This study shows that lac genes in Lactobacillus reuteri encode hydrolases and transporters that are necessary for the metabolism of GOS, as well as α-galactoside substrates. Coculture experiments with the wild-type strain and a gos mutant clearly demonstrated that GOS utilization confers a growth advantage in medium containing GOS as the sole carbohydrate source. However, the wild-type strain also outcompeted the mutant in germfree mice, suggesting that GOS genes in L. reuteri also provide a basis for utilization of other carbohydrates, including α-galactosides, ordinarily present in the diets of humans and other animals. Collectively, our work provides information on the metabolism of L. reuteri in its natural niche in the gut and may provide a basis for the development of synbiotic strategies.
KEYWORDS: Lactobacillus reuteri, carbohydrate, galactooligosaccharide, gnotobiotic, metabolism, raffinose, stachyose
ABSTRACT
Strains of Lactobacillus reuteri are commonly used as probiotics due to their demonstrated therapeutic properties. Many strains of L. reuteri also utilize the prebiotic galactooligosaccharide (GOS), providing a basis for formulating synergistic synbiotics that could enhance growth or persistence of this organism in vivo. In this study, in-frame deletion mutants were constructed to characterize the molecular basis of GOS utilization in L. reuteri ATCC PTA-6475. Results suggested that GOS transport relies on a permease encoded by lacS, while a second unidentified protein may function as a galactoside transporter. Two β-galactosidases, encoded by lacA and lacLM, sequentially degrade GOS oligosaccharides and GOS disaccharides, respectively. Inactivation of lacL and lacM resulted in impaired growth in the presence of GOS and lactose. In vitro competition experiments between the wild-type and ΔlacS ΔlacM strains revealed that the GOS-utilizing genes conferred a selective advantage in media with GOS but not glucose. GOS also provided an advantage to the wild-type strain in experiments in gnotobiotic mice but only on a purified, no sucrose diet. Differences in cell numbers between GOS-fed mice and mice that did not receive GOS were small, suggesting that carbohydrates other than GOS were sufficient to support growth. On a complex diet, the ΔlacS ΔlacM strain was outcompeted by the wild-type strain in gnotobiotic mice, suggesting that lacL and lacM are involved in the utilization of alternative dietary carbohydrates. Indeed, the growth of the mutants was impaired in raffinose and stachyose, which are common in plants, demonstrating that α-galactosides may constitute alternate substrates of the GOS pathway.
IMPORTANCE This study shows that lac genes in Lactobacillus reuteri encode hydrolases and transporters that are necessary for the metabolism of GOS, as well as α-galactoside substrates. Coculture experiments with the wild-type strain and a gos mutant clearly demonstrated that GOS utilization confers a growth advantage in medium containing GOS as the sole carbohydrate source. However, the wild-type strain also outcompeted the mutant in germfree mice, suggesting that GOS genes in L. reuteri also provide a basis for utilization of other carbohydrates, including α-galactosides, ordinarily present in the diets of humans and other animals. Collectively, our work provides information on the metabolism of L. reuteri in its natural niche in the gut and may provide a basis for the development of synbiotic strategies.
INTRODUCTION
Lactobacillus reuteri is an autochthonous member of the vertebrate gastrointestinal tract (GIT) and is considered a dominant gut symbiont in rodents, pigs, and chickens (1, 2). Although less prevalent in humans in industrialized societies (3, 4), strains of L. reuteri are widely used as probiotics due to their ability to exert health benefits through a variety of suggested mechanisms, including enhancing epithelial barrier integrity (5, 6), modulating immunity (7), increasing bone density (8, 9), reducing symptoms associated with infant colic (10), reducing incidence of necrotizing enterocolitis in premature infants (11), and improving glucose-dependent insulin release (12).
The ability of probiotics or other orally administered therapeutic bacteria to remain active in the highly competitive gut ecosystem (13–15) depends, in part, on their ability to occupy nutritional and geographic niches and sequester nutrients from competitors (16, 17). One suggested strategy to enhance the establishment and growth of specific probiotic bacteria is to provide prebiotics, such as galactooligosaccharide (GOS), together with the probiotic to generate a synbiotic (18–20). For synergistic synbiotics, in particular, the prebiotic is specifically intended to support the growth of the probiotic (18, 21).
Fermentation of GOS by L. reuteri has previously been reported (22–24), and the suggested molecular mechanisms for transport and hydrolysis in L. reuteri and other species of Lactobacillus have been described (25, 26). According to this model, GOS is transported via a LacS permease and hydrolyzed by β-galactosidase. However, commercial GOS is enzymatically synthesized from lactose via an industrial process and would not be expected to be a natural substrate for L. reuteri. Thus, the presence of a GOS utilization pathway suggests that other saccharides might serve as substrates for this pathway. Therefore, the primary goals of this study were to genetically characterize the GOS pathway and determine the role of this pathway in colonization of the murine gastrointestinal tract.
RESULTS
In silico identification of putative GOS metabolic genes in L. reuteri.
Genome analysis based on the Integrated Microbial Genomes (IMG) database confirmed that all four human-derived, GOS-fermenting strains used in this study were very similar, with few single nucleotide polymorphisms. In addition, all harbored a lacA gene that putatively encodes an intracellular β-galactosidase of the glycoside hydrolase family 42 (GH42). However, this gene was absent from the draft genome of the poor-GOS-fermenting strain ATCC 53608, which had been isolated from swine (Table 1). In the GOS-utilizing strains, the lacA gene was located within the same cluster as lacR and lacS (Fig. 1), both of which were also absent in the draft genome of strain 53608. The lacR and lacS genes were annotated in the L. reuteri genomes to encode a LacI family transcriptional regulator and a lactose symporter/permease, respectively.
TABLE 1.
Similarity among protein sequences encoded by GOS metabolic genes identified in different L. reuteri genomes
| Locus taga HMPREF0536_ | Gene symbolb | Annotated gene product | Similarityc (%) by L. reuteri strain |
||||
|---|---|---|---|---|---|---|---|
| GOS fermenting |
Poor GOS fermenting |
||||||
| ATCC PTA 6475 | DSM 20016 | ATCC PTA 4659 | ATCC 55730 | ATCC 53608 | |||
| 659 | lacR | Lacl family transcriptional regulator LacR | 100 | 100 | 100 | 95 | Absent |
| 660 | lacS | Glycoside-pentoside-hexuronide:cation symporter | 100 | 100 | 100 | 96 | Absent |
| 661 | lacA | GH42 β-galactosidase | 100 | 100 | 100 | 95 | Absent |
| 317 | lacL | GH2 β-galactosidase large subunit | 100 | 100 | 100 | 98 | 98 |
| 316 | lacM | GH2 β-galactosidase small subunit | 100 | 100 | 100 | 96 | 95 |
Locus tag as assigned in the L. reuteri ATCC 6475 (MM4-1A) genome.
lacR, lacS, and lacA as referred to by Andersen et al. (28); lacL and lacM as assigned in the Integrated Microbial Genomes (IMG) system.
Similarity to ATCC 6475 query sequences.
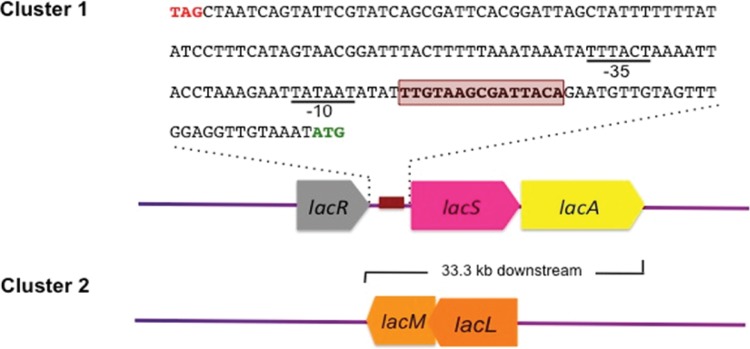
FIG 1.
Organization of two gene clusters potentially responsible for GOS metabolism in L. reuteri 6475 (MM4-1A). The predicted catabolite-responsive element (CRE)/operator region is highlighted in red, and the predicted −35 and −10 sequences of the σ70 promoter are underlined. The stop codon of lacR and the start codon of lacS are colored red and green, respectively. The lacLM gene is located 33.3 kb downstream of the lacRSA cluster in the L. reuteri ATCC 6475 genome.
In addition to the lacRSA gene cluster, another locus harboring lacL and lacM genes, encoding large and small subunits of β-galactosidase (GH2), was also detected in all studied strains, with more than 95% identity in amino acid sequences (Table 1). Thus, lacL and lacM were also considered likely GOS metabolic genes in this study.
Role of functional genes in GOS metabolism.
