Because of limited therapeutic options, infections caused by methicillin-resistant Staphylococcus aureus represent a serious problem in both civilian and military health care settings. Phages have potential as alternative antibacterial agents that can be used in combination with antibiotic drugs. For decades, phage Sb-1 has been used in former Soviet Union countries for antistaphylococcal treatment in humans. The therapeutic spectrum of activity of Sb-1 can be increased by selecting mutants of the phage with expanded host ranges. In this work, the host range of phage Sb-1 was expanded in the laboratory, and a hypervariable region in its genome was identified with a distinct allele state that correlated with this host range expansion. These results provide a genetic basis for better understanding the mechanisms of phage host range expansion.
KEYWORDS: Staphylococcus aureus, phage Sb-1, adaptation, host range expansion, hypervariable DNA region, bacteriophage adaptation, genome analysis, putative host range determinant, therapeutic bacteriophage
ABSTRACT
Staphylococci are frequent agents of health care-associated infections and include methicillin-resistant Staphylococcus aureus (MRSA), which is resistant to first-line antibiotic treatments. Bacteriophage (phage) therapy is a promising alternative antibacterial option to treat MRSA infections. S. aureus-specific phage Sb-1 has been widely used in Georgia to treat a variety of human S. aureus infections. Sb-1 has a broad host range within S. aureus, including MRSA strains, and its host range can be further expanded by adaptation to previously resistant clinical isolates. The susceptibilities of a panel of 25 genetically diverse clinical MRSA isolates to Sb-1 phage were tested, and the phage had lytic activity against 23 strains (92%). The adapted phage stock (designated Sb-1A) was tested in comparison with the parental phage (designated Sb-1P). Sb-1P had lytic activity against 78/90 strains (87%) in an expanded panel of diverse global S. aureus isolates, while eight additional strains in this panel were susceptible to Sb-1A (lytic against 86/90 strains [96%]). The Sb-1A stock was shown to be a mixed population of phage clones, including approximately 4% expanded host range mutants, designated Sb-1M. In an effort to better understand the genetic basis for this host range expansion, we sequenced the complete genomes of the parental Sb-1P and two Sb-1M mutants. Comparative genomic analysis revealed a hypervariable complex repeat structure in the Sb-1 genome that had a distinct allele that correlated with the host range expansion. This hypervariable region was previously uncharacterized in Twort-like phages and represents a novel putative host range determinant.
IMPORTANCE Because of limited therapeutic options, infections caused by methicillin-resistant Staphylococcus aureus represent a serious problem in both civilian and military health care settings. Phages have potential as alternative antibacterial agents that can be used in combination with antibiotic drugs. For decades, phage Sb-1 has been used in former Soviet Union countries for antistaphylococcal treatment in humans. The therapeutic spectrum of activity of Sb-1 can be increased by selecting mutants of the phage with expanded host ranges. In this work, the host range of phage Sb-1 was expanded in the laboratory, and a hypervariable region in its genome was identified with a distinct allele state that correlated with this host range expansion. These results provide a genetic basis for better understanding the mechanisms of phage host range expansion.
INTRODUCTION
The increasing prevalence of multidrug-resistant (MDR) bacterial pathogens, including that of methicillin-resistant Staphylococcus aureus (MRSA), is a dangerous trend that limits options for treatment and hinders the ability of physicians to treat infections (1–3). The estimated incidence of health care-associated MRSA infection in the United States in 2014 was more than 77,000 cases (4). This situation, combined with the scarcity of new antibiotics currently in the pharmaceutical development pipeline, necessitates the implementation of alternative therapeutics, including bacteriophages (phages), for patients infected with MDR bacteria. Recently, phages and their lysins were identified by the National Institute of Allergy and Infectious Diseases (NIAID) as one of the viable alternatives to antibiotic treatment (5). Phages specifically infect and kill target bacteria but leave mammalian cells intact and do not have a detrimental effect on normal microflora; thus, they are considered nontoxic and safe for clinical use (6–8). Phages have been shown to disperse S. aureus biofilms (9, 10) and to provide successful treatment against various experimental and veterinary S. aureus infections, including septicemia (11–13) and skin infection (14) in mice, tibial implant infection in rats (15), and cutaneous abscesses (16) and biofilm-infected wounds in rabbits (17). Phage therapy of S. aureus infections in silkworm larvae and in mice showed that the larval model allows for adequate evaluation of therapeutic efficacy (18). Several case reports demonstrated successful phage treatment of refractory staphylococcal infections in humans, including diabetic ulcers (19–21), otitis media (22), and infected radiation injuries (23). Despite this progress, no FDA-approved phage therapeutics are currently available for treatment of infections caused by MRSA or any other MDR bacterium. A phage cocktail was applied against Pseudomonas aeruginosa and S. aureus burn wound infections in a small clinical trial (nine patients) in Belgium and showed no adverse effects (24). Recently, AmpliPhi Biosciences Corporation reported a successful phase I clinical trial in the United States to evaluate the safety of S. aureus phage cocktail AB-SA01 applied topically to the intact skin of healthy adults (https://www.reuters.com/article/idUSFWN1BP07D).
S. aureus phages identified to date are tailed double-stranded DNA phages that belong to the Myoviridae, Siphoviridae, and Podoviridae families, characterized respectively by their long contractile, long noncontractile, or short tails (25–32). The broad-host-range S. aureus phage Sb-1 has been used extensively in former Soviet Union (FSU) countries and has shown efficacy in the treatment of various S. aureus infections, including those caused by MRSA (33–35). Phage Sb-1 is a member of the Twortlikevirus genus within the Spounavirinae subfamily of the Myoviridae family (27, 36–38) and is a close relative of phages K, G1, and Twort (31), as well as of Bacillus subtilis phage SPO1, from which the taxonomy of the subfamily is derived (39). The close genetic relationship between phages Sb-1, K, and SPO1 has been described previously, and genome architecture appears well conserved within the staphylococcal Twort-like phages (34, 38).
Phage Sb-1, produced by Eliava BioPreparations, Ltd., and referred to as “Staphylococcal Bacteriophage,” is commercially available in the country of Georgia. This phage exhibits favorable safety and efficacy profiles (35). Originally, Sb-1 was isolated in 1977 from an already available antistaphylococcal phage cocktail (40). The biology and genome of Sb-1 have been studied in detail (34, 41); it is a strictly lytic phage (it cannot become a prophage) whose genome lacks any bacterial virulence or drug resistance genes. Phage Sb-1 has a broad host range that can be further expanded by passages on phage-resistant strains (34). The broad host range has been partially explained by the lack of GATC sequences in the Sb-1 genome, which precludes the phage DNA from digestion by major S. aureus restriction endonuclease Sau3AI (42, 43). However, the molecular basis for host range expansions of both Sb-1 (34) and closely related phage K (44) have remained unknown, although it was speculated that host range expansion of phage K was due to an unidentified antirestriction property (45).
Here, we describe the isolation and characterization of Sb-1 phage mutants with expanded host ranges. Our analysis revealed the presence of a novel hypervariable repeat region in the genomes of Sb-1, K, and other Twort-like phages. This repeat region stabilized to a dominant allele state when Sb-1 was adapted to a previously phage-resistant bacterial host.
RESULTS AND DISCUSSION
Sb-1 testing on 25 military MRSA isolates and expansion of its host range.
We tested the efficiency of plating (EOP) of a Sb-1 therapeutic preparation manufactured by Eliava BioPreparations, Ltd. (production stock designated Sb-1P, with “P” for parental), on an initial panel of 25 representative MRSA strains isolated from U.S. military personnel, including wound infection isolates (Table 1, strains 1 to 25). This Sb-1P phage preparation was active against 23/25 (92%) of the strains in this initial diversity panel. In order to expand the host range of Sb-1P, a phage adaptation procedure was performed as described earlier (34; see Materials and Methods) that resulted in a phage lysate with an expanded host range, designated Sb-1A (“A” for adapted). The Sb-1A lysate was able to lyse S. aureus strain MRSN9832, which was originally resistant to the parental Sb-1 phage (Sb-1P). The efficiencies of plating of Sb-1A and Sb-1P were then tested on an expanded panel of 90 global S. aureus isolates (see Table 1). Sb-1P had lytic activity against 78/90 strains (87%), while the Sb-1A phage preparation had lytic activity against eight additional strains (86/90 [96%]). Thus, the laboratory adaptation of Sb-1 phage to infect and lyse a single previously resistant MRSA strain (MRSN9832) resulted in a significant more generalized host range expansion. Although the process of adaptation enabled the phage to infect previously resistant strains, the efficiency of plating on those strains was approximately 20 times lower than those of the previously phage-susceptible strains (data not shown). We speculated that the Sb-1A lysate was a mixed population of phage clones, of which approximately 5% were expanded-host-range mutants. To confirm this, the Sb-1A phage lysate was plated on strain MRSN8383, and 46 random single plaques were picked, treated with chloroform, and spotted both on phage-susceptible strain MRSN8383 and on previously phage-resistant strain MRSN9832. Only 2/46 (4.3%) of these subclones were able to produce plaques on both strains and thus were considered to be expanded-host-range mutants. We designated these two host range mutants derived from single plaques Sb-1M (“M” for mutant).
