Global spread of CTX-M-type extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae is a critical concern in both clinical and community settings. This dominance of CTX-M-type ESBL producers may be largely due to the successful international spread of epidemic clones, as represented by the extraintestinal pathogenic Escherichia coli (ExPEC) ST131. Our findings highlight the worrisome presence of diverse E. coli clones associated with humans, including ExPEC lineages harboring the most common blaCTX-M variants in untreated wastewater samples. Moreover, the chimeric genes blaCTX-M-64 and blaCTX-M-123, which have not yet been identified from human isolates of domestic origin in Japan, were identified. Exposure to untreated wastewater through combined sewer overflow caused by heavy rains derived from abnormal weather change could pose a risk for human health due to ingesting those antimicrobial-resistant bacteria.
KEYWORDS: wastewater, ESBL, CTX-M-64, CTX-M-123, WWTPs
ABSTRACT
The presence of antimicrobial-resistant bacteria and resistance genes in aquatic environments is a serious public health concern. This study focused on Escherichia coli possessing blaCTX-M genes in wastewater inflows. Twelve crude inflow water samples from wastewater treatment plant (WWTP) A and two samples each from three other WWTPs were collected in 2017 and 2018. A total of 73 E. coli isolates with 31 different sequence types (STs) harboring distinctive blaCTX-M gene repertoires were detected. In WWTP A influents, blaCTX-M-14 (14 isolates) was dominant, followed by blaCTX-M-15 (12 isolates) and blaCTX-M-27 (10 isolates). The chimeric blaCTX-M-64 and blaCTX-M-123 genes were each identified in one of the E. coli isolates from the same WWTP A inflow port. The blaCTX-M-27 gene was associated with five of seven B2-ST131 isolates, including three isolates of the B2-O25b-ST131-H30R/non-Rx lineage. One of the remaining two isolates belonged to the B2-O25b-ST131-H30R/Rx lineage harboring the blaCTX-M-15 gene. As for the B2-O25b-ST131-H30R/non-Rx lineage, two isolates with blaCTX-M-27 were recovered from each of the WWTP B and D influents, and one isolate with blaCTX-M-174 was also recovered from WWTP B influent. Whole-genome sequencing of chimeric blaCTX-M-harboring E. coli isolates revealed that the blaCTX-M-64 gene was integrated into the chromosome of ST10 E. coli B22 via ISEcp1-mediated transposition of a 9,467-bp sequence. The blaCTX-M-123-carrying IncI1 plasmid pB64 was 109,169 bp in length with pST108. The overall findings suggest that wastewater may act as a probable reservoir of clinically significant clonal lineages mediating antimicrobial resistance genes and chimeric genes that have not yet been identified from human isolates of domestic origin in Japan.
IMPORTANCE Global spread of CTX-M-type extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae is a critical concern in both clinical and community settings. This dominance of CTX-M-type ESBL producers may be largely due to the successful international spread of epidemic clones, as represented by the extraintestinal pathogenic Escherichia coli (ExPEC) ST131. Our findings highlight the worrisome presence of diverse E. coli clones associated with humans, including ExPEC lineages harboring the most common blaCTX-M variants in untreated wastewater samples. Moreover, the chimeric genes blaCTX-M-64 and blaCTX-M-123, which have not yet been identified from human isolates of domestic origin in Japan, were identified. Exposure to untreated wastewater through combined sewer overflow caused by heavy rains derived from abnormal weather change could pose a risk for human health due to ingesting those antimicrobial-resistant bacteria.
INTRODUCTION
The worldwide spread of extended-spectrum β-lactamase (ESBL)-producing bacteria, particularly Escherichia coli and Klebsiella pneumoniae, emerging as multidrug-resistant microbes in both clinical and community settings is a critical concern. The variations in the intestinal carriage of ESBL producers in healthy individuals have been observed among countries, especially in Africa and Asia (15 to 46%), including Japan (3 to 16%), compared to Europe (3 to 6%) and the Americas (2%) (1, 2). Asymptomatic intestinal carriage of ESBL producers could be linked to developing urinary tract and more severe invasive infections, contributing to the transmission of ESBL producers within the community as their reservoirs, as well as to the potential influx of ESBL producers into hospital settings from the community (3).
CTX-M type β-lactamases, which are the representative ESBLs, have been recognized as the most common community-derived ESBLs during the past decade (1). More than 220 different CTX-M enzymes have been deposited in GenBank to date. CTX-M-type ESBLs have been classified roughly into four groups based on their amino acid sequence similarities: CTX-M-1, CTX-M-2, CTX-M-9, and CTX-M-8/CTX-M-25. Among the CTX-M-type ESBLs exhibiting rapidly growing diversity in genotypes, the most predominant variants currently are CTX-M-15, which belongs to the CTX-M-1 group found in many countries across the world, including the United Kingdom, the United States, Canada, Africa, and India, and CTX-M-14, which belongs to the CTX-M-9 group reported especially in Asian countries, including Japan and China, as well as Spain (4–6). A high prevalence of CTX-M-1 has also been described in Italy and France. Notably, the recent rise of CTX-M-27, an Asp240Gly variant of CTX-M-14, exhibiting a higher ceftazidime MIC compared to CTX-M-14, has been documented globally (4, 5). The global dominance of CTX-M-15, CTX-M-14, and CTX-M-27 producers may be largely due to the successful international spread of epidemic clones, as represented by the extraintestinal pathogenic E. coli (ExPEC) ST131 clones belonging to phylogenetic group B2 with the specific serotype O25b:H4. This B2-O25b:H4-ST131 lineage harbors transferable plasmids mediating CTX-M-type ESBL genes, as well as other antimicrobial resistance genes, allowing their efficient dissemination among the family Enterobacteriaceae (4, 5).
There is growing concern that E. coli isolates producing CTX-M enzymes, particularly CTX-M-15 and CTX-M-14, have also been frequently detected in companion animals, food-producing animals and their meat, and environmental samples in many countries (7–11). Furthermore, a high prevalence of CTX-M-27 producers among companion animals and a relatively high incidence of CTX-M-55, an Ala77Val variant from CTX-M-15 among raw chicken meat in Japan, have been documented in our own recent studies (10, 11). The importance of the role of environmental sources in spreading ESBL producers into the community has been investigated (4); however, little is known regarding the occurrence of ESBL producers in aquatic environments in Japan.
We sought to investigate the resistance characteristics and genetic backgrounds of blaCTX-M-carrying E. coli recovered from untreated wastewater. Particularly noteworthy was the detection of blaCTX-M-64 and blaCTX-M-123 genes, encoding CTX-M-64 and CTX-M-123, respectively, chimeric genes of blaCTX-M-15 and blaCTX-M-14, despite the fact that there have been no reports so far of such chimeric genes from human isolates of domestic origin in Japan (12, 13).
RESULTS
E. coli clonal lineages and blaCTX-M gene repertoires detected from influents of municipal wastewater treatment plants (WWTPs) A, B, C, and D.