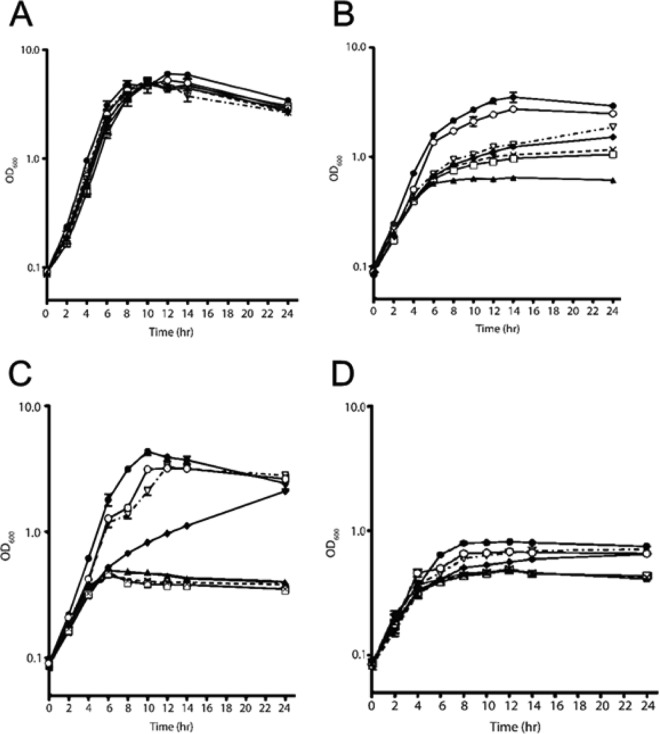
To understand the function of the lac genes, we constructed single (ΔlacS, ΔlacA, ΔlacL, ΔlacM, and ΔlacR) and double (ΔlacS ΔlacM) mutants. The recombineering method involved the introduction of in-frame stop codons so that polar effects would not occur. Growth studies in modified MRS (mMRS) supplemented with different carbohydrates were then performed to determine how each mutation affected GOS utilization. Importantly, no differences among any of the mutant strains were observed during growth in glucose-mMRS compared with the wild type (Fig. 2A), indicating the absence of pleiotropic effects of the nonsense mutations. None of the mutations affected growth on glucose (Fig. 2A). In contrast, compared with the wild type, all of the mutants showed diminished growth on GOS-mMRS, with the ΔlacS ΔlacM mutant the most affected (Fig. 2B). On lactose, the growth of the ΔlacR and ΔlacA mutants was unaffected, whereas the ΔlacS mutant grew more slowly than the wild type, but eventually reached a similar cell density (Fig. 2C). In contrast, the ΔlacL, ΔlacM, and ΔlacS ΔlacM mutants grew at markedly lower rates and reached lower cell densities than the wild-type strain. In the negative-control cultures containing only the background sugars present in GOS (mainly lactose), a slower growth of the ΔlacS, ΔlacM, and ΔlacS ΔlacM mutants was also observed (Fig. 2D).
FIG 2.
Growth of wild-type L. reuteri 6475-1A (●) and ΔlacR (○), ΔlacS (◆), ΔlacA (▽), ΔlacL (×), ΔlacM (□), and ΔlacS ΔlacM (▲) mutants on mMRS supplemented with 1% glucose (A), 1% GOS (B), 1% lactose (C), and GOS-contaminating sugars (D) (see the text for details). Results are expressed as means ± SD obtained from three independent replicates.
Because the lacS mutant was still able to grow, albeit slowly, on GOS as well as lactose, we hypothesized that a second transport system is likely present in L. reuteri. This hypothesis was supported by thin-layer chromatography (TLC) analysis of the sugar components remaining in spent mMRS medium containing purified GOS as a sole carbon source. Although the GOS constituents with a degree of polymerization (DP) of 2 were mostly consumed by the ΔlacS mutant within 24 hours, components with a DP of ≥3 remained intact (Fig. 3). This result indicated the presence of a second transporter that has an affinity for the disaccharide components of GOS.
FIG 3.
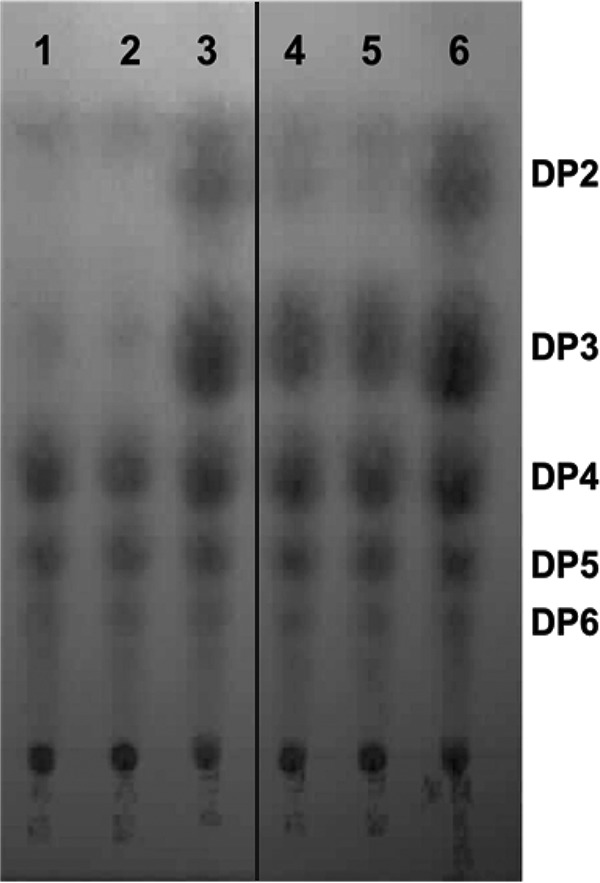
TLC analysis of GOS consumption by wild-type L. reuteri 6475 and the ΔlacS mutant. Shown are spent medium of the wild type at 12 and 24 h (lanes 1 and 2), 1% GOS-mMRS medium (lanes 3 and 6), and spent medium of the mutant at 12 and 24 h (lanes 4 and 5). DP, estimated degree of polymerization. The chromatogram is a composite of lanes (1 to 3 and 4 to 6) from the same TLC plate.
The growth of the mutants on GOS and lactose indicates the presence of two transporters, and this result also suggested distinct functional roles of the two β-galactosidases. This was demonstrated by the difference in growth profiles on GOS between the ΔlacL or ΔlacM strain and the ΔlacA mutant (Fig. 2B). A difference was also observed when the mutants were grown on lactose (Fig. 2C). Whereas the ΔlacL and ΔlacM mutants completely lost the ability to grow on lactose, the ΔlacA mutant grew almost as well as the wild type. These results indicated that the GH2 β-galactosidase LacLM was essential for lactose hydrolysis, whereas the GH42 β-galactosidase encoded by lacA was not. This finding also led us to consider if the utilization of other disaccharides present in GOS also relies primarily on the hydrolytic activity of the LacLM β-galactosidase.
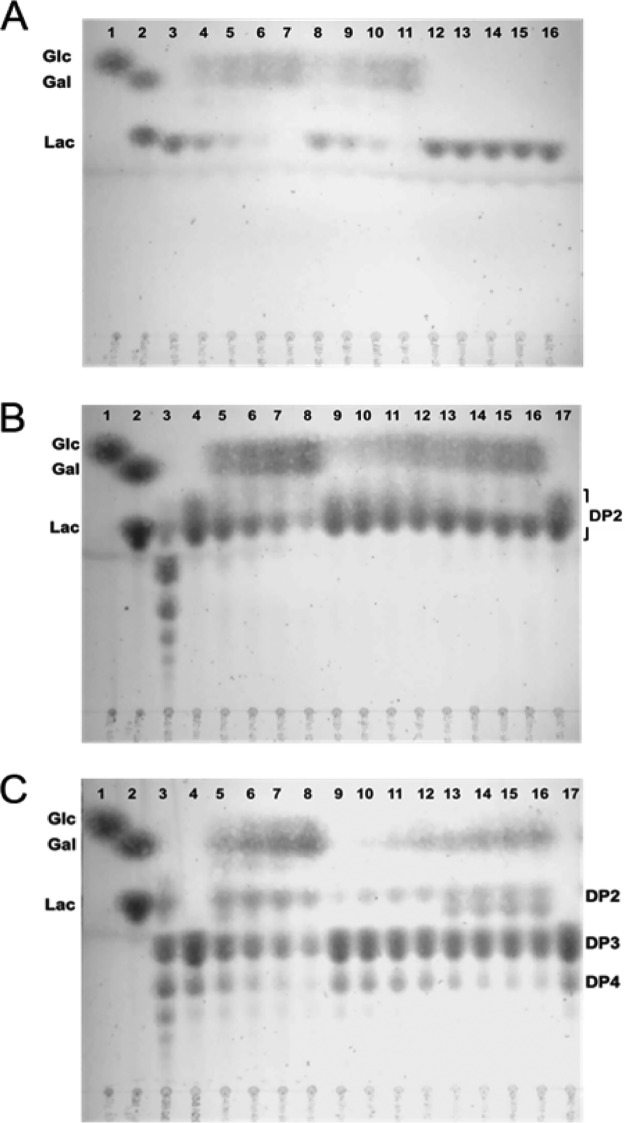
To address this question, the activity of intracellular LacA and LacLM β-galactosidases on the disaccharide components of GOS was determined using cell-free extracts separated from GOS-grown cells. A TLC analysis of lactose hydrolysis products showed that while the ΔlacA cell-free extract was capable of hydrolyzing lactose equal to that of the wild type, the ΔlacM cell-free extract was devoid of activity (Fig. 4A). This analysis confirmed that only LacLM, but not LacA, has cleavage specificity for the β(1-4) linkage between galactose and glucose. Moreover, the cleavage of terminal lactose from GOS with a DP of ≥3 could potentially be achieved by the activity of LacLM.