TABLE 1.
S. aureus strains used in this work and results of phage susceptibility testing
| Strain | Geographic location | Source | Antibiotic resistancea | Susceptibility of phageb: |
|
|---|---|---|---|---|---|
| Sb-1 | Sb-1A | ||||
| MRSN18 | Fort Bragg, NC | Wound | MRSA | S | S |
| MRSN30 | Fort Bragg, NC | Wound | MRSA | S | S |
| MRSN42 | Fort Bragg, NC | Wound | MRSA | S | S |
| MRSN214 | Bethesda, MD | Wound | MRSA | S | S |
| MRSN219 | Bethesda, MD | Urine | MRSA | S | S |
| MRSN250 | Bethesda, MD | Urine | MRSA | S | S |
| MRSN352 | Bethesda, MD | Wound | MRSA | S | S |
| MRSN549 | Fort Bragg, NC | Wound | MRSA | S | S |
| MRSN563 | Fort Bragg, NC | Wound | MRSA | S | S |
| MRSN1722 | Bethesda, MD | Sputum | MRSA | S | S |
| MRSN1732 | Bethesda, MD | Tissue | MRSA | S | S |
| MRSN1952 | Bethesda, MD | Wound | MRSA | S | S |
| MRSN2339 | Bethesda, MD | Wound | MRSA | S | S |
| MRSN2763 | Bethesda, MD | Wound | MRSA | S | S |
| MRSN3573 | Bethesda, MD | Blood | MRSA | S | S |
| MRSN3643 | Bethesda, MD | Tissue | MRSA | S | S |
| MRSN3710 | Bagram, Afghanistan | Wound | MRSA | S | S |
| MRSN3966 | Tbilisi, Georgia | Blood | MRSA | S | S |
| MRSN4109 | Bethesda, MD | Sputum | MRSA | S | S |
| MRSN4531 | Bethesda, MD | Wound | MRSA | S | S |
| MRSN4535 | El Paso, TX | Wound | MRSA | S | S |
| MRSN9832 | El Paso, TX | Nasal swab | MRSA | R | S |
| MRSN9834 | El Paso, TX | Nasal swab | MRSA | S | S |
| NAJAF22 | Najaf, Iraq | Wound | MRSA | R | R |
| NAJAF33 | Najaf, Iraq | Wound | MRSA | S | S |
| MRSN4344 | Bethesda, MD | Wound | MSSA | S | S |
| MRSN5079 | Bethesda, MD | Wound | MSSA | S | S |
| MRSN6168 | Bethesda, MD | Blood | MSSA | R | S |
| MRSN7983 | Bethesda, MD | Wound | MSSA | S | S |
| MRSN8383 | Bethesda, MD | Wound | MSSA | S | S |
| MRSN9127 | Bethesda, MD | Respiratory | MSSA | S | S |
| MRSN9287 | Bethesda, MD | Wound | MSSA | S | S |
| MRSN10110 | Bethesda, MD | Wound | MSSA | S | S |
| MRSN10185 | Bethesda, MD | Blood | MSSA | S | S |
| MRSN12239 | Bethesda, MD | Eye | MSSA | S | S |
| IS-3 | Baghdad, Iraq | Wound | MRSA | S | S |
| IS-24 | Baghdad, Iraq | Wound | MRSA | S | S |
| IS-55 | Baghdad, Iraq | Wound | MRSA | S | S |
| IS-88 | Baghdad, Iraq | Wound | MRSA | S | S |
| IS-91 | Baghdad, Iraq | Respiratory | MRSA | S | S |
| IS-99 | Baghdad, Iraq | Wound | MRSA | S | S |
| IS-105 | Baghdad, Iraq | Wound | MRSA | S | S |
| IS-111 | Baghdad, Iraq | Wound | MRSA | S | S |
| IS-122 | Baghdad, Iraq | Wound | MRSA | R | S |
| IS-125 | Baghdad, Iraq | Wound | MRSA | S | S |
| IS-157 | Baghdad, Iraq | Wound | MRSA | S | S |
| IS-160 | Baghdad, Iraq | Wound | MRSA | R | S |
| IS-189 | Baghdad, Iraq | Respiratory | MRSA | S | S |
| IS-195 | Baghdad, Iraq | Blood | MRSA | S | S |
| IS-278 | Baghdad, Iraq | Respiratory | MRSA | R | R |
| IS-M | Baghdad, Iraq | Blood | MRSA | S | S |
| CO-06 | Camp Cropper, Iraq | Abscess | MRSA | S | S |
| CO-08 | Camp Cropper, Iraq | Abscess | MRSA | S | S |
| CO-23 | Camp Cropper, Iraq | Abscess | MRSA | S | S |
| CO-30 | Camp Cropper, Iraq | Wound | MRSA | S | S |
| CO-32 | Camp Cropper, Iraq | Abscess | MRSA | R | S |
| CO-41 | Camp Cropper, Iraq | Wound | MRSA | S | S |
| CO-49 | Camp Cropper, Iraq | Abscess | MRSA | S | S |
| CO-85 | Camp Cropper, Iraq | Wound | MRSA | S | S |
| CO-86 | Camp Cropper, Iraq | Wound | MRSA | S | S |
| CO-99 | Camp Cropper, Iraq | Wound | MRSA | S | S |
| DBT1 | Japan | NAc | MSSA | S | S |
| DBT2 | USA | NA | MRSA | S | S |
| DBT3 | USA | NA | MRSA | S | S |
| DBT4 | USA | NA | MRSA | S | S |
| DBT5 | NA | NA | MRSA | R | S |
| DBT6 | Sydney, Australia | NA | MRSA | S | S |
| DBT7 | Sydney, Australia | NA | MRSA | S | S |
| DBT8 | Sydney, Australia | NA | MRSA | S | S |
| DBT9 | Sydney, Australia | NA | MRSA | S | S |
| DBT10 | Sydney, Australia | NA | MRSA | S | S |
| DBT11 | Sydney, Australia | NA | MRSA | S | S |
| DBT12 | Sydney, Australia | NA | MRSA | R | R |
| DBT13 | Sydney, Australia | NA | MRSA | S | S |
| DBT14 | Sydney, Australia | NA | MRSA | R | S |
| DBT15 | Sydney, Australia | NA | MRSA | R | R |
| DBT16 | Sydney, Australia | NA | MRSA | S | S |
| DBT17 | Sydney, Australia | NA | MRSA | S | S |
| DBT18 | Sydney, Australia | NA | MRSA | S | S |
| DBT19 | Sydney, Australia | NA | MRSA | R | S |
| DBT20 | Sydney, Australia | NA | MRSA | S | S |
| DBT21 | Sydney, Australia | NA | MRSA | S | S |
| DBT22 | St Leonards, Australia | Wound | MSSA | S | S |
| DBT23 | St Leonards, Australia | Wound | MSSA | S | S |
| DBT24 | St Leonards, Australia | Wound | MSSA | S | S |
| DBT25 | St Leonards, Australia | Wound | MSSA | S | S |
| DBT26 | St Leonards, Australia | Wound | MSSA | S | S |
| DBT27 | St Leonards, Australia | Wound | MSSA | S | S |
| DBT28 | St Leonards, Australia | Wound | MSSA | S | S |
| DBT29 | St Leonards, Australia | Wound | MSSA | S | S |
MRSA, methicillin-resistant S. aureus; MSSA, methicillin-susceptible S. aureus.
S, susceptible; R, resistant.
NA, data are not available.
Analysis of S. aureus diversity.