A total of 73 E. coli isolates comprising 31 different clonal lineages harboring distinct blaCTX-M gene repertoires were detected from WWTPs A, B, C, and D. Table 1 shows the genotypic characteristics of 50 E. coli isolates recovered from three inflow ports (A-1, A-2, and A-3) of WWTP A over 4 months. The dominant blaCTX-M genes found were blaCTX-M-14 (14 isolates), blaCTX-M-15 (12 isolates), and blaCTX-M-27 (10 isolates), which were followed by blaCTX-M-55 (5 isolates), blaCTX-M-3 (3 isolates), blaCTX-M-1 (1 isolate), blaCTX-M-2 (1 isolate), blaCTX-M-24 (1 isolate), and blaCTX-M-14 and blaCTX-M-15 (1 isolate). Of note, the chimeric genes blaCTX-M-64 and blaCTX-M-123 were each identified in one of the E. coli isolates from inflow port A-2 at 3-month intervals. The 50 E. coli isolates were assigned to 25 distinct STs, where seven isolates were found to be ST131 clone belonging to phylogroup B2. The blaCTX-M-27 gene was associated with five of these seven isolates of the clone B2-ST131, including three isolates of the B2-O25b-ST131-H30R/non-Rx lineage. As for the remaining two isolates of the clone B2-ST131, one isolate harboring blaCTX-M-15 was identified to be of the B2-O25b-ST131-H30R/Rx lineage. Six isolates belonged to one secondary dominant ST648 clone with phylogroup F though they harbored different blaCTX-M genes, blaCTX-M-14 (two isolates), and blaCTX-M-15, blaCTX-M-27, blaCTX-M-55, and blaCTX-M-14/15 (one isolate each). Six isolates of another secondary dominant ST10 clone belonged to three different phylogroups, including phylogroups A (two isolates, one isolate, and one isolate harboring blaCTX-M-15, blaCTX-M-55, and blaCTX-M-14, respectively), C (one isolate harboring blaCTX-M-64), and F (one isolate harboring blaCTX-M-14). All four isolates of the ST38 clone belonging to phylogroup D carried blaCTX-M-14. Two isolates harboring blaCTX-M-15 and blaCTX-M-3 each were identified to be the phylogroup B2-ST127 clone (Table 1). E. coli B2-ST131, F-ST648, A-ST10, D-ST38, and B2-ST127 clones were detected across different sampling months, inflow ports at WWTP A, and different WWTPs.
TABLE 1.
Genotypic characteristics of CTX-M-producing E. coli isolates recovered from WWTP A influents
| Date, no. of isolates (n = 50) | blaCTX-M gene | n | Inflow port A-1 (n = 20) |
Inflow port A-2 (n = 15) |
Inflow port A-3 (n = 15) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phylogroup | ST | Serotype O25b | Subclone/lineage | Phylogroup | ST | Serotype O25b | Subclone/lineage | Phylogroup | ST | Serotype O25b | Subclone/lineage | |||
| September 2017, 11 | blaCTX-M-27 | 3 | B2 | ST2252a | Non-O25b | Non-H30 | B2 | ST131 | Non-O25b | Non-H30 | ||||
| F | ST648 | |||||||||||||
| blaCTX-M-55 | 2 | A | ST10a,b | F | ST648 | |||||||||
| blaCTX-M-14 | 2 | F | ST648c | C | ST167c | |||||||||
| blaCTX-M-15 | 1 | B2 | ST127 | Non-O25b | Non-H30 | |||||||||
| blaCTX-M-3 | 1 | B1 | ST4720 | |||||||||||
| blaCTX-M-1 | 1 | B1 | ST2073-likea | |||||||||||
| blaCTX-M-64 | 1 | C | ST10 | |||||||||||
| October 2017, 15 | blaCTX-M-14 | 5 | D | ST38 | D | ST38 | ||||||||
| F | ST354c | F | ST354c | |||||||||||
| F | ST648 | |||||||||||||
| blaCTX-M-15 | 3 | A | ST10 | A | ST44c | |||||||||
| D | ST405 | |||||||||||||
| blaCTX-M-3 | 2 | B1 | ST156a,c | |||||||||||
| B2 | ST127 | Non-O25b | Non-H30 | |||||||||||
| blaCTX-M-55 | 2 | B1 | ST155 | D | ST5150 | |||||||||
| blaCTX-M-27 | 2 | B2 | ST131 | Non-O25b | Non-H30 | F | ST1722 | |||||||
| blaCTX-M-2 | 1 | F | ST117 | |||||||||||
| November 2017, 12 | blaCTX-M-14 | 5 | D | ST38 | F | ST393c | A | ST10c | ||||||
| B2 | ST131a | Non-O25b | Non-H30 | |||||||||||
| F | ST10c | |||||||||||||
| blaCTX-M-15 | 4 | A | ST43 | B1 | ST4681 | |||||||||
| A | ST617 | |||||||||||||
| F | ST648 | |||||||||||||
| blaCTX-M-24 | 1 | F | ST354c | |||||||||||
| blaCTX-M-27 | 1 | B2 | ST131 | O25b | H30R/non-Rx | |||||||||
| blaCTX-M-15 and blaCTX-M-14 | 1 | F | ST648a | |||||||||||
| December 2017, 12 | blaCTX-M-15 | 4 | A | ST10 | A | ST43 | A | ST43 | ||||||
| B2 | ST131a | O25b | H30R/Rx | |||||||||||
| blaCTX-M-27 | 4 | A | ST46a | B2 | ST131a | O25b | H30R/non-Rx | D | ST69a | |||||
| B2 | ST131b | O25b | H30R/non-Rx | |||||||||||
| blaCTX-M-14 | 2 | D | ST69c | D | ST38 | |||||||||
| blaCTX-M-55 | 1 | B2 | ST676 | O25b | Non-H30 | |||||||||
| blaCTX-M-123 | 1 | F | ST1674d | |||||||||||
Positive for tetA gene.
Positive for fosA3 gene.
Positive for tetB gene.
The isolate also carried the blaCMY-2 gene.
Table 2 shows genotypic characteristics of 23 E. coli isolates recovered from influents of WWTPs B, C, and D. The B2-O25b-ST131-H30R/non-Rx lineage with blaCTX-M-27 was identified in both samples from WWTPs B and D. Interestingly, despite belonging to the B2-O25b-ST131-H30R/non-Rx lineage, an isolate possessing blaCTX-M-174 was recognized. The B2-non-O25b-ST131-non-H30 lineage isolates with blaCTX-M-14 (two isolates) and blaCTX-M-27 (two isolates) were found from influents of WWTPs B/C and WWTP D, respectively. Four isolates harboring blaCTX-M-14 belonged to different sequence types (STs): ST38, ST58, ST69, and ST206.
TABLE 2.