FIG 4.
TLC analysis of cell-free extracts (CFEs) on different β-galactosides. (A) Activity on lactose. Lane 1: glucose; lane 2: lactose and galactose; lanes 3 and 16: lactose in the reaction mix; lanes 4 to 7: wild-type strain 6475 CFEs at 2, 4, 6, and 12 h; lanes 8 to 11: ΔlacA CFEs at 2, 4, 6, and 12 h; and lanes 12 to 15: ΔlacM CFEs at 2, 4, 6, and 12 h. (B) Activity on GOS disaccharides. Lane 1: glucose; lane 2: lactose and galactose; lane 3: GOS; lanes 4 and 17: disaccharides in the reaction mix; lanes 5 to 8: 6475 CFEs at 2, 4, 6, and 12 h; lanes 9 to 12: ΔlacA CFEs at 2, 4, 6, and 12 h; and lanes 13 to 16: ΔlacM CFEs at 2, 4, 6, and 12 h. (C) Activity on GOS trisaccharides and tetrasaccharides. Lane 1: glucose; lane 2: lactose and galactose; lane 3: GOS; lanes 4 and 17: oligosaccharides in the reaction mix; lanes 5 to 8: 6475 CFEs at 2, 4, 6, and 12 h; lanes 9 to 12: ΔlacA CFEs at 2, 4, 6, and 12 h; and lanes 13 to 16: ΔlacM CFEs at 2, 4, 6, and 12 h.
Despite the absence of lactose or β-galactoside hydrolysis activity, the ΔlacM cell-free extract containing intact LacA was still able to degrade other GOS species and release free glucose and galactose into the reaction mix (Fig. 4B, lanes 13 to 16). In contrast, the ΔlacA cell-free extract had diminished hydrolytic activity on the disaccharide fraction (Fig. 4B, lanes 9 to 12), emphasizing the role of this enzyme on degradation of GOS disaccharides.
Other in vitro hydrolysis assays were conducted using fractionated GOS containing larger DP (trisaccharide and tetrasaccharide) species. TLC analysis showed that although disaccharide products were detected in the reaction mix of the ΔlacA cell-free extract, trisaccharide and tetrasaccharide fractions remained intact (Fig. 4C, lanes 9 to 12). This result indicated that LacLM had minimal hydrolytic activity on GOS, with a DP of ≥3. In comparison to the ΔlacA cell-free extract, both trisaccharides and tetrasaccharides, in particular, were reduced in the ΔlacM cell-free extract (Fig. 4C, lanes 13 to 16), indicating a greater activity of LacA on these GOS components. Moreover, the accumulation of lactose as a hydrolysis product suggested that LacA cleaves trisaccharides and tetrasaccharides at Gal-Gal β-linkages and releases a lactose terminus that was resistant to LacA hydrolytic activity.
GOS genes confer a competitive advantage in vitro.
Coculture competition experiments with the wild-type and the ΔlacS ΔlacM strains were performed to determine if GOS utilization conferred a competitive advantage. As predicted, when grown in mMRS medium containing GOS, the wild-type strain quickly dominated, displacing the mutant within 21 generations (Fig. 5A). However, in MRS containing glucose, the population of the wild-type and mutant strains remained unchanged, even after 28 generations (Fig. 5B).
FIG 5.
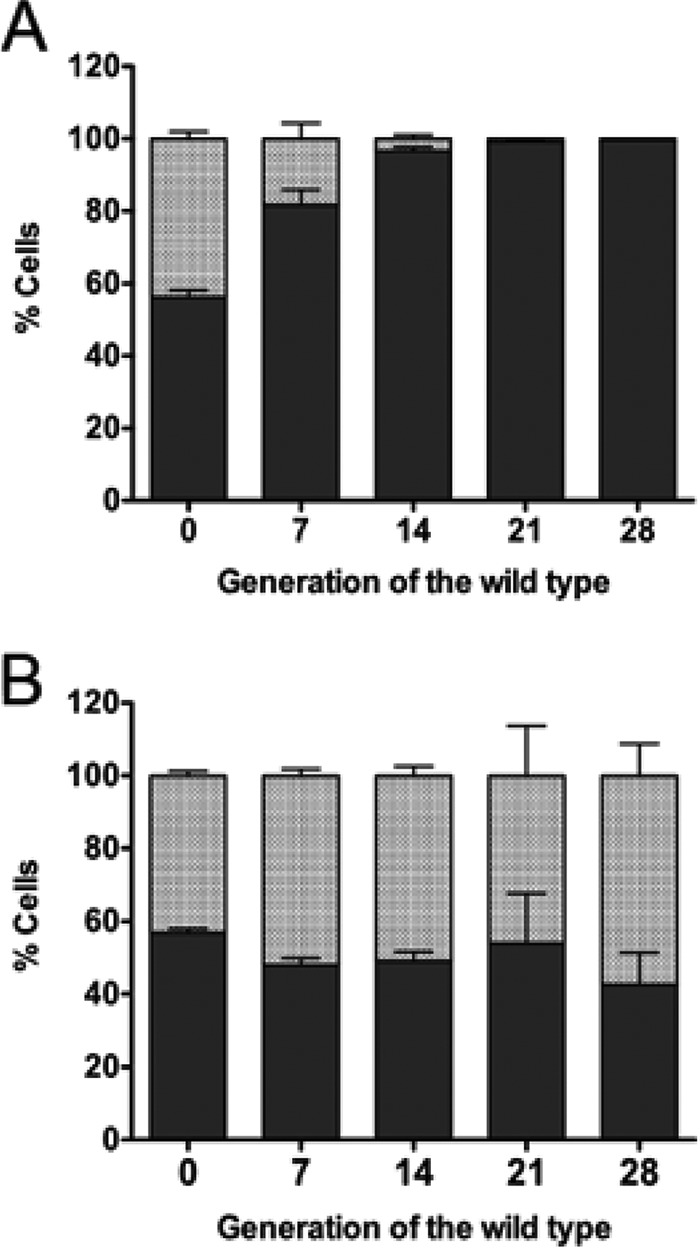
Proportions of the total L. reuteri population comprised of wild-type L. reuteri 6475 (solid bars) and the ΔlacS ΔlacM mutant (shaded bars) during in vitro coculture growth in 1% GOS-mMRS (A) and 1% Glc-mMRS (B).
Contribution of GOS genes to ecological performance in vivo is not primarily related to GOS utilization.
To determine the role of the GOS genes in vivo, we performed competition experiments in ex-germfree mice. Mice were fed a purified ingredient, no sucrose diet to minimize the amount of fermentable sugars that could serve as an alternative carbon source and obscure the impact of GOS. The wild type reached significantly higher levels in feces when drinking water was supplemented with GOS than without GOS treatment after 48 h (Fig. 6A), while the presence of GOS consumption had no effect on the mutant (Fig. 6B). In addition, the proportion of the wild type appeared to increase over time in the presence of GOS but decrease in its absence (Fig. 6C). However, the effects were small and not significant, and the mutant colonized at least as well (around 60%), indicating dietary substrates other than GOS were sufficient to support growth in vivo.
FIG 6.