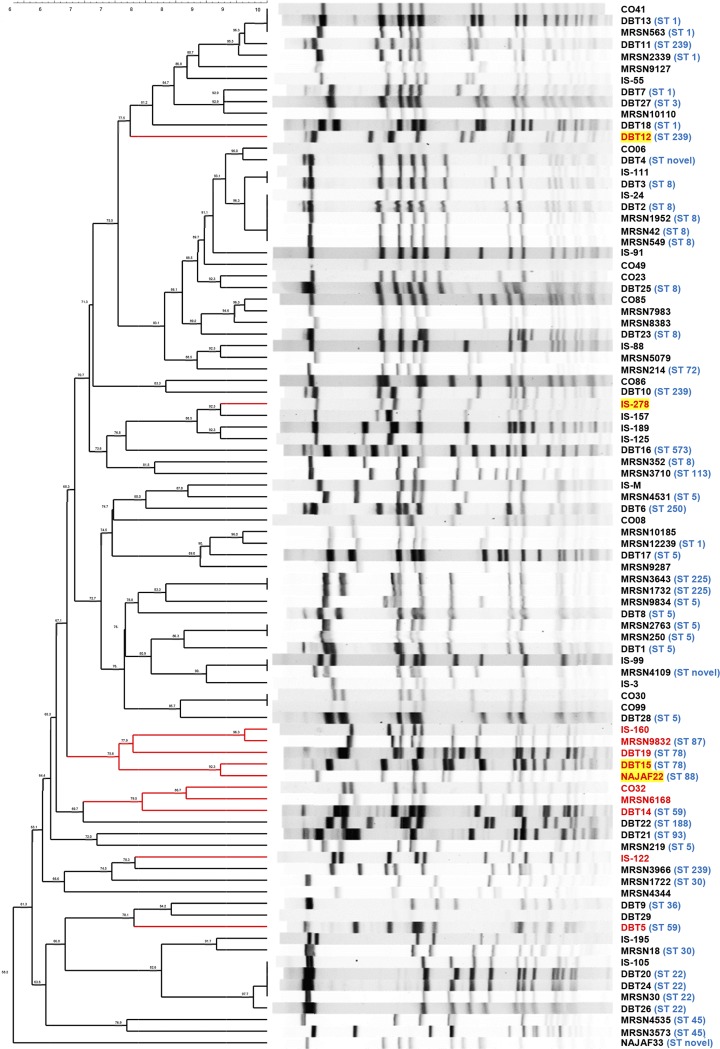
The genetic diversity of U.S. military MRSA isolates selected for this study, and also that of the expanded 90-strain global S. aureus panel used here, was analyzed using pulsed-field gel electrophoresis (PFGE) genotyping. Computer analysis of PFGE profiles identified 76 distinct restriction patterns in the expanded 90-strain diversity panel (Fig. 1). Using the Dice coefficient with BioNumerics software, isolates with ≥80% similarity were assigned to the same pulsotype. The isolates were distributed in 32 pulsotypes. Eight of the 12 strains that were resistant to Sb-1P (67%) were isolated on three different continents but were grouped into two closely related PFGE clusters (Fig. 1) suggesting some correlation between PFGE subtype and Sb-1 resistance and indicating partial association of resistance with phylogenetic grouping. Based on the extant multilocus sequence typing information available for the strains in this panel, some patterns of phage susceptibility could be observed in the S. aureus clonal complexes (CCs): strains representing CC1 (sequence type 1 [ST1]), CC5 (ST5 and ST225), CC8 (ST8 and ST250), CC22 (ST22), CC30 (ST30 and ST36), and CC45 (ST45) were susceptible. Strains in ST239 were also susceptible, with the notable exception of strain DBT12, which was resistant to both Sb-1P and Sb-1A. Clonal complexes with strains that were resistant to Sb-1P included CC59 (ST59 and ST87) and CC88 (ST78 and ST88), with CC88 strains DBT15 (ST78) and NAJAF22 (ST88) also retaining resistance to Sb-1A.
FIG 1.
Diversity of S. aureus strains according to PFGE. Strains resistant to the parental Sb-1 therapeutic preparation (Sb-1P) manufactured by Eliava Biopreparations, Ltd., are marked in red. Strains that were also resistant to the adapted Sb-1 (Sb-1A) are marked with yellow highlighting. Sequence type (ST) based on multiple locus sequence typing is also provided in blue type, if available.
Parental Sb-1 and Sb-1M adsorb to phage-resistant and phage-susceptible S. aureus strains with similar efficiency.
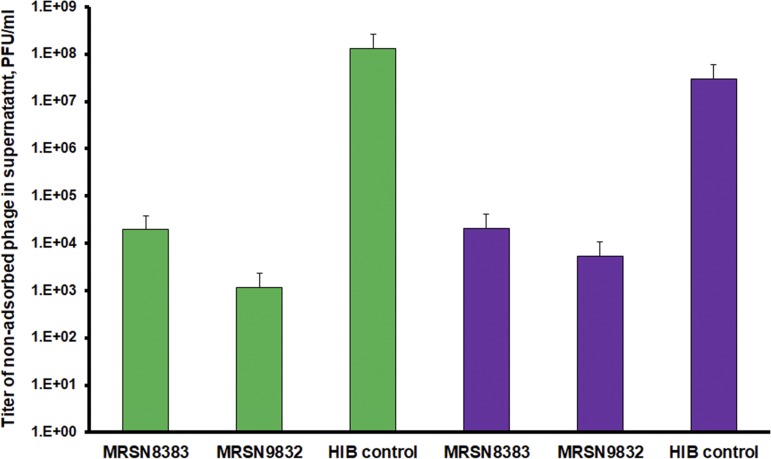
The first step of phage infection is adsorption of the virus to receptors on the host cell. In order to investigate why Sb-1P could not propagate on resistant strains, we sought to determine whether the phage was able to adsorb to the surface of phage-resistant S. aureus cells. The results of a phage adsorption assay presented in Fig. 2 suggest that both Sb-1P and Sb-1M variants were able to efficiently bind to the surface of both phage-resistant (MRSN9832) and phage-susceptible (MRSN8383) host bacteria. In all cases, more than 99% of phage particles were bound to S. aureus cells and thus lost from the supernatant after centrifugation. Therefore, phage resistance in this case is likely to be independent of phage interaction with receptors on the bacterial cell surface.
FIG 2.
Phage adsorption assay. The bars represent the titers of nonadsorbed phage particles in supernatants after precipitation of bacterial cells and phage particles adsorbed to them. Green, Sb-1P; purple, Sb-1M.
Genome comparisons of Sb-1 and its host range variants.
To explore the molecular basis of the phage Sb-1 host range expansion, we performed whole-genome sequencing of the Sb-1P phage (production stock), picked from a single plaque and propagated on phage-susceptible strain MRSN8383 (Sb-18383), and of two expanded host range mutants (Sb-1M), picked from single plaques and propagated on previously Sb-1-resistant strains MRSN6168 (Sb-1M6168) and MRSN9832 (Sb-1M9832). These whole genomes were compared to each other and to the reference genome of Sb-1 sequenced in 2010 (GenBank accession number HQ163896). The comparisons revealed the presence of multiple single-nucleotide differences, two insertion/deletions, and select unresolved regions corresponding to the positions of tandem and complex repeat structures in the genomes analyzed (see Table 2 and Table S1). Single mutations in two hypothetical proteins were observed, including an arginine-to-isoleucine mutation in open reading frame 147 (ORF147) and a lysine-to-glutamic acid mutation in ORF149, both of which were uniquely shared by both Sb1M phages (Table 2 and Table S1). We noted that the Sb-18383 phage genome encodes six unique mutations in the terminal region of ORF82 relative to the Sb-1 reference genome. Although annotated as a hypothetical protein, ORF82 has >97% amino acid similarity to capsid proteins of Staphylococcus phages JD007, phiSA039, and pSco-10. The TAT to ATA mutation observed at positions 47966 to 47968 generates a 36-amino acid extension of the ORF82 homolog in Sb-18383.
TABLE 2.
Discriminating polymorphisms among Sb-1 phage genomes sequenced in this studya
| Location | Polymorphism in phage: |
Codon | ORF | Gene | GenBank protein accession no. | |||
|---|---|---|---|---|---|---|---|---|
| Sb-1ref | Sb-18383 | Sb-1M9832 | Sb-1M6168 | |||||
| 47961 | A[E] | C[D] | Adel | Adel | 3rd | 82 | Conserved hypothetical protein | AEJ79720.1 |
| 47964 | C[G] | T[G] | C[G] | C[G] | 3rd | 82 | Conserved hypothetical protein | AEJ79720.1 |
| 47966 | T | A | T | T | 2nd | 82 | Conserved hypothetical protein | AEJ79720.1 |
| 47967 | A | T | A | A | 2nd | 82 | Conserved hypothetical protein | AEJ79720.1 |
| 47968 | T | A | T | T | 2nd | 82 | Conserved hypothetical protein | AEJ79720.1 |
| 47971 | A | G | A | A | NA | NA | NA | NA |
| 101833 | C[A] | C[A] | C[A] | A[E] | 2nd | NA | Hypothetical protein | AAX92102.1 |
| 108037 | A | G | A | A | NA | NA | NA | NA |
| 109146 | G[R] | G[R] | T[I] | T[I] | 2nd | 147 | Hypothetical protein | YP_008873656.1 |
| 109834 | G[V] | G[V] | A[I] | G[V] | 1st | 149 | Hypothetical protein | YP_008873658.1 |
| 109903 | A[K] | A[K] | G[E] | G[E] | 1st | 149 | Hypothetical protein | YP_008873658.1 |
| 123276–123740 | Region 2 allele A | Region 2 allele B | Region 2 allele C | Region 2 allele C | NA | NA | NA | NA |
NA, data are not available.