Genotypic characteristics of CTX-M-producing E. coli isolates recovered from WWTP B, C, and D influents
| WWTP (mo yr) | No. of isolates (n = 23) | blaCTX-M gene | No. of specimens | Finding in the indicated location |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phylogroup | ST | Serotype O25b | Subclone/lineage | Phylogroup | ST | Serotype O25b | Subclone/lineage | ||||
| B (July 2018) | 7 | Inflow port B-1 (n = 5) | Sampling pointa B-2 (n = 2) | ||||||||
| blaCTX-M-55 | 2 | B1 | 58b | ||||||||
| E clade | 10b,c | ||||||||||
| blaCTX-M-14 | 1 | B2 | 131b | Non-O25b | Non-H30 | ||||||
| blaCTX-M-15 | 1 | D | 70e | ||||||||
| blaCTX-M-27 | 2 | B2 | 131 | O25b | H30R/non-Rx | B2 | 131b | O25b | H30R/non-Rx | ||
| blaCTX-M-174 | 1 | B2 | 131b | O25b | H30R/non-Rx | ||||||
| C (August 2018) | 4 | Inflow port C-1 (n = 2) | Sampling pointa C-2 (n = 2) | ||||||||
| blaCTX-M-14 | 2 | B2 | 131 | Non-O25b | Non-H30 | ||||||
| D | 38 | ||||||||||
| blaCTX-M-15 | 1 | F | 648 | ||||||||
| blaCTX-M-65 | 1 | B1 | 2179 | ||||||||
| D (September 2018) | 12 | Inflow port D-1 (n= 6) | Inflow port D-2 (n = 6) | ||||||||
| blaCTX-M-27 | 5 | B2 | 131b | Non-O25b | Non-H30 | B2 | 131 | Non-O25b | Non-H30 | ||
| B2 | 131b | O25b | H30R/non-Rx | B2 | 131 | O25b | H30R/non-Rx | ||||
| D | 2003 | ||||||||||
| blaCTX-M-14 | 4 | D | 69d | D | 38 | ||||||
| B1 | 58b | ||||||||||
| A | 206b | ||||||||||
| blaCTX-M-8 | 1 | B1 | 345 | ||||||||
| blaCTX-M-15 | 1 | D | 69b | ||||||||
| blaCTX-M-55 | 1 | B1 | 767b,c | ||||||||
The sample contained inflow water and return flows from sludge treatment.
Positive for tetA gene.
Positive for fosA3 gene.
Positive for tetB gene.
The isolate also carried the blaDHA-1 gene.
Prevalence of other antimicrobial resistance genes.
Among 73 E. coli isolates harboring blaCTX-M genes, the plasmid-mediated 16S rRNA methyltransferase genes armA, rmtB, and rmtC and the plasmid-mediated colistin resistance mcr genes mcr-1, mcr-2, and mcr-3 were not detected. The frequency of the tetracycline resistance genes tetA and tetB and the plasmid-mediated fosfomycin resistance gene fosA3 were as follows: 8 isolates were positive for tetA, 10 were positive for tetB, 1 was positive for fosA3, 1 was positive for tetA and tetB, and 1 was positive for tetA and fosA3 in WWTP A influents (Table 1); 9 isolates were positive for tetA, 1 was positive for tetB, and 2 were positive for tetA and fosA3 in WWTP B, C, and D influents (Table 2).
Susceptibility testing of chimeric blaCTX-M gene-harboring E. coli isolates.
The MICs of various antimicrobials for E. coli B22 harboring blaCTX-M-64, E. coli B64 harboring plasmid pB64 with blaCTX-M-123, and E. coli χ1037 transconjugant that acquired plasmid pB64 with blaCTX-M-123 are listed in Table 3. E. coli B22 did not yield cefotaxime (CTX)-resistant transconjugant or electrotransformant in multiple experiments. E. coli B22, E. coli B64, and E. coli χ1037 transconjugant that acquired plasmid pB64 were resistant to all the third-generation cephalosporins and to aztreonam, whereas they were susceptible to carbapenems. E. coli B22 had a cefotaxime MIC of >2,048 mg/liter and a ceftazidime MIC of 64 mg/liter. E. coli B64 and the E. coli χ1037 transconjugant that acquired plasmid pB64 had cefotaxime MICs of 2,048 mg/liter and ceftazidime MICs of 32 mg/liter. Consistent results for susceptibility profiles, including a cefotaxime MIC of >2,048 mg/liter and a ceftazidime MIC of 64 mg/liter, were observed for the E. coli χ1037 transconjugant harboring pUIH-1. Levofloxacin resistance in E. coli B22 was not observed in E. coli B64 and two E. coli χ1037 transconjugants.
TABLE 3.
MICs of antimicrobials for the CTX-M-64-producing E. coli strain B22 and CTX-M-123-producing E. coli B64 strain and its transconjugants
| Antimicrobial | MIC (mg/liter) |
||||
|---|---|---|---|---|---|
| B22 (blaCTX-M-64) | B64 (blaCTX-M-123) | χ1037 Rifr transconjugant (blaCTX-M-123/pB64) | χ1037 Rifr transconjugantb (blaCTX-M-64/pUIH-1) | χ1037 Rifr | |
| Piperacillin | >64 | >64 | >64 | >64 | ≤0.25 |
| SAMc | 8/16 | 8/16 | 8/16 | 8/16 | ≤2/4 |
| Cefazolin | >16 | >16 | >16 | >16 | 1 |
| Cefotiam | >16 | >16 | >16 | >16 | ≤0.25 |
| Cefotaximea | >2,048 | 2,048 | 2,048 | >2,048 | ≤0.25 |
| Ceftazidimea | 64 | 32 | 32 | 64 | ≤0.25 |
| Cefpodoxime | >4 | >4 | >4 | >4 | ≤1 |
| Cefepime | 8 | >16 | >16 | >16 | ≤0.25 |
| Flomoxef | ≤0.5 | 16 | ≤0.5 | ≤0.5 | ≤0.5 |
| Aztreonam | >16 | >16 | 16 | >16 | ≤0.5 |
| Imipenem | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 |
| Meropenem | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 |
| Gentamicin | ≤0.25 | 0.5 | ≤0.25 | 1 | ≤0.25 |
| Amikacin | 4 | 4 | ≤1 | 2 | ≤1 |
| Minocycline | 4 | 0.5 | 1 | 1 | 1 |
| Levofloxacin | >4 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 |
| Fosfomycin | ≤32 | ≤32 | ≤32 | ≤32 | ≤32 |
| SXTc | ≤9.5/0.5 | ≤9.5/0.5 | ≤9.5/0.5 | ≤9.5/0.5 | ≤9.5/0.5 |
The antibiotic-containing plates were prepared in-house.
As described by Nagano et al. (12).
SAM, ampicillin-sulbactam; SXT, trimethoprim-sulfamethoxazole.
Genetic features of blaCTX-M-64 gene-harboring E. coli B22.
Genome assembly of E. coli B22 yielded 273 contig sequences with the total assembly length of 5,084,128 bp, GC content of 50.6%, and an N50 of 131,142 bp. Gene annotation identified 4,670 protein-coding genes, 87 tRNA genes, and 7 rRNA genes. E. coli B22 belonged to the ST10 clone, O8:H17 serotype, and fimH41 subclone and contained the blaCTX-M-64 gene and substitutions conferring quinolone resistance, Ser80Ile in ParC and Ser83Leu/Asp87Asn in GyrA (Table 4). The virulence genes astA, cma, and gad were also detected.
TABLE 4.