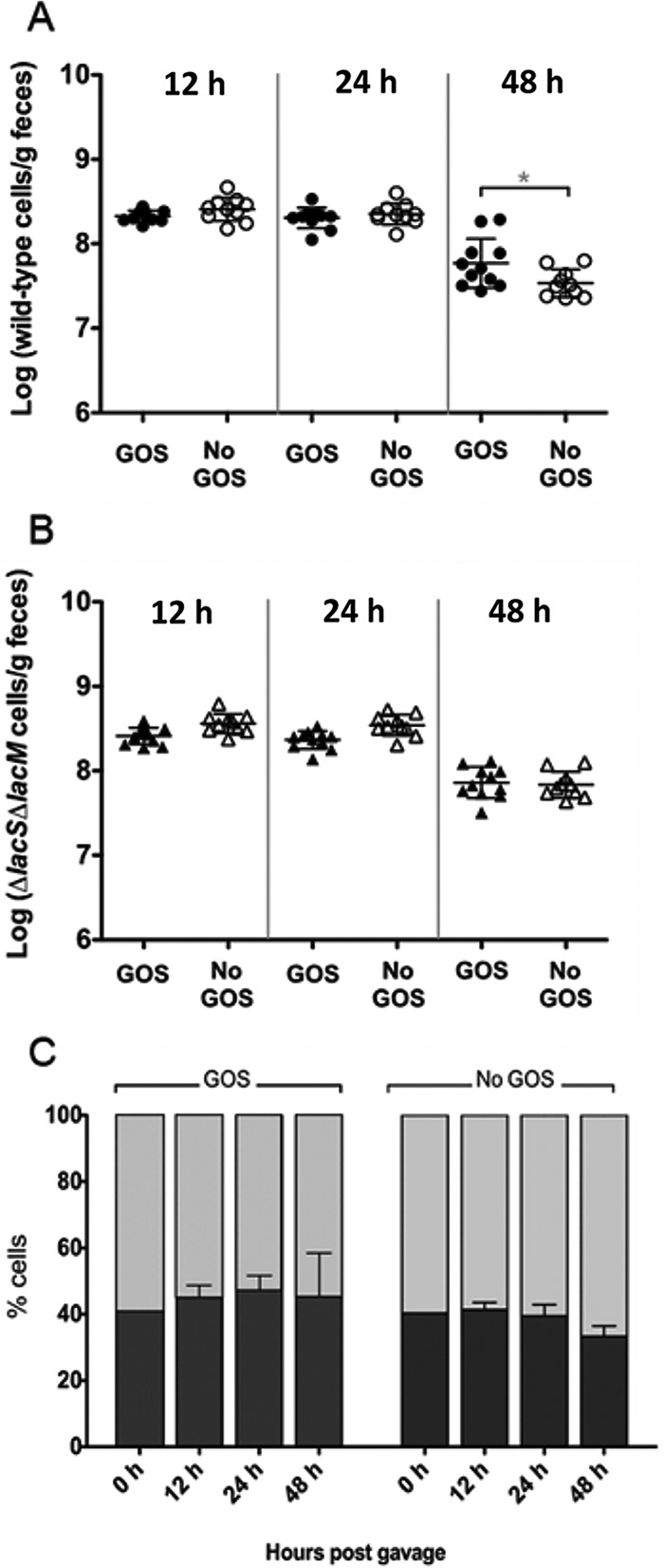
Changes in fecal populations of wild-type L. reuteri 6475 and the ΔlacS ΔlacM mutant, coinoculated into germfree C3H mice fed a purified ingredient, low sucrose diet. (A) Wild-type populations in GOS-fed mice (●) and in control mice (○). (B) Mutant populations in GOS-fed mice (▲) and in control mice (▵). Cell populations were determined by RT-qPCR using strain-specific primers. An asterisk denotes significant differences (P < 0.05) analyzed by one-way ANOVA with Tukey’s post test. (C) Proportion of the total L. reuteri population comprised of 6475 (black bars) and the ΔlacS ΔlacM mutant (gray bars) in the presence (left) or absence (right) of GOS.
In a second experiment, mice received a standard, grain-based chow, and populations of the wild-type and mutant strains in feces were again quantified by real-time PCR. After 48 hours, the wild-type strain reached about 70% of the total population in both treatment groups. However, the addition of GOS made no detectable difference (Fig. 7A). Similarly, when the two strains were cocultured in chow broth, the wild-type population reached about 90% of the total cell number after 28 generations (Fig. 7B). These experiments suggest that although GOS genes contribute to ecological performance in vivo, this is not primarily related to GOS utilization. Rather, these genes likely contribute to the metabolism of other sugars present in standard mouse chow.
FIG 7.
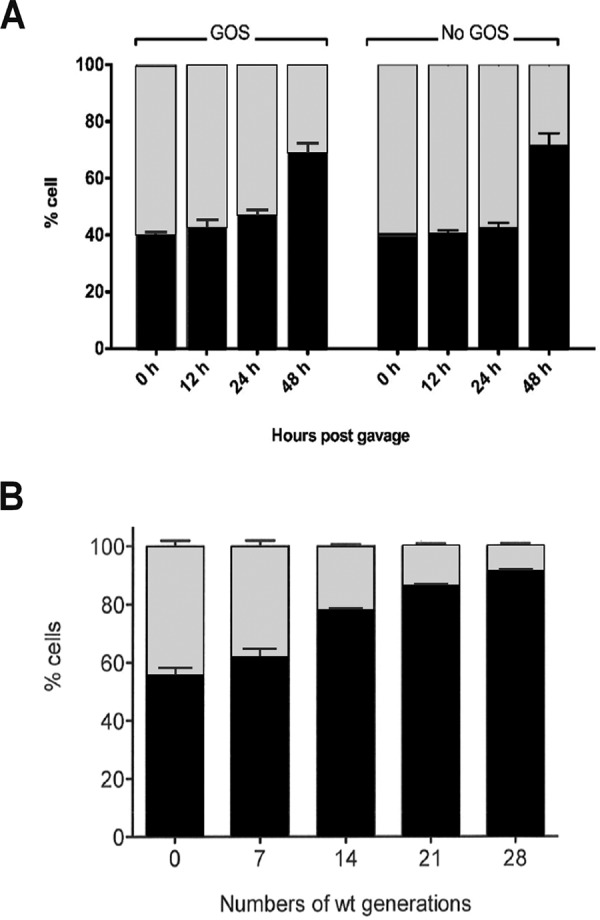
Proportions of total fecal wild-type L. reuteri (black bars) and the ΔlacS ΔlacM mutant (gray bars) after both strains were coinoculated into germfree C3H mice fed with a standard chow diet with and without GOS (A) or in a 5% standard chow broth without GOS supplementation (B).
GOS genes involved in the utilization of the α-galactosides raffinose and stachyose.
Given that the GOS strains contributed to in vivo ecological fitness on a complex mouse chow independently of GOS but not in a refined chow, we hypothesized that other carbohydrates likely present in the complex chow function as growth substrates utilized by these genes. Therefore, we tested the ability of the L. reuteri wild-type and mutant strains to grow on alternative substrates likely to be present in mouse chow and composed of galactose, such as the α-galactosides raffinose and stachyose. For these experiments, 0.5× mMRS broth was used to minimize the effect of background sugars present. Interestingly, the growth of the mutant strains on raffinose and stachyose was clearly impaired compared with the wild type (see Fig. S1 in the supplemental material). The findings are of major interest, as raffinose and stachyose are present in the human diet, and in contrast to GOS (which is rare in nature), they could constitute potential growth substrates for strains related to L. reuteri ATCC PTA 6475 that originate from humans (27).
DISCUSSION
A genome comparison between robust and weak GOS-fermenting strains of L. reuteri revealed five candidate GOS-metabolizing genes whose role in GOS transport and metabolism was confirmed by phenotypic analyses. The organization of the GOS genes was very similar to that described for Lactobacillus acidophilus NCFM (28), with structural genes encoding LacS and LacA located downstream of a predicted regulatory gene (lacR). Although LacS appears to be the main transporter for GOS in L. reuteri, the ability of the lacS mutant to grow on GOS and lactose suggested the presence of a second, as yet unidentified transporter. In contrast, in L. acidophilus NCFM, inactivation of lacS completely abolished both lactose and GOS consumption (28).
The presence of a second transport system in L. reuteri was also suggested based on the observation that lactose and disaccharide components of GOS were consumed by the lacS mutant. Genome analysis subsequently revealed that a putative transporter encoded by a gene (HMPREF0536_1595) annotated as glycoside-pentoside-hexuronide (GPH):cation symporter family was present in all studied L. reuteri strains, including ATCC 53608. Its product and LacS are similar in size (650 versus 640 amino acids) but share only 38% identity in amino acid sequences.
In addition to the suggested presence of two transport systems for GOS and other galactosides, phenotypic analyses also indicated the concerted action of two β-galactosidases, namely, LacA and LacLM, for hydrolysis of GOS. The inability of the ΔlacM mutant to grow on lactose suggested that the LacLM β-galactosidase had activity toward Galβ(1 → 4)Glc galactosidic linkages that also include the terminal lactose moiety of GOS. The accumulation of lactose in reaction mixtures containing ΔlacM cell extracts and GOS trisaccharides and tetrasaccharides further supports this suggestion.
In contrast, LacA does not possess lactose-hydrolyzing activity, as demonstrated by similar growth on lactose between the ΔlacA mutant and the wild type, as well as the degradation of lactose in the cell extract devoid of LacA. Rather, the substrates for LacA appear to be the larger GOS species. Indeed, the ΔlacM cell-free extract retained the ability to hydrolyze trisaccharides and tetrasaccharides, liberating galactose, and lactose moieties, whereas lacA cell-free extracts did not hydrolyze these substrates.