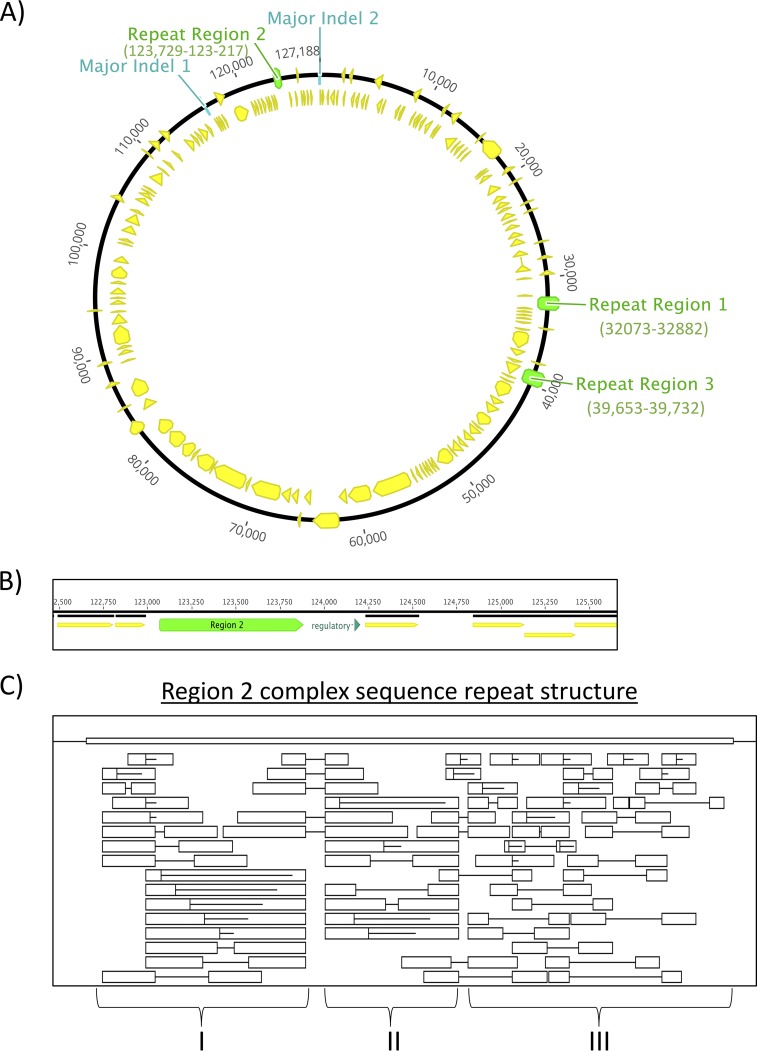
In addition, unresolved repetitive sequences were also observed among the phage genomes. Errors in reconstructing repetitive DNA sequence loci by next-generation sequence assembly algorithms challenged the accurate determination of repeat copy number. Genomic microsatellite diversity has previously been hypothesized to impact Staphylococcus phage genome evolution and host adaptation, and the need to evaluate repeat composition in the context of host range was also noted (46). Our results indicated that such sequence repeat-mediated adaptation may have contributed to the phage host range phenotypes observed in our study. Subsequent bioinformatic analyses revealed the presence of three major intergenic repetitive sequence loci in the phage genomes reported here, which we arbitrarily designated regions 1, 2, and 3. The positions of these regions are illustrated in Fig. 3. We confirmed clone-specific allele states (Table 2) in the Sb-1/Sb-1M genome using PCR fragment sizing and Sanger sequencing. No sequence variation was observed between Sb-1P and Sb-1M at the region 1 locus. Region 1 is comprised of an iteron repeat structure in the left long terminal repeat (LTR) region. We observed that the genomes of the parental and expanded-host-range Sb-1 phage genomes sequenced in this work (Sb-18383, Sb-1M6168, and Sb-1M9832) differed from the Sb-1 reference genome at this locus, in that Sb-18383, Sb-1M6168, and Sb-1M9832 display a single-copy allele state, while the repeat locus in the Sb-1 reference genome contains four copies of the repeat (Fig. 3). Surprisingly, our analysis revealed the presence of a repeat-derived mutant spectrum arising from apparent hypermutability of the region 2 locus in Sb-18383. Region 2 is an intergenic repeat locus that exhibited a complex degenerate repetitive structure comprised of tandem and indirect repeats (Fig. 3A to C), including a core repeat unit sequence of “TACTACTATTAC.” Region 2 is located upstream of the right terminus and immediately adjacent to a regulatory region that could control the transcription of the right long terminal repeat (R-LTR) genes (Fig. 3C). The region 3 locus exhibited no sequence polymorphism across host range phenotypes and the 2010 Sb-1 reference genome.
FIG 3.
Repeat structure analysis of genomic sequence repeat regions. (A) Physical location of the three repeat regions and major indels in the Sb-1 reference genome; (B) annotation of the repeat region 2 locus; (C) complex repeat structure of the region 2 locus illustrating direct and indirect repeats. Boxed regions indicate repeated sequence units. Horizontal lines designate direct repeats.
Diversity of genomic regions 1, 2, and 3.
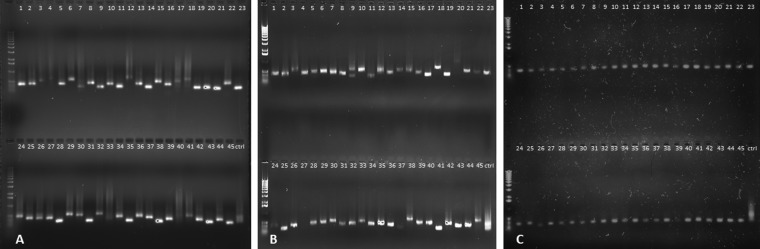
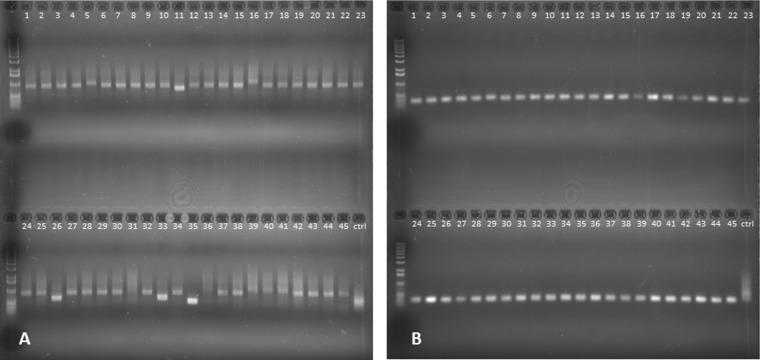
The sequence variability of Regions 1, 2 and 3 was compared in the therapeutic phage Sb-1 production stock (Sb-1P) and the Sb-1A adapted phage preparation. PCRs were performed on 45 randomly selected individual plaques from Sb-1P plated on MRSN8383 (phage-susceptible strain) and from Sb-1A plated on both MRSN8383 and on MRSN9832 (Sb-1P-resistant strain). Region 2 amplicons exhibited a high degree of length variability when Sb-1P or Sb-1A was plated on MRSN8383 (Fig. 4A and B), while Sb-1A plaques produced on previously phage-resistant strain MRSN9832 generated PCR amplicons of relatively uniform length (Fig. 4C). The control PCR product of region 2 of the Sb-1 production stock (Sb-1P) without isolation of individual plaques/clones appears as a highly diffused band that is consistent with variability in region 2 size in the Sb-1 genome. We observed no detectable length variability of PCR fragments in regions 1 and 3 in Sb-1P and Sb-1A clones plated on MRSN8383 or Sb-1A clones plated on MRSN9832 (data not shown). The results suggested that phage plaques on MRSN9832 represented Sb-1M host range mutants with a stabilized region 2 structure.
FIG 4.
Region 2 allele diversity in Sb-1P and Sb-1A phage plaques. (A) PCR products of region 2 from Sb-1P clones plated on MRSN8383. (B) PCR products of region 2 from Sb-1A clones plated on MRSN8383. (C) PCR products of region 2 from Sb-1A clones plated on MRSN9832. The control lanes (ctrl) contain the PCR product of region 2 of the Sb-1 production stock (Sb-1P) without any plaque/clone purification.
Sanger sequence analysis of regions 1, 2, and 3.
The PCR products of regions 1, 2, and 3 for each individual clone of Sb-1P production stock and Sb-1A plated on MRSN8383 (see Fig. 4A and B), and Sb-1M clones picked from previously phage-resistant strain MRSN9832 (see Fig. 4C) were sequenced by the Sanger method.