WGS-based analysis of E. coli B22 harboring blaCTX-M-64 and E. coli B64 harboring blaCTX-M-123
| Genetic characteristics | E. coli B22 | E. coli B64 | |
|---|---|---|---|
| Antimicrobial resistance genes | Chromosome(s) | blaCTX-M-64, mdfA Ser80Ile in ParC, Ser83Leu and Asp87Asn in GyrA | blaCMY-2, mdfA |
| Plasmid(s) | blaTEM-1B | blaCTX-M-123, mphA | |
| Virulence genes | Chromosome | astA, gad | air, eilA, lpfA |
| Plasmid | cma | ||
| Plasmid replicon type(s) | IncFIB, IncFII, IncI1 | IncI1 | |
| MLST | ST10 | ST1674 | |
| Serotype | O8:H17 | O11:H25 | |
| fimH type | fimH41 | fimH138 | |
Next-generation sequencing (NGS) data of blaCTX-M-64-carrying plasmid UIH-1 generated 14 assembled contigs, which were subjected to gap-closing PCR and Sanger sequencing to determine the complete nucleotide sequence. The IncI2 plasmid UIH-1 was 62,194 bp, with a GC content of 42.1% and harboring 73 protein-coding genes.
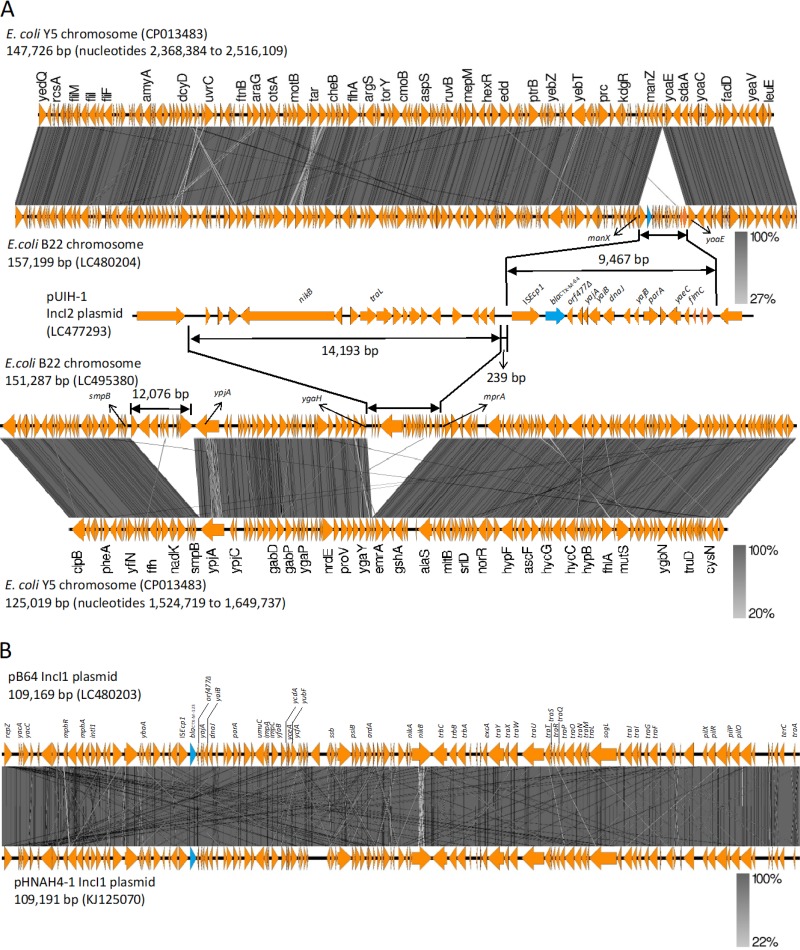
Comparison of the genetic structure of the region surrounding the blaCTX-M-64 gene revealed that a 9,467-bp sequence of ISEcp1-blaCTX-M-64-orf477Δ-yajA-yaiB-orf1-dnaJ-orf2-yajB-parA-orf3-yaeC-flmC-orf4-orf5-orf6 in pUIH-1 shared 99.99% identity with the 9,467-bp corresponding region of E. coli B22 (Fig. 1A). It is noteworthy that at least a 157-kb region of the E. coli B22 genomic sequence, with the exception of the above 9,467-bp region, exhibited a high degree of sequence identity (99.99%) with the 147.6-kb chromosomal sequence of E. coli Y5 (GenBank accession no. CP013483). The 9,467-bp region was flanked by 5-bp direct repeats (TATAA), which were integrated between open reading frames (ORFs), ManX (PTS system mannose-specific EIIAB component), and YoaE (inner membrane protein) of the E. coli B22 genomic sequence (Fig. 1A).
FIG 1.
Schematic representation of the chimeric genes. (A) Genome alignments of E. coli B22, including the 9,467-bp sequence of ISEcp1-blaCTX-M-64-orf477Δ-yajA-yaiB-orf1-dnaJ-orf2-yajB-parA-orf3-yaeC-flmC-orf4-orf5-orf6 (GenBank accession number LC480204) with E. coli Y5 (CP013483), and of E. coli B22, including 14,193-bp sequence immediately upstream region of the ISEcp1-blaCTX-M-64 (GenBank accession number LC495380) with E. coli Y5 (CP013483), and comparison of genetic environment flanking blaCTX-M-64 in E. coli B22 to that of plasmid pUHI from S. sonnei (GenBank accession number LC477293). (B) Sequence comparison between plasmid pB64 carrying blaCTX-M-123 (GenBank accession number LC480203) and pHNAH4-1 (KJ125070). The arrows show the translation orientation of the coding genes. Figure was generated using EasyFig (http://mjsull.github.io/Easyfig/).
Moreover, a 14,193-bp sequence immediately upstream region of the ISEcp1-blaCTX-M-64 in pUIH-1 shared 99.98% sequence identity with the 14,193-bp corresponding region of E. coli B22 (Fig. 1A). The 14,193-bp region was flanked by 5-bp direct repeats (TAAAT), which were integrated between ORFs MprA (negative regulator of the multidrug operon emrAB) and YgaH (uncharacterized protein) of the E. coli B22 genomic sequence (Fig. 1A).
Genetic features of the blaCTX-M-123 gene-harboring E. coli B64.
The WGS assembly of E. coli B64 contained 316 contigs, with a total length of 5,042,491 bp, a GC content of 50.3%, and an N50 of 199,993 bp. Gene annotation identified 4,544 protein-coding genes, 82 tRNA genes, and 8 rRNA genes. E. coli B64 belonged to the ST1674 clone, O11:H25 serotype, and fimH138 subclone and contained blaCTX-M-123 and blaCMY-2 resistance genes and air, eilA, lpfA virulence genes (Table 4).
NGS-based plasmid sequencing of blaCTX-M-123-carrying plasmid pB64 yielded 57 assembled contigs, which were subjected to gap-closing PCR and Sanger sequencing to determine the complete nucleotide sequence. The IncI1 plasmid pB64 with ST108 by pMLST was 109,169 bp, with a GC content of 50.6% harboring 124 protein-coding genes. The plasmid pB64 shared 99.97% sequence identity with that of pHNAH4-1 (IncI1 ST108, GenBank KJ125070) harbored by the E. coli ST746 isolate from chicken feces in China (14) (Fig. 1B).