Collectively, these observations indicate the cooperative action of LacA and LacLM for hydrolysis of the different disaccharides and β-galactosidic linkages that constitute GOS. Accordingly, we suggest that intracellular GOS species are first cleaved by LacA at their terminal β-galactosyl residue, yielding free galactose and terminal lactose moieties. LacLM then hydrolyzes the liberated terminal lactose into galactose and glucose.
The distinct hydrolytic activities of L. reuteri LacLM and LacA are congruent with the enzymatic properties between GH2 and GH42 β-galactosidases previously reported (29–32). Specifically, the GH2 β-galactosidases usually have lactose as their natural substrate (29). Enzymes in this family, including the LacLM and LacZ type, are common among lactic acid bacteria associated with dairy fermentation (30) and in bifidobacteria of human intestinal origin (31, 32). The GH42 β-galactosidases, in contrast, typically have weak affinity toward lactose and prefer to act on other galactose-containing glycosides (33–37).
The cooperative action between GH2 LacLM and GH42 LacA for hydrolyzing GOS described here in L. reuteri is consistent with other studies. Gopal et al. (38) were the first to associate the lactose permease/β-galactosidase system with the ability of lactobacillus strains to utilize GOS. Specifically, they observed that all identified GOS-fermenting strains possess a β-galactosidase. Similar results were observed by microarray transcriptome analysis of GOS metabolism in L. acidophilus NCFM, where LacS, LacA, and LacLM were identified as being responsible for GOS metabolism (28).
Evidence for similar activities of LacS, LacA, and LacLM was also reported for Lactobacillus ruminis (39). In this bacterium, two operons encode systems involved in β-galactoside utilization. One includes a GPH-family lactose permease and a GH42 β-galactosidase (LacA). A second operon, lacYZ, encodes a putative lactose permease and GH2 β-galactosidase. Both of these operons are present only in the GOS-fermenting strain but not in the strain incapable of utilizing GOS and lactose, suggesting their fundamental role in GOS metabolism (39).
In L. reuteri ATCC 53608, which grows weakly on GOS, the lacRSA cluster is absent, but lacLM is present. Not surprisingly, the absence of the former operon limits the transport and hydrolysis of most GOS components and also prevents the terminal lactose moiety from serving as a substrate of LacLM. These limitations likely account for the substantial decrease of the GOS-fermenting capacity in this strain. Gene expression analysis revealed that L. reuteri typically expresses a low basal level of LacS and LacA and that the lacSA operon was induced by lactose but not by GOS (see Fig. S2 in supplemental material). In addition, inactivation of lacR led to constitutive transcription of lacSA.
GOS utilization was not of major importance for L. reuteri growth in the mouse gastrointestinal tract. However, the GOS genes did still contribute to ecological fitness but only on a mouse chow containing the carbohydrates and not in the purified ingredient chow. This finding suggests that the GOS genes are involved in the utilization of other carbohydrates present in the complex chow, which was confirmed by competition experiments in a broth that was generated from the chow. Interestingly, the ability of the mutant strains to grow on raffinose and stachyose was also impaired, suggesting the GOS machinery may also have activity on α-galactosides, as other researchers have proposed (22). Previously, the putative galactoside transporter RafP was reported to have high homology to LacS in Streptococcus thermophilus (40). In that organism, LacS has an affinity for both α-galactosides and β-galactosides (41). More recently, investigators suggested that melibiose or raffinose transport in lactobacilli could be mediated by LacS (42).
Although β-galactosides occur primarily in milk, α-galactosides are highly abundant in nature, particularly plants (43). Thus, it is intriguing to speculate that LacS and the galactoside hydrolysis enzymes in L. reuteri may have broad substrate affinity. Consistent with this suggestion, Zhao and Gänzle recently reported that raffinose induced expression of LacS in L. reuteri 100-23 (44). Therefore, because GOS is likely not ordinarily available as a growth substrate for L. reuteri, the presence of GOS genes in this organism suggests they may also function for the utilization of other galactosidic substrates that are more commonly present in the diet of humans and other mammals.
Based on the phenotypic and gene expression analyses described above, a coherent model for GOS metabolism in L. reuteri is proposed. Accordingly, GOS is taken up into the cytosol by the activity of two different transporters—the LacS permease and a second transporter predicted to be another GPH-family symporter. Consistent with other lactobacillus lactose transporters (26, 44), the LacS permease has broad substrate specificity and is responsible for the transport of lactose, as well as a range of GOS components. In contrast, the second putative transporter appears to have affinity mainly for lactose and GOS disaccharides. In the cytosol, LacA sequentially cleaves GOS oligosaccharides from the terminal β-galactosyl residue until all galactose moieties and terminal lactose are liberated. LacLM subsequently breaks down the terminal lactose into glucose and galactose. Although the true substrates for these transporters and galactosidases remain unclear and require additional research, they may be involved in metabolism of both α-galactosides and β-galactosides. Given the fact that raffinose and stachyose are common in human food and animal feed, the ability to utilize these carbohydrates provides a more realistic explanation for the conservation of GOS genes in the species L. reuteri, other than the ability to utilize GOS, which is rare in nature. Elucidation of the true growth substrates of L. reuteri in the gastrointestinal tract will provide a basis for the development of rational synbiotic strategies.
MATERIALS AND METHODS
Cultures and growth conditions.
Lactobacillus reuteri ATCC PTA 6475 (MM4-1A) was obtained from BioGaia AB, Sweden. The strain and the isogenic GOS-metabolic-gene-deficient mutants generated in this study were routinely prepared from frozen stock cultures. Stock cultures were streaked onto De Mann, Rogosa, and Sharp (MRS; Difco Laboratories) agar plates and incubated at 37°C for 36 to 48 h under an anaerobic condition consisting of 5% CO2, 5% H2, and 90% N2. Single colonies isolated on the plates were transferred into MRS broth. Cultures were grown anaerobically at 37°C for 16 to 24 h in a Bactron anaerobic incubator and then subcultured (1%) into fresh MRS for 12 h.
GOS and fractionated GOS components.
Purimune GOS was provided by GTC Nutrition, USA (now Ingredion, Inc., Westchester, Illinois). The GOS powder was comprised of 90% to 92% GOS, 7% to 10% lactose, <1% glucose, and <0.5% galactose (45). The GOS fraction contained oligosaccharides having a degree of polymerization (DP) ranging from 2 to 7 monomers. For some experiments, the GOS mixture was fractionated and purified by size exclusion chromatography using a Sephadex G-10 (Sigma-Aldrich) column. For each separation, 5 ml of 30% (wt/vol) GOS solution was applied to the column (96 × 2.5 cm), and fractions were eluted with Nanopure water at a flow rate of 0.16 ml/min. After the void volume was eluted, 1-ml fractions were collected and assessed for the presence of carbohydrate using a refractometer (Reichert Rhino BRIX30). Approximately 70 fractions were collected from each run. Saccharide compositions were subsequently identified by thin-layer chromatography (TLC) on high-performance thin-layer chromatography (HPTLC) silica gel 60 plates (Merck KGaA, Darmstadt, Germany). The TLC plates were developed and visualized as previously described (20). Fractions were pooled and concentrated by freeze drying. Hydrated fractions were reanalyzed again by TLC to confirm the presence of the expected components. The TLC analysis of the fractionated products is shown in Fig. S3 in the supplemental material.
Genomic analysis of putative GOS genes in L. reuteri.
Genome sequences of four human-derived, GOS-fermenting L. reuteri strains, namely, L. reuteri ATCC PTA-6475, L. reuteri ATCC 55730 (SD2112), L. reuteri DSM 20016 (F275), and L. reuteri ATCC PTA-4659 (MM2-3), were compared. We specifically searched for genes functionally annotated as β-galactosidases using the Gene Search tool in the Integrated Microbial Genomes (IMG) platform (https://img.jgi.doe.gov/) (46). In addition, the genomes were also compared with that of L. reuteri ATCC 53608, a swine-derived, poor-GOS-fermenting strain. DNA loci surrounding putative β-galactosidase-encoding genes were further examined for neighboring genes annotated with functions related to carbohydrate metabolism and regulation (47).
Generation of mutants deficient in GOS metabolic genes.
We used single-stranded DNA recombineering in L. reuteri 6475 to inactivate genes predicted to be involved in GOS metabolism, as described previously (48). Briefly, for each target gene, we designed 85-mer oligonucleotides made up of 80 bases identical to the lagging strand of replication and 5 centrally located nonhomologous bases which would—when incorporated in the genome—yield an in-frame stop codon in the target gene (Table 2). L. reuteri 6475 expressing RecT, a single-stranded DNA-binding protein that facilitates incorporation of the oligomer, was transformed with 100-μg oligonucleotide. Resulting colonies were screened by mismatch amplification mutation assay (MAMA)-PCR to identify recombinant genotypes (48, 49). Colonies corresponding to recombinant genotypes were purified by streak plate, followed by a second MAMA-PCR to identify a strain with a pure recombinant genotype that was confirmed by Sanger sequencing. Recombineering oligonucleotides and the targeted gene sequences and primers for MAMA-PCR are listed in Table 2.