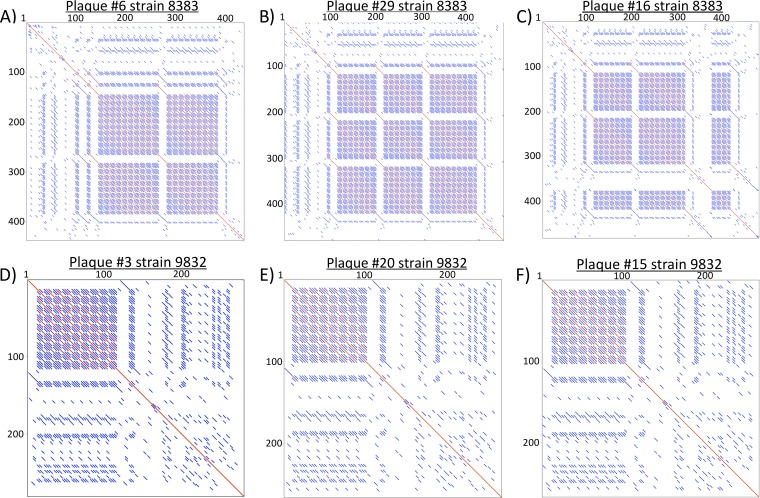
Sequencing analysis and dot plot comparisons of sequence data confirmed that Sb-1P and Sb1A clones picked from phage-susceptible strain MRSN8383 had highly diverse region 2 genotypes (see Fig. 5A to C), while the Sb-1M host range mutants selected on the previously resistant strain show a single dominant genotype (clone) and roughly four minor genotype variants (Fig. 5D to F). The extent of allele diversity at the hypervariable locus correlated with host range. Given the stability observed in this complex repeat region when grown on the previously resistant host, it is feasible that selection may occur at this particular locus that impacts viral fitness on that specific host. Regions 1 and 3 yielded uniform DNA sequence in clones from Sb-1P and Sb-1A (data not shown).
FIG 5.
Dot plot analysis. Dot plots illustrating the region 2 repeat structure of individual clones derived from the dominant genotype of Sb-1A phage plated on the phage-susceptible strain MRSN8383 (A to C) and from the dominant genotype of Sb-1M plated on the previously phage-resistant strain MRSN9832 (D to F).
Diversity generation during phage propagation from a single plaque.
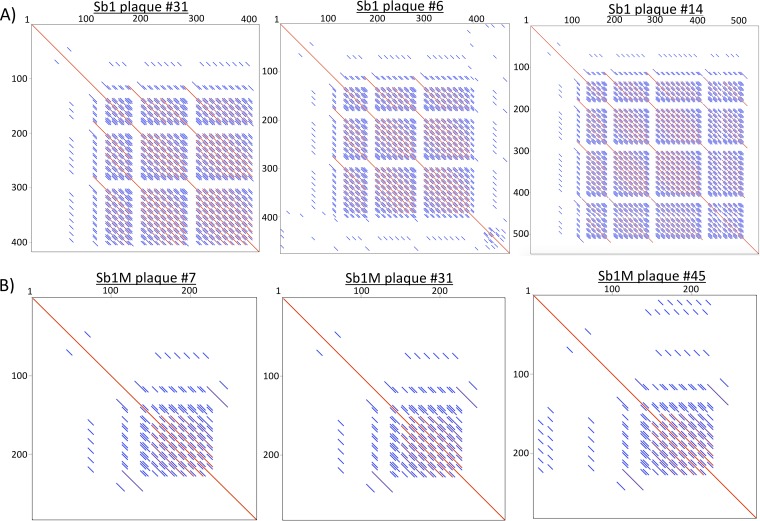
We determined whether progeny of individual clones of Sb-1P or Sb-1M generated diverse alleles in regions 1 to 3 during their propagation in broth culture, per the procedure described in Materials and Methods. For each stock, regions 1 to 3 from 45 randomly selected individual plaques were amplified by PCR, analyzed for fragment size variation, and sequenced. PCR banding patterns (Fig. 6) and dot plot analysis of sequenced amplicons (Fig. 7) consistently demonstrated that Sb-1P exhibited marked allelic diversity only in the region 2 locus and not in regions 1 or 3 (data not shown), while Sb-1M showed only a single allelic state at the region 2 repeat locus (Fig. 6B and 7B).
FIG 6.
PCR fragment size variation at the region 2 locus for isolated plaque clones of Sb-1P (A) and of Sb-1M (B). Control lanes (ctrl) show the PCR product of region 2 of the Sb-1P production stock without plaque/clone purification.
FIG 7.
Dot plot comparisons of region 2 locus sequences among Sb-1P (A) and Sb-1M (B) plaques.
Thus, the only consistent genetic variation observed between the parental Sb-1P phage and the expanded host range Sb-1M phage were the host-expansion-specific allele states observed at the region 2 locus. Whether the observed variations in the complex repeat architecture at this locus alter viral fitness is unknown. An expanded annotation search for functionality of this specific sequence did not indicate significant homologies. Region 2 is positioned upstream of the right terminus and immediately adjacent to a regulatory region that we speculate could impact the transcription of select R-LTR genes. The presence of variable LTRs that exhibit multiple tandemly repeated regions flanking the central nonrepetitive region of the genome is a shared feature of staphylococcal phages in the Spounavirinae subfamily (38). The region 2 locus is positioned at the border region of the right LTR and the nonrepetitive genome region adjacent to a palindromic sequence (36). The region 2 locus displays extensive variation among other representative Staphylococcus phage genomes characterized by the presence of repeat structure variation as well as by the presence of short hypothetical proteins (data not shown). The frequency of the singular Sb-1M region 2 allele state observed over subsequent host-specific passages argues against this being the result of neutral mutation. We speculate that the variation observed at the region 2 hypervariable repeat locus may provide an increase in fitness related to host range phenotype. The similar host adsorption efficiencies observed between Sb-1P and Sb-1M suggest a lack of differential receptor-mediated host range specificity, which would be consistent with this hypothesis. Alternatively, the observed variation in repeat structure may impart changes in gene expression of adjacent open reading frames that may participate in mediating host range. Such mechanisms are speculative, but the data reported here could guide future experimental efforts to dissect the basis of host range specificity. The basis for the novel allele variation observed in the Sb-1 phage genome that distinctly exhibits a unified allele state coinciding with host range expansion requires further investigation. Identifying potential functional consequences arising from mutations such as the ones described here that occur during host adaptation may enable the determination of the genetic requisites of host range expansion in Sb-1 and other Twort-like phages.
Phage Sb-1 has been used for decades for therapy of multiple types of S. aureus infections in Georgia and other FSU countries. The host range of Sb-1 was known to be expandable in the laboratory using a so-called adaptation procedure, but the genetic basis of this host range expansion was previously unknown. In this work, a hypervariable region (region 2) that is also present in other Twort-like phages was found in the genome of Sb-1. We showed that laboratory adaption of Sb-1 to expand host range to include previously resistant S. aureus strains produced a mixture of clones, including host range mutants (Sb-1M). The region 2 locus was variable in sequence in the parental phage preparation (Sb-1P) but exhibited a stable distinct allele in host range mutants (Sb-1M) that correlated with phenotypic host range expansion. The results reported here can inform future studies to better understand the mechanisms of host range expansion in Twort-like phages such as Sb-1.
MATERIALS AND METHODS
Bacterial strains, phage, and growth media.
S. aureus strains used in this work are presented in Table 1. The first 35 strains listed in Table 1 were received from the Multidrug-Resistant Organism Repository and Surveillance Network (MRSN; Bacterial Diseases Branch, Walter Reed Army Institute of Research), and the rest belong to the culture collection of the Department of Bacteriophage Therapeutics (in the same branch). The cultures were grown at 37°C in Bacto heart infusion broth (HIB; Becton, Dickinson and Co., Franklin Lakes, NJ). Solid medium contained 1.5% (wt/vol) Bacto agar (Becton, Dickinson). Semisolid agar overlay contained 0.7% (wt/vol) of Bacto agar (Becton, Dickinson). All S. aureus cultures were stored at −80°C in HIB containing 10% glycerol. Phage Sb-1 and its derivatives were filter sterilized through a 0.22-μm membrane and stored at 4°C in HIB or sodium chloride-magnesium sulfate (SM) buffer (Teknova, Hollister, CA).
Bacteriophage titer determination, plaque assays, and propagation.