The genetic context of blaCMY-2 was investigated by exploring ∼350-kbp fragments of the E. coli B64 genomic sequence. The results revealed that a 10,178-bp sequence, including ISEcp1-blaCMY-2-Δblc-yggR-Δtnp1-orf7-orf8-orf9-Δtnp2-ΔhsdR, sharing 100% identity with the 10,178-bp corresponding region of the E. coli CRE10 isolate from a human stool sample in Thailand (CP034404), was located on its chromosome. The macrolide resistance operon mphA-mrx-mphR located between two IS26 elements was associated with class 1 integrase gene intI1, which carried no class 1 integron gene cassettes.
DISCUSSION
Exposure to untreated wastewater could pose a risk for human health due to the ingestion of unrecognized antimicrobial-resistant bacteria. Particularly, the combined sewer overflow that has frequently occurred in recent years caused by localized heavy rains resulting from abnormal weather changes may diffuse those resistant organisms into the living environment. Thus, in the present study, the untreated wastewater samples flowing into inflow ports of WWTPs were investigated in order to gain better understanding into their probable risks for human health via exposure to clinically significant clonal lineages of ESBL-producing E. coli.
Despite the high overall genetic diversity of 31 STs among the 73 blaCTX-M-harboring E. coli isolates detected in untreated wastewater flows, clinically important lineages ST131 (16 isolates), ST10 (7 isolates), ST648 (7 isolates), ST38 (6 isolates), and ST69 (4 isolates) were noted at higher frequencies. The highly successful epidemic clone ST131, a major contributor to ExPEC infections in humans, has been broadly disseminated among nonhuman sources, such as companion animals, food animals, foods of animal origin, and environmental samples (15). Particularly, the global spread of the specific epidemic subclone B2-O25b-ST131-H30R mediating blaCTX-M-15, blaCTX-M-27, or blaCTX-M-14 poses a major public health threat because of its multidrug-resistant and potential virulent characteristics (16, 17). Recently, we have described the high prevalence of the blaCTX-M-27 gene among the pandemic B2-O25b-ST131-H30R/non-Rx lineage isolates from companion animals in Japan (10). The present study shows that this B2-O25b-ST131-H30R/non-Rx E. coli carrying blaCTX-M-27 was detected from influents of all WWTPs except those of the WWTP C, in addition to the B2-O25b-ST131-H30R/Rx E. coli carrying blaCTX-M-15 from WWTP A influent. Interestingly, blaCTX-M-174, a Glu7Leu variant of blaCTX-M-27 (only nucleotide sequence data are available in the GenBank database under the accession number KT997886), was also associated with the B2-O25b-ST131-H30R/non-Rx lineage, raising concerns regarding the future spread of this lineage carrying blaCTX-M-174 in both hospital and community settings. Moreover, in the present study, the blaCTX-M-27 was detected among several other lineages, such as B2-non-O25b-ST131-non-H30, B2-non-O25b-ST2252-non-H30, A-ST46, D-ST69, D-ST2003, F-ST648, and F-ST1722, although Matsumura et al. have reported that the blaCTX-M-27 was confined to the O25b-ST131-H30R/non-Rx lineage of human clinical isolates (17). Phylogroup D constitutes the second major ExPEC lineage after phylogroup B2. E. coli clones ST38, ST69, ST405, ST70, ST2003, and ST5150 were recognized among this phylogroup D, with ST38, which has been recognized to carry virulence determinants of both uropathogenic E. coli and enteroaggregative E. coli (18), being the most common (6 of 14 isolates).
Clinically relevant E. coli lineages, including globally circulating B2-ST131, F-ST648, A-ST10, D-ST38, and the recently emerged virulent clone B2-ST127, were detected across different sampling months and inflow ports of the WWTP A or across different WWTPs, suggesting these lineages have acquired the ability to adapt to the environment; this highlights the importance of untreated wastewater flow as an unrecognized crucial reservoir of these epidemic lineages.
The present study confirmed the overall predominance of phylogroups B2 and D among the blaCTX-M-harboring E. coli isolates derived from untreated domestic wastewater flows. Similar findings have been reported previously among ESBL-producing E. coli isolates from human commensal populations, as well as from WWTP effluents (19, 20). Nonetheless, this study newly identifies the blaCTX-M-174 gene associated with the B2-O25b-ST131-H30R/non-Rx lineage and the uncommon chimeric genes blaCTX-M-64 and blaCTX-M-123, which probably originated by recombination between blaCTX-M-14 and blaCTX-M-15. The blaCTX-M-64 was first identified by Nagano et al. as a chimeric ESBL gene in a Shigella sonnei strain UIH-1 recovered from a tourist who had returned from China (12). Subsequently, several chimeric blaCTX-M genes, including blaCTX-M-123, have been reported from pets, food animals, and humans, mostly in China (13, 21), and also recently in a German patient (22). The prevalence of the blaCTX-M-64 gene has been determined to be 6% among ESBL-producing E. coli isolates recovered from fecal samples from healthy children in Laos (23). In Japan, the occurrence of chimeric blaCTX-M genes has been associated with a travel-associated S. sonnei isolate from China for blaCTX-M-64 and with an E. coli isolate from a domestic pet dog for blaCTX-M-123; no chimeric genes from human isolates of domestic origin have yet been reported (12, 24).
CTX-M-64 and CTX-M-123, showing chimeric structures of a CTX-M-15 β-lactamase (N- and C-terminal moieties) and a CTX-M-14 β-lactamase (central portion), had amino acid sequence identities of 91.8 and 94.5%, respectively, with CTX-M-15, and of 88.0 and 85.2%, respectively, with CTX-M-14. The cefotaxime MICs for E. coli B22 producing CTX-M-64 and E. coli B64 producing CTX-M-123 were >2,048 mg/liter and 2,048 mg/liter, respectively, higher than those for E. coli transformants producing CTX-M-15 or CTX-M-14 reported previously (25). Also, it has been reported that these chimeric enzymes exhibited higher catalytic activities toward cefotaxime compared to CTX-M-15 and CTX-M-14, which was consistent with the MIC results (25).
The blaCTX-M-64 gene was integrated into the chromosome of ST10 E. coli B22 via ISEcp1-mediated transposition of a DNA fragment of 9,467-bp nucleotide sequence which showed 99.99% sequence identity to the corresponding sequence located on S. sonnei plasmid UIH-1. The distribution and high prevalence of chromosomally located blaCTX-M genes have been documented in Japan (26, 27). The stability of blaCTX-M genes integrated into the chromosome remains unclear, however, compared to the plasmid location; more stable retention of the chromosomally located antimicrobial resistance genes, allowing these organisms to live even in the absence of antimicrobial selection pressure, has been suggested. The occurrence of the chromosomal integration of blaCTX-M-64 among the clinically important ESBL lineage could pose public health concerns due to antimicrobial resistance spread from untreated wastewater through combined sewer overflow, resulting in contamination of the human environment. Intriguingly, a 14,193-bp nucleotide sequence immediately upstream region of the ISEcp1-blaCTX-M-64, exhibiting 99.98% sequence identity with the corresponding sequence located on plasmid UIH-1, was integrated into another site on the E. coli B22 chromosome. This study reports an integration of divided segments probably derived from a blaCTX-M-64-carrying IncI2 plasmid into E. coli chromosome at different sites. The mechanisms underlying this integration are unclear. Chromosome integration of the regions upstream and downstream of ISEcp1-blaCTX-M-64 may have occurred, followed by ISEcp1-mediated transposition of blaCTX-M-64 and its downstream fragments into another site on the chromosome. It is also possible that an independent integration of these 14,193- and 9,467-bp fragments occurs at different sites without significant homology on the chromosome following illegitimate recombination, as described previously (28).