TABLE 2.
Recombineering oligonucleotidesa and MAMA-PCR primersb used to generate mutants deficient in potential GOS metabolic genes
| Mutant | Recombineering oligonucleotide (5′−3′) | MAMA-PCR primers (forward; reverse) |
|---|---|---|
| ΔlacR | (230) AGTTGTTCTTTACCACTTACCCGATACAATAGAGCAAGATgctatCGGATTGGTTGCTGCTGAGAGTTTTTCTTGTGGTTGTTCC (146) | AGCAACCAATCCGatagc; ACACCAACACTTAGCGTAT |
| ΔlacS | (178) TCAACCCAATTAGTTTATTAGCAACCGTTTTACTTAGTCCctactACATTCCGCTTGTGACAAAGATAATAAAGTAAGTACTCAT (94) | CACAAGCGGAATGTagtag; CACAGATAACAGCCAATGC |
| ΔlacA | (167) TTTCCCTCCTGCGGTTCAAGCAAAGCCCATGAAAAAACATactagGTGGCGGAATTTATATCTGCCTGTTTAAATACTTTAATAT (83) | TATAAATTCCGCCACctagt; GCACAACAATCTCATTCCA |
| ΔlacL | (291) CAAAGTGTTAATGTAATTATTTTGGGCGTAATTATTTAGTagctaCTCACTAGGCACTTCGATTGTTCCAAAACTACTACTATCA (207) | AAGTGCCTAGTGAGtagct; CGAGCGTATTACCATCATTG |
| ΔlacM | (228) CGCTGCTAACCACTGAGCAGACCGAATATTAAAGCCACTTtactaGTCATTATCAGTGGTTGCCCGCCAAAACGTCGGCATTGGT (144) | GCAACCACTGATAATGACtag; GGAACGAGATACTTAGTAACC |
Recombineering oligonucleotides are identical to the lagging strand of replication except for five consecutive mismatches shown in bold lowercase. Underlined bases denote introducing stop codons (TAG), and numbers in parentheses indicate annealing sites relative to the first base of the start codon of the target genes.
MAMA-PCR forward primers are homologous to mutated sequences but harbor at the 3’ end three to five mismatches (in bold lowercase) with wild-type sequences.
Growth on different carbon sources.
The ability of the wild-type and mutant strains to utilize GOS was determined by measuring growth in modified MRS (mMRS) broth supplemented with GOS or other carbon sources. The mMRS medium was prepared without added glucose and was made at half strength (i.e., containing 50% of the normal amount of ingredients) in order to minimize carbon-source content and background growth. Twelve-hour cultures, prepared as described above, were used as the source of the inoculum. These cultures were then inoculated at 1% (vol/vol) into prewarmed mMRS broth supplemented with 1% GOS (GOS-mMRS), 1% lactose (Lac-mMRS), 1% glucose (Glc-mMRS), or a mixture of 0.076% lactose, 0.008% glucose, and 0.003% galactose. The latter was used as a negative control and represented the approximate amount of sugars present in purimune GOS. In other experiments, 1% raffinose or stachyose was added to mMRS. After inoculation, cell cultures were incubated anaerobically at 37°C. Growth on different carbon sources was determined by a measurement of the optical density at 600 nm (OD600) (Biomate3; Thermo Electron Corporation, Madison, WI).
In vitro coculture.
To determine if GOS utilization conferred a competitive advantage during growth in culture media, in vitro coculture experiments with L. reuteri 6475 and the ΔlacS ΔlacM mutant were performed. Cells were harvested from 12-hour cultures by centrifugation at 3,220 × g for 10 min, and cell pellets were washed twice with phosphate-buffered saline (PBS) buffer (pH 7.4) and resuspended in PBS buffer to obtain a cell concentration at an OD600 of 1. The cell suspensions of the two strains were mixed together at a 1:1 ratio before the cell mixtures were inoculated into either 1% GOS-mMRS or 1% Glc-mMRS. Cultures were incubated anaerobically at 37°C and then transferred (1%) into fresh media every 12 h until approximately 28 generations were obtained. One-milliliter aliquots of mix cell cultures were collected at 0, 12, 24, 36, and 48 h for quantification of each cell type. Similar experiments were performed in sterile chow broth, made by blending 5% standard chow (see below) in distilled water.
Real-time PCR quantification.
The populations of the wild-type and the ΔlacS ΔlacM mutant in cocultures were determined by real-time quantitative PCR (RT-qPCR) using primers listed in Table 3. Reaction mixes, consisting of 12.5 μl of 2× QuantiFast SYBR PCR master mix (Qiagen, Valencia, CA), 0.5 μM each primer, and 1 μl of RNA extracts, were amplified with the following program: 5 min of initial heat activation at 95°C, followed by 40 cycles of 10 s denaturation at 95°C, and 30 s combined annealing/extension at 63°C. Standard curves prepared from pure cultures of each strain were used for the absolute quantification.
TABLE 3.
Real-time PCR primers used in this study
| Expt | Primers (5′−3′) (forward; reverse) | Target |
|---|---|---|
| Gene expression analysis | TTTCTCGCGCTTCGTTTTGC; TCCTGCAAACATTCCGCTTG | lacS |
| TCCGCCATTCAAACGTTGTG; TTGCCCAATACGTTCGCTAC | recA (reference gene) | |
| GTACGCACTGGCCCAA; ACCGCAGGTCCATCCCAG | 16S rRNA gene, 16S rRNA | |
| In vitro co-culture and mouse | CACAAGCGGAATGTTTGCA; CGGGTCTTCGTATTATCAACAA | Wild-type lacS in L. reuteri MM4-1A |
| CACAAGCGGAATGTAGTAG; CGGGTCTTCGTATTATCAAC | Mutated lacS in the ΔlacS ΔlacM mutant |
GOS utilization.
GOS utilization by the ΔlacS mutant and the wild-type strain ATCC 6475 was determined by TLC analysis of spent fermentation media. Log-phase cells were prepared by inoculating 12-h cultures into prewarmed MRS at 1% and anaerobically incubating cell cultures at 37°C for 5 h or until obtaining an OD600 of 1.5 to 2.5. Cells were harvested by centrifugation, washed twice with PBS buffer (pH 7.4), and resuspended in PBS buffer to obtain a cell concentration at an OD600 of 10. Three milliliters of cell suspensions was inoculated into 27 ml of prewarmed GOS-mMRS so that an initial cell concentration of approximately 109 cells/ml (OD600 of 1) was obtained. A high cell concentration was used to ensure that the number of the mutant cells was high enough for cell activity to be observed. After an anaerobic incubation at 37°C for 0, 2, 4, 6, 9, 12, and 24 h, spent fermentation media were separated from cell cultures by centrifugation and filter sterilized through 0.22-μm membranes. Five-microliter aliquots of spent media were spotted onto HPTLC silica gel plates (Dynamic Adsorbents, Inc., Atlanta, GA) that were subsequently developed and visualized as described previously (20).
β-Galactosidase activity.
In vitro hydrolysis was performed using cell-free extracts of the ΔlacA and ΔlacM mutants to determine the hydrolytic activity of the LacA and LacLM β-galactosidase on disaccharide and oligosaccharide components of GOS, as well as lactose. Log-phase, GOS-grown cells of the ΔlacA and ΔlacM mutant were centrifuged and washed twice with PBS, as described above. Cell pellets were resuspended in PBS with 10% (wt/vol) glycerol and 1 mM dithiothreitol (DTT) to obtain a cell concentration at an OD600 of 10. One-milliliter aliquots of cell suspensions were transferred into ice-cold 2-ml microtubes containing 400 mg of 0.1-mm glass beads (zirconia/silica; BioSpec Products, Inc., Bartlesville, OK). Cells were then disrupted with a bead beater (mini-beadbeater; BioSpec Products, Inc.) at a maximum speed for three 1-min intervals, each separated by 1 min on ice. Cell-free extracts were separated from cell debris by centrifugation at 14,000 × g and 4°C for 10 min. The protein content of cell-free extracts was measured using a Qubit protein assay kit (Life Technologies, Grand Island, NY) and adjusted to 0.5 mg protein/ml. Enzyme assay reaction mixtures (200 μl in total) consisted of 20 μl of either 50 mg/ml GOS components or 10 mg/ml lactose, 160 μl PBS (pH 6.5), and 20 μl of cell-free extracts with 0.5-mg/ml protein content. Reaction mixtures were incubated in an incubator shaker (Thermomixer R; Eppendorf AG, Hamburg, Germany) at 37°C and 200 rpm for 2, 4, 6, and 12 h and then immediately heated at 95°C for 5 min to terminate the reactions. Five-microliter aliquots of reaction mixes were spotted onto TLC plates and analyzed as described previously.