Phage dilution was done in SM buffer. Phage titer determination and plaque assays were performed by the double-layer agar method as described earlier (47) with overnight incubation at 37°C. Phages were propagated on methicillin-susceptible S. aureus (MSSA) strain MRSN8383 as follows. Phage stock lysate was added to 150 ml of an early-exponential-phase bacterial culture grown in HIB at a multiplicity of infection (MOI) of 0.01 and incubated in a 250-ml plastic Erlenmeyer flask (catalog no. 07-200-668; Fisher Scientific, Hampton, NH) at 37°C at 200 rpm until visible lysis occurred (for 3 to 5 h). Phage lysate was treated with chloroform added to a final concentration of 5% (vol/vol) in order to destroy all remaining live bacteria. Bacterial debris was removed by 15-min centrifugation at 5,000 × g. The supernatant was filtered through a sterile 0.22-μm membrane.
Phage host range test.
Phage host ranges were determined using a micro-spot modification of an efficiency of plating assay. Briefly, 10-fold serial dilutions of phage were prepared in a sterile flat-bottomed 96-well plate. An aliquot (2 μl) of each phage dilution, ranging from 10−1 to 10−8, was spotted using a multichannel pipette on semisolid HIB agar overlay infused with S. aureus culture and incubated overnight at 37°C. The results were scored, and the morphology of individual plaques was evaluated. The efficiency of plating (EOP) of phage on each bacterial strain was calculated as phage titer on the test strain divided into the titer of the same phage on MSSA strain MRSN8383, which is consistently susceptible to phage Sb-1.
Host range expansion (adaptation procedure).
Phage adaptation was performed by several rounds of coincubation of phage Sb-1P with initially phage-resistant strain S. aureus MRSN9832 in liquid medium (34). Briefly, 109 PFU of phage in 0.1 ml of SM buffer and 0.1 ml of overnight culture of MRSN9832 were added to 5 ml of HIB in a 50-ml conical centrifuge tube (catalog no. 06-443-18; Fisher Scientific) and incubated for 24 h at 37°C with shaking at 100 rpm. Bacterial cells were pelleted by 15-min centrifugation at 5,000 × g, 1 ml of supernatant and 0.1 ml of overnight culture were added to 5 ml of HIB, and the second round of the procedure was performed. Slight lysis of the culture was observed after the fifth round of coincubation, and the procedure was finished after six rounds. Bacterial debris was pelleted and supernatant was filter sterilized and considered Sb-1A. This preparation contained about 4 × 109 PFU/ml of phage capable of plating on MRSN9832.
Single-plaque diversity generation assay.
A single isolated phage plaque was picked and suspended in 0.7 ml of SM buffer, filtered through a sterile 0.22-μm membrane, and diluted to a final concentration of 300 to 1,000 PFU/ml. The presence of viable phage particles was confirmed by plating on S. aureus MRSN8383. Phage suspension (100 μl) containing 30 to 100 viable phage particles was added to 10 ml of early mid-exponential-phase culture of MRSN8383 and incubated for 16 h at 37°C in a shaker incubator at 200 rpm. The resulting phage lysate was plated again on MRSN8383 to get isolated plaques. Sb-1 genome regions 1, 2, and 3 (see Results and Discussion) from 45 randomly selected individual plaques were analyzed by PCR and sequenced by the Sanger dideoxynucleotide chain termination method (48). The experiment is presented schematically in Fig. S1.
Phage adsorption assay.
A phage adsorption assay was performed as described earlier (49–51), with some variations. Briefly, test strains were grown overnight in 5 ml of HIB supplemented with 5 mM CaCl2. HIB without bacteria was used as a negative control. An aliquot (1 ml) of a tested culture was centrifuged, and the pellet was resuspended in 0.5 ml of HIB with 5 mM CaCl2. Phage (50 μl, ∼108 PFU) was added to each culture and control. Samples were incubated for 15 min at room temperature to allow phage to adsorb and then were centrifuged for 4 min at 13,000 rpm. The resulting supernatant was filter sterilized to eliminate the residual bacteria with adsorbed phage. The supernatant was diluted from 100 to 10−3, and 10 μl of each dilution was spotted on a semisolid agar overlay infused with 0.1 ml of overnight culture of indicator strain MRSN8383. The negative control was treated the same way. The percentage of phage adsorbed by bacteria of each test strain was calculated by subtracting the phage titer in the test strain from the phage titer in the negative control and dividing the result by phage titer in the negative control. Adsorption assays were done in triplicate and averaged.
Isolation of phage genomic DNA for sequencing.
Isolation of phage genomic DNA was performed as previously described (52), with minor modifications. Briefly, Sb-1P and Sb-1M phage lysates propagated from individual purified plaques were centrifuged at 13,000 × g for 4 h and resuspended in SM buffer in 1/100 of the original volume. The concentrated lysate was treated for 1 h at 37°C with RNase A (60 μg/ml) and DNase I (20 μg/ml) to remove residual S. aureus DNA and RNA. Proteinase K and SDS were added to final concentrations of 50 μg/ml and 0.5%, respectively, and the mix was incubated for 1 h at 56°C. DNA was purified by phenol-chloroform extraction followed by overnight DNA precipitation with ethanol at −20°C, centrifugation and resuspension in nuclease-free water. DNA purity was confirmed by restriction analysis with BamHI, EcoRI, and XbaI endonucleases (New England BioLabs Inc., Ipswich, MA) using horizontal agarose gel electrophoresis (47).
Whole-genome sequencing and genome assembly.
Whole-genome sequences were determined for the Sb-1P phage picked from a single plaque and propagated on strain MRSN8383 (Sb-18383) as well as for the two Sb-1M host range mutants (Sb-1M6168 and Sb-1M9832) picked from single plaques and propagated on previously resistant strains MRSN6168 and MRSN9832, respectively. DNA sequencing was performed using the Illumina MiSeq desktop sequencer (Illumina Inc., San Diego, CA) with 600-cycle v3 reagents. Sequencing libraries were prepared with the HyperPlus PCR-free next-generation sequencing (NGS) kit (Kapa Biosystems, Wilmington, MA) containing 550 bp inserts. Paired-end sequencing reads were assembled using Velvet (https://www.ebi.ac.uk/~zerbino/velvet), CLC Bio (Qiagen, Germantown, MD), and Geneious 8.0.2 (Newark, NJ). Reference mapping and variant calling was performed using the 2010 Sb-1 reference genomic sequence (accession number HQ163896) using Geneious (51) with medium sensitivity and fine tuning at 5× iterations using trimmed sequences (yielding >100-fold coverage of each genome). Annotations were performed using Geneious and Sequin and using the 2010 Sb-1 genome as the reference. Geneious software was used for partial genome alignments and to analyze the physical organization of reference genomic sequences, including ORF content and synteny. Sequence repeat structures were analyzed using DNASTAR GeneQuest v14.1.0.
Analysis of staphylococcal DNA by pulsed-field gel electrophoresis.
A single colony of each S. aureus strain was inoculated in 5 ml of BHI and grown overnight. The culture was diluted with BHI to an optical density at 600 nm (OD600) of 0.9 to 1.1. The adjusted cell suspension (200 μl) was centrifuged at 13,000 rpm on an Eppendorf 5415C microcentrifuge for 4 min, and the pellet was resuspended in 300 μl of Tris-EDTA (TE) buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8.0). The cell suspension was equilibrated at 37°C for 10 min and treated with 4 μg of lysostaphin (Sigma-Aldrich Corp., St. Louis, MO). To make plugs, 500 μl of 1.8% SeaKem Gold agarose (Lonza Inc., Allendale, NJ) in TE buffer equilibrated to 55°C was added to each cell suspension, gently mixed, and dispensed into the well of plug mold. The solid plugs were incubated at 37°C for at least 4 h with 3 ml of EC lysis buffer (6 mM Tris HCl [pH 7.6], 1 M NaCl, 100 mM EDTA, 0.5% Brij 58, 0.2% sodium deoxycholate, and 0.5% sodium lauroylsarcosine) in a shaker incubator. After incubation, EC lysis buffer was discarded, and the plugs were rinsed 4 times in 4 ml of TE buffer for 30 min at 37°C. After washing, the plugs were stored in 4 ml of TE buffer. DNA was digested with SmaI enzyme (New England Biolabs [NEB]) in NEB 4 buffer (30 units per tube) at 25°C for at least 3 h. DNA fragments were resolved using 1% SeaKem Gold agarose in 0.5× Tris-borate-EDTA (TBE) buffer (45 mM Tris-borate; 1 mM EDTA) using a Chef Mapper apparatus (Bio-Rad, Hercules, CA) for 21 h at 200 V and 14°C, with a 5 to 40 sec linear ramp. After electrophoresis, gels were stained with EnviroSafe DNA/RNA stain (Helixx Technologies, Inc., Scarborough, ON, Canada) per the manufacturer’s protocol, rinsed, and photographed using a ChemiDoc XRS+ gel documentation system (Bio-Rad). Macrorestriction patterns were analyzed using BioNumerics version 7.6 (Applied Maths, Austin, TX) software based on the Dice similarity index (optimization, 5%; band matching tolerance, 0.7%). Cluster analysis was performed using unweighted pair group method with arithmetic mean (UPGMA) with 101 total entries (taxa) and normalized to Salmonella enterica serovar Braenderup H9812 strain standards.