The blaCTX-M-123 gene has mainly been reported among food animal and human isolates, including Shigella flexneri in China, followed by isolation only among pet animals in the United States and Japan (24, 29, 30). The present study reveals that the IncI1 plasmid pB64 mediating the blaCTX-M-123 gene harbored by E. coli F-ST1674 shares 99.97% sequence identity with the blaCTX-M-123-carrying IncI1 plasmid pHNAH4-1 (GenBank accession no. KJ125070) from a chicken E. coli isolate in China. E. coli ST1674 is extremely rare in published articles, but the EnteroBase database (http://enterobase.warwick.ac.uk) reports ST1674 isolates detected in domestic water sources. E. coli ST1674 with the narrow-host-range IncI1 plasmid, which enables the development of the virulence factor type IV pili that are essential for adhesion and invasion, is more likely to be well adapted to a water environment, which thus may contribute to the horizontal transfer of blaCTX-M genes.
In conclusion, we demonstrated here the occurrence and repeated detection of blaCTX-M genes harbored by clinically significant E. coli lineages, including ST131, which is responsible for the spread of blaCTX-M-15 and blaCTX-M-27 in wastewater flow. Our present findings also show the emergence of blaCTX-M-174, a variant of blaCTX-M-27 among the B2-O25b-ST131-H30R/non-Rx lineage and chimeric genes of blaCTX-M-64 and blaCTX-M-123 characterized by enhanced catalytic activity to cephalosporins, all of which have not yet been identified in human isolates of domestic origin in Japan. Thus, wastewater may retain clinically important clonal lineages with antimicrobial resistance genes and chimeric genes spreading into human communities, including health care settings.
MATERIALS AND METHODS
Collection of wastewater inflows.
A total of 12 crude inflow water samples were collected from WWTP A serving 125,000 people in the central area of Matsumoto City, Nagano Prefecture, Japan during September and December 2017; one sample each was collected from three different inflow ports A-1, A-2, and A-3, respectively, once a month for 4 months. In order to investigate whether there were any variations among ESBL-producing E. coli lineages or ESBL genes in wastewater samples depending on time and temperature, sampling on four occasions was performed in WWTP A. In addition, two samples each collected from B-1 (inflow water) and B-2 (inflow water plus return flows from sludge treatment) at WWTP B serving 162,000 people and C-1 (inflow water) and C-2 (inflow water plus return flows from sludge treatment) at WWTP C serving 134,000 people in Nagano City, and inflow ports D-1 and D-2 at WWTP D serving 95,000 people in Azumino City during July and September 2018 were included in our analyses.
Wastewater samples (∼500 ml) collected in sterile glass bottles were transported to the laboratory under cooling conditions and were stored at 4°C until their utilization within 3 h after sampling.
Isolation of the cefotaxime-resistant E. coli and screening of ESBL production.
Portions (1 ml) of well-mixed inflow samples were added to 9 ml of brilliant green lactose bile broth (BGLB broth; Eiken Chemical Co., Tokyo, Japan). After overnight incubation at 37°C, 10 μl of the broth was plated onto MacConkey agar (Eiken) supplemented with 2 mg/liter cefotaxime (CTX; Sigma-Aldrich Japan, Tokyo, Japan) and incubated overnight at 37°C. Several different colonies showing the expected morphological appearance of E. coli were subjected to matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Bruker Daltonics Japan, Yokohama, Japan) analysis. As recommended by the manufacturer, score cutoffs of ≥2.000 were applied for species-level identification.
Phenotypic screening of ESBL production on the basis of inhibitory effect of clavulanic acid was performed on Mueller-Hinton agar (Eiken) using CTX (30 μg; Eiken) and ceftazidime (30 μg; Eiken) disks, each alone and in combination with clavulanic acid (10 μg; Eiken) according to Clinical and Laboratory Standards Institute (CLSI) guidelines (31).
Genotype analysis of blaCTX-M genes.
ESBL genes of blaCTX-M-1, blaCTX-M-2, blaCTX-M-8, and blaCTX-M-9 groups were detected by PCR with specific primers (11). The nucleotide sequences of the structural genes were determined by direct sequencing of PCR products by using BigDye Terminator v1.1 cycle sequencing kit and an ABI 3730xl genetic analyzer (Applied Biosystems, Foster City, CA) (11). The sequence results were analyzed using the NCBI BLAST program (https://www.ncbi.nlm.nih.gov/blast).
Molecular typing of E. coli.
Phylotyping was performed using the Clermont multiplex PCR method with specific primers targeting arpA, chuA, yjaA, and TspE4.C2 (32).
Multilocus sequence typing (MLST) was carried out with seven housekeeping genes—adk, fumC, gyrB, icd, mdh, purA, and recA—and the sequence type (ST) was assigned according to the E. coli MLST website (http://enterobase.warwick.ac.uk/species/ecoli/allele_st_search). If E. coli isolates sharing the same blaCTX-M genotypes, phylogroups, and STs were detected from the same samples, one representative isolate was included in the study data.
For E. coli isolates belonging to the phylogroup B2, the O25b molecular subtype was identified by PCR as described previously (33). Those isolates were further characterized; the fimH30 (H30) subclone was detected by PCR, and ciprofloxacin-resistant H30 isolates were identified as H30R (34), and the H30Rx lineage within H30R subclone was identified by sequencing of PCR product for detection of the H30Rx-specific ybbW SNP G723A (35).
PCR detection of other resistance genes.
For E. coli isolates harboring blaCTX-M genes, the plasmid-mediated 16S rRNA methyltransferase genes armA, rmtB, and rmtC and the plasmid-mediated fosfomycin resistance gene fosA3 were detected as described previously (10). Tetracycline resistance genes tetA, tetB, and tetM were detected by PCR with the following primers: tetA, 5′-GTAATTCTGAGCACTGTCGC-3′ and 5′-TCAGCGATCGGCTCGT-3′; tetB, 5′-GTTCGACAAAGATCGCATTGG-3′ and 5′-AATCCAAATCCAGCCATCCCA-3′; and tetM, 5′-TAGCTCATGTTGATGCAGGA-3′ and 5′-ATCCGACTATTTGGACGACGG-3′. The plasmid-mediated colistin resistance mcr genes mcr-1, mcr-2, and mcr-3 were detected by PCR as described previously (36–38).
Analysis of chimeric blaCTX-M-64- and blaCTX-M-123-harboring E. coli.
