KD. 2001. Genetic locus required for antigenic maturation of Rhizobium etli CE3 lipopolysaccharide. J Bacteriol 183:6054–6064. doi: 10.1128/JB.183.20.6054-6064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Díaz E, Ferrández A, Prieto MA, García JL. 2001. Biodegradation of aromatic compounds by Escherichia coli. Microbiol Mol Biol Rev 65:523–569. doi: 10.1128/MMBR.65.4.523-569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Díaz E, Ferrández A, García JL. 1998. Characterization of the hca cluster encoding the dioxygenolytic pathway for initial catabolism of 3-phenylpropionic acid in Escherichia coli K-12. J Bacteriol 180:2915–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Easom CA, Clarke DJ. 2012. HdfR is a regulator in Photorhabdus luminescens that modulates metabolism and symbiosis with the nematode Heterorhabditis. Environ Microbiol 14:953–966. doi: 10.1111/j.1462-2920.2011.02669.x. [DOI] [PubMed] [Google Scholar]
- 30.Schneider BL, Kiupakis AK, Reitzer LJ. 1998. Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J Bacteriol 180:4278–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Badri DV, Vivanco JM. 2009. Regulation and function of root exudates. Plant Cell Environ 32:666–681. doi: 10.1111/j.1365-3040.2008.01926.x. [DOI] [PubMed] [Google Scholar]
- 32.Kalir S, McClure J, Pabbaraju K, Southward C, Ronen M, Leibler S, Surette MG, Alon U. 2001. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science 292:2080–2083. doi: 10.1126/science.1058758. [DOI] [PubMed] [Google Scholar]
- 33.Hodgson MM, Day B, White DJ, Tisa LS. 2003. Effect of growth conditions on the motility of Photorhabdus temperata. Arch Microbiol 180:17–24. doi: 10.1007/s00203-003-0558-z. [DOI] [PubMed] [Google Scholar]
- 34.Lehnen D, Blumer C, Polen T, Wackwitz B, Wendisch VF, Unden G. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol Microbiol 45:521–532. doi: 10.1046/j.1365-2958.2002.03032.x. [DOI] [PubMed] [Google Scholar]
- 35.Richards GR, Herbert EE, Park Y, Goodrich-Blair H. 2008. Xenorhabdus nematophila lrhA is necessary for motility, lipase activity, toxin expression, and virulence in Manduca sexta insects. J Bacteriol 190:4870–4879. doi: 10.1128/JB.00358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Givaudan A, Lanois A. 2000. flhDC, the flagellar master operon of Xenorhabdus nematophilus: requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J Bacteriol 182:107–115. doi: 10.1128/jb.182.1.107-115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silverman M, Simon M. 1976. Operon controlling motility and chemotoxis in E. coli. Nature 264:577–580. doi: 10.1038/264577a0. [DOI] [PubMed] [Google Scholar]
- 38.Block SM, Berg HC. 1984. Successive incorporation of force-generating units in the bacterial rotary motor. Nature 309:470–472. doi: 10.1038/309470a0. [DOI] [PubMed] [Google Scholar]
- 39.Blair DF, Berg HC. 1988. Restoration of torque in defective flagellar motors. Science 242:1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]
- 40.Krikos A, Mutoh N, Boyd A, Simon MI. 1983. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell 33:615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- 41.Mutoh N, Simon MI. 1986. Nucleotide sequence corresponding to five chemotaxis genes in Escherichia coli. J Bacteriol 165:161–166. doi: 10.1128/jb.165.1.161-166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Springer MS, Goy MF, Adler J. 1977. Sensory transduction in Escherichia coli: a requirement for methionine in sensory adaptation. Proc Natl Acad Sci U S A 74:183–187. doi: 10.1073/pnas.74.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burkart M, Toguchi A, Harshey RM. 1998. The chemotaxis system, but not chemotaxis, is essential for swarming motility in Escherichia coli. Proc Natl Acad Sci U S A 95:2568–2573. doi: 10.1073/pnas.95.5.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- 45.Walker TS, Bais HP, Grotewold E, Vivanco JM. 2003. Root exudation and rhizosphere biology. Plant Physiol 132:44–51. doi: 10.1104/pp.102.019661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lesuffleur F, Paynel F, Bataillé M-P, Le Deunff E, Cliquet J-B. 2007. Root amino acid exudation: measurement of high efflux rates of glycine and serine from six different plant species. Plant Soil 294:235–246. doi: 10.1007/s11104-007-9249-x. [DOI] [Google Scholar]
- 47.Phillips DA, Fox TC, King MD, Bhuvaneswari TV, Teuber LR. 2004. Microbial products trigger amino acid exudation from plant roots. Plant Physiol 136:2887–2894. doi: 10.1104/pp.104.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carvalhais LC, Dennis PG, Fedoseyenko D, Hajirezaei M-R, Borriss R, von Wirén N. 2011. Root exudation of sugars, amino acids, and organic acids by maize as affected by nitrogen, phosphorus, potassium, and iron deficiency. J Plant Nutr Soil Sci 174:3–11. doi: 10.1002/jpln.201000085. [DOI] [Google Scholar]
- 49.Grayston SJ, Campbell CD. 1996. Functional biodiversity of microbial communities in the rhizospheres of hybrid larch (Larix eurolepis) and Sitka spruce (Picea sitchensis). Tree Physiol 16:1031–1038. doi: 10.1093/treephys/16.11-12.1031. [DOI] [PubMed] [Google Scholar]
- 50.Haichar FEZ, Santaella C, Heulin T, Achouak W. 2014. Root exudates mediated interactions belowground. Soil Biol Biochem 77:69–80. doi: 10.1016/j.soilbio.2014.06.017. [DOI] [Google Scholar]
- 51.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Easom CA, Clarke DJ. 2008. Motility is required for the competitive fitness of entomopathogenic Photorhabdus luminescens during insect infection. BMC Microbiol 8:168. doi: 10.1186/1471-2180-8-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.