Repeat region PCR.
Primers used for amplification of repeat regions of Sb-1 and Sb-1M are shown in Table 3. The primers were designed using Beacon Designer program (Premier Biosoft Int.) within unique flanking sequences for each repeat region. PCR was performed using Invitrogen Platinum PCR supermix (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s protocol. PCR fragments were resolved using a standard horizontal electrophoresis in 0.8% Tris-acetate-EDTA (TAE) agarose gel. Sb-1P and Sb-1M phages were plated on strains MRSN8383 and MRSN9832, respectively. In each case, 45 well-separated single plaques were picked up using sterile tips. Each plaque was resuspended in 1 ml of SM buffer and filter sterilized, and 1 μl of the suspension (ca. 100 PFU) was used as a template for each of three separate PCRs targeting three genome regions.
TABLE 3.
Primers used in this work
| Primer | Target region | Primer sequence |
|---|---|---|
| R1-F | 1 | CTGCTTCACTACGATTATCTTC |
| R1-R | 1 | GGAATGGATGAAACCTCTATA |
| R2-F | 2 | GAGGAAGATATAGAATGAGAACC |
| R2-R | 2 | GCTTCAGCATAGATTAACAGTAG |
| R3-R | 3 | CACAAGGCAATGAGGTAATC |
| R3-F | 3 | CAGGTAAATACTACTGATAGTGACG |
Statistical analysis.
Results of all EOP assays are presented as mean values of three independent biological replicas. Statistical significance was determined by one-way analysis of variance (ANOVA) analysis using a free online program at the website of Vassar College (53). P values of <0.05 were considered significant.
Data availability.
The nucleotide sequences of the phages reported here were submitted to GenBank under the accession numbers MN336261 (Sb-18383), MN336262 (Sb-1M6168), and MN336263 (Sb-1M9832).
Supplementary Material
ACKNOWLEDGMENTS
This material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the authors, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense.
This study was supported by the United States Defense Health Program under the administration of the Military Infectious Disease Research Program and the U.S. Army Medical Research and Materiel Command.
We thank Xiaozhe Huang and Dana M. Cash for their contributions to PFGE data for S. aureus strains and Akhil D. Reddy for his technical assistance. The Multidrug Resistant Organism Repository and Surveillance Network (MRSN) at the Walter Reed Army Institute of Research provided the initial diversity panel of 25 U.S. military MRSA isolates based on PFGE and the MSSA strains used in this work.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01209-19.
REFERENCES
- 1.Alekshun MN, Levy SB. 2007. Molecular mechanisms of antibacterial multidrug resistance. Cell 128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Chambers HF, Deleo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marston HD, Dixon DM, Knisely JM, Palmore TN, Fauci AS. 2016. Antimicrobial resistance. JAMA 316:1193–1204. doi: 10.1001/jama.2016.11764. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2014. Active bacterial core surveillance report: methicillin-resistant Staphylococcus aureus. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/abcs/reports-findings/survreports/mrsa14.html. [Google Scholar]
- 5.National Institute of Allergy and Infectious Diseases. 2014. NIAID’s antibacterial resistance program: current status and future directions. National Institute of Allergy and Infectious Diseases, North Bethesda, MD: https://www.niaid.nih.gov/sites/default/files/arstrategicplan2014.pdf. [Google Scholar]
- 6.Kutateladze M, Adamia R. 2010. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol 28:591–595. doi: 10.1016/j.tibtech.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Górski A, Międzybrodzki R, Weber-Dąbrowska B, Fortuna W, Letkiewicz S, Rogóż P, Jończyk-Matysiak E, Dąbrowska K, Majewska J, Borysowski J. 2016. Phage therapy: combating infections with potential for evolving from merely a treatment for complications to targeting diseases. Front Microbiol 7:1515. doi: 10.3389/fmicb.2016.01515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abedon ST. 2017. Bacteriophage clinical use as antibacterial “drugs”: utility and precedent. Microbiol Spectr 5. doi: 10.1128/microbiolspec.BAD-0003-2016. [DOI] [PubMed] [Google Scholar]
- 9.Alves DR, Gaudion A, Bean JE, Perez Esteban P, Arnot TC, Harper DR, Kot W, Hansen LH, Enright MC, Jenkins AT. 2014. Combined use of bacteriophage K and a novel bacteriophage to reduce Staphylococcus aureus biofilm formation. Appl Environ Microbiol 80:6694–6703. doi: 10.1128/AEM.01789-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutierrez D, Vandenheuvel D, Martinez B, Rodriguez A, Lavigne R, Garcia P. 2015. Two phages, phiIPLA-RODI and phiIPLA-C1C, lyse mono- and dual-species staphylococcal biofilms. Appl Environ Microbiol 81:3336–3348. doi: 10.1128/AEM.03560-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuzaki S, Yasuda M, Nishikawa H, Kuroda M, Ujihara T, Shuin T, Shen Y, Jin Z, Fujimoto S, Nasimuzzaman MD, Wakiguchi H, Sugihara S, Sugiura T, Koda S, Muraoka A, Imai S. 2003. Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage φMR11. J Infect Dis 187:613–624. doi: 10.1086/374001. [DOI] [PubMed] [Google Scholar]
- 12.Takemura-Uchiyama I, Uchiyama J, Osanai M, Morimoto N, Asagiri T, Ujihara T, Daibata M, Sugiura T, Matsuzaki S. 2014. Experimental phage therapy against lethal lung-derived septicemia caused by Staphylococcus aureus in mice. Microbes Infect 16:512–517. doi: 10.1016/j.micinf.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Zheng P, Ji W, Fu Q, Wang H, Yan Y, Sun J. 2016. SLPW: a virulent bacteriophage targeting methicillin-resistant Staphylococcus aureus in vitro and in vivo. Front Microbiol 7:934. doi: 10.3389/fmicb.2016.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pincus NB, Reckhow JD, Saleem D, Jammeh ML, Datta SK, Myles IA. 2015. Strain specific phage treatment for Staphylococcus aureus infection is influenced by host immunity and site of infection. PLoS One 10:e0124280. doi: 10.1371/journal.pone.0124280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yilmaz C, Colak M, Yilmaz BC, Ersoz G, Kutateladze M, Gozlugol M. 2013. Bacteriophage therapy in implant-related infections: an experimental study. J Bone Joint Surg Am 95:117–125. doi: 10.2106/JBJS.K.01135. [DOI] [PubMed] [Google Scholar]
- 16.Wills QF, Kerrigan C, Soothill JS. 2005. Experimental bacteriophage protection against Staphylococcus aureus abscesses in a rabbit model. Antimicrob Agents Chemother 49:1220–1221. doi: 10.1128/AAC.49.3.1220-1221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seth AK, Geringer MR, Nguyen KT, Agnew SP, Dumanian Z, Galiano RD, Leung KP, Mustoe TA, Hong SJ. 2013. Bacteriophage therapy for Staphylococcus aureus biofilm-infected wounds: a new approach to chronic wound care. Plast Reconstr Surg 131:225–234. doi: 10.1097/PRS.0b013e31827e47cd. [DOI] [PubMed] [Google Scholar]
- 18.Takemura-Uchiyama I, Uchiyama J, Kato S, Inoue T, Ujihara T, Ohara N, Daibata M, Matsuzaki S. 2013. Evaluating efficacy of bacteriophage therapy against Staphylococcus aureus infections using a silkworm larval infection model. FEMS Microbiol Lett 347:52–60. doi: 10.1111/1574-6968.12220. [DOI] [PubMed] [Google Scholar]
- 19.Chhibber S, Kaur T, Sandeep K. 2013. Co-therapy using lytic bacteriophage and linezolid: effective treatment in eliminating methicillin resistant Staphylococcus aureus (MRSA) from diabetic foot infections. PLoS One 8:e56022. doi: 10.1371/journal.pone.0056022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendes JJ, Leandro C, Corte-Real S, Barbosa R, Cavaco-Silva P, Melo-Cristino J, Górski A, Garcia M. 2013. Wound healing potential of topical bacteriophage therapy on diabetic cutaneous wounds. Wound Repair Regen 21:595–603. doi: 10.1111/wrr.12056. [DOI] [PubMed] [Google Scholar]
- 21.Fish R, Kutter E, Wheat G, Blasdel B, Kutateladze M, Kuhl S. 2016. Bacteriophage treatment of intransigent diabetic toe ulcers: a case series. J Wound Care 25(Suppl 7):S27–S33. doi: 10.12968/jowc.2016.25.7.S27.26949862 [DOI] [Google Scholar]