Whole-genome sequencing of blaCTX-M-64-harboring E. coli B22 and blaCTX-M-123-harboring E. coli B64, which were detected from influents at inflow port A-2 of WWTP A in September and December 2017, respectively, was performed. Genomic DNA was extracted by using a Wizard genomic DNA purification kit (Promega, Madison, WI). Sequencing libraries were prepared using the NEBNext Ultra DNA LibraryPrep kit (New England BioLabs, Ipswich, MA), and 150-bp paired-end sequencing was performed on the HiSeq platform (Illumina, San Diego, CA). De novo assembly was conducted using an A5-MiSeq Assembly pipeline with the default parameters (39). MLSTs, serotypes, fimH types, plasmid replicon types, antimicrobial resistance genes, and virulence genes were analyzed using tools available from the Center for Genomic Epidemiology (CGE; http://www.genomicepidemiology.org). Annotation of the resulting scaffolds was performed by using the DDBJ Fast Annotation and Submission Tool (DFAST; https://dfast.nig.ac.jp/) (40).
NGS-based plasmid sequencing of the plasmid pB64 carrying blaCTX-M-123 from E. coli χ1037 transconjugants (donor E. coli B64) was performed. Plasmid pUIH-1 carrying blaCTX-M-64 from E. coli χ1037 transconjugants obtained from the conjugation with Shigella sonnei strain UIH-1 was also analyzed for sequence comparison purposes (12). The plasmid DNA was extracted using the PureYield plasmid midiprep kit (Promega) and was subjected to pulsed-field gel electrophoresis. Plasmid DNA excised from the gel was used to prepare sequencing libraries using a Nextera XT DNA sample preparation kit (Illumina, San Diego, CA). Sequencing with 150-bp paired-end reads was performed on a MiSeq platform (Illumina). The contigs obtained after de novo assembly as described above were submitted to the DFAST for annotation. Plasmid MLSTs, plasmid replicon types, and antimicrobial resistance genes were analyzed using CGE web tools as described above.
Antimicrobial susceptibility testing of E. coli B22, E. coli B64, and E. coli χ1037 transconjugants that acquired plasmid pB64 carrying blaCTX-M-123 was conducted by the broth microdilution method using Dry Plate Eiken (Eiken), and the results were interpreted by using the CLSI breakpoints (31). Alternatively, for cefotaxime and ceftazidime, broth microdilution panels prepared in-house were used to provide a broader range of antimicrobial concentrations for evaluation of the MICs. For comparative purposes, E. coli χ1037 transconjugant with plasmid pUIH-1 carrying blaCTX-M-64 as described previously was also subjected to MIC determination (12).
Data availability.
Two nucleotide sequences of E. coli B22 have been deposited in GenBank under accession numbers LC480204 (157,199 bp) and LC495380 (151,287 bp). The complete nucleotide sequences of pB64 and pUIH-1 have been deposited in the GenBank database under accession numbers LC480203 and LC477293, respectively.
ACKNOWLEDGMENTS
We declare no conflicts of interest.
This study was supported by JSPS KAKENHI grant JP18K08428.
REFERENCES
- 1.Karanika S, Karantanos T, Arvanitis M, Grigoras C, Mylonakis E. 2016. Fecal colonization with extended-spectrum β-lactamase-producing Enterobacteriaceae and risk factors among healthy individuals: a systematic review and meta-analysis. Clin Infect Dis 63:310–318. doi: 10.1093/cid/ciw283. [DOI] [PubMed] [Google Scholar]
- 2.Nakane K, Kawamura K, Goto K, Arakawa Y. 2016. Long-term colonization by blaCTX-M-harboring Escherichia coli in healthy Japanese people engaged in food handling. Appl Environ Microbiol 82:1818–1827. doi: 10.1128/AEM.02929-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Ami R, Schwaber MJ, Navon-Venezia S, Schwartz D, Giladi M, Chmelnitsky I, Leavitt A, Carmeli Y. 2006. Influx of extended-spectrum β-lactamase-producing Enterobacteriaceae into the hospital. Clin Infect Dis 42:925–934. doi: 10.1086/500936. [DOI] [PubMed] [Google Scholar]
- 4.Chong Y, Shimoda S, Shimono N. 2018. Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet Evol 61:185–188. doi: 10.1016/j.meegid.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Bevan ER, Jones AM, Hawkey PM. 2017. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother 72:2145–2155. doi: 10.1093/jac/dkx146. [DOI] [PubMed] [Google Scholar]
- 6.Zhao WH, Hu ZQ. 2013. Epidemiology and genetics of CTX-M extended-spectrum β-lactamases in Gram-negative bacteria. Crit Rev Microbiol 39:79–101. doi: 10.3109/1040841X.2012.691460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang PLC, Shen X, Chalmers G, Reid-Smith RJ, Slavic D, Dick H, Boerlin P. 2018. Prevalence and mechanisms of extended-spectrum cephalosporin resistance in clinical and fecal Enterobacteriaceae isolates from dogs in Ontario, Canada. Vet Microbiol 213:82–88. doi: 10.1016/j.vetmic.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 8.Kawamura K, Nagano N, Suzuki M, Wachino J, Kimura K, Arakawa Y. 2017. ESBL-producing Escherichia coli and its rapid rise among healthy people. Food Safety 5:122–150. doi: 10.14252/foodsafetyfscj.2017011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canton R, Gonzalez-Alba JM, Galan JC. 2012. CTX-M enzymes: origin and diffusion. Front Microbiol 3:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maeyama Y, Taniguchi Y, Hayashi W, Ohsaki Y, Osaka S, Koide S, Tamai K, Nagano Y, Arakawa Y, Nagano N. 2018. Prevalence of ESBL/AmpC genes and specific clones among the third-generation cephalosporin-resistant Enterobacteriaceae from canine and feline clinical specimens in Japan. Vet Microbiol 216:183–189. doi: 10.1016/j.vetmic.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi W, Ohsaki Y, Taniguchi Y, Koide S, Kawamura K, Suzuki M, Kimura K, Wachino J-I, Nagano Y, Arakawa Y, Nagano N. 2018. High prevalence of blaCTX-M-14 among genetically diverse Escherichia coli recovered from retail raw chicken meat portions in Japan. Int J Food Microbiol 284:98–104. doi: 10.1016/j.ijfoodmicro.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Nagano Y, Nagano N, Wachino J, Ishikawa K, Arakawa Y. 2009. Novel chimeric β-lactamase CTX-M-64, a hybrid of CTX-M-15-like and CTX-M-14 β-lactamases, found in a Shigella sonnei strain resistant to various oxyimino-cephalosporins, including ceftazidime. Antimicrob Agents Chemother 53:69–74. doi: 10.1128/AAC.00227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He D, Partridge SR, Shen J, Zeng Z, Liu L, Rao L, Lv L, Liu J-H. 2013. CTX-M-123, a novel hybrid of the CTX-M-1 and CTX-M-9 group β-lactamases recovered from Escherichia coli isolates in China. Antimicrob Agents Chemother 57:4068–4071. doi: 10.1128/AAC.00541-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L, He D, Lv L, Liu W, Chen X, Zeng Z, Partridge SR, Liu J-H. 2015. blaCTX-M-1/9/1 hybrid genes may have been generated from blaCTX-M-15 on an IncI2 plasmid. Antimicrob Agents Chemother 59:4464–4470. doi: 10.1128/AAC.00501-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicolas-Chanoine MH, Bertrand X, Madec JY. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathers AJ, Peirano G, Pitout JD. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumura Y, Johnson JR, Yamamoto M, Nagao M, Tanaka M, Takakura S, Ichiyama S. 2015. CTX-M-27- and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J Antimicrob Chemother 70:1639–1649. doi: 10.1093/jac/dkv017. [DOI] [PubMed] [Google Scholar]