GOS utilization in the murine gut.
To assess the ability of L. reuteri to utilize GOS in the gut environment, experiments were conducted in germfree C3H/HeN mice. Germfree mice were born and reared in flexible film isolators and maintained under gnotobiotic conditions at the University of Nebraska—Lincoln. Mice received either an irradiated purified ingredient, no-sucrose diet (D12450K; Research Diets Inc., New Brunswick, NJ) to minimize the amount of simple carbohydrates (with 70% of calories provided as mainly starch) or a standard chow diet (LabDiet 5K67; Purina Foods, St. Louis, MO). Immediately prior to colonization with L. reuteri, mice were transferred from gnotobiotic isolators into autoclaved cages on a positive pressure, individually ventilated caging system (Allentown Inc., Allentown, NJ), maintained on autoclaved bedding, and provided autoclaved water as described previously (50). The sterile-filtered GOS solution (30 mg/ml) was provided in the drinking water 24 hours prior to inoculation with test strains. Considering an average daily water consumption of 4 ml per mouse, each mouse received approximately 120 mg of GOS per day. A 1:1 mixture of wild-type and ΔlacS ΔlacM mutant cells (each at 5 × 109 CFU/ml) was prepared immediately before inoculation, and mice were orally gavaged with 100 μl of this cell suspension. All animal protocols were approved by the Institutional Animal Care and Use Committee of the University of Nebraska—Lincoln (Project ID 731).
Fecal samples (about 30 to 60 mg) were collected from individual mice at 12, 24, and 48 h after inoculation, diluted 10-fold with PBS buffer (pH 7.4), and homogenized by vortexing. To isolate DNA, 1 ml or less of fecal suspensions was transferred into 2-ml safe-lock tubes containing 300 mg of glass beads. Fecal pellets were centrifuged at 10,000 × g for 5 min, washed twice with 1 ml ice-cold PBS buffer, and washed once with 1 ml ice-cold water. Washed fecal pellets were mixed with 500 μl of the same lysis buffer described in the in vitro coculture study and incubated at 37°C for 1 hour. Following this step, 30 μl of 20% SDS and 25 μl of 15 mg/ml proteinase K were added into cell lysates. After incubation at 60°C for 30 min, samples were cooled on ice for 1 min and mixed with 500 μl of phenol-chloroform-isoamyl alcohol (25:24:1). Cell lysates were further disrupted in a bead beater at a maximum speed for 2 min. Cell homogenates were extracted with 500 μl of phenol-chloroform-isoamyl alcohol two more times and then with 500 μl of chloroform-isoamyl alcohol (24:1) three times. Next, 450-μl aliquots of aqueous phase were mixed with 2.5 volumes of ice-cold ethanol and a 0.1 volume of 3 M sodium acetate. DNA was allowed to precipitate at –20°C for at least 1 h and then centrifuged at 14,000 × g for 20 min. DNA pellets were washed with 1 ml of ice-cold 70% ethanol, centrifuged, air-dried, and resuspended in 100 μl of Tris-Cl buffer (pH 8.0). One-microliter aliquots (containing <100 ng) were used for RT-qPCR of the wild-type and mutant population as described above.
Data availability.
Genome sequences for strains of Lactobacillus reuteri used in this study can be found under GenBank accession numbers ACGX00000000 (for ATCC PTA-6475), CP002844 (for ATCC 55730), NC_009513 (for DSM 20016), ACLB00000000 (for ATCC PTA-4659), and CACS02000000 (for ATCC 53608).
Supplementary Material
ACKNOWLEDGMENTS
We thank Robert Schmaltz and the staff at the UNL Gnotobiotic Mouse Facility for outstanding animal husbandry and technical assistance. We also thank BioGaia AB for providing Lactobacillus reuteri MM4-1A and David Gomez for technical assistance.
M.R. was supported by a fellowship from the Royal Thai Government.
REFERENCES
- 1.Walter J. 2008. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol 74:4985–4996. doi: 10.1128/AEM.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Pérez-Muñoz ME, Leulier F, Gänzle M, Walter J. 2017. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev 41:S27–S48. doi: 10.1093/femsre/fux030. [DOI] [PubMed] [Google Scholar]
- 3.Walter J, Britton RA, Roos S. 2011. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc Natl Acad Sci U S A 108:4645–4652. doi: 10.1073/pnas.1000099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martínez I, Stegen JC, Maldonado-Gómez MX, Eren AM, Siba PM, Greenhill AR, Walter J. 2015. The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Rep 11:527–538. doi: 10.1016/j.celrep.2015.03.049. [DOI] [PubMed] [Google Scholar]
- 5.Karimi S, Jonsson H, Lundh T, Roos S. 2018. Lactobacillus reuteri strains protect epithelial barrier integrity of IPEC-J2 monolayers from the detrimental effect of enterotoxigenic Escherichia coli. Physiol Rep 6:e13514. doi: 10.14814/phy2.13514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu HY, Roos S, Jonsson H, Ahl D, Dicksved J, Lindberg JE, Lundh T. 2015. Effects of Lactobacillus johnsonii and Lactobacillus reuteri on gut barrier function and heat shock proteins in intestinal porcine epithelial cells. Physiol Rep 3:e12355. doi: 10.14814/phy2.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cervantes-Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J, Cortez VS, Caparon MG, Donia MS, Gilfillan S, Cella M, Gordon JI, Hsieh C-S, Colonna M. 2017. Lactobacillus reuteri induces gut intraepithelial CD4+CD8αα+ T cells. Science 357:806–810. doi: 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins FL, Irwin R, Bierhalter H, Schepper J, Britton RA, Parameswaran N, McCabe LR. 2016. Lactobacillus reuteri 6475 increases bone density in intact females only under an inflammatory setting. PLoS One 11:e0153180. doi: 10.1371/journal.pone.0153180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nilsson AG, Sundh D, Bäckhed F, Lorentzon M. 2018. Lactobacillus reuteri reduces bone loss in older women with low bone mineral density: a randomized, placebo-controlled, double-blind, clinical trial. J Intern Med 284:307–317. doi: 10.1111/joim.12805. [DOI] [PubMed] [Google Scholar]
- 10.Savino F, Garro M, Montanari P, Galliano I, Bergallo M. 2018. Crying time and RORγ/FOXP3 expression in Lactobacillus reuteri DSM17938-treated infants with colic: a randomized trial. J Pediatr 192:171–177. doi: 10.1016/j.jpeds.2017.08.062. [DOI] [PubMed] [Google Scholar]
- 11.Olson JK, Navarro JB, Allen JM, McCulloh CJ, Mashburn-Warren L, Wang Y, Varaljay VA, Bailey MT, Goodman SD, Besner GE. 2018. An enhanced Lactobacillus reuteri biofilm formulation that increases protection against experimental necrotizing enterocolitis. Am J Physiol Liver Physiol 315:G408–G419. doi: 10.1152/ajpgi.00078.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon MC, Strassburger K, Nowotny B, Kolb H, Nowotny P, Burkart V, Zivehe F, Hwang JH, Stehle P, Pacini G, Hartmann B, Holst JJ, Mackenzie C, Bindels LB, Martinez I, Walter J, Henrich B, Schloot NC, Roden M. 2015. Intake of Lactobacillus reuteri improves incretin and insulin secretion in glucose-tolerant humans: a proof of concept. Diabetes Care 38:1827–1834. doi: 10.2337/dc14-2690. [DOI] [PubMed] [Google Scholar]
- 13.Liévin-Le Moal V, Servin AL. 2014. Anti-infective activities of Lactobacillus strains in the human intestinal microbiota: from probiotics to gastrointestinal anti-infectious biotherapeutic agents. Clin Microbiol Rev 27:167–199. doi: 10.1128/CMR.00080-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sassone-Corsi M, Raffatellu M. 2015. No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J Immunol 194:4081–4087. doi: 10.4049/jimmunol.1403169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, Raffatellu M. 2013. Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischbach MA, Sonnenburg JL. 2011. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flint HJ, Duncan SH, Scott KP, Louis P. 2007. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol 9:1101–1111. doi: 10.1111/j.1462-2920.2007.01281.x. [DOI] [PubMed] [Google Scholar]
- 18.Kolida S, Gibson GR. 2011. Synbiotics in health and disease. Annu Rev Food Sci Technol 2:373–393. doi: 10.1146/annurev-food-022510-133739. [DOI] [PubMed] [Google Scholar]
- 19.Su P, Henriksson A, Mitchell H. 2007. Prebiotics enhance survival and prolong the retention period of specific probiotic inocula in an in vivo murine model. J Appl Microbiol 103:2392–2400. doi: 10.1111/j.1365-2672.2007.03469.x. [DOI] [PubMed] [Google Scholar]