- 22.Weber-Dabrowska B, Zimecki M, Kruzel M, Kochanowska I, Lusiak-Szelachowska M. 2006. Alternative therapies in antibiotic-resistant infection. Adv Med Sci 51:242–244. [PubMed] [Google Scholar]
- 23.Jikia D, Chkhaidze N, Imedashvili E, Mgaloblishvili I, Tsitlanadze G, Katsarava R, Glenn Morris J Jr, Sulakvelidze A. 2005. The use of a novel biodegradable preparation capable of the sustained release of bacteriophages and ciprofloxacin, in the complex treatment of multidrug-resistant Staphylococcus aureus-infected local radiation injuries caused by exposure to Sr90. Clin Exp Dermatol 30:23–26. doi: 10.1111/j.1365-2230.2004.01600.x. [DOI] [PubMed] [Google Scholar]
- 24.Rose T, Verbeken G, Vos DD, Merabishvili M, Vaneechoutte M, Lavigne R, Jennes S, Zizi M, Pirnay JP. 2014. Experimental phage therapy of burn wound infection: difficult first steps. Int J Burns Trauma 4:66–73. [PMC free article] [PubMed] [Google Scholar]
- 25.O’Flaherty S, Coffey A, Edwards R, Meaney W, Fitzgerald GF, Ross RP. 2004. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J Bacteriol 186:2862–2871. doi: 10.1128/JB.186.9.2862-2871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H. 2003. Prophage genomics. Microbiol Mol Biol Rev 67:238–276. doi: 10.1128/mmbr.67.2.238-276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavigne R, Darius P, Summer EJ, Seto D, Mahadevan P, Nilsson AS, Ackermann HW, Kropinski AM. 2009. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol 9:224. doi: 10.1186/1471-2180-9-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chibani-Chennoufi S, Bruttin A, Dillmann M-L, Brüssow H. 2004. Phage-host interaction: an ecological perspective. J Bacteriol 186:3677–3686. doi: 10.1128/JB.186.12.3677-3686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ackermann HW, Krisch HM, Comeau AM. 2011. Morphology and genome sequence of phage φ1402: a dwarf myovirus of the predatory bacterium Bdellovibrio bacteriovorus. Bacteriophage 1:138–142. doi: 10.4161/bact.1.3.15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen KC, Hair BB, Wienclaw TM, Murdock MH, Hatch JB, Trent AT, White TD, Haskell KJ, Berges BK. 2015. Isolation and host range of bacteriophage with lytic activity against methicillin-resistant Staphylococcus aureus and potential use as a fomite decontaminant. PLoS One 10:e0131714. doi: 10.1371/journal.pone.0131714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwan T, Liu J, DuBow M, Gros P, Pelletier J. 2005. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci U S A 102:5174–5179. doi: 10.1073/pnas.0501140102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia G, Wolz C. 2014. Phages of Staphylococcus aureus and their impact on host evolution. Infect Genet Evol 21:593–601. doi: 10.1016/j.meegid.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 33.Kutateladze M, Adamia R. 2008. Phage therapy experience at the Eliava Institute. Med Mal Infect 38:426–430. doi: 10.1016/j.medmal.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 34.Kvachadze L, Balarjishvili N, Meskhi T, Tevdoradze E, Skhirtladze N, Pataridze T, Adamia R, Topuria T, Kutter E, Rohde C, Kutateladze M. 2011. Evaluation of lytic activity of staphylococcal bacteriophage Sb-1 against freshly isolated clinical pathogens. Microb Biotechnol 4:643–650. doi: 10.1111/j.1751-7915.2011.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merabishvili M, Pirnay JP, Verbeken G, Chanishvili N, Tediashvili M, Lashkhi N, Glonti T, Krylov V, Mast J, Van Parys L, Lavigne R, Volckaert G, Mattheus W, Verween G, De Corte P, Rose T, Jennes S, Zizi M, De Vos D, Vaneechoutte M. 2009. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS One 4:e4944. doi: 10.1371/journal.pone.0004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deghorain M, Van Melderen L. 2012. The staphylococci phages family: an overview. Viruses 4:3316–3335. doi: 10.3390/v4123316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klumpp J, Lavigne R, Loessner MJ, Ackermann HW. 2010. The SPO1-related bacteriophages. Arch Virol 155:1547–1561. doi: 10.1007/s00705-010-0783-0. [DOI] [PubMed] [Google Scholar]
- 38.Łobocka M, Hejnowicz MS, Dąbrowski K, Gozdek A, Kosakowski J, Witkowska M, Ulatowska MI, Weber-Dąbrowska B, Kwiatek M, Parasion S, Gawor J, Kosowska H, Głowacka A. 2012. Genomics of staphylococcal Twort-like phages – potential therapeutics of the post-antibiotic era. Adv Virus Res 83:143–216. doi: 10.1016/B978-0-12-394438-2.00005-0. [DOI] [PubMed] [Google Scholar]
- 39.Stewart CR, Casjens SR, Cresawn SG, Houtz JM, Smith AL, Ford ME, Peebles CL, Hatfull GF, Hendrix RW, Huang WM, Pedulla ML. 2009. The genome of Bacillus subtilis bacteriophage SPO1. J Mol Biol 388:48–70. doi: 10.1016/j.jmb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andriashvili IA, Kvachadze LI, Adamiia R, Tushishvili DG, Kalandarishvili LL. 1981. Characteristics of the primary and secondary DNA structure of staphylococcal phage Sb-1. Vopr Virusol 1:100–103. [PubMed] [Google Scholar]
- 41.Tevdoradze E, Kvachadze L, Kutateladze M, Stewart CR. 2014. Bactericidal genes of staphylococcal bacteriophage Sb-1. Curr Microbiol 68:204–210. doi: 10.1007/s00284-013-0456-9. [DOI] [PubMed] [Google Scholar]
- 42.Kvachadze LI, Andriashvili IA, Chanishvili TG, Arutiunian EE, Nikol’skaia II. 1985. Modification of the modification-restriction system in staphylococci. Vopr Med Khim 31:121–125. [PubMed] [Google Scholar]
- 43.Andriashvili IA, Kvachadze LI, Bashakidze RP, Adamiia R, Chanishvili TG. 1986. Molecular mechanism of phage DNA protection from the restriction endonucleases of Staphylococcus aureus cells. Mol Gen Mikrobiol Virusol 8:43–45. [PubMed] [Google Scholar]
- 44.O’Flaherty S, Ross RP, Meaney W, Fitzgerald GF, Elbreki MF, Coffey A. 2005. Potential of the polyvalent anti-Staphylococcus bacteriophage K for control of antibiotic-resistant staphylococci from hospitals. Appl Environ Microbiol 71:1836–1842. doi: 10.1128/AEM.71.4.1836-1842.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelly D, McAuliffe O, Ross RP, O’Mahony J, Coffey A. 2011. Development of a broad-host-range phage cocktail for biocontrol. Bioeng Bugs 2:31–37. doi: 10.4161/bbug.2.1.13657. [DOI] [PubMed] [Google Scholar]
- 46.Alam CM, Iqbal A, Tripathi D, Sharfuddin C, Ali S. 2017. Microsatellite diversity and complexity in eighteen Staphylococcus phage genomes. Gene Cell Tissue 4:e14543. doi: 10.5812/gct.14543. [DOI] [Google Scholar]
- 47.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 48.Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu Z, Breidt F Jr, Fleming HP, Altermann E, Klaenhammer TR. 2003. Isolation and characterization of a Lactobacillus plantarum bacteriophage, φJL-1, from a cucumber fermentation. Int J Food Microbiol 84:225–235. doi: 10.1016/S0168-1605(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 50.Shao Y, Wang IN. 2008. Bacteriophage adsorption rate and optimal lysis time. Genetics 180:471–482. doi: 10.1534/genetics.108.090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwiatek M, Parasion S, Mizak L, Gryko R, Bartoszcze M, Kocik J. 2012. Characterization of a bacteriophage isolated from a cow with mastitis that is lytic against Staphylococcus aureus strains. Arch Virol 157:225–234. doi: 10.1007/s00705-011-1160-3. [DOI] [PubMed] [Google Scholar]
- 52.Farlow J, Filippov AA, Sergueev KV, Hang J, Kotorashvili A, Nikolich MP. 2014. Comparative whole genome analysis of six diagnostic brucellaphages. Gene 541:115–122. doi: 10.1016/j.gene.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 53.Lowry R. 2019. One-way analysis of variance for independent or correlated samples. http://vassarstats.net/anova1u.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nucleotide sequences of the phages reported here were submitted to GenBank under the accession numbers MN336261 (Sb-18383), MN336262 (Sb-1M6168), and MN336263 (Sb-1M9832).