- 18.Chattaway MA, Jenkins C, Ciesielczuk H, Day M, DoNascimento V, Day M, Rodríguez I, van Essen-Zandbergen A, Schink A-K, Wu G, Threlfall J, Woodward MJ, Coldham N, Kadlec K, Schwarz S, Dierikx C, Guerra B, Helmuth R, Mevius D, Woodford N, Wain J. 2014. Evidence of evolving extraintestinal enteroaggregative Escherichia coli ST38 clone. Emerg Infect Dis 20:1935–1937. doi: 10.3201/eid2011.131845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura A, Komatsu M, Noguchi N, Ohno Y, Hashimoto E, Matsutani H, Abe N, Fukuda S, Kohno H, Nakamura F, Matsuo S, Kawano S. 2016. Analysis of molecular epidemiologic characteristics of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli colonizing feces in hospital patients and community dwellers in a Japanese city. J Infect Chemother 22:102–107. doi: 10.1016/j.jiac.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Gomi R, Matsuda T, Matsumura Y, Yamamoto M, Tanaka M, Ichiyama S. 2017. Occurrence of clinically important lineages, including the sequence type 131 C1-M27 subclone, among extended-spectrum-β-lactamase-producing Escherichia coli in wastewater. Antimicrob Agents Chemother 61:e00564-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia S, Fan X, Huang Z, Xia L, Xiao M, Chen R, Xu Y, Zhuo C. 2014. Dominance of CTX-M-type extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolated from patients with community-onset and hospital-onset infection in China. PLoS One 9:e100707. doi: 10.1371/journal.pone.0100707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeifer Y, Werner G, Körber-Irrgang B, Jonas D, Kresken M. 2018. Escherichia coli ST1421 harboring the hybrid extended-spectrum β-lactamase CTX-M-64 from a German patient. J Glob Antimicrob Resist 12:167–168. doi: 10.1016/j.jgar.2018.01.025. [DOI] [PubMed] [Google Scholar]
- 23.Stoesser N, Xayaheuang S, Vongsouvath M, Phommasone K, Elliott I, Del Ojo Elias C, Crook DW, Newton PN, Buisson Y, Lee SJ, Dance DAB. 2015. Colonization with Enterobacteriaceae producing ESBLs in children attending pre-school childcare facilities in the Lao People’s Democratic Republic. J Antimicrob Chemother 70:1893–1897. doi: 10.1093/jac/dkv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawamura K, Sugawara T, Matsuo N, Hayashi K, Norizuki C, Tamai K, Kondo T, Arakawa Y. 2017. Spread of CTX-type extended-spectrum β-lactamase-producing Escherichia coli isolates of epidemic clone B2-O25-ST131 among dogs and cats in Japan. Microb Drug Resist 23:1059–1066. doi: 10.1089/mdr.2016.0246. [DOI] [PubMed] [Google Scholar]
- 25.He D, Chiou J, Zeng Z, Liu L, Chen X, Zeng L, Chan EWC, Liu J-H, Chen S. 2015. Residues distal to the active site contribute to enhanced catalytic activity of variant and hybrid β-lactamases derived from CTX-M-14 and CTX-M-15. Antimicrob Agents Chemother 59:5976–5983. doi: 10.1128/AAC.04920-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirai I, Fukui N, Taguchi M, Yamauchi K, Nakamura T, Okano S, Yamamoto Y. 2013. Detection of chromosomal blaCTX-M-15 in Escherichia coli O25b-B2-ST131 isolates from the Kinki region of Japan. Int J Antimicrob Agents 42:500–506. doi: 10.1016/j.ijantimicag.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Hamamoto K, Hirai I. 2019. Characterization of chromosomally located blaCTX-M and its surrounding sequence in CTX-M-type extended-spectrum β-lactamase-producing Escherichia coli isolates. J Glob Antimicrob Resist 17:53–57. doi: 10.1016/j.jgar.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Richardson PT, Park SF. 1997. Integration of heterologous plasmid DNA into multiple sites on the genome of Campylobacter coli following natural transformation. J Bacteriol 179:1809–1812. doi: 10.1128/jb.179.5.1809-1812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu G, Qian H, Tang B, Chen Y, Kang H, Jiang F, Ma P, Gu B, Huo X. 2018. Prevalence and characterization of third-generation cephalosporin-resistant Shigella flexneri isolates from Jiangsu Province, China, 2013-2015. J Glob Antimicrob Resist 15:283–287. doi: 10.1016/j.jgar.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Thungrat K, Boothe DM. 2016. Occurrence of OXA-48 carbapenemase and other β-lactamase genes in ESBL-producing multidrug resistant Escherichia coli from dogs and cats in the United States, 2009-2013. Front Microbiol 7:1057. doi: 10.3389/fmicb.2016.01057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing, 28th ed CLSI supplement M100 CLSI, Wayne, PA. [Google Scholar]
- 32.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylotyping method revisited: improvement of specificity and detection of new phylogroups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 33.Clermont O, Lavollay M, Vimont S, Deschamps C, Forestier C, Branger C, Denamur E, Arlet G. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J Antimicrob Chemother 61:1024–1028. doi: 10.1093/jac/dkn084. [DOI] [PubMed] [Google Scholar]
- 34.Colpan A, Johnston B, Porter S, Clabots C, Anway R, Thao L, Kuskowski MA, Tchesnokova V, Sokurenko EV, Johnson JR. 2013. Escherichia coli sequence type 131 (ST131) as an emergent multidrug-resistant pathogen among U.S. veterans. Clin Infect Dis 57:1256–1265. doi: 10.1093/cid/cit503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banerjee R, Robicsek A, Kuskowski MA, Porter S, Johnston BD, Sokurenko E, Tchesnokova V, Price LB, Johnson JR. 2013. Molecular epidemiology of Escherichia coli sequence type 131 and its H30 and H30-Rx subclones among extended-spectrum-β-lactamase-positive and -negative E. coli clinical isolates from the Chicago region, 2007 to 2010. Antimicrob Agents Chemother 57:6385–6388. doi: 10.1128/AAC.01604-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 37.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21:30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 38.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, Zhang R, Walsh TR, Shen J, Wang Y. 2017. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e00543-17. doi: 10.1128/mBio.01166-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 40.Tanizawa Y, Fujisawa T, Kaminuma E, Nakamura Y, Arita M. 2016. DFAST and Daga: web-based integrated genome annotation tools and resources. Biosci Microbiota Food Health 35:173–184. doi: 10.12938/bmfh.16-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Two nucleotide sequences of E. coli B22 have been deposited in GenBank under accession numbers LC480204 (157,199 bp) and LC495380 (151,287 bp). The complete nucleotide sequences of pB64 and pUIH-1 have been deposited in the GenBank database under accession numbers LC480203 and LC477293, respectively.