- 20.Rattanaprasert M, Roos S, Hutkins RW, Walter J. 2014. Quantitative evaluation of synbiotic strategies to improve persistence and metabolic activity of Lactobacillus reuteri DSM 17938 in the human gastrointestinal tract. J Funct Foods 10:85–94. doi: 10.1016/j.jff.2014.05.017. [DOI] [Google Scholar]
- 21.Krumbeck JA, Walter J, Hutkins RW. 2018. Synbiotics for improved human health: recent developments, challenges, and opportunities. Annu Rev Food Sci Technol 9:451–479. doi: 10.1146/annurev-food-030117-012757. [DOI] [PubMed] [Google Scholar]
- 22.Gänzle MG, Follador R. 2012. Metabolism of oligosaccharides and starch in lactobacilli: a review. Front Microbiol 3:340. doi: 10.3389/fmicb.2012.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watson D, O'Connell Motherway M, Schoterman MHC, van Neerven RJJ, Nauta A, van Sinderen D. 2013. Selective carbohydrate utilization by lactobacilli and bifidobacteria. J Appl Microbiol 114:1132–1146. doi: 10.1111/jam.12105. [DOI] [PubMed] [Google Scholar]
- 24.Mao B, Li D, Zhao J, Liu X, Gu Z, Chen YQ, Zhang H, Chen W. 2014. In vitro fermentation of lactulose by human gut bacteria. J Agric Food Chem 62:10970–10977. doi: 10.1021/jf503484d. [DOI] [PubMed] [Google Scholar]
- 25.Andersen JM, Barrangou R, Abou Hachem M, Lahtinen SJ, Goh YJ, Svensson B, Klaenhammer TR. 2012. Transcriptional analysis of prebiotic uptake and catabolism by Lactobacillus acidophilus NCFM. PLoS One 7:e44409. doi: 10.1371/journal.pone.0044409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thongaram T, Hoeflinger JL, Chow JM, Miller M. 2017. Prebiotic galactooligosaccharide metabolism by probiotic lactobacilli and bifidobacteria. J Agric Food Chem 65:4184–4192. doi: 10.1021/acs.jafc.7b00851. [DOI] [PubMed] [Google Scholar]
- 27.Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, Walter J. 2010. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J 4:377–387. doi: 10.1038/ismej.2009.123. [DOI] [PubMed] [Google Scholar]
- 28.Andersen JM, Barrangou R, Hachem MA, Lahtinen S, Goh YJ, Svensson B, Klaenhammer TR. 2011. Transcriptional and functional analysis of galactooligosaccharide uptake by lacS in Lactobacillus acidophilus. Proc Natl Acad Sci U S A 108:17785–17790. doi: 10.1073/pnas.1114152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juers DH, Matthews BW, Huber RE. 2012. LacZ β-galactosidase: structure and function of an enzyme of historical and molecular biological importance. Protein Sci 21:1792–1807. doi: 10.1002/pro.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Vos WM, Vaughan EE. 1994. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol Rev 15:217–237. doi: 10.1111/j.1574-6976.1994.tb00136.x. [DOI] [PubMed] [Google Scholar]
- 31.van den Broek LAM, Hinz SWA, Beldman G, Vincken J-P, Voragen A. 2008. Bifidobacterium carbohydrases—their role in breakdown and synthesis of (potential) prebiotics. Mol Nutr Food Res 52:146–163. doi: 10.1002/mnfr.200700121. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, Ashida H, Hirose J, Katayama T, Yamamoto K, Kumagai H. 2012. Bifidobacterium longum subsp. infantis uses two different β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology 22:361–368. doi: 10.1093/glycob/cwr116. [DOI] [PubMed] [Google Scholar]
- 33.Hinz SWA, van den Broek LAM, Beldman G, Vincken J-P, Voragen A. 2004. β-Galactosidase from Bifidobacterium adolescentis DSM20083 prefers β(1,4)-galactosides over lactose. Appl Microbiol Biotechnol 66:276–284. doi: 10.1007/s00253-004-1745-9. [DOI] [PubMed] [Google Scholar]
- 34.Schwab C, Sørensen KI, Gänzle MG. 2010. Heterologous expression of glycoside hydrolase family 2 and 42 β-galactosidases of lactic acid bacteria in Lactococcus lactis. Syst Appl Microbiol 33:300–307. doi: 10.1016/j.syapm.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Goulas T, Goulas A, Tzortzis G, Gibson GR. 2009. Expression of four β-galactosidases from Bifidobacterium bifidum NCIMB41171 and their contribution on the hydrolysis and synthesis of galactooligosaccharides. Appl Microbiol Biotechnol 84:899–907. doi: 10.1007/s00253-009-2009-5. [DOI] [PubMed] [Google Scholar]
- 36.Viborg AH, Katayama T, Abou Hachem M, Andersen MCF, Nishimoto M, Clausen MH, Urashima T, Svensson B, Kitaoka M. 2014. Distinct substrate specificities of three glycoside hydrolase family 42 β-galactosidases from Bifidobacterium longum subsp. infantis ATCC 15697. Glycobiology 24:208–216. doi: 10.1093/glycob/cwt104. [DOI] [PubMed] [Google Scholar]
- 37.Shigehisa A, Sotoya H, Sato T, Hara T, Matsumoto H, Matsuki T. 2015. Characterization of a bifidobacterial system that utilizes galacto-oligosaccharides. Microbiology 161:1463–1470. doi: 10.1099/mic.0.000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gopal PK, Sullivan PA, Smart JB. 2001. Utilisation of galacto-oligosaccharides as selective substrates for growth by lactic acid bacteria including Bifidobacterium lactis DR10 and Lactobacillus rhamnosus DR20. Int Dairy J 11:19–25. doi: 10.1016/S0958-6946(01)00026-7. [DOI] [Google Scholar]
- 39.O’ Donnell MM, Forde BM, Neville B, Ross PR, O’ Toole PW. 2011. Carbohydrate catabolic flexibility in the mammalian intestinal commensal Lactobacillus ruminis revealed by fermentation studies aligned to genome annotations. Microb Cell Fact 10:S12. doi: 10.1186/1475-2859-10-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silvestroni A, Connes C, Sesma F, De Giori GS, Piard JC. 2002. Characterization of the melA locus for α-galactosidase in Lactobacillus plantarum. Appl Environ Microbiol 68:5464–5471. doi: 10.1128/aem.68.11.5464-5471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poolman B. 2002. Transporters and their roles in LAB cell physiology. Antonie Van Leeuwenhoek 82:147–164. doi: 10.1023/A:1020658831293. [DOI] [PubMed] [Google Scholar]
- 42.Petrova P, Petrov K. 2017. Prebiotic-probiotic relationship: the genetic fundamentals of polysaccharides conversion by Bifidobacterium and Lactobacillus genera, p 237–278. In Grumezescu AM and Holban AM (ed), Food bioconversion. Elsevier, Philadelphia, PA. [Google Scholar]
- 43.Martínez-Villaluenga C, Frias J, Vidal-Valverde C. 2008. Alpha-galactosides: antinutritional factors or functional ingredients? Crit Rev Food Sci Nutr 48:301–316. doi: 10.1080/10408390701326243. [DOI] [PubMed] [Google Scholar]
- 44.Zhao X, Gänzle MG. 2018. Genetic and phenotypic analysis of carbohydrate metabolism and transport in Lactobacillus reuteri. Int J Food Microbiol 272:12–21. doi: 10.1016/j.ijfoodmicro.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 45.Davis LMG, Martínez I, Walter J, Hutkins R. 2010. A dose dependent impact of prebiotic galactooligosaccharides on the intestinal microbiota of healthy adults. Int J Food Microbiol 144:285–292. doi: 10.1016/j.ijfoodmicro.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 46.Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Pillay M, Ratner A, Huang J, Woyke T, Huntemann M, Anderson I, Billis K, Varghese N, Mavromatis K, Pati A, Ivanova NN, Kyrpides NC. 2014. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res 42:D560–D567. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miwa Y, Nakata A, Ogiwara A, Yamamoto M, Fujita Y. 2000. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res 28:1206–1210. doi: 10.1093/nar/28.5.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Pijkeren J-P, Britton RA. 2012. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res 40:e76. doi: 10.1093/nar/gks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cha RS, Zarbl H, Keohavong P, Thilly WG. 1992. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. Genome Res 2:14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- 50.Gomes-Neto JC, Mantz S, Held K, Sinha R, Segura Munoz RR, Schmaltz R, Benson AK, Walter J, Ramer-Tait AE. 2017. A real-time PCR assay for accurate quantification of the individual members of the Altered Schaedler Flora microbiota in gnotobiotic mice. J Microbiol Methods 135:52–62. doi: 10.1016/j.mimet.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequences for strains of Lactobacillus reuteri used in this study can be found under GenBank accession numbers ACGX00000000 (for ATCC PTA-6475), CP002844 (for ATCC 55730), NC_009513 (for DSM 20016), ACLB00000000 (for ATCC PTA-4659), and CACS02000000 (for ATCC 53608).